Abstract
Summary: SimBoolNet is an open source Cytoscape plugin that simulates the dynamics of signaling transduction using Boolean networks. Given a user-specified level of stimulation to signal receptors, SimBoolNet simulates the response of downstream molecules and visualizes with animation and records the dynamic changes of the network. It can be used to generate hypotheses and facilitate experimental studies about causal relations and crosstalk among cellular signaling pathways.
Availability: SimBoolNet package (with manual) is freely available at http://www.ncbi.nlm.nih.gov/CBBresearch/Przytycka/SimBoolNet
Contact: przytyck@ncbi.nlm.nih.gov; zhengj@ncbi.nlm.nih.gov
1 INTRODUCTION
Signal transduction is one of the most important cellular processes, and defects in signaling pathways are associated with many serious diseases such as cancer. It is therefore highly desirable to understand the mechanisms of cell signaling for therapeutic purposes. The availability of high-throughput data combined with the complexity of signaling mechanisms calls for a system-level understanding of signaling networks. An important systems biology approach to the analysis of signaling networks is computer simulation. Many existing tools for the simulation of signal transduction use kinetic model based on differential equations (Gilbert et al., 2006). However, some of the kinetic parameters may not be available for newly discovered pathways, since they usually require many experimental data and long-time accumulation of knowledge. Alternatively, one can obtain insight of qualitative relations using much less data and simpler models. Boolean networks are one of such simple models that have been applied to systems biology. A Boolean network is a directed graph in which the nodes represent elements (e.g. genes, proteins) in the network and the edges represent interactions (e.g. gene regulation, phosphorylation) between two types of elements. Every node is assigned a state of ON/OFF, and at each time point the state of a node is determined by the states of its upstream neighbors via a transfer logical function. Recently, it was shown that simple models combining Boolean networks with fuzzy logic can capture most features of the signaling data (Aldridge et al., 2009).
Here, we present SimBoolNet, a simulation tool based on an extended Boolean network model, in which the state of each node is a variable between 0 and 1 representing the probability (or percentage) of activation (e.g. phosphorylation). A similar existing simulation tool is BooleanNet (Albert et al., 2008), which is also based on Boolean networks. However, since BooleanNet is a library of Python code, despite its flexibility it requires users to write their own Python scripts for simulation, which may imply a steep learning curve for users without any programming experience. In contrast, SimBoolNet does not require any programming. As a Java plugin for Cytoscape, an open source framework for analyzing biological networks (Shannon et al., 2003), SimBoolNet allows users to take advantage of the powerful functionalities and friendly graphic user interface of Cytoscape. Given the user-specified levels of stimulation to a signaling network, SimBoolNet simulates the response of downstream molecules, which can be visualized with animations and recorded for further analyses. Despite its simplicity, SimBoolNet is able to capture the general trends in signaling networks and recapitulate experimental results in our previous study of crosstalk among three signaling pathways (Zielinski et al., 2009). It is a useful tool for exploratory analysis of signaling network, e.g. hypothesizing causal relation and crosstalks among signaling molecules, elucidating mechanisms behind signaling abnormalities in diseases, etc.
2 PROGRAM OVERVIEW
The input to SimBooNet is a text file specifying a directed graph in which each node represents a specific molecule and has a variable denoting the percentage of activity of the molecule, and each directed edge (u, v) models the transduction of signals from u to v of type A (activation) or B (blockage). Each edge is associated with a weight between 0 and 1, an important parameter representing the efficiency of signals passing along a given edge. Starting from signal receptors, we iteratively traverse the whole network by a depth-first search and update the state of every node. Each iterative step simulates a small change of the molecules in the process of signal transduction, approximating the continuous process with discrete steps. The state of each node u is determined from its own previous state and the amounts of signals received by upper-stream nodes, according to the formula X(t)=X(t − 1)+[1 − ∏(1 − Ai)]×∏ (1 − Bj)×[1 − X(t −1)], where X(t) is the activity of u at time t, Ai (Bj) is the product of signal received by the i-th activating (blocking) node in t-th step times the edge weight, and ∏ is the product over all incoming edges of type A or B from the neighboring nodes of u. After a user-specified number of iterations the simulation stops.
