Abstract
Recently, we reported the discovery of several potential rodent reservoirs of hantaviruses in western (Holochilus chacarius) and eastern Paraguay (Akodon montensis, Oligoryzomys chacoensis, and O. nigripes). Comparisons of the hantavirus S- and M-segments amplified from these four rodents revealed significant differences from each another and from other South American hantaviruses. The ALP strain from the semiarid Chaco ecoregion clustered with Leguna Negra and Rio Mamore (LN/RM), whereas the BMJ-ÑEB strain from the more humid lower Chaco ecoregion formed a clade with Oran and Bermejo. The other two strains, AAI and IP37/38, were distinct from known hantaviruses. With respect to the S-segment sequence, AAI from eastern Paraguay formed a clade with ALP/LN/RM, but its M-segment clustered with Pergamino and Maciel, suggesting a possible reassortment. AAI was found in areas experiencing rapid land cover fragmentation and change within the Interior Atlantic Forest. IP37/38 did not show any strong association with any of the known hantavirus strains.
Introduction
Hantaviruses are enzootic viruses that can maintain persistent infections in their natural hosts without apparent disease symptoms in these rodents.1 More than 20 different hantavirus species are recognized, and each is predominantly associated with one rodent species or a few very closely related species.2,3 In nature, transmission of hantavirus between rodents within a species is thought to be primarily through aggressive behavior and exposure to saliva and excreta.4–6 Laboratory studies of virus transmission among rodents of the same species show that an infected rodent can transmit virus horizontally to another rodent within the same cage or through infected bedding.7–11 However, to our knowledge it has not been shown that hantavirus can be transmitted between different rodent species in the laboratory, which suggests that the coevolution of each virus with its rodent host may be the reason that observations of reassortments in nature are rare12,13 and are difficult to obtain in the laboratory.14,15 However, it also plausible that the geographic and habitat restrictions of these rodents may simply limit overlap and interactions among hantavirus strains and their rodent reservoirs.16 Very little is known about potential routes of transmission of hantavirus from rodent to human outside of inhalation of aerosolized virus from rodent excreta, which is thought to be the main route.17,18 In Argentina, person to person transmission has been reported.19 Give the distribution of hantaviruses and that human cases are most often linked to outbreaks, it would seem that additional factors, perhaps environmental, contribute to effective transmission. When transmitted to humans, several members of the genus cause deadly illnesses such as hemorrhagic fever with renal syndrome (HFRS)20 and hantavirus pulmonary syndrome (HPS).21 Old World hantaviruses cause HFRS in Asia and Europe, whereas New World hantaviruses cause HPS in the Americas. The pattern of disease follows the geographical distribution of the habitat preference of the rodent.
Since the initial discovery in 1993 of Sin Nombre (SN) virus, an etiologic agent of HPS in the United States,22 several new hantaviruses and hantavirus sequences have been identified from Sigmodontinae throughout the Americas.22,23 In contrast with North America, studies of hantaviruses in Latin America have revealed an increasingly complex picture of their ecology. Strikingly, numerous rodent species have been identified to harbor unique strains of Hantavirus in Argentina,24,25 Bolivia,26 Brazil,27 Chile,28 Costa Rica,29 Panama,30 Peru,31 and Venezuela.32 In Paraguay, Calomys laucha was discovered as the rodent reservoir for Laguna Negra (LN) virus, which was responsible for the outbreak of HPS in the western region of Paraguay or Chaco.33 We have recently reported additional rodent species with serological evidence of a hantavirus infection in Paraguay34 in Akodon azarae, A. montensis, Bibimys chacoensis, Graomys griseoflavus, Holochilus chacarius, Nectomys squamipes, Oligoryzomys chacoensis, O. fornesi, O. nigripes, and an unidentified Oryzomys species. The discovery of hantaviruses in rodents in three areas of eastern Paraguay was surprising, because cases of HPS have only been reported in western Paraguay. However, HPS cases have been confirmed in eastern Paraguay in 2005 and 2006, suggesting the rodents in this area can transmit a HPS to humans.
To map the phylogenetic and geographical relationships of these hantaviruses and their rodent hosts in Paraguay, we amplified hantaviral cDNA from the lung tissues of four species, including H. chacarius collected in the Chaco region in western Paraguay and A. montensis, O. chacoensis, and O. nigripes collected in eastern Paraguay. Hantaviruses contain three negative-sense single-stranded RNA genome segments L, M, and S, which encode the RNA-dependent RNA polymerase, the G1 and G2 envelope glycoproteins, and the nucleocapsid protein, respectively. Phylogenetic relationships were reconstructed from S- and M-segment sequences. Herein, we present the deduced amino acid homologies and compare them with those from all available North and South American hantavirus sequences. Furthermore, we report phylogenetic relationships of the sequences that were amplified and cloned from the S- and M-segments within the context of the genetic diversity of the known Paraguayan and South American hantaviruses. To assist with the phylogenetic analysis, cDNA was made from the M-segment of Rio Mamore (RM) virus from Bolivia26 and Cano Delgadito (CD) virus,32 which have not been previously reported. The phylogenetic and geographical relationships of these Paraguayan hantavirus sequences shed light into their evolution in Paraguay and suggest that host switching and reassortment along with Paraguay's geographic features may have played a role in their evolution and distribution.
Materials and Methods
Rodent samples, RNA isolation, and nested reverse transcriptase-polymerase chain reaction
As reported previously,34 rodent samples were obtained from an inventory of small mammals of Paraguay. Five RNA-positive rodent lung tissues were provided by the Museum of Texas Tech University (TTU). We isolated RNA from H. chacarius (TK62276), which harbored the Alto Paraguay strain (ALP) in western Paraguay. In eastern Paraguay in the department of Ñeembucu, we isolated RNA from O. chacoensis (TK64399), which harbored the Bermejo-Ñeembucu strain (BMJ-ÑEB), and in the department of Itapúa, we isolated RNA from A. montensis (TK65816), which harbored the Ape Aime-Itapúa strain (AAI) and from two O. nigripes (TK65937, TK65938), which harbored the Itapúa strains (IP37 strain; IP38 strain). TK numbers are the catalog numbers for the individual rodents in the TTU frozen tissue collections. Hereafter, we will refer to each of these strains identified by their location rather than by their strain identification.
Partial M- and S-genome segments were amplified by nested reverse transcriptase-polymerase chain reaction (RT-PCR).35 We amplified complete open reading frame of the S-segment of ALP, ÑEB, IP37, IP38, and 1038 nt from AAI. The regions cloned for each strain were 1944 nt of ALP (TK62276, aligned to LN from 28 to 1881), 1887 nt of BMJ-ÑEB (TK64399, aligned to LN from 28 to 1881), 1038 nt of AAI (TK65816 aligned to LN from 28 to 1065), 1853 nt of IP37 (TK65937, aligned to LN from 28 to 1881), and 1853 nt of IP38 (TK65938, aligned to LN from 28 to 1881). For the M-segment, identical regions were cloned for each sample, 439 nt from the G1 region (1236-1674 nt based on LN virus), and 575 nt from the G2 region of the M genome segment (2234-2808 nt based on LN virus). These regions were chosen because they are the most commonly reported sequences in GenBank and thus this region allows for the maximal number of comparisons. Briefly, 0.1 g of lung tissue was ground in a 1.5-mL microfuge tube containing 1 mL of Trizol (Invitrogen, Bethesda, MD) with a disposable tissue grinder (Fisher, Atlanta, GA). Total RNA was extracted from Vero E6 cells, infected with CD or RM virus in a T25 flask, with 2 mL of Trizol. The extracted RNA pellet was diluted with 10 μL of RNase-free distilled water and subjected to RT-PCR with S- and M-segment outer generic primers (available on request) using the One-Step RT-PCR kit (Invitrogen, San Diego, CA). Degenerate S-segment outer generic primers were selected from a consensus region chosen from an alignment of seven different American hantaviruses as reported previously.34 Degenerate M-segment outer generic primers to the G1 and G2 regions of the M segment were selected from the conserved region identified by an alignment of Andes (AND) virus, Bayou (BAY) virus, Blue River (BR) virus, LN virus, Lechiguanas (LEC) virus, Oran (ORN) virus, and Sin Nombre (SN) virus (strain CC107) using Vector NTI software package, version 7.0 (Informax, Bethesda, MD). Two microliters of each amplicon from the first RT-PCR was further amplified by PCR, using the PCR Core Kit (Roche, Indianapolis, IN), with the generic inner primers that were specific for the S, G1, and G2 regions of the M-segment. The initial and second rounds of amplification were performed for one cycle of 45 minutes at 45°C, followed by 35 cycles at 94°C for 30 seconds; at 50°C for 30 seconds; and at 72°C for 45 seconds. PCR was conducted for 35 cycles at 94°C for 30 seconds and at 72°C for 30 seconds. Amplicons were separated in 1.2% agarose gels in Tris-Acetate-EDTA (TAE) buffer and visualized by ethidium bromide staining.
Cloning, nucleotide sequencing, and genetic analysis
The cDNA from each sample was extracted from an agarose gel, purified (Bio101, La Jolla, CA), and ligated into pGEM-T (Promega, Madison, WI). T7 and SP6 primers were used to sequence three clones from each amplicon in both directions using the BigDye 3.0 terminator sequencing system (ABI, Foster City, CA) as described by the manufacturer. Nucleotide and deduced amino acids sequences of the S- and M-segment were obtained for all available American hantaviruses from GenBank (Table 1). Sequence alignments and comparisons were performed with the AlignX programs from the Vector NTI software package, version 7.0. Phylogenetic analysis was performed using the maximum likelihood program from PAUP software version 4.0 b10 (Sinauer Associates, Sunderland, MA). These analyses excluded the primer regions for PCR amplification. Phylogenetic trees were obtained using the heuristic search method with transversions weighted four times transitions. Gaps were treated as missing data or as a fifth character. Bootstrap confidence limits were obtained by 1,000 heuristic search repetitions.
Table 1.
North and South American hantavirus sequences analyzed in this study
| Virus | Abbreviation | Source of virus | Origin | GenBank accession number | References |
|---|---|---|---|---|---|
| Sin Nombre CC107 | SN | Peromyscus maniculatis | S.W. United States | L33474 [M], L33683 [S] | 45 |
| New York 1 | NY | Peromyscus leucopus | E. United States | U36802 [M], U29210 [S] | 46, 47 |
| Blue River | BR | Peromyscus leucopus | Cen. United States | AF030551 [M] | |
| Black Creek Canal | BCC | Sigmodon hispidus | S.E. United States | L39950 [M], L39949 [S] | 49 |
| Bayou | BAY | Human | S.E. United States | L36930 [M], L36929 [S] | 50 |
| Mule Shoe | MUL | Sigmodon spp | S. United States | U54575 [S] | 51 |
| El Moro Canyon | EMC | Reithrodontomys megalotis | S.W. United States | U26828 [M], U11427 [S] | 53 |
| Rio Segundo | RS | Reithrodontomys mexicanus | Costa Rica | U18100 [S] | |
| Laguna Negra | LN | Calomys laucha | Paraguay | AF005728 [M], AF005727 [S] | 33 |
| Rio Mamore | RM | Oligoryzomys microtis | Bolivia | AY953443 [M], AY953445 [M], U52136 [S] | 26 |
| Cano Delgadito | CD | Sigmodon alstoni | Venezuela | AY953442 [M], AY953444 [M], AF000140 [S] | 32 |
| Maciel | MAC | Necromys benefactus | Argentina | AF028027 [M], AF482716 [S] | 24, 36 |
| Pergamino | PRG | Akodon azarae | Argentina | AF028028 [M], AF482717 [S] | 24, 36 |
| Bermejo | BMJ | Oligoryzomys chacoensis | Argentina | AF028025 [M], AF482713 [S] | 24, 36 |
| Lechiguanas | LEC | Oligoryzomys flavescens | Argentina | AF028022 [M], AF482714 [S] | 24, 36 |
| Andes Chile-9717869 | CHL | Oligoryzomys longicaudatus | Chile | AF291703 [M], AF291702 [S] | 54 |
| Andes AH-1 | AH1 | Human | Argentina | AF324901 [M], AF324902 [S] | 55 |
| Oran (“Andes Norte”) | ORN | Oligoryzomys longicaudatus | Argentina | AF028023 [M], AF482715 [S] | 24, 36 |
| Andes | AND | Human | Argentina | AF004660 [S] | 56 |
| Castelo dos Sonho | CAS | Human | Brazil | AF307326 [M] | 57 |
| Araraquara | ARA | Human | Brazil | AF307327 [M] | 57 |
| Alto Paraguay (TK62276) | ALP | Holochilus chacoensis | W. Paraguay | AY515597 [M], AY515602 [M], [S] | |
| Bermejo-Ñeembucu (TK64399) | BMJ-ÑEB | Oligoryzomys chacoensis | E. Paraguay | AY515598 [M], AY515603 [M], [S] | |
| Ape Aime Itapua (TK65816) | AAI | Akodon montensis | E. Paraguay | AY515599 [M], AY515604 [M], [S] | |
| Itapua 37 (TK65937) | IP37 | Oligoryzomys nigripes | E. Paraguay | AY515600 [M], AY515605 [M], [S] | |
| Itapua 38 (TK65938) | IP38 | Oligoryzomys nigripes | E. Paraguay | AY515601 [M], AY515606 [M], [S] |
Results
Sequence similarity of the G1 and G2 regions of the M-segment
Nucleotide sequence and amino acid similarities of the G1 (Table 2) and G2 (Table 3) regions for the five Paraguayan hantavirus sequences from the four different Paraguayan rodent species (Alto Paraguay [ALP], Bermejo-Ñeembucú [BMJ-ÑEB], Ape Aime-Itapúa [AAI], Itapúa37 [IP37], and Itapúa38 [IP38]) were compared with all North and South American hantavirus sequences available from GenBank (Table 1). IP37 and IP38 sequences were amplified from RNA extracted from lung tissues of O. nigripes, whereas each of the others came from unique rodent species (H. chacarius: ALP, O. chacoensis: ÑEB, A. montensis: AAI strain). LN virus showed 74–81% nucleotide (nt) and 88–93% amino acid (aa) sequence similarity to the five sequences from the G1 segment (Table 2). The nucleotide sequence and amino acid sequence similarity between IP37 and IP38 viruses were 98% and 99%, respectively. This is not surprising given that both were obtained from O. nigripes collected in the same locale in Itapúa. The other hantavirus sequence identified in Itapúa, AAI from A. montenis, was distinct from IP37 and IP38 (77% nt; 89–90% aa). Nucleotide and deduced amino acid sequence similarities between hantaviruses from Paraguay and other South American hantaviruses showed a wide range in nucleotide (71–88%) and amino acid (82–99%) similarity (Table 2). In contrast, nucleotide and deduced amino acid sequence similarities of the hantavirus sequences from Paraguay with the North American hantaviruses were 72–75% (nt) and 81–88% (aa). For comparison, the nucleotide and deduced amino acid sequence similarities between North American hantaviruses were 72–79% and 84–97%, respectively.
Table 2.
Sequence similarity of a G1 region within the M-segment among American hantaviruses
| Paraguayan | South American | North American | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALP | ÑEB | AAI | IP37 | IP38 | LN | RM | CHL | AH1 | ORN | LEC | CD | BR | EMC | NY | SN | BAY | BCC | |
| ALP | – | 75 | 75 | 76 | 76 | 81 | 76 | 77 | 76 | 77 | 76 | 73 | 73 | 72 | 72 | 73 | 72 | 73 |
| BMJ ÑEB | 89 | – | 78 | 80 | 79 | 74 | 76 | 78 | 78 | 82 | 88 | 71 | 75 | 72 | 73 | 76 | 75 | 74 |
| AAI | 90 | 90 | – | 77 | 77 | 75 | 74 | 78 | 77 | 78 | 78 | 74 | 75 | 72 | 72 | 72 | 73 | 74 |
| IP37 | 92 | 94 | 90 | – | 98 | 75 | 76 | 78 | 78 | 80 | 77 | 76 | 75 | 73 | 74 | 74 | 74 | 76 |
| IP38 | 91 | 93 | 89 | 99 | – | 75 | 76 | 78 | 78 | 78 | 77 | 75 | 74 | 73 | 74 | 73 | 73 | 76 |
| LN | 93 | 88 | 90 | 89 | 88 | – | 78 | 73 | 74 | 76 | 75 | 74 | 75 | 73 | 77 | 75 | 72 | 75 |
| RM | 95 | 90 | 88 | 92 | 91 | 94 | – | 77 | 78 | 77 | 74 | 74 | 75 | 74 | 75 | 74 | 71 | 72 |
| CHL | 90 | 93 | 90 | 92 | 92 | 87 | 89 | – | 93 | 76 | 77 | 74 | 72 | 70 | 71 | 72 | 73 | 72 |
| AH1 | 90 | 94 | 90 | 93 | 92 | 88 | 90 | 99 | – | 76 | 79 | 74 | 72 | 70 | 73 | 73 | 73 | 72 |
| ORN | 89 | 98 | 90 | 94 | 93 | 88 | 90 | 92 | 92 | – | 80 | 73 | 74 | 71 | 72 | 72 | 73 | 74 |
| LEC | 89 | 99 | 90 | 94 | 93 | 88 | 90 | 93 | 94 | 96 | – | 71 | 75 | 71 | 73 | 74 | 75 | 73 |
| CD | 86 | 82 | 83 | 85 | 84 | 84 | 86 | 82 | 83 | 82 | 82 | – | 74 | 73 | 72 | 73 | 75 | 72 |
| BR | 86 | 87 | 85 | 88 | 87 | 88 | 87 | 86 | 87 | 86 | 87 | 86 | – | 72 | 78 | 79 | 73 | 78 |
| EMC | 84 | 81 | 82 | 81 | 82 | 83 | 83 | 82 | 83 | 79 | 80 | 82 | 87 | – | 72 | 76 | 73 | 73 |
| NY | 86 | 85 | 87 | 86 | 85 | 86 | 86 | 86 | 86 | 84 | 85 | 84 | 93 | 88 | – | 77 | 74 | 76 |
| SN | 86 | 86 | 86 | 88 | 87 | 87 | 86 | 88 | 88 | 85 | 86 | 86 | 94 | 87 | 97 | – | 74 | 77 |
| BAY | 86 | 84 | 82 | 86 | 86 | 86 | 85 | 84 | 85 | 84 | 83 | 88 | 86 | 86 | 87 | 88 | – | 76 |
| BCC | 84 | 84 | 84 | 86 | 85 | 84 | 83 | 83 | 84 | 84 | 83 | 84 | 86 | 84 | 89 | 88 | 90 | – |
Table 3.
Sequence similarity of a G2 region within the M-segment among American hantaviruses
| Paraguayan | South American | North American | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALP | ÑEB | AAI | IP37 | IP38 | LN | RM | CHL | AH1 | ORN | LEC | MAC | PRG | BMJ | ARA | CAS | CD | BAY | BCC | BR | EMC | NY | SN | |
| ALP | – | 77 | 78 | 79 | 80 | 79 | 81 | 79 | 79 | 79 | 77 | 78 | 78 | 76 | 77 | 78 | 76 | 76 | 76 | 76 | 77 | 78 | 75 |
| BMJ ÑEB | 94 | – | 79 | 82 | 82 | 78 | 79 | 82 | 82 | 83 | 89 | 80 | 82 | 91 | 81 | 81 | 76 | 75 | 76 | 78 | 74 | 75 | 79 |
| AAI | 93 | 94 | – | 78 | 79 | 80 | 77 | 78 | 78 | 79 | 81 | 80 | 84 | 80 | 79 | 79 | 75 | 76 | 77 | 76 | 76 | 76 | 78 |
| IP37 | 96 | 98 | 94 | – | 99 | 78 | 78 | 81 | 80 | 82 | 80 | 77 | 82 | 82 | 79 | 81 | 75 | 74 | 76 | 80 | 77 | 79 | 77 |
| IP38 | 96 | 98 | 94 | 100 | – | 78 | 78 | 80 | 80 | 82 | 80 | 76 | 82 | 83 | 79 | 81 | 75 | 75 | 76 | 80 | 76 | 79 | 77 |
| LN | 94 | 94 | 92 | 93 | 93 | – | 81 | 80 | 79 | 79 | 78 | 78 | 82 | 78 | 77 | 81 | 75 | 76 | 77 | 78 | 75 | 75 | 78 |
| RM | 95 | 94 | 91 | 94 | 94 | 96 | – | 79 | 80 | 80 | 79 | 76 | 79 | 78 | 77 | 79 | 72 | 77 | 77 | 78 | 75 | 77 | 73 |
| CHL | 94 | 98 | 93 | 98 | 98 | 93 | 94 | – | 94 | 81 | 80 | 80 | 79 | 80 | 82 | 80 | 75 | 76 | 77 | 79 | 74 | 79 | 77 |
| AH1 | 94 | 98 | 93 | 98 | 98 | 93 | 93 | 99 | – | 79 | 80 | 81 | 78 | 79 | 81 | 81 | 73 | 77 | 76 | 77 | 74 | 78 | 76 |
| ORN | 94 | 99 | 94 | 98 | 98 | 94 | 94 | 98 | 98 | – | 82 | 80 | 79 | 83 | 80 | 82 | 73 | 75 | 76 | 77 | 76 | 75 | 76 |
| LEC | 95 | 99 | 94 | 99 | 99 | 94 | 95 | 99 | 98 | 99 | – | 79 | 81 | 88 | 80 | 80 | 75 | 75 | 75 | 78 | 75 | 77 | 80 |
| MAC | 93 | 93 | 95 | 93 | 93 | 93 | 92 | 92 | 93 | 93 | 93 | – | 78 | 80 | 79 | 80 | 75 | 75 | 76 | 77 | 74 | 72 | 75 |
| PRG | 94 | 96 | 97 | 96 | 96 | 93 | 93 | 95 | 95 | 96 | 96 | 94 | – | 81 | 79 | 80 | 75 | 73 | 76 | 80 | 76 | 77 | 76 |
| BMJ | 92 | 99 | 93 | 98 | 98 | 93 | 94 | 98 | 97 | 98 | 99 | 92 | 95 | – | 80 | 80 | 75 | 74 | 77 | 78 | 75 | 75 | 77 |
| ARA | 96 | 95 | 94 | 95 | 95 | 91 | 91 | 95 | 94 | 95 | 96 | 93 | 95 | 95 | – | 81 | 74 | 76 | 76 | 77 | 75 | 75 | 75 |
| CAS | 96 | 98 | 94 | 97 | 97 | 94 | 94 | 97 | 97 | 98 | 98 | 93 | 96 | 97 | 95 | – | 76 | 75 | 78 | 80 | 76 | 77 | 78 |
| CD | 87 | 84 | 86 | 85 | 85 | 84 | 83 | 84 | 85 | 84 | 85 | 87 | 86 | 84 | 85 | 86 | – | 72 | 75 | 75 | 76 | 74 | 76 |
| BAY | 88 | 87 | 86 | 87 | 87 | 87 | 87 | 87 | 87 | 87 | 87 | 87 | 87 | 86 | 85 | 88 | 84 | – | 80 | 76 | 76 | 76 | 76 |
| BCC | 86 | 86 | 85 | 86 | 86 | 87 | 86 | 85 | 85 | 85 | 86 | 86 | 85 | 85 | 85 | 86 | 82 | 93 | – | 78 | 78 | 76 | 75 |
| BR | 90 | 92 | 90 | 91 | 91 | 89 | 89 | 91 | 91 | 92 | 92 | 89 | 93 | 91 | 89 | 92 | 86 | 90 | 88 | – | 77 | 81 | 83 |
| EMC | 87 | 87 | 87 | 87 | 87 | 87 | 86 | 86 | 87 | 87 | 87 | 87 | 88 | 86 | 88 | 86 | 84 | 87 | 86 | 87 | – | 73 | 74 |
| NY | 90 | 90 | 90 | 90 | 90 | 88 | 88 | 89 | 89 | 90 | 90 | 87 | 91 | 89 | 87 | 91 | 86 | 88 | 86 | 96 | 86 | – | 81 |
| SN | 92 | 91 | 89 | 91 | 91 | 89 | 89 | 90 | 91 | 91 | 91 | 89 | 92 | 90 | 88 | 92 | 87 | 91 | 89 | 98 | 88 | 97 | – |
Nucleotide sequence comparisons of the G2 region among the five hantavirus strains from Paraguay showed a slightly higher similarity than those of the G1 region (77–82% nt and 93–98% aa; Table 3). The nucleotide sequence similarity and amino acid sequence similarities between IP37 and IP38 were 99% and 100%, respectively, clearly supporting the idea that these sequences were from one virus circulating in the same rodent species, O. nigripes. The amino acid similarities of AAI with these two strains were 94%, which suggests this sequence is from a distinct strain of hantavirus. Nucleotide and deduced amino acid sequence similarities of hantaviruses from Paraguay to other South American hantaviruses were 75–91% (nt) and 84–99% (aa), respectively. Nucleotide and deduced amino acid sequence similarities between hantaviruses from Paraguay and North America were 74–80% and 85–92%. Nucleotide and deduced amino acids sequence similarity among North American hantaviruses ranged from 73–83% to 85–98%, respectively.
Sequence similarity of S-segment
Nucleotide sequence similarity of the S-segment among the five sequences ranged from 76% to 82%, whereas the level of amino acid sequence similarity ranged from 86% to 95% (Table 4). As noted for G1 and G2, the nucleotide and amino acid sequence similarities between IP37 and IP38 were 99% and 100%, respectively. The nucleotide and deduced amino acid sequence similarities of the hantaviruses from Paraguay to other South American hantaviruses were 69–93% and 82–99%, respectively. Nucleotide and deduced amino acid sequence similarities of Paraguayan hantaviruses to the North American hantaviruses were 68–78% and 80–88%, respectively, whereas the similarities among North American hantaviruses were 63–81% and 81–93%.
Table 4.
Sequence similarity of the S-segment among American hantaviruses
| Virus | Paraguayan | South American | North American | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALP | ÑEB | AAI | IP37 | IP38 | LN | RM | ORN | AH1 | CHL | BMJ | LEC | MAC | PRG | CD | BAY | BCC | MUL | RS | SN | |
| ALP | – | 79 | 78 | 79 | 79 | 79 | 82 | 78 | 79 | 79 | 78 | 79 | 77 | 78 | 69 | 77 | 77 | 76 | 71 | 76 |
| BMJ-ÑEB | 88 | – | 76 | 82 | 82 | 81 | 80 | 87 | 83 | 83 | 93 | 90 | 82 | 82 | 69 | 77 | 78 | 77 | 69 | 76 |
| AAI | 89 | 86 | – | 77 | 78 | 77 | 78 | 76 | 77 | 77 | 75 | 77 | 77 | 76 | 75 | 77 | 76 | 75 | 73 | 75 |
| IP37 | 89 | 95 | 87 | – | 99 | 79 | 79 | 84 | 82 | 82 | 83 | 83 | 80 | 81 | 72 | 78 | 77 | 76 | 71 | 76 |
| IP38 | 89 | 95 | 87 | 100 | – | 79 | 80 | 84 | 83 | 82 | 83 | 83 | 80 | 82 | 73 | 78 | 77 | 76 | 71 | 77 |
| LN | 92 | 89 | 86 | 90 | 90 | – | 82 | 79 | 79 | 79 | 79 | 80 | 78 | 78 | 66 | 78 | 77 | 76 | 68 | 75 |
| RM | 96 | 90 | 89 | 90 | 90 | 93 | – | 81 | 80 | 81 | 80 | 80 | 78 | 79 | 68 | 77 | 77 | 77 | 68 | 76 |
| ORN | 89 | 98 | 86 | 96 | 97 | 90 | 91 | – | 84 | 84 | 87 | 87 | 82 | 83 | 70 | 78 | 76 | 76 | 68 | 76 |
| AH1 | 89 | 96 | 87 | 95 | 95 | 90 | 91 | 97 | – | 94 | 84 | 84 | 81 | 82 | 72 | 77 | 76 | 77 | 70 | 77 |
| CHL | 89 | 96 | 87 | 95 | 95 | 90 | 91 | 96 | 100 | – | 84 | 84 | 81 | 82 | 69 | 77 | 76 | 76 | 69 | 76 |
| BMJ | 89 | 99 | 87 | 96 | 96 | 90 | 90 | 99 | 97 | 96 | – | 92 | 82 | 82 | 70 | 77 | 78 | 77 | 67 | 77 |
| LEC | 89 | 99 | 87 | 95 | 96 | 90 | 91 | 98 | 96 | 96 | 100 | – | 82 | 83 | 72 | 77 | 78 | 77 | 68 | 77 |
| MAC | 89 | 94 | 86 | 93 | 93 | 88 | 89 | 95 | 94 | 94 | 94 | 94 | – | 83 | 66 | 76 | 77 | 76 | 66 | 77 |
| PRG | 89 | 95 | 85 | 93 | 93 | 90 | 90 | 96 | 95 | 95 | 96 | 96 | 96 | – | 68 | 77 | 76 | 76 | 68 | 75 |
| CD | 83 | 83 | 82 | 83 | 84 | 82 | 83 | 83 | 84 | 84 | 84 | 84 | 82 | 82 | – | 75 | 73 | 74 | 74 | 74 |
| BAY | 88 | 88 | 86 | 87 | 88 | 87 | 88 | 89 | 88 | 88 | 89 | 88 | 88 | 88 | 81 | – | 81 | 80 | 68 | 76 |
| BCC | 87 | 86 | 84 | 85 | 86 | 86 | 87 | 86 | 86 | 87 | 86 | 86 | 86 | 86 | 80 | 92 | – | 81 | 67 | 75 |
| MUL | 86 | 84 | 82 | 84 | 84 | 85 | 86 | 85 | 86 | 86 | 85 | 85 | 86 | 86 | 80 | 93 | 90 | – | 63 | 76 |
| RS | 81 | 82 | 80 | 81 | 81 | 81 | 82 | 82 | 82 | 82 | 82 | 82 | 82 | 81 | 79 | 84 | 82 | 81 | – | 69 |
| SN | 84 | 86 | 85 | 86 | 86 | 85 | 84 | 86 | 86 | 86 | 87 | 86 | 85 | 85 | 82 | 86 | 83 | 82 | 83 | – |
Phylogenetic relationships of Paraguayan hantaviruses to other American hantaviruses
The phylogenetic relationships among the five newly identified Paraguayan hantavirus M sequences and the previously characterized North and South American hantaviruses were examined using a maximum likelihood model. The resulting cladogram (Figure 1) showed the BMJ-ÑEB strain from O. chacoensis formed a subclade with Lechiguanas virus (LEC) from O. flavescens and Bermejo virus (BMJ) from O. chacoensis, which were both discovered in Argentina. AAI from A. montensis also fell within the clade that encompassed BMJ-ÑEB and all of the reported Argentinean and Chilean hantaviruses, and Chile-9717869, and AH-1, LEC, and ORN. IP37 and IP38 strains from O. nigripes formed a separate clade within the South American hantaviruses. The ALP strain from H. chacarius, a rodent captured in the northern part of the Chaco, formed a clade with the LN and RM viruses, which were originally isolated from C. laucha in the Chaco and O. microtis in Bolivia. Together they form a third South American clade in this phylogenetic tree. The M-segment sequence of AAI formed a strong subclade with Pergamino (PRG).
Figure 1.
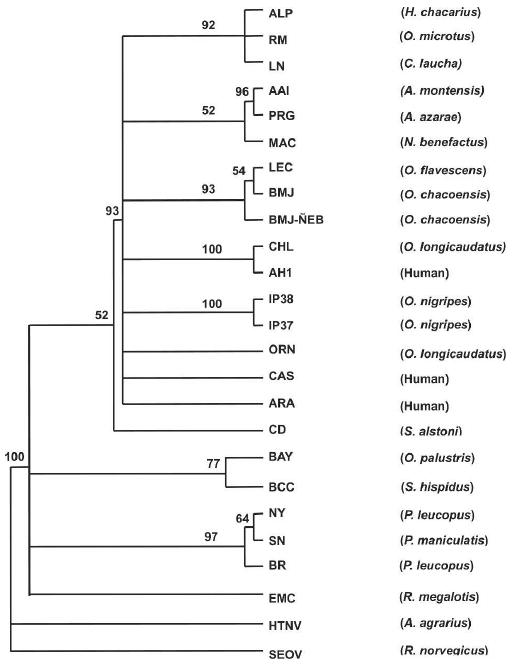
Phylogenetic tree based on maximum likelihood analysis of 438 and 575 nucleotides from G1 and G2, respectively. Nucleotide sequences were analyzed by maximum likelihood analysis of PAUP version 4 b10, using heuristic search option and weighting transversions four times transitions. Bootstrap values of > 50%, obtained from 1,000 replicates of the analysis, are shown at the appropriate branch points. See Table 1 for the definition of the abbreviations used to identify each sequence.
The phylogenetic tree based on the S-segment (Figure 2) showed that the BMJ-ÑEB sequence made a strong subclade with the BMJ virus and LEC and was part of a larger clade with other viruses from Argentina: Andes and ORN viruses. ALP formed a clade with LN and RM as observed for the M-segment. Strikingly, AAI was also part of this clade, in contrast to the M-segment cladogram that showed a strong association with PRG. Finally, as shown in the M-segment cladogram, the S-segments of IP37 and 38 were not closely associated with other viruses. El Moro Canyon (ELMC) virus from Reithrodontomys megalotis (southwestern United States) and the Rio Segundo (RIOS) virus from R. mexicanus (Costa Rica) formed one clade. SN and NY viruses formed a clade, distinct from a group of BAY, BCC, and MUL viruses, and a group of EMC and RS viruses.
Figure 2.
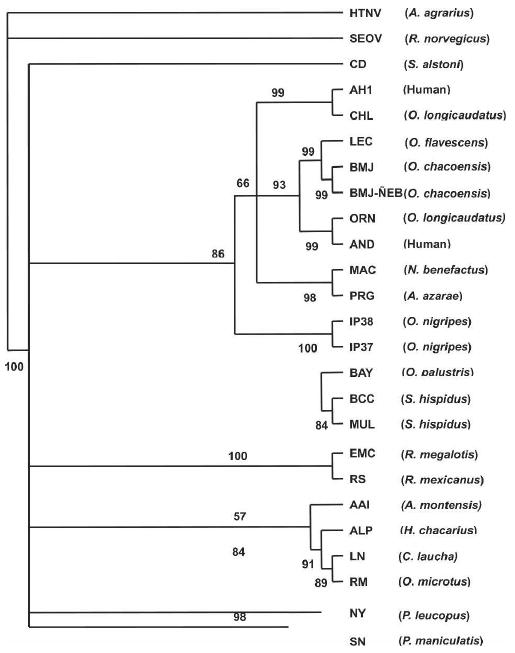
Phylogenetic tree based on maximum likelihood analysis of the partial S-segment. Nucleotide sequences were analyzed by maximum likelihood analysis of PAUP version 4 b10, using heuristic search option and weighting transversions four times transitions. Bootstrap values of > 50%, obtained from 1,000 replicates of the analysis, are shown at the appropriate branch points. See Table 1 for the definition of the abbreviations used to identify each sequence.
Discussion
Considering the general paradigm of “one rodent host species—one hantavirus” in hantavirus evolution, the South American hantaviruses are phylogenetically intriguing. For example, numerous strains of hantaviruses have been have been isolated or identified from various Oligoryzomys species by a number of laboratories. In Argentina alone, there are at least six distinct hantavirus strains, Andes virus, Oran or Andes Norte virus, Lechiguanas virus, Bermejo virus, Maciel virus, and Pergamino virus, which form a single clade that also includes the hantaviruses from Chile.24,36,37
In nature, hantaviruses maintain persistent infections in one or a few closely related rodent species, which reside within a specific geographical region. The ecological constraints of the rodents' habitat have played a primary role in Hantavirus evolution.2 Phylogenetic analysis of the relationships among hantaviruses and their rodent hosts suggests that hantaviruses have a long-standing co-evolutionary history with their predominant rodent carriers.38,39 Therefore, the existing relationships among hantaviruses are most likely derived from adaptation to the distinct genetic environment of their rodent hosts, which evolved from a complicated history of co-speciation events40 and the geographic constraints of the landscape that have influenced rodent migration patterns and habitats.
The diversity of sigmodontine rodents in South America is one of the most impressive examples of broad adaptive radiation of an evolutionary lineage after invading a previously unoccupied region.41 Moreover, this phylogenetic diversity is also reflected as ecological diversity, and a complex biogeographic history within continental South America.42 It is therefore not surprising that South American hantaviruses seem to be highly diverse and biogeographically complex, because they follow their rodent hosts through speciation events and changing landscapes.
We show herein that the South American hantaviruses, including those we identified from Paraguay, share 87–92% and 92–99% amino acid sequence homology in the G1 and G2 region and that their phylograms were concordant as noted previously.2,37 In contrast to the M-segment, amino acid sequence homologies of the S-segment among South American hantaviruses varied from 82% to 99%. The greater homology of the M-segment compared with the S-segment among South American hantaviruses may suggest that the South American Sigmodontinae rodents share similar receptors for viral entry. In the development of effective diagnostics for hantaviruses, these ranges should be considered. In our studies, we obtained a greater sensitivity in the detection of antibody in rodent blood when we use both antigens.34
The BMJ-ÑEB strain, which we identified from O. chacoensis in eastern Pargauay in Ñeembucu, seems to be a strain of the BMJ virus that was identified from the same rodent species in Argentina. Nucleotide sequence differences of the G2 region of the M- and complete ORF region of the S-segments of BMJ-ÑEB and BMJ was 9% and 7%, respectively, whereas their amino acid difference was 1% in both segments. BMJ was originally isolated from the northern part of Argentina in Oran, which borders with the southern part of Bolivia. The location of these two strains suggests that the habitat of O. chacoensis and this strain of hantavirus extends along the Argentine–Paraguayan border in the Gran Chaco and into parts of eastern Paraguay (Figure 3). The other three hantavirus strains, ALP, AAI, and IP37/IP38, were identified from rodent species that have not been detected previously; H. chacarius, A. montensis, and O. nigripes.34 IP37/IP38 from O. nigripes did not form a clade with any of the known American hantaviruses. The ALP strain from H. chacarius, which was collected from the northern Paraguayan Chaco, formed a clade with RM virus (Bolivia) and LN virus (Paraguayan Chaco). The amino acid sequence differences in G1 and G2 regions of the M-segment of the ALP strain with RM and LN were 5% and 7%, respectively, and the amino acid difference of the S-segment of ALP strain to RM and LN was 5% and 6%, respectively, which would suggest they are all the same strain. Nucleotide and amino acid sequence homology of ALP was higher to RM than LN. Thus, one might conclude that ALP, RM, and LN are all closely related species based on both M- and S-segments. The AAI strain from A. montensis clustered with the PRG strain from A. azarae in Argentina when comparing the M-segment, but when comparing the S-segment, it clustered with ALP, RM, and LN viruses. Natural genetic reassortment of genomic RNA segments has been reported within the SN virus among Peromyscus maniculatis12,16 and within the DOB virus among two different rodent host species, Apodemus flavicollis and A. agrarius.43 In an in vitro study with mixed infection using AND and SN viruses, genetic reassortants were isolated with the large (L) and small (S) segments of SNV and M-segment of ANDV.15 Last, phylogenetic analyses of CD virus from Sigmodon alstoni in Venezuela has shown that its M- and S-segment clusters with different clades within the cladogram of North and South American hantaviruses. Further efforts in sequencing of the L-segments from these hantaviruses will help to substantiate the reassorment relationships of the virus strains of Paraguay.
Figure 3.
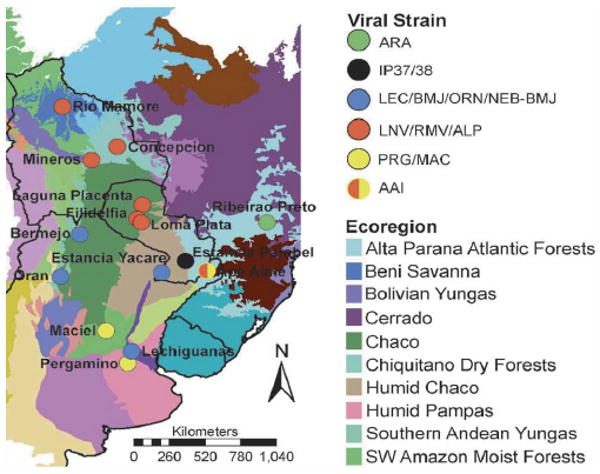
Distribution of hantavirus strains in Paraguay and surrounding countries. Symbols indicate viral strain occurring at each geographic location. Map also shows the distribution of viral strains relative to major South American ecoregions.44
Although there is no entirely consistent relationship between the virus strains that we and others have identified, and the ecoregions in which they occur, there are some apparent biogeographic patterns of note (Figure 3). The group of viruses that include ALP, LN, and RM occur within the Gran Chaco ecoregion, which is characterized by semiarid climate pattern. Several habitats occur in the region, with savanna and thorn forest dominating. Quebracho woodlands, more open than thorn forests, are characterized by thorny bushes, shrubs, and cactuses among scattered trees. The group of viral strains associated with the BMJ-ÑEB strain in Argentina (ORN and BMJ) also occur in the Chaco ecoregion, but this group of interrelated viral strains occurs in the Lower Chaco, a humid subregion of the Gran Chaco. The Lower Chaco encompasses a variety of landscapes, creating a transition zone between the arid Upper Chaco ecoregion to the west and the humid subtropical forests to the east. The region encompasses the flooded savannas of southwestern Paraguay, including the site where the BMJ-ÑEB strain was identified. The third related group of hantaviruses, including the AAI and PRG strains, were found in rodents that exist in very different ecoregions. The AAI strain, identified at Ape Aime in Akodon montensis in Itapúa, occurs in the Upper Paraná Atlantic Forest biome. This biome is characterized by a humid continental climate, and is dominated by semi-deciduous broadleaf forest. In contrast, the rodent that harbors the PRG strain, Akodon azarae, lives in a region dominated by the Espinal, a semiarid temperate grassland ecoregion. Although these two regions are distinct ecologically, they are both experiencing rapid anthropogenic land cover change and are increasingly characterized by fragmentation and habitat disturbance. Comparable and full length sequence information on all genomic segments for each viral isolate is especially important for ascertaining both the degree of reassortment among segments and the geographic diffusion of viral strains. While we have been able to detect a potential reassortment event between the M- and S-segments and have been able to determine relationships suggesting complex geographical differentiation within South America, our ability to test appropriate hypotheses quantitatively is limited by the available sequence data. Future efforts will focus on isolation of Paraguayan hantaviruses, and obtaining full length clones so that comprehensive phylogenetic comparisons among segments and among isolates can be made.
Acknowledgments
The authors thank Robert J. Baker and Heath Garner of the Museum of Texas Tech University for approving and facilitating loans of rodent tissues; the Secretaría del Ambiente (Paraguay) for permits to collect and export rodents and tissues; and the field crew, led by Ismael Mora, for dedication to their work, regardless of circumstances.
Financial support: This work was supported by a grant from the Fogarty International Center 1 R01 TW006986-01 to CBJ under the NIH-NSF Ecology of Infectious Diseases initiative. In addition, this work was supported by a grant from the NIH to B.M., S06 GM08136, project 2-S2. Rodents and tissues were collected under NSF Grants DEB-9400926, DEB-9741543, and DEB-9741134 to R.D.O. and Michael R. Willig.
Contributor Information
Yong-Kyu Chu, Email: Chu@sri.org, Southern Research Institute, 2000 9th Avenue S, Birmingham, AL 35205, Telephone: 205-581-2693, Fax: 205-581-2093.
Brook Milligan, Email: brook@nmsu.edu, Department of Biology, New Mexico State University, Las Cruces, NM 88003, Telephone: 505-646-7980, Fax: 505-646-5665.
Robert D. Owen, Department of Biological Sciences, Texas Tech University, Lubbock, TX, 79409-3131, Telephone: 806-742-3232, Fax: 806-742-2963
Douglas G. Goodin, Department of Geography, Kansas State University, Manhattan, KS 66506, Telephone: 785-532-3411, Fax: 785-532-7310
Colleen B. Jonsson, Email: jonsson@sri.org, Southern Research Institute, 2000 9th Avenue S, Birmingham, AL 35205, Telephone: 205-581-2681, Fax: 205-581-2093.
References
- 1.Hjelle B, Yates T. Modeling hantavirus maintenance and transmission in rodent communities. Curr Top Microbiol Immunol. 2001;256:77–90. doi: 10.1007/978-3-642-56753-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr Top Microbiol Immunol. 2001;256:47–75. doi: 10.1007/978-3-642-56753-7_4. [DOI] [PubMed] [Google Scholar]
- 3.Plyusnin A. Genetics of hantaviruses: Implications to taxonomy. Arch Virol. 2002;147:665–682. doi: 10.1007/s007050200017. [DOI] [PubMed] [Google Scholar]
- 4.Glass GE, Childs JE, Korch GW, LeDuc JW. Association of intraspecific wounding with hantaviral infection in wild rats (Rattus norvegicus) Epidemiol Infect. 1988;101:459–472. doi: 10.1017/s0950268800054418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinson ER, Shone SM, Zink MC, Glass GE, Klein SL. Wounding: The primary mode of Seoul virus transmission among male Norway rats. Am J Trop Med Hyg. 2004;70:310–317. [PubMed] [Google Scholar]
- 6.Root JJ, Black WCt, Calisher CH, Wilson KR, Beaty BJ. Genetic relatedness of deer mice (Peromyscus maniculatus) infected with Sin Nombre virus. Vector Borne Zoonotic Dis. 2004;4:149–157. doi: 10.1089/1530366041210747. [DOI] [PubMed] [Google Scholar]
- 7.Lee HW, Lee PW, Baek LJ, Song CK, Seong IW. Intraspecific transmission of Hantaan virus, etiologic agent of Korean hemorrhagic fever, in the rodent Apodemus agrarius. Am J Trop Med Hyg. 1981;30:1106–1112. doi: 10.4269/ajtmh.1981.30.1106. [DOI] [PubMed] [Google Scholar]
- 8.Kariwa H, Fujiki M, Yoshimatsu K, Arikawa J, Takashima I, Hashimoto N. Urine-associated horizontal transmission of Seoul virus among rats. Arch Virol. 1998;143:365–374. doi: 10.1007/s007050050292. [DOI] [PubMed] [Google Scholar]
- 9.Bernshtein AD, Apekina NS, Mikhailova TV, Myasnikov YA, Khlyap LA, Korotkov YS, Gavrilovskaya IN. Dynamics of Puumala hantavirus infection in naturally infected bank voles (Clethrinomys glareolus) Arch Virol. 1999;144:2415–2428. doi: 10.1007/s007050050654. [DOI] [PubMed] [Google Scholar]
- 10.Hutchinson KL, Rollin PE, Shieh WJ, Zaki S, Greer PW, Peters CJ. Transmission of Black Creek Canal virus between cotton rats. J Med Virol. 2000;60:70–76. doi: 10.1002/(sici)1096-9071(200001)60:1<70::aid-jmv12>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 11.Iwasa MA, Kariwa H, Cui BZ, Lokugamage K, Lokugamage N, Hagiya T, Mizutani T, Takashima I. Modes of hantavirus transmission in a population of Clethrionomys rufocanus bedfordiae inferred from mitochondrial and microsatellite DNA analyses. Arch Virol. 2004;149:929–941. doi: 10.1007/s00705-003-0255-x. [DOI] [PubMed] [Google Scholar]
- 12.Li D, Schmaljohn AL, Anderson K, Schmaljohn CS. Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: Evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology. 1995;206:973–983. doi: 10.1006/viro.1995.1020. [DOI] [PubMed] [Google Scholar]
- 13.Plyusnin A, Cheng Y, Vapalahti O, Pejcoch M, Unar J, Jelinkova Z, Lehvaslaiho H, Lundkvist A, Vaheri A. Genetic variation in Tula hantaviruses: Sequence analysis of the S and M segments of strains from Central Europe. Virus Res. 1995;39:237–250. doi: 10.1016/0168-1702(95)00086-0. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez LL, Owens JH, Peters CJ, Nichol ST. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology. 1998;242:99–106. doi: 10.1006/viro.1997.8990. [DOI] [PubMed] [Google Scholar]
- 15.McElroy AK, Smith JM, Hooper JW, Schmaljohn CS. Andes virus M genome segment is not sufficient to confer the virulence associated with Andes virus in Syrian hamsters. Virology. 2004;326:130–139. doi: 10.1016/j.virol.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Henderson WW, Monroe MC, St Jeor SC, Thayer WP, Rowe JE, Peters CJ, Nichol ST. Naturally occurring Sin Nombre virus genetic reassortants. Virology. 1995;214:602–610. doi: 10.1006/viro.1995.0071. [DOI] [PubMed] [Google Scholar]
- 17.Lee HW, Johnson KM. Laboratory-acquired infections with Hantaan virus, the etiologic agent of Korean hemorrhagic fever. J Infect Dis. 1982;146:645–651. doi: 10.1093/infdis/146.5.645. [DOI] [PubMed] [Google Scholar]
- 18.Tsai TF. Hemorrhagic fever with renal syndrome: Mode of transmission to humans. Lab Anim Sci. 1987;37:428–430. [PubMed] [Google Scholar]
- 19.Padula PJ, Edelstein A, Miguel SD, Lopez NM, Rossi CM, Rabinovich RD. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology. 1998;241:323–330. doi: 10.1006/viro.1997.8976. [DOI] [PubMed] [Google Scholar]
- 20.LeDuc JW, Childs JE, Glass GE. The Hantaviruses, etiologic agents of hemorrhagic fever with renal syndrome: a possible cause of hypertension and chronic renal disease in the United States. Annu Rev Public Health. 1992;13:79–98. doi: 10.1146/annurev.pu.13.050192.000455. [DOI] [PubMed] [Google Scholar]
- 21.Peters CJ, Khan AS. Hantavirus pulmonary syndrome: the new American hemorrhagic fever. Clin Infect Dis. 2002;34:1224–1231. doi: 10.1086/339864. [DOI] [PubMed] [Google Scholar]
- 22.Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 23.Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levis S, Morzunov SP, Rowe JE, Enria D, Pini N, Calderon G, Sabattini M, St Jeor SC. Genetic diversity and epidemiology of hantaviruses in Argentina. J Infect Dis. 1998;177:529–538. doi: 10.1086/514221. [DOI] [PubMed] [Google Scholar]
- 25.Calderon G, Pini N, Bolpe J, Levis S, Mills J, Segura E, Guthmann N, Cantoni G, Becker J, Fonollat A, Ripoll C, Bortman M, Benedetti R, Enria D. Hantavirus reservoir hosts associated with peridomestic habitats in Argentina. Emerg Infect Dis. 1999;5:792–797. doi: 10.3201/eid0506.990608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bharadwaj M, Botten J, Torrez-Martinez N, Hjelle B. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am J Trop Med Hyg. 1997;57:368–374. doi: 10.4269/ajtmh.1997.57.368. [DOI] [PubMed] [Google Scholar]
- 27.Figueiredo LT, Moreli ML, Campos GM, Sousa RL. Hantaviruses in Sao Paulo State, Brazil. Emerg Infect Dis. 2003;9:891–892. doi: 10.3201/eid0907.030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toro J, Vega JD, Khan AS, Mills JN, Padula P, Terry W, Yadon Z, Valderrama R, Ellis BA, Pavletic C, Cerda R, Zaki S, Shieh WJ, Meyer R, Tapia M, Mansilla C, Baro M, Vergara JA, Concha M, Calderon G, Enria D, Peters CJ, Ksiazek TG. An outbreak of hantavirus pulmonary syndrome, Chile, 1997. Emerg Infect Dis. 1998;4:687–694. doi: 10.3201/eid0404.980425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hjelle B, Anderson B, Torrez-Martinez N, Song W, Gannon WL, Yates TL. Prevalence and geographic genetic variation of hantaviruses of New World harvest mice (Reithrodontomys): identification of a divergent genotype from a Costa Rican Reithrodontomys mexicanus. Virology. 1995;207:452–459. doi: 10.1006/viro.1995.1104. [DOI] [PubMed] [Google Scholar]
- 30.Vincent MJ, Quiroz E, Gracia F, Sanchez AJ, Ksiazek TG, Kitsutani PT, Ruedas LA, Tinnin DS, Caceres L, Garcia A, Rollin PE, Mills JN, Peters CJ, Nichol ST. Hantavirus pulmonary syndrome in Panama: Identification of novel hantaviruses and their likely reservoirs. Virology. 2000;277:14–19. doi: 10.1006/viro.2000.0563. [DOI] [PubMed] [Google Scholar]
- 31.Powers AM, Mercer DR, Watts DM, Guzman H, Fulhorst CF, Popov VL, Tesh RB. Isolation and genetic characterization of a hantavirus (Bunyaviridae: Hantavirus) from a rodent, Oligoryzomys microtis (Muridae), collected in northeastern Peru. Am J Trop Med Hyg. 1999;61:92–98. doi: 10.4269/ajtmh.1999.61.92. [DOI] [PubMed] [Google Scholar]
- 32.Fulhorst CF, Monroe MC, Salas RA, Duno G, Utrera A, Ksiazek TG, Nichol ST, de Manzione NM, Tovar D, Tesh RB. Isolation, characterization and geographic distribution of Cano Delgadito virus, a newly discovered South American hantavirus (family Bunyaviridae) Virus Res. 1997;51:159–171. doi: 10.1016/s0168-1702(97)00091-9. [DOI] [PubMed] [Google Scholar]
- 33.Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- 34.Chu YK, Owen RD, Gonzalez LM, Jonsson CB. The complex ecology of hantavirus in Paraguay. Am J Trop Med Hyg. 2003;69:263–268. [PubMed] [Google Scholar]
- 35.Chu YK, Jennings G, Schmaljohn A, Elgh F, Hjelle B, Lee HW, Jenison S, Ksiazek T, Peters CJ, Rollin P, Schmaljohn C. Cross-neutralization of hantaviruses with immune sera from experimentally infected animals and from hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome patients. J Infect Dis. 1995;172:1581–1584. doi: 10.1093/infdis/172.6.1581. [DOI] [PubMed] [Google Scholar]
- 36.Bohlman MC, Morzunov SP, Meissner J, Taylor MB, Ishibashi K, Rowe J, Levis S, Enria D, St Jeor SC. Analysis of hantavirus genetic diversity in Argentina: S segment-derived phylogeny. J Virol. 2002;76:3765–3773. doi: 10.1128/JVI.76.8.3765-3773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padula PJ, Colavecchia SB, Martinez VP, Gonzalez Della Valle MO, Edelstein A, Miguel SD, Russi J, Riquelme JM, Colucci N, Almiron M, Rabinovich RD. Genetic diversity, distribution, and serological features of hantavirus infection in five countries in South America. J Clin Microbiol. 2000;38:3029–3035. doi: 10.1128/jcm.38.8.3029-3035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doyle TJ, Bryan RT, Peters CJ. Viral hemorrhagic fevers and hantavirus infections in the Americas. Infect Dis Clin North Am. 1998;12:95–110. doi: 10.1016/s0891-5520(05)70411-6. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez AJ, Abbott KD, Nichol ST. Genetic identification and characterization of limestone canyon virus, a unique Peromyscus-borne hantavirus. Virology. 2001;286:345–353. doi: 10.1006/viro.2001.0983. [DOI] [PubMed] [Google Scholar]
- 40.Engel SR, Hogan KM, Taylor JF, Davis SK. Molecular systematics and paleobiogeography of the South American sigmodontine rodents. Mol Biol Evol. 1998;15:35–49. doi: 10.1093/oxfordjournals.molbev.a025845. [DOI] [PubMed] [Google Scholar]
- 41.Pardiñas U, D'Elía G, Ortiz P. Sigmodontinos fosiles (Rodentia, Muroidea, Sigmodontinea) de América del Sur: Estado actual de conocimiento y prospectivo. Mastozoología Neotropical. 2002;9:209–252. [Google Scholar]
- 42.D'Elía G. Phylogenetics of Sigmodontinae (Rodentia, Muroidea, Cricetidae), with special reference to the akodont group, and with additional comments on biogeography. Cladistics. 2003;19:307–323. [Google Scholar]
- 43.Klempa B, Schmidt HA, Ulrich R, Kaluz S, Labuda M, Meisel H, Hjelle B, Kruger DH. Genetic interaction between distinct Dobrava hantavirus subtypes in Apodemus agrarius and A. flavicollis in nature. J Virol. 2003;77:804–809. doi: 10.1128/JVI.77.1.804-809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D'Amico JA, Itoua I, Strand HE, Morrinson JC, Loucks CJ, Allnutt TF, Ricketts TH, Kura Y, Lamoreux JF, Wettengel WW, Hedao P, Kassem KR. Terrestrial ecoregions of the world: A new map of life on earth. Bioscience. 2001;51:933–938. [Google Scholar]
- 45.Schmaljohn AL, Li D, Negley DL, Bressler DS, Turell MJ, Korch GW, Ascher MS, Schmaljohn CS. Isolation and initial characterization of a newfound hantavirus from California. Virology. 1995;206:963–972. doi: 10.1006/viro.1995.1019. [DOI] [PubMed] [Google Scholar]
- 46.Hjelle B, Lee SW, Song W, Torrez-Martinez N, Song JW, Yanagihara R, Gavrilovskaya I, Mackow ER. Molecular linkage of hantavirus pulmonary syndrome to the white-footed mouse, Peromyscus leucopus: Genetic characterization of the M genome of New York virus. J Virol. 1995;69:8137–8141. doi: 10.1128/jvi.69.12.8137-8141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song JW, Baek LJ, Gavrilovskaya IN, Mackow ER, Hjelle B, Yanagihara R. Sequence analysis of the complete S genomic segment of a newly identified hantavirus isolated from the white-footed mouse (Peromyscus leucopus): Phylogenetic relationship with other sigmodontine rodent-borne hantaviruses. Virus Genes. 1996;12:249–256. doi: 10.1007/BF00284645. [DOI] [PubMed] [Google Scholar]
- 48.Morzunov SP, Rowe JE, Ksiazek TG, Peters CJ, St Jeor SC, Nichol ST. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J Virol. 1998;72:57–64. doi: 10.1128/jvi.72.1.57-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravkov EV, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. Genetic and serologic analysis of Black Creek Canal virus and its association with human disease and Sigmodon hispidus infection. Virology. 1995;210:482–489. doi: 10.1006/viro.1995.1366. [DOI] [PubMed] [Google Scholar]
- 50.Morzunov SP, Feldmann H, Spiropoulou CF, Semenova VA, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. J Virol. 1995;69:1980–1983. doi: 10.1128/jvi.69.3.1980-1983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rawlings JA, Torrez-Martinez N, Neill SU, Moore GM, Hicks BN, Pichuantes S, Nguyen A, Bharadwaj M, Hjelle B. Cocirculation of multiple hantaviruses in Texas, with characterization of the small (S) genome of a previously undescribed virus of cotton rats (Sigmodon hispidus) Am J Trop Med Hyg. 1996;55:672–679. doi: 10.4269/ajtmh.1996.55.672. [DOI] [PubMed] [Google Scholar]
- 52.Torrez-Martinez N, Song W, Hjelle B. Nucleotide sequence analysis of the M genomic segment of El Moro Canyon hantavirus: Antigenic distinction from four corners hantavirus. Virology. 1995;211:336–338. doi: 10.1006/viro.1995.1413. [DOI] [PubMed] [Google Scholar]
- 53.Hjelle B, Chavez-Giles F, Torrez-Martinez N, Yates T, Sarisky J, Webb J, Ascher M. Genetic identification of a novel hantavirus of the harvest mouse Reithrodontomys megalotis. J Virol. 1994;68:6751–6754. doi: 10.1128/jvi.68.10.6751-6754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meissner JD, Rowe JE, Borucki MK, St Jeor SC. Complete nucleotide sequence of a Chilean hantavirus. Virus Res. 2002;89:131–143. doi: 10.1016/s0168-1702(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 55.Padula PJ, Sanchez AJ, Edelstein A, Nichol ST. Complete nucleotide sequence of the M RNA segment of Andes virus and analysis of the variability of the termini of the virus S, M and L RNA segments. J Gen Virol. 2002;83:2117–2122. doi: 10.1099/0022-1317-83-9-2117. [DOI] [PubMed] [Google Scholar]
- 56.Lopez N, Padula P, Rossi C, Miguel S, Edelstein A, Ramirez E, Franze-Fernandez MT. Genetic characterization and phylogeny of Andes virus and variants from Argentina and Chile. Virus Res. 1997;50:77–84. doi: 10.1016/s0168-1702(97)00053-1. [DOI] [PubMed] [Google Scholar]
- 57.Johnson AM, de Souza LT, Ferreira IB, Pereira LE, Ksiazek TG, Rollin PE, Peters CJ, Nichol ST. Genetic investigation of novel hantaviruses causing fatal HPS in Brazil. J Med Virol. 1999;59:527–535. doi: 10.1002/(sici)1096-9071(199912)59:4<527::aid-jmv17>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]


