Abstract
Since the function of the spinal cord depends on the proteins found there, better defing the normal Spinal Cord Proteome is an important and challenging task. Although brain and cerebrospinal fluid samples from patients with different central nervous system (CNS) disorders have been studied, a thorough examination of specific spinal cord proteins and the changes induced by injury or associated to conditions such as neurodegeneration, spasticity and neuropathies has yet to be performed. In the present study, we aimed to describe total protein content in the spinal cord of healthy rats, employing different proteomics tools. Accordingly, we have developed a fast, easy, and reproducible sequential protocol for protein extraction from rat spinal cords. We employed conventional two dimensional electrophoresis (2DE) in different pH ranges (eg. 4–7, 3–11 NL) combined with identification by mass spectrometry (MALDI-TOF/TOF), as well as first dimension protein separation combined with Liquid Chromatography Mass Spectrometry/Mass Spectrometry (LC-MS/MS) to maximise the benefits of this technology. The value of these techniques is demonstrated here by the identification of several proteins known to be associated with neuroglial structures, neurotransmission, cell survival and nerve growth in the central nervous system. Furthermore this study identified many spinal proteins that have not previously been described in the literature and which may play an important role as either sensitive biomarkers of dysfunction or of recovery after Spinal Cord Injury.
Keywords: proteomics, two dimensional electrophoresis (2-DE), Liquid Chromatography Mass Spectrometry/Mass Spectrometry (LC-MS MS), spinal cord, proteome
Introduction
Spinal cord injury (SCI) has a significant disabling and lifelong effect on many people and as such, it represents a major challenge for successful health care management. SCI is a devastating neurotrauma insult that can lead to the loss of sensory and motor function below the level of injury.1, 2 The progressive pathological changes initiated by SCI include complex and evolving molecular cascades whose interrelationships are not fully understood, and many molecules involved in these processes remain to be discovered.3–7 To date, brain and cerebrospinal fluid samples from patients with different central nervous system (CNS) disorders have been studied extensively using different biochemical assays.8–12 However, relatively few studies have focused on spinal cord protein content, and the changes induced after spinal neurotrama or in association with symptoms such as spasticity or neuropathic pain. Indeed, recent studies have been conducted to screen for a wide range of proteins following SCI using comparative proteomic technologies.13–17
The tremendous advances in molecular biology, mainly in the field of genomics and proteomics, open the possibility to understand the mechanisms underlying many neuropathologies. After genomics, proteomics is often considered the next logical step to study biological systems, with the added capacity to describe the spatiotemporal differences in protein expression, both in normal and pathological tissue.18–20 The proteome represents all the proteins expressed by a genome, cell, tissue or organism at a given time under defined physiological conditions. Since most physiological body functions reflect the integrity of their proteins, understanding the complex biological processes active in the spinal cord during pathological conditions like SCI requires the key proteins involved at an early stage of the neurotrauma21, 22 (acute phase) and during injury progression to be identified.
Proteomic analysis is now a key biomedical tool to establish protein maps that can assist in biomarker discovery and in the identification of therapeutic targets. In this respect, an important and challenging task is to develop protocols designed to extend our knowledge of the spinal cord (SC) protein profile that combine mass spectrometry with two dimensional gels (2-DE). Until now most studies have focussed on one protein or on a small number of proteins using standard techniques such as Western blotting, immunohistochemistry or RT-PCR, which fail to provide complete information regarding the general physiological state of the SC. In contrast, proteomic analysis is useful as multiple molecules can be assayed simultaneously using separation techniques combined with the powerful new mass spectrometry technologies, such as MALDI-TOF/TOF (Matrix Assisted Laser Desorption Ionization-Time of Flight/Time of Flight Mass Spectrometry), SELDI-TOF (Surface Enhanced Laser Desorption Ionization Time Of Flight Mass Spectrometry), Protein Arrays, LCM (Laser Capture Microdissection), MS-Imaging, LC-MS (Liquid Chromatography Mass Spectrometry), TOF-SIMS (Time of Flight Secondary Ion Mass Spectrometry).23–29
However, the development of global protein analysis using proteomic technologies needs to address several limitations and challenges. An important tool applied to study the proteome is 2-DE, whereby proteins are first separated by isoelectric focusing (IEF) and then based on their molecular weight by SDS-PAGE (sodium dodecyl sulphate polyacrylamide gel electrophoresis).30–32 However, this technique presents some important limitations that could be resolved by the application of other proteomics tools such as LC-MS/MS.33 In addition, there is a need to develop efficient protocols to extract most of the proteins present in the spinal cord, given the limitations of each technique and the complexity of the proteome.
In this technical report, we present a fast, easy and reproducible protocol to extract SC proteins and analyze its proteome (Fig. 1). The aim of this study is to describe the majority of the proteins extracted from the rat SC proteome by employing conventional 2-DE spot maps over different pH ranges and MALDI-TOF/TOF for their identification, in combination with LC-MS/MS to maximise the utility of this technology. The application of this newly developed optimal protein extraction protocol compatible with 2-DE and LC-MS/MS will permit future translational studies to identify the main pathophysiological mechanisms associated with SCI.
Figure 1.
The proteomic platforms used in this study and a flowchart demonstrating the strategy for the rat spinal cord analysis. Schematic illustration of the proteomics methods used to characterise the rat spinal cord proteome.
Materials and Methods
Collection of rat spinal cords
Thoracico-lumbar spinal cord tissue was obtained from 12 week old male adult Wistar rats (n = 6: Harlan SA, Milano, Italy) weighing between 300–400 g sacrificed with an intraperitoneal overdose of Sodium Pentobarbital (Dolethal, Norman SA). Shortly afterwards, the spinal cord tissue was extracted using hydraulic pressure applied to the caudal vertebral canal, whereupon the tissue was cleaned with a saline solution (0.9%). The thoracico-lumbar segments were carefully dissected out and then frozen and stored at −20 °C until analyses.
Rat spinal cord processing: protein extraction
After removal from −20 °C storage, the tissue was maintained at 4 °C in PBS solution and all the following steps in the protocol were performed at 4 °C (Fig. 2).
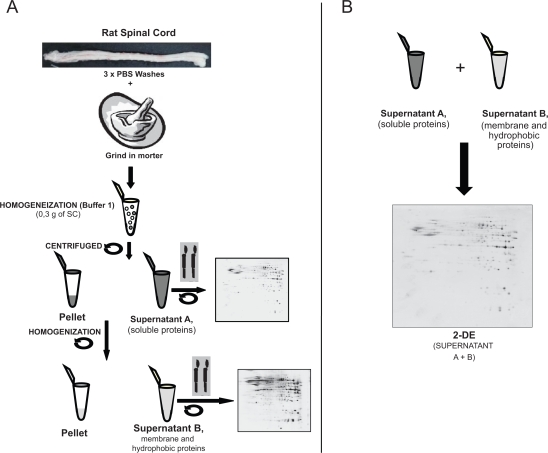
Figure 2.
The protocol to extract proteins from the rat spinal cord. A) After surgery the spinal cord tissue was washed in saline buffer to eliminate blood contaminants and tissue was homogenized (Buffer 1) and later a new extraction of proteins was realized using buffer 2. Supernatant A, containing most of the soluble proteins and supernatant B, containing membrane and hydrophobic proteins were analysed separately in 2-DE in order to check the efficiency of the protein extraction protocol. B) Supernatant A and B were mixed and analysed by 2-DE.
Firstly, the tissue was washed 3 times in PBS to remove blood contaminants and it was then ground into a powder with a mortar in Liquid Nitrogen. This powder (0.3 g) was resuspended in 300 μL of protein extraction buffer 1 (Tris 10 mM [pH 7.5], 500 mM NaCl 0.1%, Triton x-100, 1% β-mercaptoethanol and 1 mM PMSF).34 The homogenate was sonicated for 5 minutes and centrifuged at 21,000 g (5840 R Eppendorf) for 15 minutes at 4 °C to precipitate the membrane and tissue debris. The supernatant (supernatant A), containing most of the soluble proteins was collected and stored at 4 °C. The pellet was then dissolved in a buffer containing 7 M Urea, 2 M Thiourea, 5% CHAPS,35, 36 and it was again centrifuged at 21,000g to obtain a second supernatant separated from the pellet of tissue debris (Supernatant B), mainly composed of membrane proteins. The tissue debris was then resuspended in protein loading buffer (Tris 0.5 M [pH 8.0], SDS 10%, Glycerol, β-mercaptoethanol and bromophenol blue 0.02%) and the protein concentration was determined by the Bradford-Lowry method using the Bio-Rad protein assay commercial Kit.37 Finally, the protein composition was analyzed by resolving 25 μg of total protein content from each sample by SDS-PAGE 12% (Acrylamide/Bisacrylamide 30%/0.8% v/v).
Two-dimensional electrophoresis (2-DE)
All chemicals and instruments used for 2-DE gels have been described previously.35, 36 Both the soluble and hydrophobic protein extracts were mixed and dialysed against 2 mM Tris buffer using Mini dialysis Kit 1 kDa cut-off (GE Healthcare). Subsequently, 300 μg of each protein extract was cleaned with the 2 D Clean up Kit (GE-Healthcare) and resuspended in rehydration buffer (7 M Urea, 2 M Thiourea, 4% CHAPS, 1%–2% Ampholites and 1% TBP: Bio-Rad). Isoelectric focusing (IEF) was performed in an IPGphor unit (GE Healthcare). The strips (17 cm and pH 4–7: Bio-Rad, or 24 cm pH 3–11 NL—non-lineal: GE Healthcare) were actively rehydrated at 20 °C for 12 h at 50 V to enhance protein uptake, and the voltage was then increased according to the following program: 500 V for 30 minutes, 1000 V for 1 h, 1000–2000 V in 1 h (gradient), 2000–5000 V in 2 h (gradient), 5000–8000 V in 1 h (gradient), 8000 V to a total 88,000 V/h.
Subsequently, the strips IEF were equilibrated as described previously35, 36 and the second dimension (SDS-PAGE) was run according to Laemmli’s method,38 using a Protean II system (Bio-Rad) at 1 W/gel at 20 °C overnight. Gels were fixed and stained by Silver Staining (GE Healthcare, according to the manufacturer’s instructions) and they were then scanned with a GS-800 Calibrated Densitometer (Bio-Rad). Evaluation of the 2-DE gels was performed using PDQuest 2DE Gel Analysis Software version 8.0.1 (Bio-Rad). Reproducibility was tested comparing the variation within the different gels in the same group using the same software.
In gel digestion
Spots (200) were manually excised, automatically digested with “Ettan Digester” (GE Healthcare) and identified at the HNP Proteomic Unit according to Schevchenko et al39 with minor modifications.40 Gel plugs were reduced with 10 mM dithiothreitol (Sigma Aldrich) in 50 mM ammonium bicarbonate (99% purity; Scharlau) and by alkylation with 55 mM iodoacetamide (Sigma Aldrich) in 50 mM ammonium bicarbonate. The gel fragments were then rinsed with 50 mM ammonium bicarbonate in 50%. Methanol (gradient, HPLC grade, Scharlau) and acetonitrile (gradient, HPLC grade, Scharlau), and they were dried in a Speedvac. Modified porcine trypsin (sequencing grade; Promega, Madison, WI, USA) was added to the dry gel pieces at a final concentration of 20 ng/μl in 20 mM ammonium bicarbonate and the digestion proceeded at 37 °C overnight. Finally, 70% aqueous acetonitrile and 0.1% formic acid (99.5% purity; Sigma Aldrich) was added for peptide extraction.
Protein identification by MALDI-TOF/TOF
An aliquot of each digestion was mixed with an aliquot of the matrix solution (3 mg/mL α-cyano-4-Hydroxycinnamic acid: Sigma Aldrich) in 30% ACN, 15% 2-propanol and 0.1% TFA. This mixture was pipetted directly onto the stainless steel sample plate of the mass spectrometer (384 Opti-TOF 123 × 81 mm MALDI: Applied Biosystem) and dried at room temperature.
The MALDI-MS/MS data were obtained in an automated analysis loop using a 4800 Plus MALDI TOF/TOF Analyzer (Applied Biosystems). Spectra were acquired in the reflector positive-ion mode with a Nd:YAG laser (355 nm wavelength at a frequency of 200 Hz), and between 100 and 2000 individual spectra were averaged. The experiments were acquired in a uniform mode with a fixed laser intensity. For the MS/MS 1 kV analysis mode, precursors were accelerated to 8 kV in source 1, and they were selected at a relative resolution of 350 (FWHM) with metastable suppression. Fragment ions generated by collision with air in a CID chamber were further accelerated at 15 kV in source 2. Mass data was analysed automatically with the 4000 Series Explorer Software version 3.5.3 (Applied Biosystems). Internal calibration of MALDI-TOF mass spectra was performed using two trypsin autolysis ions with m/z = 842.510 and m/z = 2211.105. For calibration in the MS/MS mode, the fragment ion spectra obtained from Glub-fibrinopeptide were used (4700 Cal Mix, Applied Biosystems). MALDI-MS and MS/MS data were combined through the GPS Explorer Software Version 3.6 to search a nonredundant protein database (Swissprot 56.7) using the Mascot software (version 2.2, Matrix Science), employing the following parameters: 50 ppm precursor tolerance; 0.6 Da MS/MS fragment tolerance; and allowing 1 missed cleavage, carbamidomethyl cysteines and methionine oxidation as a modification. The MALDI-MS/MS spectra and database search results were manually inspected in detail using the aforementioned software.
LC-MS/MS and database searching
Sample preparation
Total spinal cord proteins (50 μg) were resolve by one dimensional (1-D) SDS-PAGE 12%. Each lane in the 1-D gel was divided into 24 gel slices that were manually excised and then digested automatically using the Ettan Digester (GE Healthcare). The digestion was performed according to Schevchenko et al39 with minor modifications40 and using Modified porcine trypsin (sequencing grade; Promega, Madison, WI, USA) diluted to a final concentration of 20 ng/μl in 20 mM ammonium bicarbonate. The gel slices were incubated with 10 mM dithiothreitol (Sigma Aldrich) in 50 mM ammonium bicarbonate (99% purity; Scharlau) for 30 minutes at 56 °C and after reduction, they were alkylated with 55 mM iodoacetamide (Sigma Aldrich) in 50 mM ammonium bicarbonate for 20 minutes at RT. Gel plugs were washed with 50 mM ammonium bicarbonate in 50% methanol (gradient, HPLC grade, Scharlau), rinsed in acetonitrile (gradient, HPLC grade, Scharlau) and dried in a Speedvac. Dry gel pieces were then embedded in sequencing grade modified porcine trypsin (20 ng/μL: Promega, Madison, WI, USA) and after digestion at 37 °C overnight, the peptides were extracted with 70% acetonitrile (ACN) in 0.1% formic acid (99.5% purity; Sigma Aldrich). Finally, the samples were dried in a speedvac and resuspended in 98% water with 0.1% formic acid (FA) and 2% ACN.
LC-MS/MS and database searching
The LC/MSMS system was comprised of a TEMPO nano LC system (Applied Biosystems) combined with a nano LC Autosampler. Each sample was injected in three replicates (3 μL) using mobile phase A (2% ACN/98% water, 0.1% FA) at a flow rate of 10 μL/minute for 10 minutes. Peptides were loaded onto a μ-Precolumn Cartridge (Acclaim Pep Map 100 C18, 5 μm, 100Å; 300 μm i.d. × 5 mm, LC Packings) to preconcentrate and desalt samples. The RPLC was performed on a C18 column (Acclaim Pep Map 100 C18, 3 μm, 100Å; NAN75-15-03-C18PM, 75 μm I.D. × 15 cm, LC Packings) using mobile phase A (2% ACN/98% water, 0.1% FA) and mobile phase B (98% ACN/2% water, 0.1% FA). Peptides were eluted at a flow rate of 300 nL/minute over the following gradient: initial conditions of 5% B that increased to 50% B over 70 minutes, 50 to 95% B for 1 minute and then 95% B for 3 minutes, returning to the initial conditions (5% B) over 2 minutes and maintaining these conditions for a further 14 minutes.
The LC-MS/MS analysis was performed on an AB/MDS Sciex 4000 Q TRAP System with NanoSprayII Source (Applied Biosystems). The TEMPO nano LC system and 4000 QTRAP were both controlled by Analyst Software v.1.4.2.
All the MS and MS/MS data were obtained in positive ion mode, with an ion spray voltage of 2800 V and a declustering of 85V. Nanoflow interface was heated at 150 °C, and the source gas 1 and curtain gas were set to 20 and 10, respectively. Nitrogen was applied as both curtain and collision gas. An Information Dependent Acquisition (IDA) method was programmed, with a full scan Enhanced MS (EMS) experiment at 4000 amu/s for ion profiling that was followed by an enhanced resolution (ER) MS experiment at 250 amu/s. The ER experiment permitted charge state recognition that was further submitted to IDA criteria to select precursor ions, and to estimate the collision energy to fragment them. These IDA criteria were set to select the 8 most intense double, triple or quadruple charged ions from 400–1200 m/z that exceed 100,000 counts for fragmentation in the LINAC collision cell. Isotopes within a 4.0 amu window and with a mass tolerance of 1,000,000 mmu were excluded. These 8 ions were submitted to 8 independent Enhanced Product Ion (EPI) MS/MS experiments at 4000 amu/s with Dynamic Fill Time (DFT). The total number of MS and MS/MS experiments per cycle was 10 (1 EMS, 1 ER and 8 EPI), resulting in a total cycle time of 5.0058 s.
Analyst software creates wiff format files including all the spectra data that were batch-processed with ProteinPilot™ Software 2.0.1 (Applied Biosystems/MDS Sciex). This software automatically generated peak lists that were searched against the Swissprot database version 56.7 using Paragon Algorithm (Applied Biosystems). Settings in the Paragon Algorithm included a detected protein threshold >1.0 (90%), Iodoacetamide was selected for Cys alkylation and Gel-based ID was selected as a special factor.
Results
Rat spinal cord processing and protein extraction
To describe the complete proteome of an organ or tissue, it is necessary to establish an efficient extraction protocol to maximize protein recovery. Here, we present a flowchart to explain our approach to the proteomic study of rat SC (Fig. 1) and a schedule of the consecutive extraction protocol (Fig. 2). This method was based on two consecutive steps using two distinct extraction buffers, the first of which extracted the more soluble proteins, while the second was designed to dissolve the membrane and hydrophobic proteins that were assumed to be abundant in SC tissue.
Sample preparation and conventional 2-DE
In order to reduce the presence of lipids and other interfering substances, samples were sonicated, filtered with a micro spin-filter (SIGMA) and cleaned with the Clean-up Kit (GE Healthcare). We tested different pH ranges (pH 4–7; pH 3–11 NL) in order to select that which was optimal to detect the maximal number of spots with the greatest resolution. Spinal Cord protein extracts were quantified and approximately 300 μg was loaded onto each 2-DE gel. After analysis with the PD-Quest software (Bio-Rad), around 300 spots were detected by 2-DE in the 4–7 pH range (Fig. 3A). However, these gels did not present an homogeneous spot distribution due to the fact that most of them co-localized in the same area.
Figure 3.
2-DE gel images. 2-DE was performed with IPG strips at different pH ranges: A) pH 4–7 (left) and B) pH 3–11 NL (right). C) 2-DE gel performed with 3–11 NL IPG strip and 9%–16% acrylamide/bisacrylamide.
For this reason, we performed 2-DE gels with 24 cm pH 3–11 NL IPG strips. We obtained a good distribution, definition and a large number of spots under these conditions, although some streaking in the 53–96 kDa molecular weight region could be due to the high concentration of these abundant proteins. This problem did not arise in the same region of the pH 4–7 2 D gels. Hence, the use of the two types of gels with complementary pH ranges (pH 4–7 and 3–11 NL) helped improve the overall spot resolution, as reported previously.35
Thus, more than 1000 spots were detected after PD-Quest software analysis, improving the resolution and permitting the subsequent identification of the spots (Fig. 3B). Reproducibility was tested by comparing the variation within the different gels in the same group using the PD Quest 8.0 software. An analysis of 1126 spots revealed a coefficient of variation (CV) < 50% for 90.4% of the spots in same group of gels. Among these, a CV < 30% was obtained for 67.1% of the spots. These data confirmed the high reproducibility of the gels obtained with the method used.
Protein identification (MALDI-TOF/TOF)
In order to verify the effectiveness of our methodology, 200 spots were chosen at random, they were excised from the stained 2-DE gels, digested and the resultant tryptic peptides were deposited an a MALDI plaque and applied in a 4800 Plus MALDI-TOF/TOF Analyzer (Applied Biosystem). Proteins were identified by Peptide mass fingerprinting using the “MASCOT” search engine (www.matrixscience.com). All the spots were identified and they corresponded to 128 proteins (Fig. 4), as summarized in Table 1 where their molecular weight, isoelectric point, cellular sublocalization and function are shown.
Figure 4.
Preparative 2-DE Gel (700 μg). Spot Map of the proteins identified. The characterization of the spots identified is shown in Table 1.
Table 1.
Spots identified with 2-DE gel (pH: 3-11 NL). The data indicates accession number, the isoelectric point (theoretical and experimental), molecular weight (theoretical and experimental), subcellular localization and recognised function.
| Protein name | Accession no. | MALDI-TOF/Spot Nº | LC-MS Q-TRAP | MW Da theorical | MW Da experimental | pI theorical | pI experimental | Subcellular localization | Function |
|---|---|---|---|---|---|---|---|---|---|
| Myelin basic protein S | MBP_RAT | Spot Nº 2 | Identified | 21,489.00 | 15 | 11.25 | 10.8 | Cell mb. | Structural… |
| Tubulin polymerization-promoting protein family | TPPP3_BOVIN | Spot Nº 3 | Identified | 18,931.00 | 24.5 | 9.18 | 10.8 | Myelin mb. | Structural… |
| Protein NipSnap homolog 1 | NIPS1_MOUSE | Spot Nº 6 | No | 33,342.00 | 35 | 9.48 | 10.8 | Mit inn mb. | Others. |
| Prohibitin-2 | PHB2_MOUSE | Spot Nº 8 | No | 33,276.00 | 40 | 9.83 | 10.8 | Mit inn mb; Cp; N. | Others. |
| Aspartate aminotransferase, mitochondrial | AATM_RAT | Spot Nº 10 | Identified | 47,284.00 | 51 | 9.13 | 10.8 | Mit; Cell mb. | Metabolism. |
| Elongation factor 1-alpha 1 | EF1A1_CRIGR | Spot Nº 11 | No | 50,109.00 | 56 | 9.10 | 10.8 | Cp. | Prot regulation. |
| Profilin-1 | PROF1_RAT | Spot Nº 13 | Identified | 14,948.00 | 13 | 8.46 | 9.6 | Cp. | Structural… |
| Peptidyl-prolyl cis-trans isomerase A | PPIA_RAT | Spot Nº 16 | Identified | 17,863.00 | 18 | 8.34 | 9.5 | Cp. | Protregulation. |
| Destrin OS | DEST_RAT | Spot Nº 17 | Identified | 18.522,00 | 20 | 8.19 | 9.2 | Structural… | |
| Cofilin-1 | COF1_RAT | Spot Nº 18 | Identified | 18,521.00 | 23 | 8.22 | 9.2 | N; Cp. | Structural… |
| Peroxiredoxin-1 | PRDX1_RAT | Spot Nº 19 | Identified | 22,095.00 | 31 | 8.27 | 9.5 | Cp; Mel. | Stress Resp... |
| Peroxiredoxin-1 | PRDX1_RAT | Spot Nº 20 | Identified | 22,095.00 | 31 | 8.27 | 9.2 | Cp; Mel. | Stress Resp... |
| Proteasome subunit beta type-1 | PSB1_MOUSE | Spot Nº 21 | Identified | 26,355.00 | 33 | 7.67 | 9.4 | Cp; N. | Prot regulation. |
| Glutathione S-transferase alpha-3 | GSTA3_RAT | Spot Nº 22 | No | 25,303.00 | 35 | 8.78 | 9.6 | Cp. | Stress Resp... |
| Glutathione S-transferase Mu 1 | GSTM1_RAT | Spot Nº 23 | No | 25,897.00 | 34.2 | 8.27 | 9.4 | Cp. | Stress Resp... |
| Proteasome subunit alpha type-7 | PSA7_MOUSE | Spot Nº 24 | Identified | 27,838.00 | 35.5 | 8.59 | 9.6 | Cp; N. | Prot regulation. |
| ATP synthase subunit alpha liver isoform, mitochondrial | ATPA2_BOVIN | Spot Nº 25 | No | 38,852.00 | 37 | 9.57 | 9.6 | Mit inn mb. | Metabolism. |
| ATP synthase subunit gamma, mitochondrial | ATPG_RAT | Spot Nº 26 | Identified | 30,172.00 | 38 | 8.87 | 9.5 | Mit inn mb. | Metabolism. |
| Voltage-dependent anion-selective channel protein 1 | VDAC1_RABIT | Spot Nº 27 | Identified | 30,722.00 | 39 | 8.62 | 9.5 | Mit out mb; Cell mb. | Stress Resp... |
| Malate dehydrogenase, mitochondrial | MDHM_RAT | Spot Nº 28 | Identified | 35,661.00 | 45 | 8.93 | 9.9 | Mit. | Metabolism. |
| Malate dehydrogenase, mitochondrial | MDHM_RAT | Spot Nº 29 | Identified | 35,661.00 | 45 | 8.93 | 9.65 | Mit. | Metabolism. |
| L-lactate dehydrogenase A chain | LDHA_RAT | Spot Nº 30 | Identified | 36,427.00 | 45 | 8.45 | 9.5 | Cp. | Metabolism. |
| Malate dehydrogenase, mitochondrial | MDHM_RAT | Spot Nº 31 | Identified | 35,661.00 | 49 | 8.93 | 9.5 | Mit. | Metabolism. |
| Malate dehydrogenase, mitochondrial | MDHM_RAT | Spot Nº 32 | Identified | 35,661.00 | 45 | 8.93 | 9.4 | Mit. | Metabolism. |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P_RAT | Spot Nº 33 | Identified | 35,787.00 | 49 | 8.44 | 9.4 | Cp. | Metabolism. |
| Fructose-bisphosphate aldolase A | ALDOA_RAT | Spot Nº 34 | Identified | 39,327.00 | 51 | 8.31 | 9.4 | Mit. | Metabolism. |
| Cytochrome b-c1 complex subunit 2, mitochondrial | QCR2_RAT | Spot Nº 35 | Identified | 48,366.00 | 53 | 9.16 | 9.5 | Mit inn mb. | Metabolism. |
| 2’,3’-cyclic-nucleotide 3’-phosphodiesterase | CN37_RAT | Spot Nº 36 | Identified | 47,239.00 | 60 | 9.03 | 9.4 | Cell mb; Mel. | Metabolism. |
| Septin-7 | SEPT7_RAT | Spot Nº 37 | No | 50,518.00 | 66 | 8.73 | 9.4 | Cp. | Structural… |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P_RAT | Spot Nº 38 | Identified | 35,787.00 | 49 | 8.44 | 9.2 | Cp. | Metabolism. |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P_RAT | Spot Nº 39 | Identified | 35,787.00 | 50 | 8.44 | 9.2 | Cp. | Metabolism. |
| Fumarate hydratase, mitochondrial | FUMH_RAT | Spot Nº 41 | No | 54,429.00 | 60 | 9.06 | 9.2 | Mit; Cp. | Metabolism. |
| ATP synthase subunit alpha, mitochondrial | ATPA_RAT | Spot Nº 42 | Identified | 59,717.00 | 75 | 9.22 | 9.1 | Mit inn mb. | Metabolism. |
| T-complex protein 1 subunit eta | TCPH_PONAB | Spot Nº 43 | No | 59,329.00 | 80 | 7.55 | 9.3 | Cp. | Prot regulation. |
| Phosphoglycerate kinase 1 | PGK1_RAT | Spot Nº 44 | Identified | 44,510.00 | 53 | 8.02 | 9.1 | Cp. | Metabolism. |
| Fumarate hydratase, mitochondrial | FUMH_RAT | Spot Nº 45 | No | 54,429.00 | 60 | 9.06 | 8.9 | Mit; Cp. | Metabolism. |
| Citrate synthase, mitochondrial | CISY_RAT | Spot Nº 46 | Identified | 51,833.00 | 53 | 8.53 | 8.88 | Mit. | Metabolism. |
| Isocitrate dehydrogenase [NAD] subunit beta, | IDH3B_RAT | Spot Nº 47 | Identified | 42,327.00 | 51 | 8.89 | 8.87 | Mit. | Metabolism. |
| Fructose-bisphosphate aldolase A | ALDOA_RAT | Spot Nº 48 | Identified | 39,327.00 | 51 | 8.31 | 8.8 | Mit. | Metabolism. |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P_RAT | Spot Nº 49 | Identified | 35,787.00 | 49 | 8.44 | 8.9 | Cp. | Metabolism. |
| Thiosulfate sulfurtransferase | THTR_RAT | Spot Nº 50 | Identified | 33,386.00 | 45 | 7.71 | 9.2 | Mit. | Carrier. |
| Hydroxyacyl-coenzyme A dehydrogenase, | HCDH_RAT | Spot Nº 51 | No | 34,426.00 | 40 | 8.83 | 9.2 | Mit. | Metabolism. |
| Voltage-dependent anion-selective channel protein 1 | VDAC1_RABIT | Spot Nº 52 | Identified | 30,722.00 | 39 | 8.62 | 9.2 | Mit out mb; Cell mb. | Stress Resp... |
| Superoxide dismutase [Mn], mitochondrial | SODM_RAT | Spot Nº 56 | Identified | 24,659.00 | 30 | 8.96 | 8.88 | Mit. | Stress Resp... |
| Peptidyl-prolyl cis-trans isomerase A | PPIA_RAT | Spot Nº 57 | Identified | 17,863.00 | 18.3 | 8.34 | 8.9 | Cp. | Prot regulation. |
| Peptidyl-prolyl cis-trans isomerase A | PPIA_RAT | Spot Nº 58 | Identified | 17,863.00 | 18.3 | 8.34 | 8.7 | Cp. | Prot regulation. |
| Cofilin-2 OS=Homo sapiens | COF2_HUMAN | Spot Nº 59 | Identified | 18,725.00 | 24 | 7.66 | 8.2 | N; Cp. | Structural… |
| Superoxide dismutase [Mn], mitochondrial | SODM_RAT | Spot Nº 60 | Identified | 24,659.00 | 30 | 8.96 | 8.3 | Mit. | Stress Resp... |
| Adenylate kinase isoenzyme 1 | KAD1_RAT | Spot Nº 60 | No | 21,570.00 | 31.5 | 7.66 | 8.5 | Cp. | Metabolism. |
| Glutathione S-transferase P | GSTP1_RAT | Spot Nº 61 | Identified | 23,424.00 | 31.3 | 6.89 | 8.6 | Stress Resp... | |
| Peroxiredoxin-1 | PRDX1_RAT | Spot Nº 62 | Identified | 22,095.00 | 30 | 8.27 | 8.7 | Cp; Mel. | Stress Resp... |
| Superoxide dismutase [Mn], mitochondrial | SODM_RAT | Spot Nº 63 | Identified | 24,659.00 | 34 | 8.96 | 8.7 | Mit. | Stress Resp... |
| Cytochrome b-c1 complex subunit Rieske | UCRI_RAT | Spot Nº 64 | Identified | 29,427.00 | 35 | 9.04 | 8.7 | Mit inn mb. | Metabolism. |
| Dihydropteridine reductase | DHPR_RAT | Spot Nº 65 | No | 25,536.00 | 36 | 7.67 | 8.7 | Stress Resp... | |
| Electron transfer flavoprotein subunit beta | ETFB_RAT | Spot Nº 66 | Identified | 27,670.00 | 39 | 7.60 | 8.8 | Mit. | Metabolism. |
| Voltage-dependent anion-selective channel protein 1 | VDAC1_RABIT | Spot Nº 67 | Identified | 30,722.00 | 49 | 8.62 | 8.8 | Mit out mb; Cell mb. | Stress Resp... |
| Phosphoserine aminotransferase | SERC_HUMAN | Spot Nº 69 | No | 40,397.00 | 51 | 7.56 | 8.6 | mb, mithoc | Metabolism. |
| Isocitrate dehydrogenase [NAD] subunit beta, | IDH3B_RAT | Spot Nº 70 | Identified | 42,327.00 | 53 | 8.89 | 8.6 | Mit. | Metabolism. |
| Fructose-bisphosphate aldolase C | ALDOC_RAT | SpotNº 70 | Identified | 39,259.00 | 60 | 6.67 | 8.6 | Metabolism. | |
| Creatine kinase, ubiquitous mitochondrial | KCRU_MOUSE | Spot Nº 71 | Identified | 46,974.00 | 57 | 8.39 | 8.7 | Mit inn mb. | Metabolism. |
| Phosphoglycerate kinase 1 | PGK1_HORSE | Spot Nº 71 | No | 42,327.00 | 57 | 8.89 | 8.5 | Metabolism. | |
| Fumarate hydratase, mitochondrial | FUMH_RAT | Spot Nº 72 | No | 54,429.00 | 64 | 9.06 | 8.3 | Mit; Cp. | Metabolism. |
| Pyruvate kinase isozymes M1/M2 | KPYM_RAT | Spot Nº 73 | Identified | 57,781.00 | 90 | 6.63 | 8.2 | Metabolism. | |
| Transketolase | TKT_RAT | Spot Nº 74 | Identified | 67,601.00 | 84 | 7.23 | 8.2 | Prot regulation. | |
| Aconitate hydratase, mitochondrial | ACON_RAT | Spot Nº 75 | Identified | 85,380.00 | 80 | 7.87 | 8.2 | Mit. | Metabolism. |
| Aconitate hydratase, mitochondrial | ACON_RAT | Spot Nº 76 | Identified | 85,380.00 | 78 | 7.87 | 8.3 | Mit. | Metabolism. |
| Transketolase | TKT_RAT | Spot Nº 77 | Identified | 67,601.00 | 75 | 7.23 | 8.1 | Prot regulation. | |
| Pyruvate kinase isozymes M1/M2 | KPYM_RAT | Spot Nº 78 | Identified | 57,781.00 | 75 | 6.63 | 8.0 | Metabolism. | |
| Glucose-6-phosphate isomerase | G6PI_RAT | Spot Nº 79 | No | 62,787.00 | 65 | 7.38 | 8.1 | Cp. | Metabolism. |
| Glutamate dehydrogenase 1, mitochondrial | DHE3_RAT | Spot Nº 80 | Identified | 61,298.00 | 60 | 8.05 | 8.2 | Mit. | Metabolism. |
| Glutamate dehydrogenase 1, mitochondrial | DHE3_RAT | Spot Nº 81 | Identified | 61,298.00 | 74 | 8.05 | 7.5 | Mit. | Metabolism. |
| Vesicle-fusing ATPase | NSF_MOUSE | Spot Nº 82 | Identified | 82,561.00 | 52 | 6.52 | 8.2 | Cp. | Prot regulation. |
| Platelet-activating factor acetylhydrolase IB subunit | LIS1_MOUSE | Spot Nº 83 | Identified | 46,670.00 | 53 | 6.97 | 8.0 | Cp; N. | Structural… |
| Creatine kinase, ubiquitous mitochondrial | KCRU_MOUSE | Spot Nº 84 | Identified | 46,974.00 | 50 | 8.39 | 8.1 | Mit inn mb. | Metabolism. |
| Aspartate aminotransferase, cytoplasmic | AATC_RAT | Spot Nº 85 | Identified | 46,400.00 | 49 | 6.73 | 8.2 | Cp. | Metabolism. |
| Glutamine synthetase | GLNA_RAT | Spot Nº 86 | Identified | 42,240.00 | 49 | 6.64 | 8.05 | Cp. | Metabolism. |
| Fructose-bisphosphate aldolase C | ALDOC_RAT | Spot Nº 87 | Identified | 39,259.00 | 50 | 6.67 | 8.0 | Metabolism. | |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P_RAT | Spot Nº 88 | Identified | 35,787.00 | 51 | 8.44 | 7.9 | Cp. | Metabolism. |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P_RAT | Spot Nº 89 | Identified | 35,787.00 | 53 | 8.44 | 7.7 | Cp. | Metabolism. |
| Alcohol dehydrogenase [NADP+] | AK1A1_RAT | Spot Nº 90 | Identified | 36,483.00 | 51 | 6.84 | 7.7 | Stress Resp... | |
| Isocitrate dehydrogenase [NADP] cytoplasmic | IDHC_RAT | Spot Nº 92 | No | 46,705.00 | 49 | 6.53 | 7.5 | Cp. | Metabolism. |
| NAD-dependent deacetylase sirtuin-2 | SIRT2_RAT | Spot Nº 95 | Identified | 39,294.00 | 36 | 6.67 | 8.1 | Cp. | Structural… |
| Ribose-phosphate pyrophosphokinase 1 | PRPS1_HUMAN | Spot Nº 96 | No | 34,812.00 | 36 | 6.51 | 7.8 | Metabolism. | |
| Electron transfer flavoprotein subunit alpha | ETFA_RAT | Spot Nº 97 | No | 34,929.00 | 35.5 | 8.62 | 7.7 | Mit. | Metabolism. |
| Carbonic anhydrase 2 | CAH2_RAT | Spot Nº 98 | No | 29,096.00 | 35 | 6.89 | 8.3 | Cp. | Stress Resp... |
| Hydroxyacylglutathione hydrolase | GLO2_RAT | Spot Nº 99 | Identified | 28,878.00 | 34.1 | 6.46 | 8.2 | Metabolism. | |
| Phosphoglycerate mutase 1 | PGAM1_MOUSE | Spot Nº 100 | No | 28,786.00 | 34 | 6.67 | 8.25 | N. | Metabolism. |
| Triosephosphate isomerase | TPIS_RAT | Spot Nº 101 | Identified | 26,832.00 | 34 | 6.89 | 8.3 | Metabolism. | |
| Protein-L-isoaspartate (D-aspartate) | PIMT_RAT | Spot Nº 103 | No | 24,619.00 | 34 | 7.10 | 7.8 | Cp. | Metabolism. |
| Glutathione S-transferase Yb-3 | GSTM4_RAT | Spot Nº 104 | No | 25,664.00 | 35 | 6.84 | 7.75 | Cp. | Stress Resp... |
| GTP-binding nuclear protein Ran | RAN_CANFA | Spot Nº 105 | No | 24,408.00 | 32 | 7.01 | 7.8 | Cp; N; Mel. | Prot regulation. |
| Proteasome subunit alpha type-2 | PSA2_RAT | Spot Nº 106 | No | 25,909.00 | 28 | 8.39 | 8.0 | Cp; N. | Prot regulation. |
| Alpha-crystallin B chain | CRYAB_RAT | Spot Nº 109 | Identified | 20,076.00 | 18 | 6.76 | 8.2 | Stress Resp... | |
| Nucleoside diphosphate kinase B | NDKB_RAT | Spot Nº 110 | No | 17,272.00 | 8 | 6.92 | 8.3 | Cp; Cell mb. | Metabolism. |
| Peroxiredoxin-5, mitochondrial | PRDX5_RAT | Spot Nº 111, 112 | Identified | 22,165.00 | 10 | 8.94 | 7.5 | Mit; Cp; Per. | Stress Resp... |
| Peroxiredoxin-5, mitochondrial | PRDX5_RAT | Spot Nº 111, 112 | Identified | 22,165.00 | 11 | 8.94 | 7.0 | Mit; Cp; Per. | Stress Resp... |
| Macrophage migration inhibitory factor | MIF_RAT | Spot Nº 113 | Identified | 12,496.00 | 15 | 6.79 | 7.1 | Cp; ES; N. | Stress Resp... |
| Cytochrome c oxidase polypeptide 6A1, mitochondrial | CX6A1_CANFA | Spot Nº 114 | No | 2,109.00 | 15 | 6.48 | 7.3 | Mit inn mb. | Metabolism. |
| D-dopachrome decarboxylase OS | DOPD_RAT | Spot Nº 115 | Identified | 13,125.00 | 15 | 6.09 | 6.8 | Cp. | Stress Resp... |
| Histidine triad nucleotide-binding protein 1 | HINT1_MOUSE | Spot Nº 116, 118 | Identified | 13,768.00 | 13.5 | 6.38 | 6.8 | Cp. | Others. |
| Fatty acid-binding protein, epidermal OS | FABP5_RAT | Spot Nº 117 | No | 15,050.00 | 12.5 | 6.73 | 6.1 | Cp. | Metabolism. |
| Histidine triad nucleotide-binding protein 1 | HINT1_MOUSE | Spot Nº 116, 118 | Identified | 13,768.00 | 14 | 6.38 | 6.3 | Cp. | Others. |
| Prefoldin subunit 1 | PFD1_HUMAN | Spot Nº 119 | Identified | 14,202.00 | 20 | 6.32 | 6.3 | ??? | Prot regulation. |
| Profilin-2 | PROF2_RAT | Spot Nº 121 | No | 14,992.00 | 18.5 | 6.55 | 6.5 | Cp. | Structural… |
| Nucleoside diphosphate kinase A | NDKA_RAT | Spot Nº 122 | Identified | 17,182.00 | 17 | 5.96 | 4.0 | Cp; N. | Metabolism. |
| Superoxide dismutase [Cu-Zn] | SODC_RAT | Spot Nº 122 | Identified | 15,912.00 | 19 | 5.88 | 4.3 | Cp. | Stress Resp... |
| Gamma-synuclein | SYUG_MOUSE | Spot Nº 125 | Identified | 13,152.00 | 19.5 | 4.63 | 5.2 | Cp. | Structural… |
| Beta-synuclein | MNME_XYLFT | Spot Nº 126 | Identified | 14,268.00 | 22 | 4.43 | 4.7 | Cp. | Structural… |
| Calmodulin | CALM_BOVIN | Spot Nº 127 | Identified | 16,827.00 | 31 | 4.09 | 3.6 | Spindle. | Prot regulation. |
| Phosphatidylethanolamine-binding protein 1 | PEBP1_RAT | Spot Nº 128 | Identified | 20,788.00 | 35 | 5.48 | 4.9 | Cp; Cell mb. | Metabolism. |
| Peroxiredoxin-2 | PRDX2_RAT | Spot Nº 129 | Identified | 21,765.00 | 35 | 5.20 | 4.7 | Cp. | Stress Resp... |
| Peroxiredoxin-2 | PRDX2_RAT | Spot Nº 129 | Identified | 21,765.00 | 32 | 5.20 | 4.3 | Cp. | Stress Resp... |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1_MOUSE | Spot Nº 131 | Identified | 24,822.00 | 32 | 5.14 | 4.5 | Cp. | Prot regulation. |
| Rho GDP-dissociation inhibitor 1 | GDIR1_RAT | Spot Nº 132 | Identified | 23,393.00 | 36 | 5.12 | 4.3 | Cp. | Prot regulation. |
| Translationally-controlled tumor protein | TCTP_MOUSE | Spot Nº 133 | Identified | 19,450.00 | 38 | 4.76 | 4.2 | Cp. | Structural… |
| Lactoylglutathione lyase | LGUL_RAT | Spot Nº 134 | Identified | 20,806.00 | 40 | 5.12 | 4.4 | Stress Resp... | |
| 14-3-3 protein gamma | 1433G_HUMAN | Spot Nº 135 | Identified | 28,235.00 | 40 | 4.80 | 4.5 | Cp. | Prot regulation. |
| Calretinin | CALB2_RAT | Spot Nº 135 | Identified | 31,384.00 | 43 | 4.94 | 4.3 | Carrier. | |
| 14-3-3 protein epsilon | 1433E_BOVIN | Spot Nº 136 | Identified | 29,155.00 | 51 | 4.63 | 4.3 | Cp; Mel. | Prot regulation. |
| Annexin A5 | ANXA5_RAT | Spot Nº 137 | Identified | 35,722.00 | 53 | 4.93 | 4.4 | Others. | |
| Annexin A5 | ANXA5_RAT | Spot Nº 138 | Identified | 35,722.00 | 75 | 4.93 | 3.88 | Others. | |
| Ubiquitin thioesterase OTUB1 | OTUB1_RAT | Spot Nº 139 | No | 31,250.00 | 80 | 4.85 | 4.6 | Prot regulation. | |
| Glial fibrillary acidic protein | GFAP_RAT | Spot Nº 140 | Identified | 49,927.00 | 47 | 5.35 | 4.5 | Cp. | Structural… |
| 40S ribosomal protein SA | RSSA_RAT | Spot Nº 140 | Identified | 32,803.00 | 47 | 4.80 | 4.3 | Cp. | Structural… |
| Glial fibrillary acidic protein | GFAP_RAT | Spot Nº 141 | Identified | 49,927.00 | 51 | 5.35 | 4.3 | Cp. | Structural… |
| Calreticulin | CALR_RAT | Spot Nº 142 | No | 47,966.00 | 63 | 4.33 | 4.6 | ER. | Prot regulation. |
| Rab GDP dissociation inhibitor alpha | GDIA_RAT | Spot Nº 143 | Identified | 50,504.00 | 64 | 5.00 | 5.0 | Cp. | Carrier. |
| Heat shock protein HSP 90-beta | HS90B_RAT | Spot Nº 144 | Identified | 83,229.00 | 97 | 4.97 | 5.2 | Cp; Mel. | Prot regulation. |
| Neurofilament medium polypeptide | NFM_RAT | Spot Nº 145 | Identified | 95,734.00 | 115 | 4.77 | 5.18 | Structural… | |
| Neurofilament medium polypeptide | NFM_RAT | Spot Nº 145 | Identified | 95,734.00 | 115 | 4.77 | 5.5 | Structural… | |
| Neurofilament heavy polypeptide | NFH_RAT | Spot Nº 146 | Identified | 115,308.00 | 160 | 5.74 | 5.2 | Structural… | |
| Gamma-enolase | ENOG_RAT | Spot Nº 147 | Identified | 47,111.00 | 66 | 5.03 | 5.3 | Cp; Cell mb. | Metabolism. |
| Actin, cytoplasmic 1 | ACT5_CHICK | Spot Nº 148 | No | 41,809.00 | 58 | 5.30 | 5.3 | Cp. | Structural… |
| Tropomodulin-2 | TMOD2_MOUSE | Spot Nº 149 | Identified | 39,487.00 | 57 | 5.28 | 5.6 | Cp. | Structural… |
| Tubulin alpha-1 chain (Fragment) | TBA1_CHICK | Spot Nº 150 | Identified | 50,104.00 | 50 | 4.96 | 6.0 | Structural… | |
| L-lactate dehydrogenase B chain | LDHB_RAT | Spot Nº 151 | Identified | 36,589.00 | 51 | 5.70 | 5.7 | Cp. | Metabolism. |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | ODPB_MOUSE | Spot Nº 152 | Identified | 38,912.00 | 49 | 6.41 | 6.0 | Mit. | Metabolism. |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | ODPB_MOUSE | Spot Nº 154 | Identified | 38,912.00 | 45 | 6.41 | 6.4 | Mit. | Metabolism. |
| L-lactate dehydrogenase B chain | LDHB_RAT | Spot Nº 155 | Identified | 36,589.00 | 45 | 5.70 | 6.8 | Cp. | Metabolism. |
| L-lactate dehydrogenase B chain | LDHB_RAT | Spot Nº 158 | Identified | 36,589.00 | 36 | 5.70 | 5.5 | Cp. | Metabolism. |
| Malate dehydrogenase, cytoplasmic | MDHC_RAT | Spot Nº 159 | Identified | 36,460.00 | 52 | 6.16 | 5.9 | Cp. | Metabolism. |
| Malate dehydrogenase, cytoplasmic | MDHC_RAT | Spot Nº 160 | Identified | 36,460.00 | 33 | 6.16 | 5.7 | Cp. | Metabolism. |
| Prohibitin | PHB_RAT | Spot Nº 162 | Identified | 29,802.00 | 34 | 5.57 | 6.2 | Mit inn mb. | Others. |
| 6-phosphogluconolactonase | 6PGL_RAT | Spot Nº 164 | No | 27,217.00 | 34 | 5.54 | 6.7 | Metabolism. | |
| Peroxiredoxin-6 | PRDX6_RAT | Spot Nº 165 | Identified | 24,803.00 | 32.5 | 5.64 | 6.5 | Cp; Lys. | Stress Resp... |
| Peroxiredoxin-6 | PRDX6_RAT | Spot Nº 165 | Identified | 24,803.00 | 32.5 | 5.64 | 6.7 | Cp; Lys. | Stress Resp... |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | NDUS3_MOUSE | Spot Nº 166 | Identified | 30,131.00 | 31.5 | 6.67 | 6.65 | Mit inn mb. | Metabolism. |
| Protein DJ-1 | PARK7_RAT | Spot Nº 167 | Identified | 19,961.00 | 25 | 6.32 | 6.65 | N; Cp. | Stress Resp... |
| Dynactin subunit 3 | DCTN3_BOVIN | Spot Nº 168 | Identified | 21,178.00 | 30 | 5.39 | 6.6 | Cp. | Structural… |
| Glutathione S-transferase A6 | GSTA6_RAT | Spot Nº 170 | No | 25,791.00 | 32 | 5.90 | 7.3 | Cp. | Stress Resp... |
| Thioredoxin-dependent peroxide reductase, mitochondrial | PRDX3_RAT | Spot Nº 171 | Identified | 28,277.00 | 32 | 7.14 | 7.6 | Mit. | Stress Resp... |
| Thioredoxin-dependent peroxide reductase, mitochondrial | PRDX3_RAT | Spot Nº 171 | Identified | 28,277.00 | 35.5 | 7.14 | 7.3 | Mit. | Stress Resp... |
| Protein DJ-1 | PARK7_RAT | Spot Nº 173 | Identified | 19,961.00 | 34 | 6.32 | 7.2 | N; Cp. | Stress Resp... |
| ATP synthase subunit d, mitochondrial | ATP5H_RAT | Spot Nº 175 | Identified | 18,752.00 | 34 | 6.17 | 6.9 | Mit inn mb. | Metabolism. |
| Protein-L-isoaspartate (D-aspartate) O-methyltransferase OS=Macaca fascicularis GN=PCMT1 PE=2 SV=3 | PIMT_MACFA | Spot Nº 180 | Identified | 24,622.00 | 34 | 6.23 | 7.0 | Cp. | Metabolism. |
| Flavin reductase | BLVRB_MOUSE | Spot Nº 181 | No | 22,183.00 | 35 | 6.49 | 7.15 | Cp. | Stress Resp... |
| Protein-L-isoaspartate (D-aspartate) O-methyltransferase OS=Macaca fascicularis GN=PCMT1 PE=2 SV=3 | PIMT_MACFA | Spot Nº 182 | Identified | 24,622.00 | 34 | 6.23 | 6.8 | Cp. | Metabolism. |
| V-type proton ATPase subunit E 1 | VATE1_BOVIN | Spot Nº 183 | Identified | 26,123.00 | 39 | 8.45 | 7.2 | Metabolism. | |
| Pirin | PIR_RAT | Spot Nº 184 | No | 32,158.00 | 40 | 6.22 | 7.5 | Others. | |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | NDUAA_RAT | Spot Nº 187 | No | 40,468.00 | 41 | 7.64 | 6.7 | Mit. | Metabolism. |
| Elongation factor Tu, mitochondrial | EFTU_RAT | Spot Nº 190 | Identified | 49,491.00 | 75 | 7.23 | 7.1 | Mit. | Prot regulation. |
| Alpha-enolase | ENOA_MOUSE | Spot Nº 191 | Identified | 47,111.00 | 80 | 6.37 | 7.8 | Cp; Cell mb. | Metabolism. |
| Alpha-enolase | ENOA_MOUSE | Spot Nº 192 | Identified | 47,111.00 | 80 | 6.37 | 8.0 | Cp; Cell mb. | Metabolism. |
| Beta-centractin | ACTY_MOUSE | Spot Nº 193 | No | 42,255.00 | 86 | 5.98 | 7.4 | Cp. | Structural… |
| Alpha-enolase | ENOA_MOUSE | Spot Nº 194 | Identified | 47,111.00 | 80 | 6.37 | 6.5 | Cp; Cell mb. | Metabolism. |
| Alpha-enolase | ENOA_MOUSE | Spot Nº 195 | Identified | 47,111.00 | 80 | 6.37 | 6.3 | Cp; Cell mb. | Metabolism. |
| Syntaxin-binding protein 1 | STXB1_BOVIN | Spot Nº 197 | Identified | 67,526.00 | 82 | 6.49 | 6.8 | Cp; Cell mb. | Prot regulation. |
Abbreviations: Cp, Cytoplasm; N, nucleus; Mit inn mb, mitochondrial inner membrane; Mit, mitochondrion; cell mb, cellular membrane; ES, extra cellular space; Mit out mb, mitochondrial outer membrane; Mel, Melanosome; Cp-sec-syn ves, Cytoplasmic-secreted-synaptic vesicle; Per mb, peroxisomal membrane; Lys, lysosome; Gol app, golgi apparatus; ER, endoplasmic reticulum; EM, extracellular matrix.
Our data show the broad range of proteins identified by 2-DE from Macrophage migration inhibitory factor 12.5 kDa up to the Neurofilament heavy polypeptide with a molecular weight of 115.31 kDa. Furthermore we identified the Myelin basic protein, as the most basic protein (pI 11.25) and Calreticulin as the most acidic (pI 4.33).
Liquid-Chromatography Mass Spectrometry (LC-MS/MS)
To improve the number of proteins identified by MALDI, a LC-MS analysis was carried out. Total rat SC protein (50 μg) was resolved by SDS-PAGE and after Coomassie staining (PageBlue™ Protein Staining Solution, Fermentas), the gel was divided and cut into 24 pieces, each of which was subjected to in-gel tryptic digestion. After digestion, the peptide samples were analyzed by HPLC (TEMPO, Applied Biosystem) and the peptides eluted were analyzed on a Q-TRAP ion trap MS workstation (Applied Biosystem).
These analyses identified a total of 18,734 peptides that corresponded to 41,481 spectra. After data grouping and filtration, 387 proteins were identified (cut off > 1 and 90% of confident) and their theoretical MW, pI, subcellular localization and function are shown in Table 2, excluding the proteins previously identified by 2-DE. Many acidic proteins were identified, such as Acidic leucine-rich-nuclear phosphoprotein 32 family member B with a pI of 3.87, and basic proteins such as Myelin basic protein with a pI of 11.25. The molecular weights of these proteins ranged from 299.53 kDa for the Microtubule-associated protein 1A to 7850.14 Da for the gamma-2 subunit of the Guanine nucleotide-binding protein G(I)/G(S)/G(O).
Table 2.
Proteins identified with 1-D gel and LC-MS/MS analysis.
| Protein name | Accession no. | MW Da | pI | Subcellular localization | Function |
|---|---|---|---|---|---|
| Slice 2, 3 | |||||
| Protein S100-B | |P50114|S100B_MOUSEN | 10728.05 | 4.52 | Cp; N. | Carrier. |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4 | |Q62425|NDUA4_MOUSE | 9326.79 | 9.52 | Mit inn mb. | Metabolism. |
| 10 kDa heat shock protein, mitochondrial | s|P26772|CH10_RAT | 10901.67 | 8.89 | Mit. | Prot regulation. |
| Cytochrome c oxidase polypeptide 7A2, mitochondrial | sp|P35171|CX7A2_RAT | 9352.97 | 10.28 | Mit inn mb. | Metabolism. |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-12 | |Q9DAS9|GBG12_MOUSE | 7997.23 | 9.14 | Cell mb; Cp. | Carrier. |
| Cytochrome c oxidase subunit VIb isoform 1 | |P56391|CX6B1_MOUSE | 10071.45 | 8.96 | Mit intermb sp. | Metabolism. |
| Slice 4 | |||||
| ATP synthase subunit e, mitochondrial | sp|P29419|ATP5I_RAT | 8254.65 | 9.34 | Mit inn mb. | Metabolism. |
| Histone H2A type 1-A | |Q96QV6|H2A1A_HUMAN | 14233.51 | 10.86 | N. | Structural… |
| Acyl-CoA-binding protein | |P11030|ACBP_RAT | 10027.46 | 8.78 | Carrier. | |
| Dynein light chain roadblock-type 1 | |P62628|DLRB1_RATE | 10989.68 | 6.58 | Cp. | Structural… |
| Glutaredoxin-1 | sp|Q9ESH6|GLRX1_RAT | 11878.88 | 8.93 | Cp. | Stress Resp... |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | |P63213|GBG2_MOUSE | 7850.14 | 7.78 | Cell mb; Cp. | Carrier. |
| Glycogen phosphorylase, brain form | |P11216|PYGB_HUMAN | 96695.96 | 6.40 | Metabolism. | |
| Mitochondrial import inner membrane translocase subunit Tim13 | |Q9Y5L4|TIM13_HUMAN | 10500.02 | 8.42 | Mit inn mb. | Prot regulation. |
| Neurofilament light polypeptide | sp|P19527|NFL_RAT | 61335.28 | 4.63 | Structural… | |
| Slice 5 | |||||
| Galectin-1 | |P16045|LEG1_MOUSE | 14865.85 | 5.32 | ES. | Others. |
| Cytochrome b-c1 complex subunit 7 | |Q9D855|QCR7_MOUSE | 13527.47 | 9.10 | Mit inn mb. | Metabolism. |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | sp|Q63362|NDUA5_RAT | 13411.79 | 6.84 | Mit inn mb. | Metabolism. |
| Ribonuclease UK114 | |P52759|UK114_RAT | 14303.46 | 7.80 | Mit; Cp; N. | Prot regulation. |
| Myotrophin | |P62775|MTPN_RAT | 12860.77 | 5.27 | Cp. | Structural… |
| Thioredoxin | |P11232|THIO_RAT | 11673.47 | 4.80 | Cp. | Stress Resp... |
| Fatty acid-binding protein, brain | |P55051|FABP7_RAT | 14863.98 | 5.46 | Cp. | Carrier. |
| Slice 6 | |||||
| Cytochrome c, somatic | |P62898|CYC_RAT | 11605.44 | 9.61 | Mit. | Metabolism. |
| Glial fibrillary acidic protein | lP47819 |GFAP_RAT | 49957.09 | 5.35 | Cp. | Structural… |
| CDGSH iron sulfur domain-containing protein 1 | |Q9NZ45|CISD1_HUMAN | 12199.05 | 9.20 | Mit out mb. | Metabolism. |
| Parvalbumin alpha | |P02625|PRVA_RAT | 11925.52 | 5.00 | Carrier. | |
| Astrocytic phosphoprotein PEA-15 | |Q5U318|PEA15_RAT | 15040.10 | 4.93 | Cp. | Stress Resp... |
| Slice 7 | |||||
| 60S acidic ribosomal protein P2 | sp|P02401|RLA2_RAT | 11691.96 | 4.44 | Prot regulation. | |
| Myosin light polypeptide 6 | sp|Q64119|MYL6_RAT | 16975.15 | 4.46 | Structural… | |
| V-type proton ATPase subunit G 2 | |Q9TSV6|VATG2_PIG | 13579.34 | 10.26 | Mel. | Carrier. |
| V-Vesicle-associated membrane protein 2 | |P63045|VAMP2_RAT | 12690.78 | 7.84 | Cp-sec-syn ves. | Carrier. |
| Ubiquitin-conjugating enzyme E2 N | |Q9EQX9|UBE2N_RAT | 17123.79 | 6.13 | Prot regulation. | |
| Histone H2A.J | |A9UMV8|H2AJ_RAT | 14045.45 | 11.05 | N. | Structural… |
| ATP synthase subunit delta, mitochondrial | |P35434|ATPD_RAT | 17595.07 | 5.16 | Mit inn mb. | Metabolism. |
| Calcineurin subunit B type 1 | |P63100|CANB1_RAT | 19299.91 | 4.64 | Carrier. | |
| Histone H2B type 2-E | |Q64524|H2B2E_MOUSE | 13993.26 | 10.31 | N. | Structural… |
| Vesicle-associated membrane protein 3 | |Q4R8T0|VAMP3_MACFA | 11319.13 | 8.89 | Cell mb. | Prot regulation. |
| Single-stranded DNA-binding protein, mitochondrial | sp|P28042|SSB_RAT | 17454.93 | 9.84 | Mit. | Others. |
| Tubulin alpha-1A chain | |Q6AYZ1|TBA1C_RAT | 50135.63 | 4.94 | Structural… | |
| Creatine kinase B-type | sp|Q04447|KCRB_MOUSE; sp|P07335|KCRB_RAT | 42725.27 | 5.39 | Cp. | Metabolism. |
| Fibrous sheath-interacting protein 1 | |Q66H16|FSIP1_RAT | 49568.06 | 5.02 | Others. | |
| Mitochondrial fission 1 protein | |Q9CQ92|FIS1_MOUSE | 17008.65 | 8.55 | Mit out mb; Per mb. | Stress Resp... |
| Slice 8 | |||||
| Low molecular weight phosphotyrosine protein phosphatase | |Q5REM7|PPAC_PONAB | 18086.50 | 6.29 | Cp. | Prot regulation. |
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | |Q9QUR7|PIN1_MOUSE | 18370.46 | 8.93 | N. | Structural… |
| Protein S100-A16 | |Q96FQ6|S10AG_HUMAN | 11801.40 | 6.28 | Carrier. | |
| Vesicle-associated membrane protein 1 | |Q63666|VAMP1_RAT | 12796.81 | 6.24 | Cp-sec-syn ves. | Prot regulation. |
| Histone H2B type 1-K | |Q8CGP1|H2B1K_MOUSE | 13920.17 | 10.31 | Nucleus. | Structural… |
| Visinin-like protein 1 | |Q5RD22|VISL1_PONAB | 22338.24 | 5.32 | Prot regulation. | |
| Thrombospondin type-1 domain-containing protein 7B | |Q6P4U0|THS7B_MOUSE | 179309.14 | 8.01 | Cell mb. | Others. |
| Slice 9 | |||||
| ADP-ribosylation factor 3 | |P61206|ARF3_RAT | 20456.51 | 6.74 | Gol app. | Prot regulation. |
| Prefoldin subunit 2 | |B0BN18|PFD2_RAT | 16579.73 | 6.20 | Prot regulation. | |
| Stathmin | |Q6DUB7|STMN1_PIG | 17302.51 | 5.76 | Cp. | Structural… |
| Vesicle-associated membrane protein-associated protein B | sp|A5GFS8|VAPB_PIG | 27053.25 | 6.85 | Cp ves. | Prot regulation. |
| Nucleoside diphosphate kinase A | |Q05982|NDKA_RAT | 17192.74 | 5.96 | Cp; N. | Metabolism. |
| Ubiquitin-conjugating enzyme E2 variant 2 | |Q7M767|UB2V2_RAT | 16352.71 | 7.79 | Prot regulation. | |
| Transgelin-3 | |Q5R6R2|TAGL3_PONAB | 22472.64 | 6.84 | Others. | |
| Ubiquitin-conjugating enzyme E2 L3 | |P68037|UB2L3_MOUSE | 17861.58 | 8.68 | Prot regulation. | |
| Endothelin-1 | sp|P22388|EDN1_RAT | 23134.93 | 9.77 | Sec. | Stress Resp... |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 | sp|Q95KV7|NDUAD_BOVIN | 16673.39 | 9.22 | Mit inn mb; N. | Stress Resp... |
| Slice 10 | |||||
| Tubulin beta chain | sp|P02554|TBB_PIG | 49860.95 | 4.78 | Structural… | |
| Actin-related protein 2/3 complex subunit 5-like protein | |Q9BPX5|ARP5L_HUMAN | 17010.32 | 6.31 | Cp. | Structural… |
| Hippocalcin-like protein 1 | |P37235|HPCL1_HUMAN | 22338.24 | 5.32 | Others. | |
| Neurocalcin-delta | |Q5PQN0|NCALD_RAT | 22245.23 | 5.23 | Others. | |
| Transcription factor BTF3 | sp|Q64152|BTF3_MOUSE | 22030.81 | 9.52 | N. | Others. |
| Ferritin heavy chain | |P19132|FRIH_RAT | 21126.66 | 5.86 | Others. | |
| Cell division control protein 42 homolog | |Q8CFN2|CDC42_RAT | 21258.61 | 6.16 | Cell mb. | Stress Resp... |
| Phospholipid hydroperoxide glutathione peroxidase, nuclear | |Q91XR8|GPX42_RAT | 29184.69 | 10.83 | N. | Stress Resp... |
| 60S ribosomal protein L12 | |P35979|RL12_MOUSE | 17804.56 | 9.48 | Others. | |
| Slice 11 | |||||
| Ferritin light chain 1 | sp|P02793|FRIL1_RAT | 20748.50 | 5.99 | Others. | |
| Cysteine and glycine-rich protein 1 | sp|P47875|CSRP1_RAT | 20613.48 | 8.90 | N. | Others. |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | |Q9DCS9|NDUBA_MOUSE | 21023.81 | 8.19 | Mit inn mb. | Metabolism. |
| Tubulin beta-2C chain | |Q6P9T8|TBB2C_RAT | 49800.98 | 4.79 | Structural… | |
| Slice 12 | |||||
| ATP synthase subunit O, mitochondrial | sp|Q06647|ATPO_RAT | 23397.55 | 10.03 | Mit inn mb. | Metabolism. |
| Glutathione S-transferase P | sp|P47954|GSTP1_CRIMI | 23469.00 | 7.64 | Stress Resp... | |
| Adenylate kinase isoenzyme 1 | sp|P39069|KAD1_RAT | 21583.76 | 7.66 | Cp. | Metabolism. |
| Ras-related protein Rab-1B | |Q9H0U4|RAB1B_HUMAN | 22163.10 | 5.55 | Cell mb; Cp. | Prot regulation. |
| Glycolipid transfer protein | |B0BNM9|GLTP_RAT | 23703.65 | 6.90 | Cp. | Carrier. |
| Ras-related protein Rab-11B | |O35509|RB11B_RAT | 24488.50 | 5.64 | Cell mb. | Carrier. |
| Histone H2A.x | sp|P27661|H2AX_MOUSE | 15142.60 | 10.74 | N. | Structural… |
| Ras-related protein Rab-7a | |P51150|RAB7A_MOUSE | 23489.75 | 6.39 | Mel. | Prot regulation. |
| Cell cycle exit and neuronal differentiation protein 1 | |Q5FVI4|CEND_RAT | 15043.01 | 9.01 | Cell mb. | Structural… |
| UMP-CMP kinase | |Q9DBP5|KCY_MOUSE | 22165.33 | 5.68 | N. Cp. | Metabolism. |
| Apolipoprotein D | |P23593|APOD_RAT | 21634.86 | 4.93 | Sec. | Carrier. |
| Microtubule-actin cross-linking factor 1, isoform 4 | |Q96PK2|MACF4_HUMAN | 670150.80 | 5.20 | Cp. | Structural… |
| GrpE protein homolog 1, mitochondrial | |Q99LP6|GRPE1_MOUSE | 24307.02 | 8.58 | Mit. | Prot regulation. |
| Transgelin-2 | |Q9WVA4|TAGL2_MOUSE | 22393.42 | 8.41 | Others. | |
| Ras-related protein Rap-1b | |Q62636|RAP1B_RAT | 20824.79 | 5.65 | Cell mb; Cp. | Prot regulation. |
| Glutathione S-transferase P 1 | |P19157|GSTP1_MOUSE | 23609.18 | 7.69 | Stress Resp... | |
| Slice 13 | |||||
| Heat shock protein beta-1 | |P14602|HSPB1_MOUSE | 23013.85 | 6.12 | Stress Resp... | |
| Myelin-oligodendrocyte glycoprotein | |Q63345|MOG_RAT | 27881.56 | 8.61 | Cell mb. | Structural… |
| Glutathione S-transferase Y1 | |Q00285|GSTMU_CRILO | 25818.98 | 8.74 | Cp. | Stress Resp... |
| Tumor protein D52 | |Q62393|TPD52_MOUSE | 20059.41 | 4.87 | Structural… | |
| Osteoclast-stimulating factor 1 | |Q62422|OSTF1_MOUSE | 23782.74 | 5.46 | Cp. | Others. |
| UPF0568 protein C14orf166 homolog | |Q9CQE8|CN166_MOUSE | 28152.21 | 6.40 | N; Cp. | Others. |
| NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | |P19234|NDUV2_RAT | 27378.34 | 6.23 | Mit inn mb. | Metabolism. |
| 3-hydroxyacyl-CoA dehydrogenase type-2 | |O02691|HCD2_BOVIN | 27140.29 | 8.45 | Mit. | Others. |
| Glutathione S-transferase alpha I | |Q08863|GSTA1_RABIT | 25691.11 | 8.92 | Cp. | Stress Resp... |
| Ras-related protein Rab-5A | |P20339|RAB5A_HUMAN | 23658.68 | 8.23 | Cell mb; Mel. | Prot regulation. |
| Slice 14 | |||||
| 14-3-3 protein zeta/delta | |P63102|1433Z_RAT; sp|P63101|1433Z_MOUSE | 27771.14 | 4.73 | Cp; Mel. | Prot regulation. |
| Dihydropteridine reductase | |P11348|DHPR_RAT | 25552.20 | 7.67 | Stress Resp... | |
| Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | |P21913|DHSB_RAT | 31829.94 | 8.96 | Mit inn mb. | Metabolism. |
| Gamma-enolase | |P09104|ENOG_HUMAN | 47268.58 | 4.91 | Cp; Cell mb. | Metabolism. |
| Tumor protein D54 | |Q6PCT3|TPD54_RAT | 23991.85 | 5.80 | Prot regulation. | |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 3, mitochondrial | |Q9CRB9|CHCH3_MOUSE | 26334.52 | 8.56 | Others. | |
| 14-3-3 protein eta | |P68511|1433F_RAT; sp|P68510|1433F_MOUSE; sp|P68509|1433F_BOVIN | 28211.74 | 4.81 | Cp. | Carrier. |
| Endoplasmic reticulum protein ERp29 | |P52555|ERP29_RAT | 28574.83 | 6.23 | ER. | Prot regulation. |
| 14-3-3 protein theta | |Q5ZMD1|1433T_CHICK | 27782.28 | 4.68 | Cp. | Prot regulation. |
| Proteasome subunit alpha type-5 | |Q9Z2U1|PSA5_MOUSE | 26411.03 | 4.74 | Cp; N. | Prot regulation. |
| Electron transfer flavoprotein subunit beta | sp|Q68FU3|ETFB_RAT | 27687.42 | 7.61 | Mit. | Metabolism. |
| 14-3-3 protein beta/alpha | |P35213|1433B_RAT | 28054.39 | 4.81 | Cp; Mel. | Prot regulation. |
| Myelin P0 protein | |Q6WEB5|MYP0_HORSE | 27570.67 | 9.40 | Cell mb. | Structural… |
| ADP/ATP translocase 1 | |P48962|ADT1_MOUSE | 32904.27 | 9.73 | Mit inn mb. | Carrier. |
| Hypoxanthine-guanine phosphoribosyltransferase (Fragment) | |P00493|HPRT_MOUSE | 24081.78 | 5.74 | Cp. | Metabolism. |
| Acidic leucine-rich nuclear phosphoprotein 32 family member A | |P49911|AN32A_RAT | 28564.59 | 3.99 | N; Cp. | Stress Resp... |
| Cytochrome c1, heme protein, mitochondrial | sp|P00125|CY1_BOVIN | 35296.75 | 9.14 | Mit inn mb. | Metabolism. |
| Microtubule-associated protein 1A | |Q9QYR6|MAP1A_MOUSE | 300139.96 | 4.92 | Structural… | |
| N(G),N(G)-dimethylarginine dimethylaminohydrolase 2 | |Q6MG60|DDAH2_RAT | 29687.91 | 5.66 | Metabolism. | |
| Myelin proteolipid protein | |P60203|MYPR_RAT | 30077.17 | 8.71 | Cell mb. | Structural… |
| Tropomyosin alpha-3 chain | |P06753|TPM3_HUMAN | 32818.79 | 4.68 | Cp. | Structural… |
| Peflin | |Q641Z8|PEF1_RAT | 30012.40 | 5.67 | Cp; Cell mb. | Others. |
| Rap guanine nucleotide exchange factor-like 1 | |Q68EF8|RPGFL_MOUSE | 73695.33 | 5.84 | Carrier. | |
| Calcyclin-binding protein | |Q6AYK6|CYBP_RAT | 26541.19 | 7.64 | N; Cp. | Prot regulation. |
| Slice 15 | |||||
| Tropomyosin alpha-1 chain | |P42639|TPM1_PIG | 32680.56 | 4.69 | Cp. | Structural… |
| Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | |P49432|ODPB_RAT | 38982.13 | 6.20 | Mit. | Metabolism. |
| 3-hydroxyisobutyrate dehydrogenase, mitochondrial | |P29266|3HIDH_RAT | 35302.71 | 8.73 | Mit. | |
| Carbonyl reductase [NADPH] 1 | |P47727|CBR1_RAT | 30578.12 | 8.21 | Cp. | Stress Resp... |
| EF-hand domain-containing protein D2 | |A5D7A0|EFHD2_BOVIN | 26918.43 | 5.26 | Others. | |
| Charged multivesicular body protein 4b | |Q9D8B3|CHM4B_MOUSE | 24936.13 | 4.76 | Cp. | Prot regulation. |
| Elongation factor 1-beta | |O70251|EF1B_MOUSE | 24693.68 | 4.53 | Prot regulation. | |
| Clathrin light chain B | |P08082|CLCB_RAT | 25117.44 | 4.56 | Cp ves. | Prot regulation. |
| Complement component 1 Q subcomponent-binding protein, mitochondrial | |O35796|C1QBP_RAT | 30996.92 | 4.77 | Mit. | Others. |
| Methylglutaconyl-CoA hydratase, mitochondrial | |Q9JLZ3|AUHM_MOUSE | 33394.99 | 9.56 | Mit. | Metabolism. |
| Tropomyosin alpha-4 chain | |P09495|TPM4_RAT | 28509.70 | 4.66 | Cp. | Structural… |
| Coiled-coil-helix-coiled-coil-helix domain-containing protein 6 | |Q91VN4|CHCH6_MOUSE | 29798.81 | 8.41 | Others. | |
| Polymerase I and transcript release factor | |Q6NZI2|PTRF_HUMAN | 43476.14 | 5.51 | Cell mb; ER; Cp; Mit; N. | Others. |
| Drebrin-like protein | |Q9JHL4|DBNL_RAT | 48612.51 | 4.89 | Cp. | Structural… |
| Tubulin alpha-1B chain | |Q6P9V9|TBA1B_RAT | 50151.63 | 4.94 | Structural… | |
| Syntaxin-1B | |P61265|STX1B_RAT | 33244.69 | 5.25 | Cell mb. | Carrier. |
| Clathrin light chain A | |P08081|CLCA_RAT | 26980.50 | 4.41 | Cp ves. | Prot regulation. |
| Phosphoglycerate kinase 2 | |Q8MIF7|PGK2_HORSE | 44879.16 | 8.62 | Cp. | Metabolism. |
| Annexin A3 | |P14669|ANXA3_RAT | 36363.20 | 5.96 | Others. | |
| Acidic leucine-rich nuclear phosphoprotein 32 family member B | |Q9EST6|AN32B_RAT | 31060.63 | 3.87 | N. | Stress Resp... |
| Alpha-S1-casein | |O62823|CASA1_BUBBU | 24326.77 | 4.87 | Sec. | Carrier. |
| Tubulin beta chain | lP02554lTBB_PIG | 49860.95 | 4.78 | Structural… | |
| Slice 16 | |||||
| Apolipoprotein E | |P02650|APOE_RAT | 35753.46 | 5.23 | Sec. | Stress Resp... |
| Breast carcinoma-amplified sequence 1 homolog (Fragment) | |Q3ZB98|BCAS1_RAT | 58623.87 | 5.58 | Cp. | Others. |
| Heterogeneous nuclear ribonucleoproteins A2/B1 | |O88569|ROA2_MOUSE | 37402.67 | 8.97 | N; Cp. | Others. |
| 60S acidic ribosomal protein P0 | sp|P19945|RLA0_RAT | 34215.47 | 5.91 | Prot regulation. | |
| Adaptin ear-binding coat-associated protein 1 | |P69682|NECP1_RAT | 29792.40 | 5.97 | Cp ves; Cell mb. | Prot regulation. |
| Alpha-internexin | |P23565|AINX_RAT | 56115.38 | 5.20 | Structural… | |
| Gamma-soluble NSF attachment protein | |Q9CWZ7|SNAG_MOUSE | 34732.33 | 5.31 | Cell mb. | Prot regulation. |
| Heterogeneous nuclear ribonucleoprotein H3 | |P31942|HNRH3_HUMAN | 36926.49 | 6.37 | N. | Others. |
| Elongation factor 1-delta | |P57776|EF1D_MOUSE | 31293.03 | 4.91 | Prot regulation. | |
| RNA-binding protein Musashi homolog 2 | |Q96DH6|MSI2H_HUMAN | 39133.53 | 7.71 | Cp; N. | Others. |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | |P54311|GBB1_RAT | 37376.97 | 5.60 | Carrier. | |
| NSFL1 cofactor p47 | p|O35987|NSF1C_RAT | 40679.96 | 5.04 | N; Gol app. | Others. |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | |Q9DC69|NDUA9_MOUSE | 42509.15 | 9.75 | Mit. | Metabolism. |
| F-actin-capping protein subunit alpha-1 | |B2GUZ5|CAZA1_RAT | 32909.77 | 5.34 | Structural… | |
| Putative heterogeneous nuclear ribonucleoprotein A1-like protein 3 | |P0C7M2|RA1L3_HUMAN | 34223.28 | 9.23 | N; Cp. | Carrier. |
| Translocon-associated protein subunit alpha | |Q9CY50|SSRA_MOUSE | 32065.01 | 4.36 | ER: | Carrier. |
| Annexin A2 | |Q07936|ANXA2_RAT | 38678.24 | 7.55 | Sec; EM; Mel. | Others. |
| Palmitoyl-protein thioesterase 1 | |P45479|PPT1_RAT | 34455.01 | 7.09 | Lys. | Stress Resp... |
| Dihydropyrimidinase-related protein 2 | |P47942|DPYL2_RAT | 62277.57 | 5.95 | Cp. | Others. |
| Putative L-aspartate dehydrogenase | |Q9DCQ2|ASPD_MOUSE | 30269.63 | 6.45 | Metabolism. | |
| Transcriptional activator protein Pur-alpha | |P42669|PURA_MOUSE | 34883.73 | 6.07 | N. | Stress Resp... |
| Slice 17 | |||||
| Tubulin beta-4 chain | |B4F7C2|B4F7C2_RAT | 49585.77 | 4.78 | Structural… | |
| Tubulin alpha chain | |P68370|TBA1A_RAT | 50630.14 | 4.93 | Structural… | |
| Vimentin | |P08670|VIME_HUMAN | 53651.68 | 5.06 | Structural… | |
| Tubulin beta-3 chain | |Q4QRB4|TBB3_RAT | 50418.65 | 4.82 | Structural… | |
| Citrate synthase, mitochondrial | |Q8VHF5|CISY_RAT | 51866.75 | 8.54 | Mit. | Metabolism. |
| Reticulocalbin-2 | |Q8BP92|RCN2_MOUSE | 37432.96 | 4.27 | ER. | Others. |
| Septin-4 | |Q4R4X5|SEPT4_MACFA | 55147.26 | 5.64 | Structural… | |
| Protein kinase C and casein kinase substrate in neurons protein 1 | |Q5R411|PACN1_PONAB | 50921.58 | 5.15 | Cp. | Structural… |
| Obg-like ATPase 1 | |Q9NTK5|OLA1_HUMAN | 44743.57 | 7.64 | Metabolism. | |
| Sodium/potassium-transporting ATPase subunit beta-1 | |P07340|AT1B1_RAT | 35201.59 | 8.83 | Cell mb. | Carrier. |
| Hsc70-interacting protein | |P50503|F10A1_RAT | 41279.50 | 5.28 | Cp. | Prot regulation. |
| Dynactin subunit 2 | |Q99KJ8|DCTN2_MOUSE | 44116.88 | 5.14 | Cp; Cell mb. | Structural… |
| Hsp90 co-chaperone Cdc37 | |Q61081|CDC37_MOUSE | 44510.36 | 5.24 | Cp. | Prot regulation. |
| Creatine kinase, ubiquitous mitochondrial | |P30275|KCRU_MOUSE | 47003.72 | 8.39 | Mit inn mb. | Metabolism. |
| Slice 18 | |||||
| Endophilin-A1 | |O35179|SH3G2_RAT | 39899.28 | 5.26 | Cp; Cell mb. | Carrier. |
| F-box only protein 2 | |Q80UW2|FBX2_MOUSE | 33675.95 | 4.21 | Prot regulation. | |
| Septin-2 | |Q91Y81|SEPT2_RAT | 41592.55 | 6.15 | Cp. | Structural… |
| Acetyl-CoA acetyltransferase, mitochondrial | |P17764|THIL_RAT | 44695.00 | 8.92 | Mit. | Metabolism. |
| Stomatin-like protein 2 | |Q99JB2|STML2_MOUSE | 38413.95 | 8.74 | Cp; Cell mb. | Structural… |
| Fumarylacetoacetase | |A5PKH3|FAAA_BOVIN | 45975.54 | 6.67 | Metabolism. | |
| NAD-dependent deacetylase sirtuin-2 | |Q5RJQ4|SIRT2_RAT | 39319.27 | 6.67 | Cp. | Structural… |
| Acetyl-CoA acetyltransferase, cytosolic | |Q5XI22|THIC_RAT | 41108.41 | 6.86 | Cp. | Metabolism. |
| Thioredoxin-dependent peroxide reductase, mitochondrial | |P20108|PRDX3_MOUSE | 28127.03 | 7.15 | Mit. | Stress Resp... |
| Microtubule-associated protein 6 | |Q63560|MAP6_RAT | 100484.89 | 9.45 | Cp; Gol app. | Structural… |
| Macrophage-capping protein | |Q6AYC4|CAPG_RAT | 38798.86 | 6.11 | N; Cp; Mel. | Structural… |
| Tubulin alpha-1D chain | |Q2HJ86|TBA1D_BOVIN | 50282.78 | 4.91 | Structural… | |
| Heterogeneous nuclear ribonucleoprotein A3 | |Q6URK4|ROA3_RAT | 39651.99 | 9.10 | N. | Others. |
| Neuromodulin | |P07936|NEUM_RAT | 23603.34 | 4.61 | Cell mb; | Structural… |
| Proto-oncogene C-crk | |Q63768|CRK_RAT | 33844.72 | 5.39 | Cp; Cell mb. | Others. |
| Sodium/potassium-transporting ATPase subunit beta-3 | |Q63377|AT1B3_RAT | 31829.68 | 8.08 | Cell mb; Mel. | Others. |
| Slice 19 | |||||
| 60 kDa heat shock protein, mitochondrial | |P18687|CH60_CRIGR | 60955.49 | 5.91 | Mit. | Stress Resp... |
| Tubulin alpha-1C chain | |P68373|TBA1C_MOUSE | 49937.37 | 4.96 | Structural… | |
| Peripherin | |P21807|PERI_RAT | 53549.76 | 5.37 | Structural… | |
| Myc box-dependent-interacting protein 1 | |O08839|BIN1_RAT | 64533.21 | 4.95 | Cp; N. | Structural… |
| Dihydrolipoyl dehydrogenase, mitochondrial | |Q6P6R2|DLDH_RAT | 54038.09 | 7.96 | Mit. | Metabolism. |
| Septin-8 | |Q8CHH9|SEPT8_MOUSE | 49812.39 | 5.68 | Structural… | |
| Slice 20 | |||||
| Stress-70 protein, mitochondrial | |O35501|GRP75_CRIGR | 73857.70 | 5.97 | Mit. | Prot regulation. |
| Actin, cytoplasmic 2 | |P63259|ACTG_RAT | 41792.84 | 5.31 | Cp. | Structural… |
| V-type proton ATPase catalytic subunit A | |P50516|VATA_MOUSE | 68326.08 | 5.42 | Metabolism. | |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | |P08461|ODP2_RAT | 67165.84 | 8.76 | Mit. | Metabolism. |
| 78 kDa glucose-regulated protein | |P06761|GRP78_RAT | 72346.99 | 5.07 | ER; Mel. | Carrier. |
| Annexin A6 | |P48037|ANXA6_RAT | 75754.16 | 5.39 | Cp; Mel. | Others. |
| Lamin-A/C | |P48678|LMNA_MOUSE | 74237.57 | 6.54 | N. | Structural… |
| Myristoylated alanine-rich C-kinase substrate | |P29966|MARCS_HUMAN | 29794.51 | 4.32 | Cp; Cell mb. | Structural… |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | |Q66HF1|NDUS1_RAT | 79412.33 | 5.65 | Mit inn mb. | Metabolism. |
| Synaptotagmin-2 | |P29101|SYT2_RAT | 47209.57 | 8.18 | Cp ves; syn ves. | Structural… |
| Heat shock-related 70 kDa protein 2 | |P14659|HSP72_RAT | 69641.66 | 5.51 | Stress Resp... | |
| Heat shock 70 kDa protein 12A | |Q8K0U4|HS12A_MOUSE | 74978.38 | 6.32 | Carrier. | |
| Slice 21 | |||||
| Heat shock protein HSP 90-alpha | sp|P46633|HS90A_CRIGR | 84814.91 | 4.93 | Stress Resp... | |
| Mitochondrial inner membrane protein | sp|Q8CAQ8|IMMT_MOUSE | 83900.08 | 6.18 | Mit inn mb. | Others. |
| Slice 22 | |||||
| Spectrin alpha chain, brain | |P16086|SPTA2_RAT | 284637.50 | 5.20 | Cp. | Structural… |
| Heat shock cognate 71 kDa protein | |P63018|HSP7C_RAT | 70871.07 | 5.37 | Cp. Mel. | Stress Resp... |
| Microtubule-associated protein 2 | |P11137|MAP2_HUMAN | 202410.75 | 4.77 | Cp. | Structural… |
| Neural cell adhesion molecule 1 | |P13595|NCAM1_MOUSE | 94658.31 | 4.83 | Cell mb. | Structural… |
| Rab GDP dissociation inhibitor alpha | |Q7YQM0|GDIA_PONPY | 50536.64 | 5.00 | Cp. | Carrier. |
| Dihydropyrimidinase-related protein 5 | |Q9JHU0|DPYL5_RAT | 61540.39 | 6.60 | Cp. | Others. |
| Neurofascin | |Q810U3|NFASC_MOUSE | 138004.21 | 5.79 | Cell mb. | Structural… |
| Slice 23 | |||||
| Tubulin beta-2A chain | |Q7TMM9|TBB2A_MOUSE | 49906.97 | 4.78 | Structural… | |
| Tubulin alpha-1B chain | |Q6P9V9|TBA1B_RAT | 50151.63 | 4.94 | Structural… | |
| Neuroblast differentiation-associated protein AHNAK | Q09666|AHNK_HUMAN | 629101.22 | 5.80 | N. | Others. |
| Regulating synaptic membrane exocytosis protein 1 | |Q86UR5|RIMS1_HUMAN | 179654.84 | 9.62 | Cell mb. | Carrier. |
| Slice 24 | |||||
| Sodium/potassium-transporting ATPase subunit alpha-3 | |P06687|AT1A3_RAT | 111691.53 | 5.26 | Cell mb. | Carrier. |
| Aconitate hydratase, mitochondrial | |Q9ER34|ACON_RAT | 85433.44 | 7.87 | Mit. | Metabolism. |
| Tubulin beta-5 chain | |P09653|TBB5_CHICK | 49670.82 | 4.78 | Structural… | |
| Myelin-associated glycoprotein | |P20917|MAG_MOUSE | 69352.86 | 4.96 | Cell mb. | Structural… |
| 6-phosphofructokinase type C | |P47860|K6PP_RAT | 85720.28 | 6.94 | Metabolism. | |
| Microtubule-associated protein 1B | |P15205|MAP1B_RAT | 269499.65 | 4.74 | Structural… | |
| Plasma membrane calcium-transporting ATPase 2 | |P11506|AT2B2_RAT | 136811.20 | 5.70 | Cell mb. | Metabolism. |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | |P08461|ODP2_RAT | 67165.84 | 8.76 | Mit. | Metabolism. |
| Microtubule-associated protein 1A | |P34926|MAP1A_RAT | 299530.68 | 4.87 | Structural… | |
| Spectrin beta chain, brain 1 | |Q62261|SPTB2_MOUSE | 274223.06 | 5.40 | Cp. | Structural… |
| Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | |Q76MZ3|2AAA_MOUSE | 65322.61 | 5.00 | Prot regulation. | |
| Hexokinase-1 | |P27595|HXK1_BOVIN | 102408.01 | 6.29 | Mit out mb. | Metabolism. |
Characterization and classification of the proteins identified
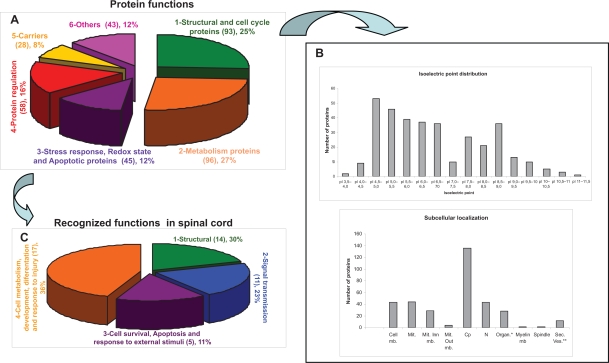
The proteins identified by MALDI-TOF/TOF and LC-MS/MS were characterized according to their molecular weight (MW), isoelectric point (pI), subcellular localization and recognized function. In total 367 unique proteins were identified with the different techniques employed. On the basis of Swiss-Prot and NCBI database information, the proteins were classified into six functional groups (Fig. 5A): Structural and Cell Cycle Proteins; Metabolic Proteins; Stress Response, Redox State and Apoptosis Proteins; Regulation proteins; Carriers and Other proteins. The different types of protein functions assigned to the proteins identified and the relative proportion of each group were represented (Fig. 5A represents), and a graph of the distribution of pI’s and cellular localization was generated (Fig. 5B). In addition, similar graphs were generated to represent the same features of those proteins recognized to be active in the nervous system.
Figure 5.
Characterization of the spinal cord proteins identified. A) The functional grouping of all the proteins identified using 2-DE and MALDI-TOF/TOF together with LC-MS/MS are presented. B) Isoelectric point distribution and subcellular localization of the proteins identified. C) Additional classification of the proteins with recognized function in spinal cord.
Discussion
To understand the complex biological processes at play in the central nervous system the key proteins involved must be identified. The exploration of the proteome has attracted increasing interest in recent years, particularly to establish reference maps designed to assist in biomarker discovery. In this regard, defining the complete spinal cord proteome is still an important challenge. This proteome may represent a fundamental key to better understand normal spinal cord physiology, as well as providing important clues to discover the molecular basis of neurodegeneration after spinal cord injury.
In the present study, we have described the proteins present in the rat spinal cord by employing different proteomic tools. Accordingly, we have defined a fast, easy and reproducible protein extraction protocol for the spinal cord. Efficient protein extraction is an essential step in proteomic studies, and the development of this specific sequential extraction augmented the number of proteins isolated, focusing mainly on membrane and hydrophobic proteins. As expected, we identified many mitochondrial and membrane proteins, as well as many soluble proteins, further supporting the efficiency of this methodology.
One of the major problems associated with proteomic analyses are the contaminants in the sample that could interfere with the isoelectrofocusing of spinal proteins (salts, DNA, lipids …). To diminish the effect of this interference, a filter step was included before initiating the 2-DE gel protocol. We employed conventional 2-DE over different pH ranges (e.g. 4–7 and 3–11 NL) to generate different maps that could help search for potential biomarkers. Furthermore, the high degree of 2-DE gel reproducibility and the resolution obtained is necessary to generate good quality maps from the rat spinal cord and for future differential expression analyses. The gels focused with 17 cm pH 4–7 IPG strips did not resolve a large number of spots, and some proteins with a high isoelectric point were not focused correctly with a line of precipitated proteins appearing at the basic extreme of the gel. This distribution in 2-DE gels pH 4–7 could present problems for posterior spot identification, and even for future differential expression analyses between healthy individuals and patients. Accordingly, better resolution was obtained with 2-DE gels with non-linear pH3–11 24 cm IPG strips, avoiding the precipitation of basic proteins. These quality of these gels was relatively high and with a good protein spot distribution, leading to the identification of 200 different spots by MALDI-TOF/TOF.
It is important to note that 2-DE gels cannot resolve proteins below 10 kDa and above 100 kDa, including the more acidic or basic proteins. To maximize the number of proteins identified and to complement the results obtained for 2-DE MALDI-MS/MS, LC-MS/MS analyses identified a further 367 unique proteins. Interestingly both proteomic tools could detect proteins with a broad range of molecular weights and isoelectric points, reflecting the efficiency of the methods employed. We found many proteins in the rat spinal cord with theoretical isoelectric points between 4.0–6.0 and 8.0–9.5, although less were obtained between 6.5 and 7.5.
The spinal proteins were classified into 6 different functional groups: Structural and Cell Cycle Proteins (25%), Metabolic Proteins (30%), Stress Response, Redox State and Apoptosis Proteins (16%), Regulation proteins (8%), Carriers and Other proteins Structural Proteins 12%. Structural and cell cycle proteins constituted a complex and heterogeneous group of cytoskeleton proteins, such as Microtubule-associated protein 1A, myelin sheet, or extracellular matrix and attachment proteins. In addition, DNA scaffold proteins and other structural proteins implicated in mitotic division and cell cycle regulation were characterized, making up around 25% of the total proteins identified. The second category, metabolic proteins, was also very broad and it reached nearly 30% of the total protein content, mainly containing hydrolytic and glucolytic enzymes. The third group, Stress Response, Redox State and Apoptosis proteins, was also a complex group made up of different proteins implicated in stress and injury response (Heat Shock Proteins). Furthermore, we included other proteins here associated with reducing oxidative damage and apoptosis. This group contained around 12% of the total proteins identified. Regulatory proteins related to protein synthesis, including transcription and translation, protein folding and degradation, made up about 16% of the proteins identified. Protein carriers were comprised of transporters and other metabolite binding molecules that represented approximately 8% of the total. Finally, a category of proteins that could not be classified into any of the above groups was denominated as “other” and contributed up to 12% to the complete proteome described here.
The proteins identified with a recognized function in the SC were organized into four functional groups. The numerous proteins in each functional group suggests that the technique developed in this report will be extremely useful to identify possible therapeutic targets for spinal cord injury, and pathways that may arrest the development of associated pathologies such as neuropathic pain and spasticity. Furthermore this technique will be important to develop future regenerative strategies.
Structural proteins were defined that included many common neuronal and glial proteins normally present in central nervous system tissue such as: Neurofilament (NF), Glial fibrillary acidic protein (GFAP), Myelin basic protein (MBP), Myelin-associated glycoprotein (MAG), Neural cell adhesion molecule (NCAM) and Macrophage migration inhibitory factor (MIF). Several of these proteins have a clear role during acute SCI such as GFAP in gliogenesis41 or MIF in astrocyte proliferation,42 while an increase in MAG would suggest the presence of a spinal environment that is inhibitory to nerve growth.43
The second group of proteins were related to neurotransmission. Several Vesicle-associated membrane proteins (VAMPs) were identified but only some of these are thought to be upregulated in the pathological state following SCI, although similar changes may have been identified following peripheral nerve injury axotomy.44 Many others were related to glutamatergic communication such as Glutamine synthetase (GS) and Glutamate dehydrogenase (GDH). These two proteins are known to be therapeutic targets for the successful treatment of spinal cord ischemia.45
Among the proteins responsible for cell survival and combating apoptosis, the presence of Gamma-enolase, Glucose-6-phosphate isomerase, Peroxiredoxin-2 (possible anti-oxidant protein) and Protein DJ-1 in the normal spinal cord should be highlighted, as opposed to only one protein (Glyceraldehyde-3-phosphate dehydrogenase) associated with a pro-apoptotic profile. The upregulation of neuron-specific enolase has been previously described as a potential biomarker of acute SCI.46 An increase in glyceraldehyde-3-phosphate dehydrogenase in spinal cord tissue has been demonstrated after contusion injury,47 while previous proteomic analysis has highlighted the upregulation of peroxiredoxin 2 protein after experimental SCI.48
Lastly, numerous proteins associated with cell metabolism, development, and response to injury were identified, including those associated with neuronneuron interactions (Neural cell adhesion molecule 1) and neuron-glial cell interactions (Neurofascin), neurogenesis (Lyssencephaly-1 homologue A, Alpha-Internexin, Stathmin, Dihydropyrimidinase-related protein), neurite outgrowth (Neural cell adhesion molecule 1, Neurofascin), neuronal precursor proliferation (Lyssencephaly-1 homologue A), synaptogenesis and synaptic plasticity (Neurofascin and 14-3-3 protein gamma), axonal guidance (Neurofascin), axonal regeneration (Macrophage migration inhibitory factor) and myelination (Neurofascin). Significantly, the induction of a serine-threonine kinase stathmin after SCI has already been demonstrated and it was associated with an increase in glial proliferation.49
In addition, several proteins with no known spinal function were identified (following a NCBI bibliographic database search), as well as Protein S100-B that has been proposed as a marker of SCI severity46 and Ubiquitin carboxyl-terminal hydrolase isozyme L1 that may be related to axon degradation.50 An upregulation of Gamma-synuclein has been described in the SC51 and the spinal dorsal horn45 although its precise role during acute SCI is not known. Moreover, both Thioredoxin-dependent peroxide reductase and Palmitoyl-protein thioesterase 1 have been linked to the negative regulation of neuron apoptosis (Swiss-prot Database). Finally, the Platelet-activating factor acetylhydrolase IB subunit alpha may promote the proliferation of neuronal precursors (Swiss-prot Database).
Taken together these data help highlight the change in the spinal cord proteome during acute and chronic SCI, as well helping to define the different profiles associated with symptoms such as neuropathic pain, spasticity, they will serve to benchmark future neuro-regenerative therapies. Despite the promising results obtained in these studies, it will be necessary to define more of the proteins present in the spinal cord proteome. We hope that by continuing these studies and complementing them, the characterization of the complete protein profile of the rat spinal cord will be possible, and differential expression analyses can be carried out in human and/or other animal models.
Acknowledgments
We thank Carmen Bermudez and Ana Isabel Carrasco for their technical support.
This work was supported by grants from the Instituto de Salud Carlos III (FIS PI070537), Fondo de Investigación Sanitaria de Castilla la Mancha (FISCAM, PI2008/08), Fondo de Investigación Sanitaria de Castilla la Mancha (FISCAM, PI2008/28), REDES TEMATICAS DE INVESTIGACION COOPERATIVA (RD06/0014/1015).
Footnotes
Disclosures
The authors report no conflicts of interest.
References
- 1.Bonita R. Epidemiology of Stroke. Lancet. 1992;339:342–4. doi: 10.1016/0140-6736(92)91658-u. [DOI] [PubMed] [Google Scholar]
- 2.Alonso I, Regidor E, Rodriguez C, Gutierrez-Fisac JL. The main causes of death in Spain. Med Clin (Barc) 1996;12:441–5. [PubMed] [Google Scholar]
- 3.Beattie MS, Hermann GE, Rogers RC, Bresnahan JC. Cell death in models of spinal cord injury. Prog Brain Res. 2002;137:37–47. doi: 10.1016/s0079-6123(02)37006-7. [DOI] [PubMed] [Google Scholar]
- 4.Profyris C, Cheema SS, Zang D, Azari MF, Boyle K, Petratos S. Degenerative and regenerative mechanisms governing spinal cord injury. Neurobiol Dis. 2004;3:415–36. doi: 10.1016/j.nbd.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Sindrup SH, Jensen TS. Efficacy of pharmacological treatments of neuropathic pain: An update and effect related to mechanism of drug action. Pain. 1999;83:389–400. doi: 10.1016/S0304-3959(99)00154-2. [DOI] [PubMed] [Google Scholar]
- 6.Hansson PT, Dickenson AH. Pharmacological treatment of peripheral neuropathic pain conditions based on shared commonalities despite multiple etiologies. Pain. 2005;113:251–4. doi: 10.1016/j.pain.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Lamer TJ, Rho RH, et al. Contemporary management of neuropathic pain for the primary care physician. Mayo Clin Proc. 2004;79:1533–45. doi: 10.4065/79.12.1533. [DOI] [PubMed] [Google Scholar]
- 8.Lanzrein AS, Johnston CM, Perry VH, et al. Longitudinal study of inflammatory factors in serum, cerebrospinal fluid, and brain tissue in Alzheimer disease: interleukin-1beta, interleukin-6, interleukin-1 receptor antagonist, tumor necrosis factor-alpha, the soluble tumor necrosis factor receptors I and II and alpha1-antichymotrypsin. Alzheimer Dis Assoc Disord. 1998 Sep;12(3):215–27. doi: 10.1097/00002093-199809000-00016. [DOI] [PubMed] [Google Scholar]
- 9.Rachel Pardes Berger, Mary Clyde Pierce, Stephen R, Wisniewski, et al. Neuron-Specific Enolase and S100B in Cerebrospinal Fluid After Severe Traumatic Brain Injury in Infants and Children. Pediatrics. 2002 Feb 2;109(1):e31. doi: 10.1542/peds.109.2.e31. [DOI] [PubMed] [Google Scholar]
- 10.Brown P, Kenney K, Little B, et al. Gajdusek. Intracerebral distribution of infectious amyloid protein in spongiform encephalopathy. Annals of Neurology. 38(2):245–53. doi: 10.1002/ana.410380218. [DOI] [PubMed] [Google Scholar]
- 11.Allan Butterfield D. Debra Boyd-Kimball and Alessandra Castegna. Proteomics in Alzheimer’s disease: insights into potential mechanisms of neurodegeneration. Journal of Neurochemistry. 2003;86:1313–27. doi: 10.1046/j.1471-4159.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 12.Maja Puchadesa, Sara Folkesson Hanssona, Carol L, et al. Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer’s disease. Molecular Brain Research. 2003 Oct 21;:118, 1–2, 140–6. doi: 10.1016/j.molbrainres.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Kunz S, Tegeder I, Coste O, et al. Comparative proteomic analysis of the rat spinal cord in inflammatory and neuropathic pain models. Neuroscience Letters. 2005;381:289–93. doi: 10.1016/j.neulet.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Ding Q, Wu Z, Guo Y, et al. Proteome analysis of up-regulated proteins in the rat spinal cord induced by transection injury. Proteomics. 2006;6:505–18. doi: 10.1002/pmic.200500296. [DOI] [PubMed] [Google Scholar]
- 15.Yan X, Liu T, Yang S, et al. Proteomic profiling of the insoluble pellets of the transected rat spinal cord. J Neurotrauma. 2009;26:179–93. doi: 10.1089/neu.2008.0533. [DOI] [PubMed] [Google Scholar]
- 16.Ka Wan Li, Martin P, Hornshaw Roel C, Van der Schors. Proteomics Analysis of Rat Brain Postsynaptic Density. Implications of the Diverse Protein Functional Groups for the Integration Ofsynaptic Physiology. Journal of Biological Chemistry. 2004 Jan 9;279(2):987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- 17.Michael Fountoulakis, Sophia Kossida. Proteomics-driven progress in neurodegeneration research. Electrophoresis. 2006;27:1556–73. doi: 10.1002/elps.200500738. [DOI] [PubMed] [Google Scholar]
- 18.Orth AP, Batalov S, Perrone M, Chanda SK. The promise of genomics to identify novel therapeutic targets. Expert Opin Ther Targets. 2004;8:587–96. doi: 10.1517/14728222.8.6.587. [DOI] [PubMed] [Google Scholar]
- 19.Choudhary J, Grant SG. Proteomics in postgenomic neuroscience: the end of the beginning. Nat Neurosci. 2004;7:440–5. doi: 10.1038/nn1240. [DOI] [PubMed] [Google Scholar]
- 20.Bermudez-Crespo J, Lopez JL. A better understanding of molecular mechanisms underlying human diseases. