SUMMARY
The gastrointestinal tract of mammals is inhabited by hundreds of distinct species of commensal microorganisms that exist in a mutualistic relationship with the host. How commensal microbiota influence the host immune system is poorly understood. We show here that colonization of the small intestine of mice with a single commensal microbe, segmented filamentous bacterium (SFB), is sufficient to induce the appearance of CD4+ T helper cells that produce IL-17 and IL-22 (Th17 cells) in the lamina propria. SFB adhere tightly to the surface of epithelial cells in the terminal ileum of mice with Th17 cells but are absent from mice that have few Th17 cells. Colonization with SFB was correlated with increased expression of genes associated with inflammation and anti-microbial defenses, and resulted in enhanced resistance to the intestinal pathogen Citrobacter rodentium. Thus, manipulation of this commensalregulated pathway may provide new opportunities for enhancing mucosal immunity and treating autoimmune disease.
INTRODUCTION
The vertebrate intestine is typically colonized by hundreds of distinct species of microorganisms that have a mutually beneficial relationship with the host. Intestinal microbiota are known to influence the development and balance of the host immune system, and have been implicated in prevention of damage induced by opportunistic microbes, in repair of damage to the mucosal barrier, and in influencing systemic autoimmune diseases (Backhed et al., 2005; Macpherson and Harris, 2004; Rakoff-Nahoum and Medzhitov, 2006). CD4+ T cells acquire distinct functional properties in response to signals conveyed by commensal and pathogenic microbe-activated cells of the innate immune system (Seder and Paul, 1994). T-helper type 1 (Th1) and Th2 cells control intracellular microorganisms and helminths, respectively (Abbas et al., 1996; Glimcher and Murphy, 2000), whereas the induced regulatory T cells (iTreg) suppress excessive immune responses (Gavin and Rudensky, 2003). Th17 cells secrete IL-17, IL-17F, and IL-22, and have significant roles in protecting the host from bacterial and fungal infections, particularly at mucosal surfaces. Th17 cells also have potent inflammatory potential, and thus are key mediators of autoimmune disease (Aujla et al., 2007; Bettelli et al., 2007). Th17 and Treg cells are both dependent on TGF-β for their differentiation and are defined by the expression of the lineage-specific transcription factors RORγt and Foxp3, respectively (Fontenot et al., 2003; Hori et al., 2003; Ivanov et al., 2006; Khattri et al., 2003; Mangan et al., 2006)(Veldhoen et al., 2006). At appropriate concentrations of TGF-β and IL-6, antigen-activated CD4+ T cells up-regulate RORγt and express Th17 cell cytokines (Zhou et al., 2008).
Th17 cells are most abundant at steady state in gut-associated tissues, particularly the small intestinal lamina propria (SI LP) (Ivanov et al., 2008; Ivanov et al., 2006), where they accumulate only in the presence of luminal commensal microbiota (Atarashi et al., 2008; Hall et al., 2008; Ivanov et al., 2008). Germ-free (GF) mice, which lack Th17 cells in the SI LP (and also in the colon), acquired them following colonization with conventional microbiota. Treatment of newborn mice with antibiotics, particularly vancomycin, resulted in marked reduction in the number of Th17 cells in the SI LP. Most strikingly, C57BL/6 (B6) mice obtained from different commercial vendors displayed marked differences in the proportion of Th17 cells in the SI LP (Ivanov et al., 2008). Thus, mice from the Jackson Laboratory had very low numbers of SI LP Th17 cells compared to mice of the same strain obtained from Taconic Farms. Transfer into GF mice of intestinal contents of Taconic B6 mice, but not Jackson B6 mice, induced Th17 cell accumulation, and Jackson mice acquired Th17 cells within weeks of co-housing with mice from Taconic Farms. GF mice colonized only with a defined cocktail of bacteria (Altered Schaedler Flora, or ASF) lacked intestinal Th17 cells (Ivanov et al., 2008). These results demonstrated that the induction of Th17 cells in the SI LP is controlled not by the presence of bacteria per se, but by the composition of the intestinal microbiota and, presumably, the presence of specific bacterial taxa. Intriguingly, Treg cells, which, like Th17 cells, are abundant in the intestine, were increased in proportion in the SI LP in GF mice, and their numbers were inversely correlated to the proportion of Th17 cells. Signals derived from microbiota may thus influence the differentiation potential of multipotent CD4+ T cells in the lamina propria (Zhou et al., 2008).
Here we report that specific members of the commensal microbiota known as segmented filamentous bacteria (SFB), with the candidate name Arthromitus, are potent inducers of Th17 cells in the SI LP of mice. SFB, spore-forming gram-positive bacteria most closely related to the genus Clostridium, have been reported to colonize the intestines of numerous species, including humans (Davis and Savage, 1974; Klaasen et al., 1993a). They typically adhere tightly to epithelium in the ileum, where their abundance has been noted to correlate with reduced colonization and growth of pathogenic bacteria (Garland et al., 1982; Heczko et al., 2000). SFB were present in large numbers in conventionally raised B6 mice from Taconic Farms, but were undetectable in the same strain of mice obtained from the Jackson Laboratory. Introduction of SFB, but not other bacteria, into Th17 cell-deficient mouse models induced IL-17 and IL-22 expression in CD4+ T cells in the SI LP. Upon colonization, SFB induced a pro-inflammatory gene program that was similar to that induced in Jackson B6 mice co-housed with Taconic animals, suggesting that SFB are major modulators of immune responses in conventional mice. SFB colonization induced production of serum amyloid A (SAA) in the terminal ileum, and SAA acted on lamina propria dendritic cells (LP DCs) to promote Th17 cell differentiation in vitro. SFB colonization resulted in reduced growth of an intestinal pathogen, suggesting that intestinal commensal microbes can contribute to Th17 cell-mediated mucosal protection.
RESULTS
Comparative analysis of the intestinal microbiota of Jackson and Taconic B6 mice
To identify the bacterial species that induce Th17 cells in the small intestine, we compared the bacterial content in B6 mice purchased from Taconic Farms and Jackson Laboratory. We previously showed that transfer of cecal contents could induce Th17 cells in recipient mice (Ivanov et al., 2008). Because we find more Th17 cells in the small intestine than in the large intestine (LI), we surmised that the Th17 cell-inducing bacterial species are present also in the small intestinal microbiota. Indeed, colonization of GF and Jackson B6 mice with the contents of the small intestines of Taconic B6 mice induced numbers of Th17 cells similar to those in specific-pathogen-free (SPF) Taconic B6 mice (Figure 1A). We therefore chose to investigate the bacterial composition of the small intestine in detail. To provide an in-depth profile of the bacterial communities present in the small intestine of Taconic and Jackson mice, we analyzed these samples using the 16S rRNA PhyloChip (Brodie et al., 2006), a high-density microarray with over 300,000 probes targeting the sequence polymorphisms in the 16S rRNA gene, permitting detection of approximately 8,500 bacterial taxa. Overall 1,164 taxa were detected across all samples, 509±32 taxa were detected in Jackson B6 mice and 828±82 taxa in Taconic B6 mice (Figure S1). Our previous analysis by FISH had demonstrated a correlation between a probe for the Cytophaga-Flavobacter-Bacteroides (CFB) phylum and the presence of Th17 cells in Taconic mice (Ivanov et al., 2008). However, closer analysis of recently published 16S rRNA sequences revealed that this probe also matches perfectly to a number of non-CFB taxa, including bacteria in the Firmicutes, Actinobacteria, and Verrucomicrobia phyla. The PhyloChip analysis demonstrated that, indeed, the overall representation of the major bacterial phyla, including CFB, was not statistically different between the two mouse strains (Figure S2).