SimBoolNet provides both single- and batch running modes.
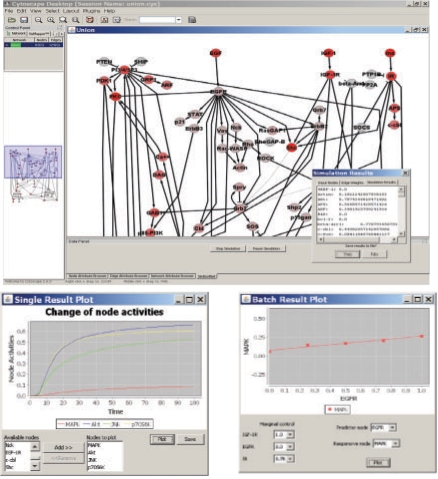
In single mode, users can specify the input activity (e.g. percentages of phosphorylation) of signal receptors. The activities of nodes will change step by step according to the above algorithm. This process is visualized both by the dynamic change of node colors from gray (less active) to red (active). A batch-mode run consists of multiple single-mode runs, each with a different combination of activities of input nodes. Using JFreeChart (Gilbert, 2009), SimBoolNet draws the time series of node changes in a single-mode simulation, and a scatter plot with a regression line between an input node and a downstream node in a batch-mode simulation (Fig. 1). All configurations of simulation model and results can be exported to hard disk for further analysis. Precise description of SimBoolNet is given in the user's manual provided with the software.
Fig. 1.
Screenshots of SimBoolNet. The top is the result of a single-mode simulation, where red color represents high percentage of phosphorylation. The bottom-left is a chart visualizing the changes of node activities with time. The bottom-right is a scatter plot and regression line between a signal receptor and a down-stream node from a batch simulation.
In our recent work (Zielinski et al., 2009), we used SimBoolNet to study the crosstalk in a combined signaling network of three cancer-related signaling pathways: epidermal growth factor receptor, insulin-like growth factor-1 receptor and insulin receptor. We first predicted the response of the output molecules using SimBoolNet, and then carried out real experiments to measure the phosphorylation levels of the selected molecules in SKOV3 (human ovary carcinoma) cells. Most of the trends suggested by SimBoolNet simulation have been confirmed by the experimental studies. Allowing for dynamic visualization and interactive analysis of simulated states of the network, SimBoolNet provided valuable insight about the mechanism of crosstalk between the three signaling pathways.
3 CONCLUSIONS
Here, we present SimBoolNet, a Cytoscape plugin for the dynamic simulation of signaling networks based on Boolean networks with probabilistic transitions. SimBoolNet facilitated our study of crosstalks among three cancer-related signaling pathways with promising results.
ACKNOWLEDGEMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organization imply endorsement by the US Government.
Funding: Intramural Research Program of the National Institutes of Health (in part); National Library of Medicine (in part); National Cancer Institute, Center for Cancer Research (in part).
Conflict of Interest: none declared.
REFERENCES
- Albert I. Boolean network simulations for life scientists. Source Code Biol. Med. 2008;3:16. doi: 10.1186/1751-0473-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge BB, et al. Fuzzy logic analysis of kinase pathway crosstalk in TNF/EGF/insulin-induced signaling. PLoS Comput. Biol. 2009;5:e1000340. doi: 10.1371/journal.pcbi.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D. The JFreeChart Class Library. 2009 Available: http://www.jfree.org/jfreechart/ (last accessed date September 18, 2009) [Google Scholar]
- Gilbert D, et al. Computational methodologies for modelling, analysis and simulation of signalling networks. Brief. Bioinform. 2006;7:339–353. doi: 10.1093/bib/bbl043. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski R, et al. The crosstalk between EGF, IGF, and insulin cell signaling pathways – computational and experimental analysis. BMC Syst. Biol. 2009;3 doi: 10.1186/1752-0509-3-88. [DOI] [PMC free article] [PubMed] [Google Scholar]