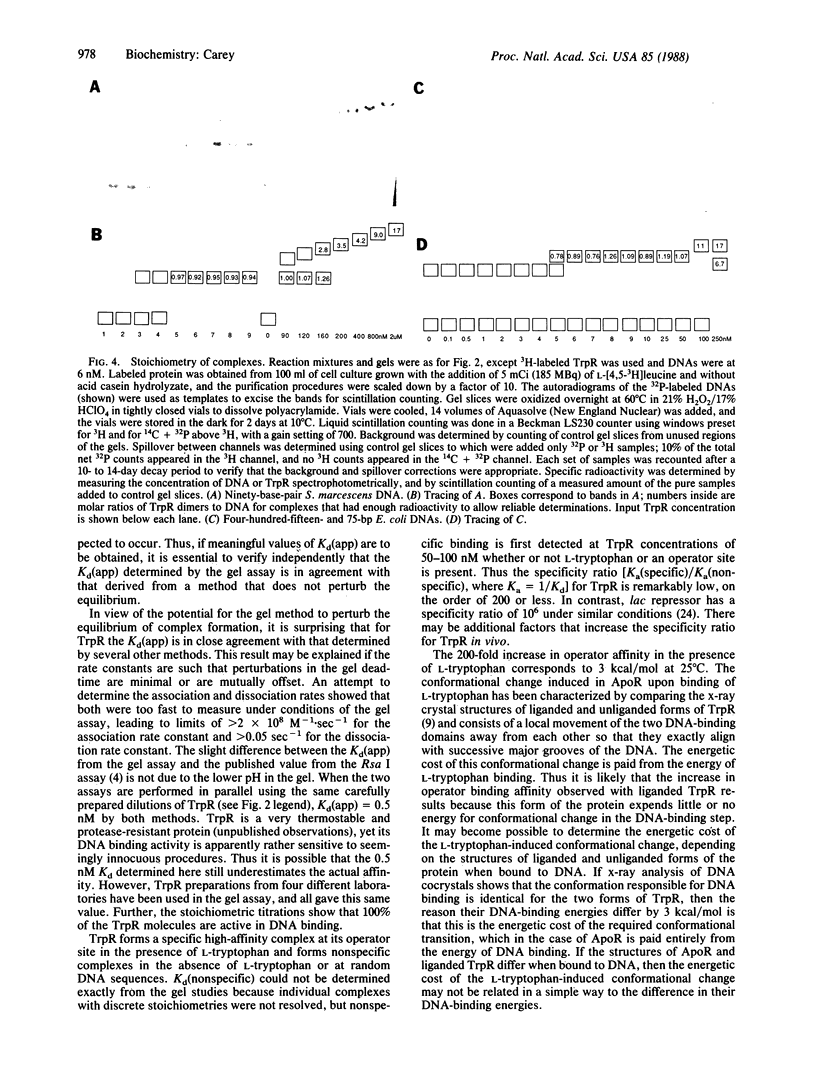
Abstract
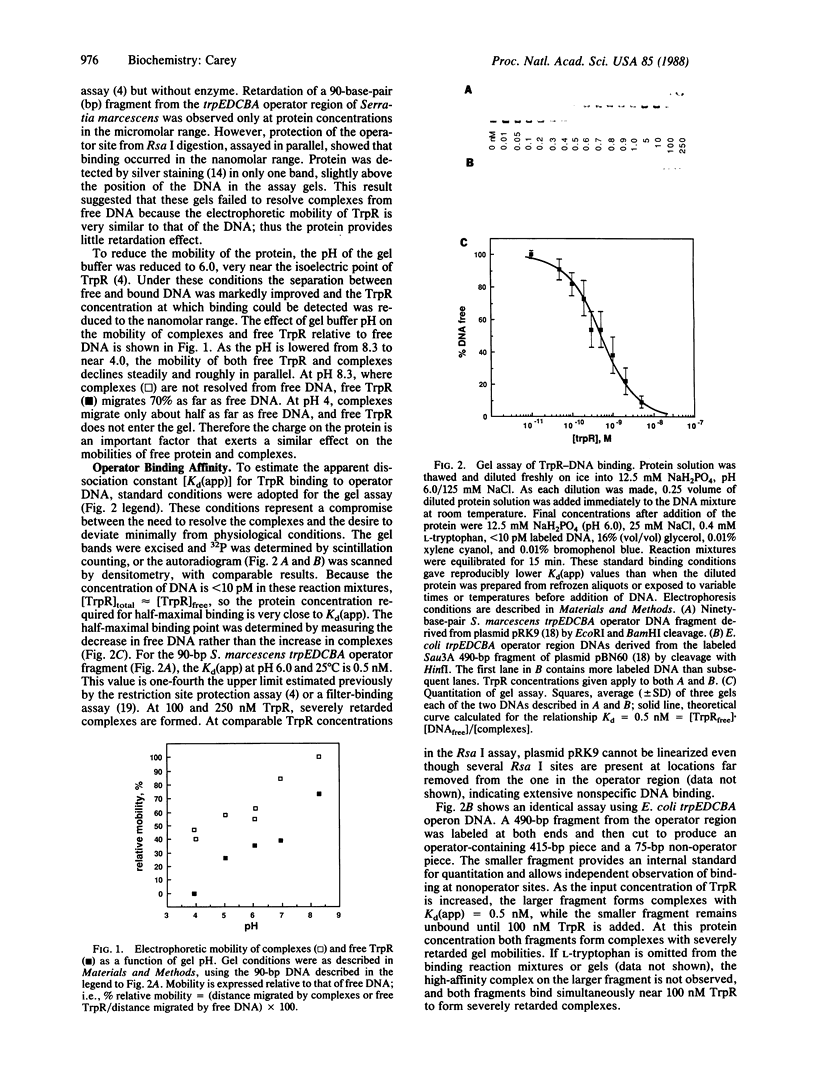
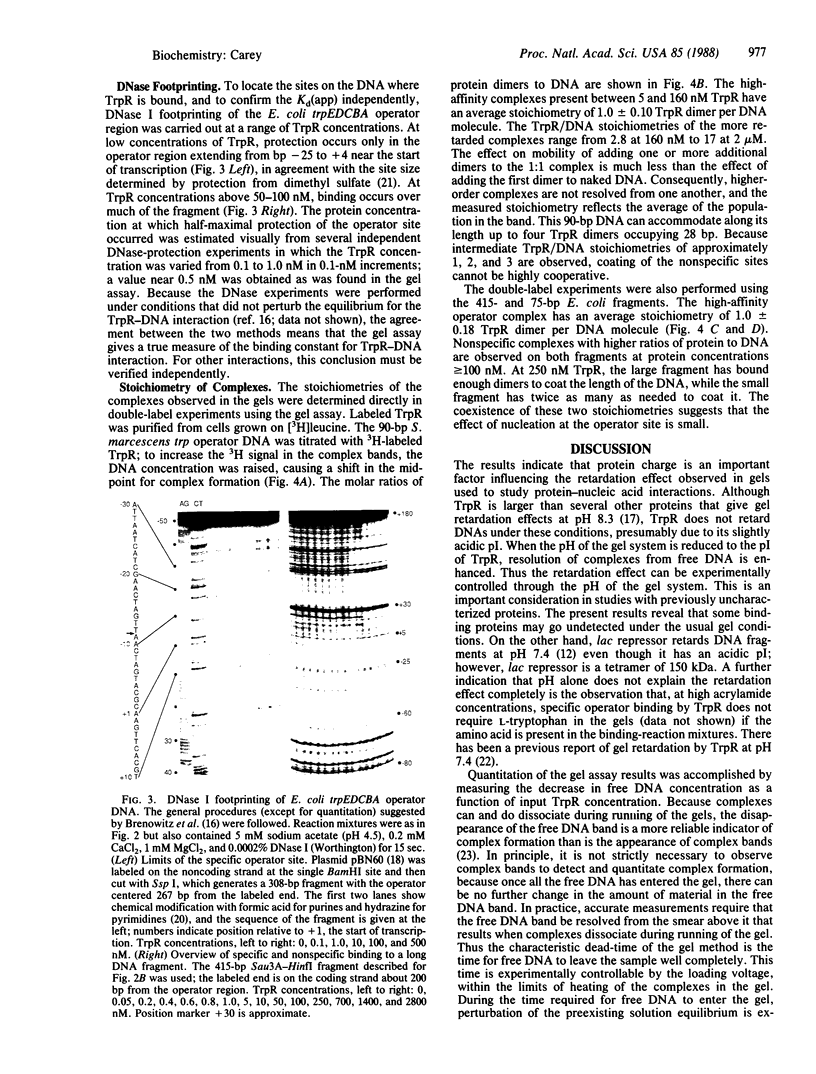
The affinity and stoichiometry of DNA binding by Escherichia coli trp repressor were studied by electrophoresis in nondenaturing gels. The ability of trp repressor to retard the electrophoretic mobility of an operator DNA fragment depends on the pH of the gel system. Above the pI of the protein, little retardation of DNA is observed, although complex formation can be detected by other assays. As the pH of the gel is lowered, retardation is enhanced. The apparent dissociation constant for the interaction between trp repressor and trpEDCBA operator fragments is 0.5 nM under the conditions used here. Nonspecific binding occurs with only about 200-fold weaker affinity. The stoichiometries of specific and nonspecific complexes were determined directly by using trp repressor labeled in vivo. High-affinity operator binding requires a single dimer of trp repressor. DNase I-protection analysis ("footprinting") was used to confirm the dissociation constants and to locate the binding site.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arvidson D. N., Bruce C., Gunsalus R. P. Interaction of the Escherichia coli trp aporepressor with its ligand, L-tryptophan. J Biol Chem. 1986 Jan 5;261(1):238–243. [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Dingman C. W., Peacock A. C. Electrophoretic characterization of bacterial polyribosomes in agarose-acrylamide composite gels. J Mol Biol. 1969 Apr 14;41(1):139–147. doi: 10.1016/0022-2836(69)90131-4. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Miguel A. G., Gunsalus G. L. Intracellular Trp repressor levels in Escherichia coli. J Bacteriol. 1986 Jul;167(1):272–278. doi: 10.1128/jb.167.1.272-278.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Yanofsky C. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7117–7121. doi: 10.1073/pnas.77.12.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydock P. V., Somerville R. L. Trp holorepressor-trp operator interaction studied by protein distribution analysis. Biochem Biophys Res Commun. 1984 Mar 30;119(3):926–932. doi: 10.1016/0006-291x(84)90862-3. [DOI] [PubMed] [Google Scholar]
- Joachimiak A., Kelley R. L., Gunsalus R. P., Yanofsky C., Sigler P. B. Purification and characterization of trp aporepressor. Proc Natl Acad Sci U S A. 1983 Feb;80(3):668–672. doi: 10.1073/pnas.80.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Yanofsky C. Mutational studies with the trp repressor of Escherichia coli support the helix-turn-helix model of repressor recognition of operator DNA. Proc Natl Acad Sci U S A. 1985 Jan;82(2):483–487. doi: 10.1073/pnas.82.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Yanofsky C. Trp aporepressor production is controlled by autogenous regulation and inefficient translation. Proc Natl Acad Sci U S A. 1982 May;79(10):3120–3124. doi: 10.1073/pnas.79.10.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klig L. S., Crawford I. P., Yanofsky C. Analysis of trp repressor-operator interaction by filter binding. Nucleic Acids Res. 1987 Jul 10;15(13):5339–5351. doi: 10.1093/nar/15.13.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. P., Yanofsky C. Plasmids containing the trp promoters of Escherichia coli and Serratia marcescens and their use in expressing cloned genes. Methods Enzymol. 1983;101:155–164. doi: 10.1016/0076-6879(83)01011-3. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Oppenheim D. S., Bennett G. N., Yanofsky C. Escherichia coli RNA polymerase and trp repressor interaction with the promoter-operator region of the tryptophan operon of Salmonella typhimurium. J Mol Biol. 1980 Dec 5;144(2):133–142. doi: 10.1016/0022-2836(80)90029-7. [DOI] [PubMed] [Google Scholar]