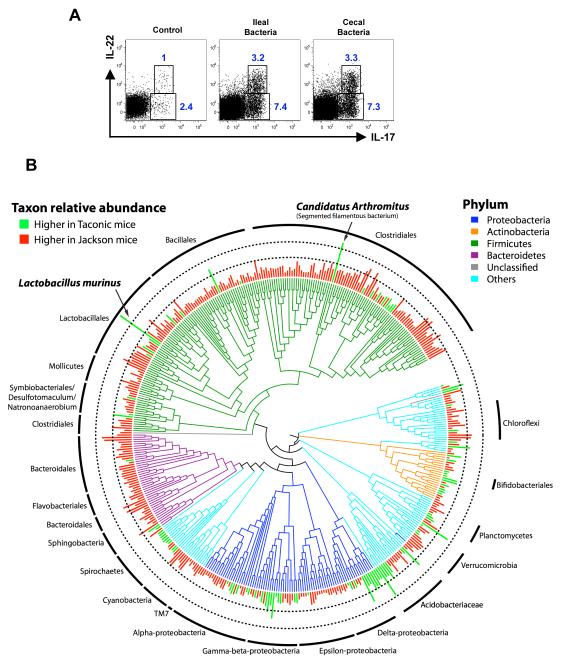
Figure 1. Comparative analysis of the microbiota in the terminal ileum of C57BL/6 (B6) mice from Jackson Laboratory versus Taconic Farms.
A. Lumenal bacteria from both cecum and terminal ileum can induce Th17 cell differentiation upon transfer into Jackson B6 mice. Jackson B6 mice were gavaged with water (control) or with intestinal luminal contents from cecum or terminal ileum of Taconic B6 mice. LPL from small intestine were isolated 10 days later and analyzed for intracellular cytokines. Representative plots from one experiment with 3 mice per group. Plots gated on TCRβ+CD4+ LPL.
B. Phylogenetic tree based on 16S rRNA gene sequences of bacterial taxa detected in the terminal ileum showing significantly different relative abundances (PhyloChip fluorescence intensity) between the suppliers, Taconic and Jackson. Branches of the tree are color coded according to phylum while green and red bars display taxa with significantly greater relative abundance in Taconic and Jackson mice, respectively. The inner and outer dotted rings represents intensities corresponding to 5-fold and 25-fold differences in 16S copy number. The two taxa with the greatest difference between Taconic and Jackson mice, Lactobacillus murinus (~94-fold difference) and Candidatus Arthromitus (~40-fold difference), are noted by arrows.
The depth of coverage and phylogenetic breadth of the PhyloChip allowed us to assay the microbial community at multiple phylogenetic levels, while its sensitivity permitted detection of less abundant organisms even in dominated communities (DeSantis et al., 2007). Comparative analysis of 766 bacterial taxa detected in at least 3 out of 4 replicates from either strain of mice demonstrated that the relative abundance of 479 taxa was significantly different (p≤0.05) between the two mouse strains, with 372 taxa having greater abundance in Jackson mice and 107 taxa overrepresented in the Taconic group. However, of the 479 significantly different taxa, most differences were subtle, with only 52 being above 5-fold (17 greater in Taconic and 35 greater in Jackson) and only two taxa were >25-fold more abundant. These were identified as members of the Lactobacillaceae and Clostridiaceae families – Lactobacillus murinus ASF361 and a segmented filamentous species of the candidate genus Arthromitus (Figure 1B). Both were of significantly greater (p<0.001) relative abundance in Taconic mice (~94-fold for Lactobacillus murinus and ~40-fold for Candidatus Arthromitus); however, since both were below the PhyloChip threshold of detection in the Jackson mice, these fold changes should be considered a minimum. For both taxa, overrepresentation of close phylogenetic relatives was not observed, suggesting a species-specific increase in relative abundance (Figure 1B).
Lactobacillus murinus ASF361 is a component of the ASF (Dewhirst et al., 1999). ASF is used by Taconic Farms as a basal inoculum introduced into all Taconic re-derived strains, but is not intentionally introduced into Jackson Laboratory animals. Because of these differences, we previously tested if L. murinus ASF361, in the context of ASF, induces Th17 cell differentiation. Colonization of germ-free mice with ASF, including L. murinus ASF361, did not induce any Th17 cells in the SI LP (Ivanov et al., 2008). We therefore concluded that L. murinus ASF 361 is not involved in the induction of Th17 cell differentiation.
Presence of SFB correlates with the presence of Th17 cells
We next examined the representation of Candidatus Arthromitus in Th17 cell-sufficient and Th17 cell-deficient mice. Arthromitus is an unofficial candidate genus name for the group of so-called segmented filamentous bacteria (Snel et al., 1995). SFB are yet to be cultured, commensal, gram-positive, anaerobic, spore-forming bacteria that are resident in the terminal ileum under steady state conditions (Davis and Savage, 1974). SFB have a characteristic long filamentous morphology, are comprised of multiple segments with well-defined septa, and often span the length of several villi. They colonize the gastrointestinal tract of mice at weaning time and adhere tightly to epithelial cells (Koopman et al., 1987). SFB are present in a many vertebrate species, including rodents (Davis and Savage, 1974), fish, chicken, dogs, and primates (Klaasen et al., 1993a; Ley et al., 2008). A phylogenetic tree based on an alignment of the available SFB 16S rRNA gene sequences according to their sequence origin is presented in Figure S3. SFB are known to actively interact with the immune system (Klaasen et al., 1993b). Colonization of germ-free animals with SFB leads to stimulation of secretory IgA (SIgA) production and recruitment of intraepithelial lymphocytes (IELs) to the gut (Talham et al., 1999; Umesaki et al., 1999). Mice lacking the activation-induced cytidine deaminase (AID) required for antibody diversification had outgrowth of SFB in their small intestine (Suzuki et al., 2004).
We validated the abundance of SFB in the gut of Taconic and Jackson B6 mice by quantitative real-time PCR (qPCR) for 16S rDNA sequences. SFB were present in fecal material from cecum as well as small and large intestine of Taconic B6 mice, but could not be detected in Jackson B6 mice (Figure 2A and data not shown). Scanning electron microscopy revealed a thick network of SFB present in the terminal ileum of 6-8 week old Taconic B6 mice (Figure 2B). In contrast we could not detect any bacteria with SFB morphology in age- and sex-matched Jackson B6 mice, even after equilibration of housing conditions and diet (Figure 2B). Despite similar numbers of total bacteria in the feces of both strains, only non-SFB bacteria were evident in the terminal ileum of mice from Jackson Laboratory (Figure S4). Transmission electron microscopy confirmed typical SFB morphology with well-defined segments in tight contact with the epithelial cells of ileum from Taconic but not Jackson B6 mice (Figure 2B). To confirm that SFB can be horizontally transferred, we co-housed female mice obtained from the two sources and observed Th17 cells in the lamina propria of Jackson B6 mice within 10 days ((Ivanov et al., 2008) and Figure S5). qPCR analysis of fecal material and microscopy of terminal ileum confirmed the appearance of SFB in the co-housed Jackson B6 mice (Figure 2C and 2D).
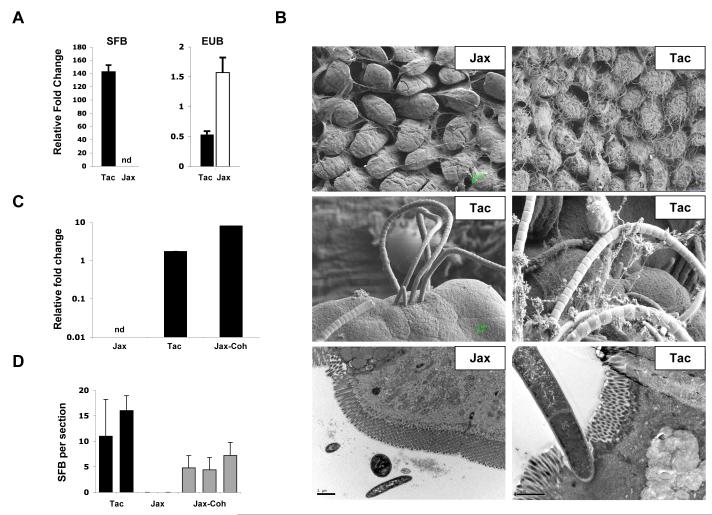
Figure 2. Segmented Filamentous Bacteria (SFB) in the intestinal tract of Th17 cell-sufficient and Th17 cell-deficient mice.
A. Quantitative PCR (qPCR) analysis of SFB and total bacterial (EUB) 16S rRNA genes in mouse feces from Taconic (Tac) and Jackson (Jax) B6 mice. Genomic DNA was isolated from combined fecal pellets from 4 animals from each strain. The experiment was repeated numerous times with similar results.
B. Scanning (SEM) and transmission (TEM) electron microscopy of terminal ileum of 8 week-old Jackson (Jax) and Taconic (Tac) C57BL/6 mice housed under similar conditions and diet for at least one week. Note the presence of long filamentous bacteria with SFB morphology in Taconic, but not Jackson mice
C. qPCR analysis for SFB presence in Jackson B6 mice after 14 days of co-housing with Taconic B6 mice (Jax-Coh). Genomic DNA was isolated from pooled feces from 3-4 mice per group.
D. SFB colonization of terminal ileum of Jackson B6 mice after 14 days of co-housing with Taconic B6 mice (Jax-Coh). Toluidine-blue sections were prepared from 0.5 cm piece of the terminal ileum as described in Methods and examined by light microscopy. Adherent bacteria with SFB morphology were counted in 4-5 sections from each sample. Each column represents a separate animal.
SFB specifically induce Th17 cells in the intestinal lamina propria
To test if SFB are sufficient to induce Th17 cells, we colonized germ-free (GF) Swiss-Webster mice with fecal material obtained from mice mono-colonized with SFB (SFB-mono mice) (Umesaki et al., 1995) and examined lamina propria CD4+ T cells for Th17 cell differentiation 10 days later. Non-colonized control GF mice housed under separate but similar conditions had no Th17 cells (Figure 3A). In contrast, SFB colonization induced robust accumulation of Th17 cells in both the SI and LI LP (Figure 3A and Figure S6). SFB induced production of both IL-22 and IL-17 in CD4+ T cells (Figure 3A and 3B). The effect of SFB on Th17 cell differentiation was similar in Swiss-Webster and IQI GF mice housed at different institutions (Figure 3B and 3C). Moreover, the effect of SFB on inducing IL-17 production in LP T cells is bacterial species specific, because colonization with Bacteroides species as well as with a defined mix of Clostridium species, which are closely related to SFB, did not induce Th17 cells in GF mice (Figure 3C). Finally, SFB had no effect on IFN-γ production, indicating that they specifically influence Th17 and not Th1 cell differentiation (Figure 3D). Colonization of GF mice with SFB restored RORγt+ T cells to the levels observed in mice kept under SPF conditions (Figure 3E). By contrast, the number of RORγt+ non-T cells, which include lymphoid tissue inducer-like cells and NK-like cells, was similar in GF mice, SFB-mono mice, and mice kept in SPF conditions (Figure 3E), and there was no significant difference in IL-17 and IL-22 production by these cells (Figure S7). Notably, SFB colonization and induction of Th17 cells did not reverse the elevated proportion of Foxp3+ cells among the CD4+ T cells in the SI LP and the peritoneal cavity of GF mice. (Figure S8).
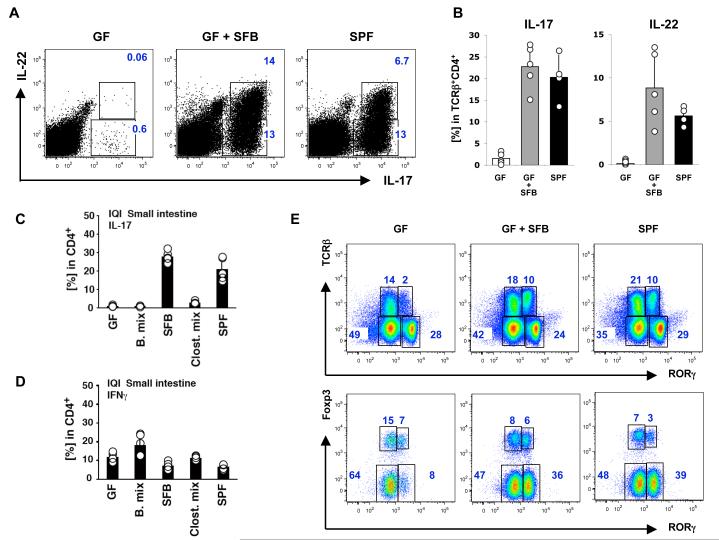
Figure 3. SFB specifically induce Th17 cell differentiation in germ-free mice.
A-B. 6-week old Swiss-Webster (SW) germ-free mice (GF) were colonized with SFB (GF+SFB) as described in Methods and small intestinal lamina propria lymphocytes (SI LPL) were isolated 10 days later. Representative plots in (A) and combined data in (B) of IL-17 and IL-22 expression in TCRβ+CD4+ LPL. Data are from one of three separate experiments with similar results. Error bars (SD). SPF – mice raised under conventional specific pathogen free conditions. Each circle in (B) represents a separate animal
C-D. IL-17 (C) and IFNγ (D) expression in TCRβ+CD4+ SI LPL from mice colonized with different commensal bacteria. IQI germ-free (GF) mice were colonized with 16 strains of Bacteroidaceae (B. mix), SFB, 46 strains of Clostridium sp mixture (Clost. mix), or microbiota from conventionally raised mice (SPF). Intracellular cytokine production in SI LP CD4 T cells was analyzed 3 weeks later by flow cytometry. Circles represent separate animals
E. SFB colonization induces RORγt expression only in CD4+ T cells. RORγt expression in total SI LPL (top panels) and RORγt and Foxp3 expression in TCRβ+CD4+ SI LPL (bottom panels) in GF mice, GF mice colonized with SFB (GF+SFB) and conventionally raised mice (SPF).
To determine whether SFB can also induce Th17 cell differentiation in conventionally raised mice, we introduced fecal material from SFB-mono mice by oral gavage into 6 week-old Jackson B6 mice and analyzed colonization and cytokine production in the SI LP. By 10 days, SFB were detected by scanning electron microscopy in the terminal ileum (Figure 4A) and by qPCR in the feces (data not shown), and robust Th17 cell differentiation was observed in the SI LP (Figures 4B and 4C). In contrast, control untreated Jackson B6 mice or Jackson B6 mice gavaged with bacterial suspensions from their littermates did not show an increase in Th17 cells (Figure 4B and 4C). Similarly, introduction of Jackson microbiota into GF animals did not induce Th17 cells, unless the microbiota were supplemented with SFB (Figure 4D and 4E). Th17 cell induction by SFB was also demonstrated by the expression of a number of Th17 cell effector cytokine mRNAs, including those for IL-17 and IL-21 (Figure 4F). We therefore conclude that SFB are members of the commensal microbiota that specifically induce the accumulation of Th17 cells in the SI LP.
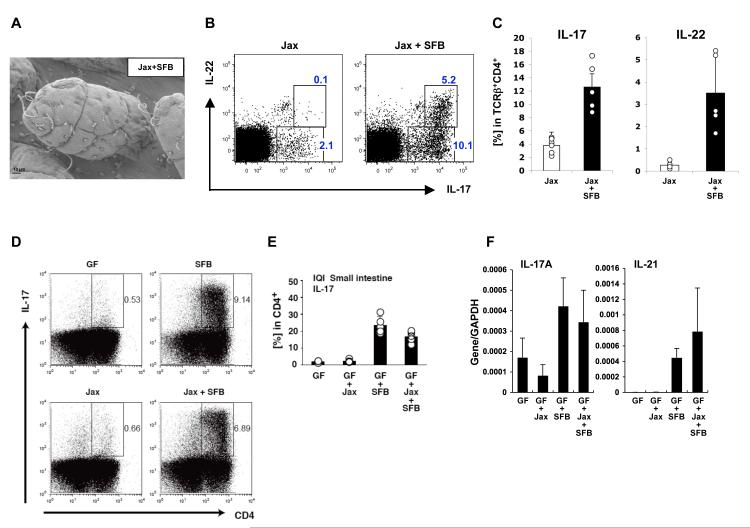
Figure 4. SFB induce Th17 cell differentiation upon colonization of Jackson C57BL/6 mice.
A. Colonization of the terminal ileum by SFB 10 days after transfer of fecal homogenates from SFB-mono mice into Jackson B6 mice
B-C. IL-17 and IL-22 expression in TCRβ+CD4+ SI LPL in Jackson B6 mice colonized with SFB (Jax+SFB) compared to controls (Jax). Data from one of two experiments
D-E. Jackson microbiota induces Th17 cells only when complemented with SFB. Germ-free IQI mice were colonized with SFB, Jackson microbiota isolated from fecal pellets by itself (Jax), or a mixture of both (Jax + SFB). Th17 cell proportions were analyzed in the LP 3 weeks later by flow cytometry. Plots in (D) gated on total lymphocytes. Data in (E) represent percentage of IL-17+ cells in the CD4+ gate
F. RT-PCR for Th17 cell effector cytokines in total RNA from terminal ileum of the mice in (E)
SFB induce an immune response program in the gut
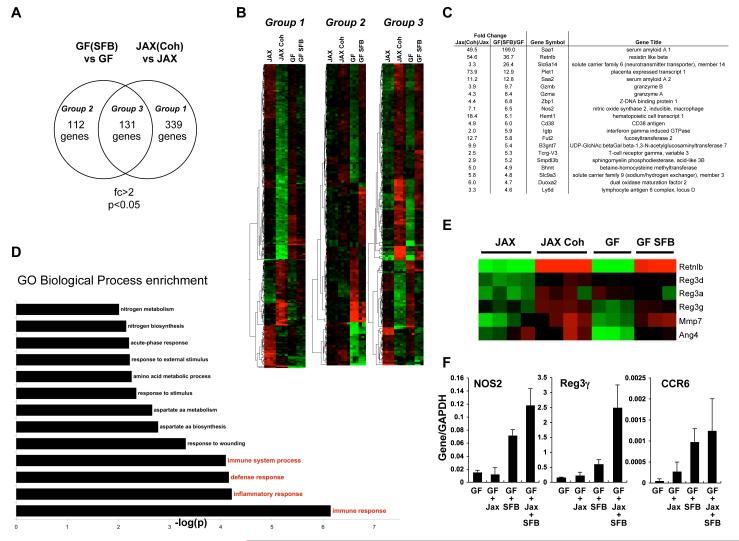
To identify specific effects of SFB, we compared the gene expression profiles in the terminal ileum of Swiss-Webster GF mice before and after colonization with SFB and in Jackson B6 mice before and after co-housing with Taconic B6 animals. Colonization of GF mice with SFB induced at least a two-fold change in expression of 253 genes while co-housing of Jackson B6 mice with Taconic B6 mice induced a similar change in 470 genes (Figure 5A). More importantly, there was a high degree of overlap between the two groups, with expression of 131 genes affected by both treatments. We could therefore distinguish three groups of genetic profiles. Group 1 includes genes whose expression was affected only in Jackson mice by co-housing, but was not statistically different after SFB colonization. This group most likely includes genes whose expression is influenced by microbiota other than SFB that differs between the mice from the different vendors, as well as strain-specific changes. Group 2 consists of genes whose expression only changed in GF mice upon colonization with SFB, but not in Jackson B6 mice following co-housing. A subset of these genes is expected to reflect changes induced in GF animals upon general intestinal colonization with bacteria. Group 3 includes the genes with expression differences following both SFB colonization and co-housing with Taconic mice (Figure 5A) and thus contains genes specifically induced by SFB and associated with Th17 cell induction.
Figure 5. Transcriptional programs induced by SFB colonization.
A. Venn diagrams showing the overlap between genes affected by either SFB colonization of SW GF mice only (Group 2), introduction of Taconic microbiota into Jackson B6 mice by co-housing (Group 1), or both (Group 3). Total RNA was prepared from terminal ileum of the corresponding mice after 10 days of colonization and Affymetrix gene chip analysis was performed as described in Methods.
B. Heat-map analysis of the three groups in (A). Each line represents a single Affymetrix probe and each column a single mouse. Green, probes that were at least 2 fold down-regulated. Red, probes that were at least 2 fold up-regulated.
C. Top up-regulated genes in Group 3 in (A) arranged by fold change in GF+SFB mice.
D. Biological processes specifically induced by SFB (genes in Group 3 in (A)). Gene ontology analysis was performed as described in Methods.
E. Changes in anti-microbial peptide related genes upon SFB colonization and Th17 cell induction by co-housing. Each column represents an individual mouse.
F. RT-PCR analysis of selected genes, induced by SFB colonization. IQI GF mice (GF) were colonized with fecal homogenates from SFB-mono mice (SFB), Jackson B6 mice (Jackson), or a mixture of both (Jackson+SFB). Total RNA from terminal ileum was prepared 3 weeks later and RT-PCR performed as described in Methods.
SFB exerted an inductive effect in the host, which was demonstrated by the finding that most (>70%) of the genes in Group 3 were up-regulated after SFB colonization (Figure 5B). By comparison, most genes in Group 1 (>70%) were down-regulated, which suggests that the rest of the Taconic microbiota has a suppressive effect that may possibly restrain the inductive effect of SFB (Figure 5B). Group 2, on the other hand, consisted of roughly equal numbers of up-regulated and down-regulated genes.
To evaluate changes specifically associated with Th17 cell-inducing SFB, we next concentrated on the genes in Group 3. A list of the top up-regulated genes is presented in Figure 5C. A GO Biological Pathway analysis of up-regulated genes in Group 3 showed that immune system pathways were among the programs most significantly induced by SFB (Figure 5D) and raised the possibility that at least some of the observed gene expression changes were mediated by Th17 cells or their effector cytokines. Because IL-17 and IL-22 have been associated with induction of anti-microbial peptides (AMP) (Curtis and Way, 2009; Kolls et al., 2008; Zheng et al., 2008), we compared the induction of AMP-related genes in our arrays. Multiple AMP genes were induced specifically by colonization with SFB, consistent with an up-regulated Th17 cell response (Figure 5E). Upregulation of Th17 cell-associated genes (Il17, Il21, Ccr6, Nos2) and AMPs (Reg3g) following SFB colonization was confirmed by quantitative RT-PCR (Figures 4F and 5F).
Serum Amyloid A is induced by SFB colonization and influences Th17 cell differentiation
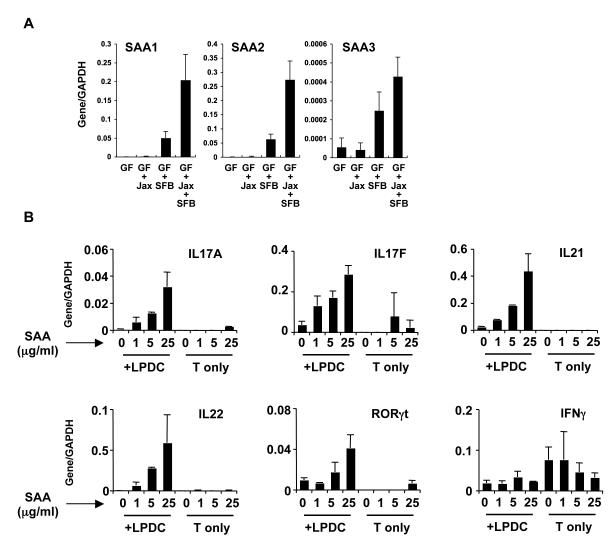
The top up-regulated transcript upon SFB colonization of GF mice encoded an isoform of SAA – Saa1, a member of the family of acute-phase response proteins induced during infection, tissue damage, or inflammatory disease (Uhlar and Whitehead, 1999). This transcript was also up-regulated upon co-housing of Jackson B6 mice with Taconic B6 animals (Figure 5C). Transcripts for the other SAA isoforms, Saa2 and Saa3, were also among the most highly up-regulated genes upon colonization with SFB or co-housing (Figure 5C).
Real-time PCR confirmed that all three SAA isoforms were induced in the terminal ileum of GF mice upon colonization with SFB or SFB + Jackson microbiota, but not by Jackson microbiota alone (Figure 6A). Recent studies have demonstrated that SAA may act as a cytokine that induces IL-8, TNFα, and IL-1β in neutrophils and IL-23 in monocytes (Furlaneto and Campa, 2000; He et al., 2006). We therefore investigated the effect of SAA on Th17 cell differentiation in vitro. Addition of recombinant SAA to co-cultures of naïve CD4+ T cells and LP DCs induced a Th17 cell differentiation program in a concentration-dependent manner, including Th17 cell effector cytokines and RORγt (Figure 6B). In addition, SAA induced production of IL-17 in CD4+ splenic OT-II T cells co-cultured with LP DCs in vitro (Figure S9). Addition of SAA to cultures containing only T cells, without DCs, did not induce Th17 cell cytokines (Figure 6B) and SAA induced production of IL-6 and IL-23 by LP DCs in vitro (Figure S10). We conclude that SFB colonization results in the production of SAA, which in turn acts on gut DCs to stimulate a Th17 cell-inducing environment.
Figure 6. SFB colonization induces SAA expression that influences Th17 differentiation.
A. Relative mRNA expression levels of SAA1-3 genes by real-time RT-PCR in the terminal ileum of IQI GF mice (GF) colonized with fecal homogenates from SFB-mono mice (SFB), Jackson B6 mice (Jackson), or a mixture of both (Jackson+SFB).
B. Splenic naïve CD4+ T cells were cocultured with or without LP CD11c+ cells in the presence of an anti-CD3 antibody with the indicated concentration of recombinant Apo-SAA for 4 days. T cells were collected, restimulated with PMA and ionomycin, and real-time RT-PCR performed. Results were normalized to expression of GAPDH mRNA. The data are representative of four independent experiments with similar results.
SFB colonization reduces growth of an intestinal pathogen
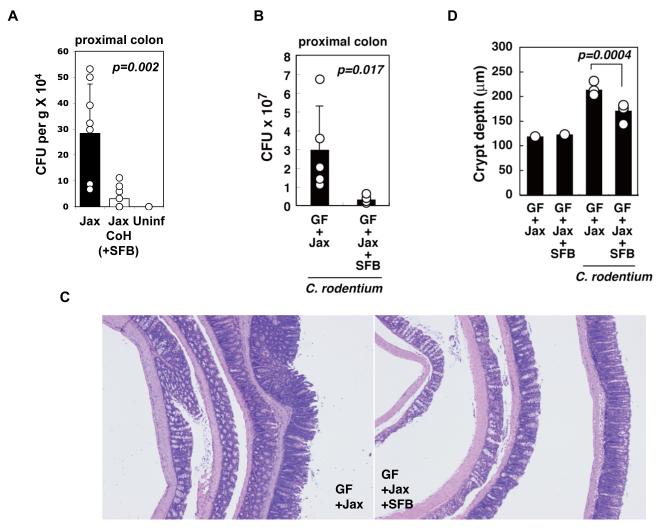
We next examined the effect of Th17 cell-inducing microbiota and SFB on oral infection with Citrobacter rodentium, an intestinal pathogen whose clearance by the host requires an immune response dependent on IL-23, IL-22, and RegIIIγ (Mangan et al., 2006; Torchinsky et al., 2009; Zheng et al., 2008). Jackson B6 mice that had been co-housed with Taconic B6 mice and hence were colonized with SFB were significantly more resistant to growth of C. rodentium compared to non-co-housed mice, as demonstrated by recovery of infectious units from the wall of the colon (Figure 7A).
Figure 7. SFB colonization confers protection from infection by Citrobacter rodentium.
A. Jackson B6 mice (Jax) were co-housed with Taconic B6 mice (Jax CoH) for 10 days to induce colonization with SFB and Th17 cells. Both groups were infected with ~ 1 × 109 CFU of C. rodentium/mouse and pathogen colonization of the colon was examined at day 8 of infection (n = 9/group). Uninf – uninfected controls.
B-D. IQI GF mice colonized with Jackson microbiota (Jax) or Jax+SFB were orally infected with 2 × 109 CFU of C. rodentium, and colons were harvested at day 8 of infection (n= 5/group). C. rodentium CFUs in proximal colon (B), histopathology (C) and crypt length (D) in the distal colon (H&E). Data represent means ± SD and circles represent separate animals. Similar effects of SFB on C. rodentium colonization were observed in a separate experiment with C.B17 mice colonized with ASF with and without SFB.
To assess specifically the ability of SFB to provide protection, we colonized GF IQI mice with Jackson microbiota with or without SFB for 14 days. The mice were then infected orally with C. rodentium and pathogen colonization and disease were assessed at day 8 post-infection. Although some infection and disease were observed in both experimental groups, the presence of SFB in the gut prevented infiltration of the pathogen into the colonic wall (Figure 7B). In addition, SFB colonization ameliorated colonic inflammation as demonstrated by reduced epithelial hyperplasia and colon shortening in its presence (Figure 7C, 7D, S11, and data not shown). We thus conclude that the presence of SFB as a component of the commensal microbiota increases mucosal protection to infection with C. rodentium.
DISCUSSION
Commensal intestinal bacteria and regulation of Th17 cell differentiation
Commensal intestinal bacteria influence multiple metabolic and physiological functions of the host (Backhed et al., 2005; Turnbaugh et al., 2006), but they also have profound effects on the host immune system (Cash et al., 2006; Macpherson and Harris, 2004). For example, most rodent colitis models are dependent on the presence of microbiota (Elson et al., 2005; Sartor, 2008), whose products can also influence systemic immune responses (Mazmanian et al., 2005; Turnbaugh et al., 2006). The effects of intestinal bacteria on the immune system are considered to be the result of stimulation of innate immune “pattern recognition receptors”, but we are limited in our understanding of how individual bacteria influence the type and location of immune responses in gnotobiotic models or in the presence of other commensal microorganisms (Macpherson and Harris, 2004; Umesaki et al., 1999) (Kim et al., 2005). A notable exception was the recent demonstration that a polysaccharide product from the commensal bacterium Bacteroides fragilis can specifically induce systemic Th1 and mucosal regulatory T cell responses and protect mice from pathogen-induced colitis (Mazmanian et al., 2005; Mazmanian et al., 2008).
We recently discovered that the homeostasis of effector helper T cell populations in the gut is dependent on the composition of intestinal bacteria (Ivanov et al., 2008). Th17 cell induction was not controlled simply by the presence of high numbers of diverse bacterial species that activate major bacterial pattern-recognition pathways. Thus, colonization of mice with several defined bacterial species as well as with diverse microbiota from Jackson Laboratory B6 mice did not induce Th17 cell differentiation, in sharp contrast to the induction observed with bacteria from Th17 cell-sufficient Taconic Farms B6 mice (Ivanov et al., 2008). The identity of microorganisms that induce Th17 cells and the signaling mechanisms involved had remained important unresolved issues.
In this study, we have identified SFB as the first commensal bacterium that can induce accumulation in the gut of CD4+ T cells with a defined effector function. Colonization with a number of other species, including members of the SFB-related Clostridiaceae family, failed to induce Th17 cells. Our results are most consistent with a mechanism that requires unique features of a specific commensal species to trigger Th17 cell differentiation and/or accumulation in the lamina propria. Pathways activated by common bacterial patterns and shared by large classes of bacteria appear to be dispensable or redundant, as both MyD88/TRIF double deficient animals and RIP-2 mutant mice still possessed mucosal Th17 cells [(Atarashi et al., 2008; Ivanov et al., 2008) and Figure S12]. We previously reported that adenosine 5′-triphosphate (ATP) derived from commensal bacteria led to the differentiation of Th17 cells in the colonic LP (Atarashi et al., 2008). However, the ATP concentrations in the ileal and colonic luminal contents of SFB-mono mice were lower than those in mice gavaged with SPF feces (data not shown). Thus, SFB-mediated Th17 cell differentiation is likely to occur through a mechanism independent of TLR-, NOD-, and ATP-signaling.
SFB associate closely with epithelial cells in the terminal ileum. This interaction was reflected in the host genes induced after SFB colonization. Multiple epithelial cell-specific genes, as well as inflammatory response host genes, were up-regulated by the bacteria. Among these were the three inducible or “acute-phase” isoforms of SAA (A-SAA). SAA is highly induced during both acute and chronic inflammation. A-SAA expression is induced in hepatocytes in the liver and in macrophages and other cells in extrahepatic sites, including the intestine, by bacterial products and inflammatory cytokines, such as IL-6 and IL-1β (Uhlar and Whitehead, 1999). In addition to its role in the acute-phase response, SAA can induce IL-23 production by monocytes at concentrations that are orders of magnitude lower than the peak plasma concentration during an acute-phase response (He et al., 2006). In accordance with this, SAA induced transient production of IL-23 by LP DCs in vitro. We further demonstrated that SAA can act on LP DCs in vitro to induce Th17 cell differentiation, suggesting that an acute phase inflammation-like response, including induction of A-SAAs, is responsible for the SFB-mediated accumulation of Th17 cells in the intestine. Although the signaling pathways induced by A-SAA are currently unknown, it most likely acts on DCs and contributes to the establishment of a Th17 cell-inducing cytokine environment.
SFB and Th17 cell-mediated protection from pathogenic microorganisms
The identification of SFB as Th17 cell inducers in the intestine may have important implications for a better understanding of how components of the commensal microbiota contribute to host protection from microbial pathogens. It is well known that treatment with broad-spectrum antibiotics can result in outgrowth of intestinal pathogens, such as vancomycin-resistant Enterococcus (VRE) or Clostridium difficile, resulting in severe colitis. SFB colonization has been suggested to reduce replication in rabbits of enteropathogenic Escherichia coli (EPEC) and in rats of Salmonella enteritidis (Garland et al., 1982; Heczko et al., 2000). In mice, Th17 cell effector cytokines, such as IL-17 and IL-22, as well as IL-23, which is required for Th17 cell function, have been proposed to play protective roles in infections with Salmonella and Citrobacter rodentium (Curtis and Way, 2009). We found that colonization with SFB reduced the capacity of orally inoculated C. rodentium to grow and/or invade colonic tissue. Although we cannot at this point formally demonstrate that this protection is a direct result of Th17 cell induction, our data, taken together with results of recent studies (Kolls et al., 2008; Zheng et al., 2008), strongly suggest that SFB-induced Th17 cytokines, particularly IL-22, limit the growth of C. rodentium, at least in part through production of AMP’s such as RegIIIγ. While IL-22, IL-23, and RegIIIγ are required for host survival after C. rodentium infection (Mangan et al., 2006; Zheng et al., 2008), mice lacking SFB and Th17 cells survive despite increased bacterial growth. This may be because intestinal γδT cells, CD4+CD3− lymphoid tissue inducer (LTi)-like cells, and NK22 cells that also produce Th17 cytokines are present even in the absence of SFB and other microbiota. Contribution of these cells to SFB-independent anti-microbial defense may hence protect the host from lethal outgrowth of the pathogenic bacteria. Our results are also consistent with the report that a vancomycin-sensitive component of the commensal microbiota induces RegIIIγ in the mouse small intestine, thus reducing colonization by VRE and enhancing killing of the pathogen (Brandl et al., 2008). Future studies will be required to determine if vancomycin-sensitive SFB enhance mucosal protection from pathogenic VRE and other bacteria through the up-regulation of Th17 cells and anti-microbial peptides. Such studies will further test the hypothesis that specific commensal microbiota, by regulating the host immune system rather than by direct microbial competition, enhance protection from potentially harmful microbes.
Do SFB influence Th17 cell-mediated inflammatory disease?
Th17 cells are recognized to have significant roles in multiple mouse models of autoimmune disease, and there is accumulating evidence that they likewise contribute to human autoimmune disease pathogenesis (Hue et al., 2006; Langrish et al., 2005; Murphy et al., 2003; Yen et al., 2006). Mice with almost complete loss of Th17 cells due to the absence of RORγt are resistant to experimental autoimmune encephalomyelitis and colitis (Ivanov et al., 2006; Leppkes et al., 2009). In humans, polymorphisms in the gene encoding the IL-23 receptor are associated with both increased resistance and susceptibility to Crohn’s disease, and inhibition of the Th17 cell differentiation pathway has been reported to be an effective therapy for psoriasis (Duerr et al., 2006; Krueger et al., 2007).
Although Th17 cells are involved in multiple organ-specific inflammatory diseases, they are not normally present in such organs, and they are relatively scarce in secondary lymphoid tissues. However, Th17 cells are abundant in the intestinal lamina propria, and, as described in this study, their differentiation within and/or migration to this lymphoid-rich site is dependent on commensal microbes with specialized properties. There is evidence that the course of certain autoimmune diseases in humans and in animal models can be altered by treatment with antibiotics and probiotics and by restricting the complexity of the microbiota (O’Dell et al., 2006; Sartor, 2008). Indeed, in rodents, differential arthritogenic potential of different commensal microbiota components and dependence of spontaneous arthritis models on “cleanliness” of housing conditions have been reported (Severijnen et al., 1989; Simelyte et al., 2003). Moreover, K/BxN mice that have a genetic predisposition to spontaneous arthritis (Monach et al., 2008) fail to develop disease when kept in GF conditions, but do progress to arthritis when colonized with SFB (Wu et al., submitted). Our results thus raise the possibility that manipulation of the number of SFB that colonize the terminal ileum may alter the course of Th17 cell-associated autoimmune diseases.
If Th17 cells involved in organ-specific autoimmunity originate in the gut, then the question arises as to what is the antigenic specificity of such cells. It is not yet known if Th17 cells in the lamina propria are specific for intestinal microbiota. If they are mostly reactive with microbial products, then it may be surprising that similar numbers of Th17 cells are observed in mice monocolonized with SFB and in mice with a broad distribution of microbiota. Th17 cells specific for bacterial products may constitute a sufficiently broad repertoire to provide subsets that are cross-reactive with self-antigen. Alternatively, intestinal Th17 cells may be broadly specific for self-antigen, rather than bacterial products, but may normally be kept in check by mechanisms of peripheral tolerance. Signals from bacteria such as SFB may provide an adjuvant effect that polarizes such self-reactive T helper cells towards the Th17 lineage without tissue damage under the immune suppressive environment in the gut. Further studies on the repertoire and antigen specificity of Th17 cells and on the role of SFB in autoimmune disease models will be necessary to resolve these issues.
SFB represent the first example of a specific component of the commensal microbiota that induces a particular helper T cell population in the lamina propria. The elucidation of additional commensal bacteria involved in this or other immune pathways and of the mechanisms employed will undoubtedly lead to further understanding of the complex host-commensal interactions that shape our immunity and will allow for tailored therapeutic manipulation of these processes.
EXPERIMENTAL PROCEDURES
Mice and Bacterial Strains
B6 mice were obtained from Taconic Farms or Jackson Laboratory. Swiss-Webster germ-free and conventionally raised (SPF) mice were purchased from Taconic Farms. Germ-free IQI mice were purchased from Japan CLEA Inc. Mice mono-colonized with SFB or 46 strains of Clostridia were developed previously (Itoh and Mitsuoka, 1985; Umesaki et al., 1995). To generate Bacteroides-associated mice, 16 strains of Bacteroides (six strains of B. vulgatus, seven of B. acidifaciens group 1, and three of B. acidifaciens group 2), which were originally isolated from murine intestinal commensal bacteria (Miyamoto and Itoh, 2000), were cultured on Eggerth-Gangon agar (Nissui) in an anaerobic stainless steel jar and inoculated orally into germ-free IQI mice.
PhyloChip Analysis
Six week old Jackson B6 and Taconic B6 mice were purchased from the corresponding vendor and housed for 3 weeks in separate microisolator cages at the NYUSOM animal facility to equilibrate housing conditions, including bedding and diet. Sample collection, processing and PhyloChip analysis are described in detail in the Supplemental section.
16S rRNA Gene Quantitative PCR Analysis
Bacterial genomic DNA was isolated from fecal pellets as described in the Supplemental section. Quantitative PCR analysis was carried out as described in (Barman et al., 2008). Primer sequences for SFB and bacterial 16S rRNA genes as well as PCR conditions were as described in (Barman et al., 2008). For SFB, relative quantity was calculated by the ΔCt method and normalized by the presence of total bacteria (EUB primers), dilution and weight of the sample and presented as relative fold change to an external sample. Typical Ct values for SFB were ~20 cycles and for EUB ~11 cycles. Samples that were negative after 40 cycles were considered “not detected” (n.d.).
Gene Expression Analysis
RNA was prepared from terminal ileum as described (Ivanov et al., 2008). For microarray analysis, RNA was labeled and hybridized to GeneChip Mouse Genome 430 2.0 arrays following the Affymetrix protocols. Data were analyzed in GeneSpring GX10. Significant genes were selected based on p values smaller than 0.05 and fold change greater than 2. For enrichment analysis of biological process ontology, probe lists were analyzed in DAVID (Dennis et al., 2003; Huang da et al., 2009) and processes were selected based on p values smaller than 0.01.
Real-time RT-PCR (Q-PCR)
cDNAs were synthesized from RNA samples prepared with a RNeasy Mini Kit (QIAGEN) using M-MLV Reverse Transcriptase (Promega). Real-time RT-PCR was performed using the ABI 7300 real time PCR system. Serial dilutions of a standard were included for each gene to generate a standard curve and allow calculation of the input amount of cDNA for each gene. Values were then normalized by the amount of GAPDH in each sample. Primer sequences are reported in the Supplemental section.
Co-housing and Microbiota Reconstitution
Co-housing and microbiota reconstitutions were performed as described before (Ivanov et al., 2008). For inoculation of germ-free mice with SFB, fecal pellets were collected from SFB-monocolonized mice using sterilized test tubes in the vinyl-isolator and were preserved frozen under dry ice until immediately before oral administration. SFB colonizations were performed by oral gavage with 300-400 μl of suspension obtained by homogenizing the fecal pellets from SFB-mono donor mice in water. Control mice were gavaged with water or homogenates prepared from their own feces.
Cell Isolation and Flow Cytometry
Lamina propria lymphocyte (LPL) isolation and intracellular cytokine staining were performed as described before (Ivanov et al., 2008). Naive CD4+ T cells were purified from spleens using a CD4+CD62L+ T cell isolation kit II (Miltenyi Biotec; purity 95%). Anti-mouse RORγ monoclonal antibody conjugated to PE was purchased from eBioscience.
In vitro T-cell Differentiation
Naïve CD4+ T cells were cultured in 24-well plates at 2 × 105 cells/well for 4 days with MACS-purified LP CD11c+ cells (1 × 105/ well) and 1 μg/ml anti-CD3 antibody (BD Biosciences) in the presence or absence of recombinant human Apo-SAA (Peprotech). The cultured cells were harvested and restimulated with PMA and ionomycin for 3 h before analysis.
Electron Microscopy
Electron micrscopy was performed on 0.5-1 cm pieces from terminal ileum (immediately proximal to the ileal-cecal junction). Tissue processing for EM is described in the Supplemental section. The analysis was performed with a Zeiss Supra 55 FESEM.
C. rodentium Infection
IQI mice were inoculated with 200 μl of a bacterial suspension (1-2 × 109 CFU) by way of oral gavage. For the colony formation assays, proximal colons and MLNs were harvested and homogenized, and serially diluted homogenates were plated on MacConkey agar (Difco). For histological analysis, distal colons were fixed with 4% paraformaldehyde and analyzed after haematoxylin and eosin staining. For assessment of crypt depth, only crypts visible along the entire length of the distal colon were analyzed (20~30 crypts/mouse).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Littman laboratory for valuable discussions, Takeshi Egawa and Homer Boushey for their contribution to establishing the collaborative study, and Junichi Nishimura for technical assistance. We also thank Feng-Xia (Alice) Liang and Eric Roth from the NYU imaging core facility for performing transmission electron microscopy and for preparing samples for SEM. We also thank Jiri Zavadil and Agnes Viale and the genomic core facilities of NYU and Memorial Sloan Ketterring Cancer Center, respectively, for performing the array studies. We thank the staff at the Yakult Central Institute for gnotobiotic handling of the mice. The work was supported by Crohn’s and Colitis Foundation of America (I.I.I.) and Cancer Research Institute (N.M.) fellowships and by the Howard Hughes Medical Institute (D.R.L.), the Helen and Martin Kimmel Center for Biology and Medicine (D.R.L.), NIH grant AI33856 (D.R.L.), Grants-in-Aid for Scientific Research from PRESTO, the Ministry of Education, Culture, Sports, Science and Technology (K.H.), Senri Life Science Foundation and the Naito Foundation (K.H.). Part of this work was performed under the auspices of the US Department of Energy by the University of California, Lawrence Berkeley National Laboratory, under contract DE-AC02-05CH11231.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377–382. doi: 10.1016/j.smim.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie EL, Desantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect Immun. 1974;10:948–956. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, Fox JG. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elson CO, Cong Y, McCracken VJ, Dimmitt RA, Lorenz RG, Weaver CT. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Furlaneto CJ, Campa A. A novel function of serum amyloid A: a potent stimulus for the release of tumor necrosis factor-alpha, interleukin-1beta, and interleukin-8 by human blood neutrophil. Biochemical and biophysical research communications. 2000;268:405–408. doi: 10.1006/bbrc.2000.2143. [DOI] [PubMed] [Google Scholar]
- Garland CD, Lee A, Dickson MR. Segmented Filamentous Bacteria in the Rodent Small Intestine: Their Colonization of Growing Animals and Possible Role in Host Resistance to Salmonella. Microb Ecol. 1982:181–190. doi: 10.1007/BF02010451. [DOI] [PubMed] [Google Scholar]
- Gavin M, Rudensky A. Control of immune homeostasis by naturally arising regulatory CD4+ T cells. Curr Opin Immunol. 2003;15:690–696. doi: 10.1016/j.coi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Shepard LW, Chen J, Pan ZK, Ye RD. Serum amyloid A is an endogenous ligand that differentially induces IL-12 and IL-23. J Immunol. 2006;177:4072–4079. doi: 10.4049/jimmunol.177.6.4072. [DOI] [PubMed] [Google Scholar]
- Heczko U, Abe A, Finlay BB. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. The Journal of infectious diseases. 2000;181:1027–1033. doi: 10.1086/315348. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, Powrie F, Maloy KJ. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Mitsuoka T. Characterization of clostridia isolated from faeces of limited flora mice and their effect on caecal size when associated with germ-free mice. Laboratory animals. 1985;19:111–118. doi: 10.1258/002367785780942589. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Laboratory animals. 1993a;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Van der Heijden PJ, Stok W, Poelma FG, Koopman JP, Van den Brink ME, Bakker MH, Eling WM, Beynen AC. Apathogenic, intestinal, segmented, filamentous bacteria stimulate the mucosal immune system of mice. Infect Immun. 1993b;61:303–306. doi: 10.1128/iai.61.1.303-306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolls JK, McCray PB, Jr., Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat Rev Immunol. 2008;8:829–835. doi: 10.1038/nri2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman JP, Stadhouders AM, Kennis HM, De Boer H. The attachment of filamentous segmented micro-organisms to the distal ileum wall of the mouse: a scanning and transmission electron microscopy study. Laboratory animals. 1987;21:48–52. doi: 10.1258/002367787780740743. [DOI] [PubMed] [Google Scholar]
- Krueger GG, Langley RG, Leonardi C, Yeilding N, Guzzo C, Wang Y, Dooley LT, Lebwohl M. A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. The New England journal of medicine. 2007;356:580–592. doi: 10.1056/NEJMoa062382. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppkes M, Becker C, Ivanov II, Hirth S, Wirtz S, Neufert C, Pouly S, Murphy AJ, Valenzuela DM, Yancopoulos GD, et al. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of Mammals and Their Gut Microbes. Science. 2008 doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Itoh K. Bacteroides acidifaciens sp. nov., isolated from the caecum of mice. International journal of systematic and evolutionary microbiology. 2000;50(Pt 1):145–148. doi: 10.1099/00207713-50-1-145. [DOI] [PubMed] [Google Scholar]
- Monach PA, Mathis D, Benoist C. The K/BxN arthritis model. In: Coligan John E, et al., editors. Current protocols in immunology. 2008. Chapter 15, Unit 15 22. [DOI] [PubMed] [Google Scholar]
- Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dell JR, Elliott JR, Mallek JA, Mikuls TR, Weaver CA, Glickstein S, Blakely KM, Hausch R, Leff RD. Treatment of early seropositive rheumatoid arthritis: doxycycline plus methotrexate versus methotrexate alone. Arthritis Rheum. 2006;54:621–627. doi: 10.1002/art.21620. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Medzhitov R. Role of the innate immune system and host-commensal mutualism. Curr Top Microbiol Immunol. 2006;308:1–18. doi: 10.1007/3-540-30657-9_1. [DOI] [PubMed] [Google Scholar]
- Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- Seder RA, Paul WE. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu Rev Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- Severijnen AJ, van Kleef R, Hazenberg MP, van de Merwe JP. Cell wall fragments from major residents of the human intestinal flora induce chronic arthritis in rats. The Journal of rheumatology. 1989;16:1061–1068. [PubMed] [Google Scholar]
- Simelyte E, Rimpilainen M, Zhang X, Toivanen P. Role of peptidoglycan subtypes in the pathogenesis of bacterial cell wall arthritis. Annals of the rheumatic diseases. 2003;62:976–982. doi: 10.1136/ard.62.10.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. International journal of systematic bacteriology. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchinsky MB, Garaude J, Martin AP, Blander JM. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature. 2009;458:78–82. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. European journal of biochemistry / FEBS. 1999;265:501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.