- Paluh J. L., Yanofsky C. High level production and rapid purification of the E. coli trp repressor. Nucleic Acids Res. 1986 Oct 24;14(20):7851–7860. doi: 10.1093/nar/14.20.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Rose J. K., Squires C. L., Yanofsky C., Yang H. L., Zubay G. Regulation of in vitro transcription of the tryptophan operon by purified RNA polymerase in the presence of partially purified repressor and tryptophan. Nat New Biol. 1973 Oct 3;245(144):133–137. doi: 10.1038/newbio245133a0. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Otwinowski Z., Joachimiak A., Lawson C. L., Sigler P. B. The three-dimensional structure of trp repressor. 1985 Oct 31-Nov 6Nature. 317(6040):782–786. doi: 10.1038/317782a0. [DOI] [PubMed] [Google Scholar]
- Squires C. L., Lee F. D., Yanofsky C. Interaction of the trp repressor and RNA polymerase with the trp operon. J Mol Biol. 1975 Feb 15;92(1):93–111. doi: 10.1016/0022-2836(75)90093-5. [DOI] [PubMed] [Google Scholar]
- Takeda Y., Ohlendorf D. H., Anderson W. F., Matthews B. W. DNA-binding proteins. Science. 1983 Sep 9;221(4615):1020–1026. doi: 10.1126/science.6308768. [DOI] [PubMed] [Google Scholar]
- Zhang R. G., Joachimiak A., Lawson C. L., Schevitz R. W., Otwinowski Z., Sigler P. B. The crystal structure of trp aporepressor at 1.8 A shows how binding tryptophan enhances DNA affinity. Nature. 1987 Jun 18;327(6123):591–597. doi: 10.1038/327591a0. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Gunsalus R. P., Brown K. D., Yanofsky C. Structure and regulation of aroH, the structural gene for the tryptophan-repressible 3-deoxy-D-arabino-heptulosonic acid-7-phosphate synthetase of Escherichia coli. J Mol Biol. 1981 Jan 5;145(1):47–73. doi: 10.1016/0022-2836(81)90334-x. [DOI] [PubMed] [Google Scholar]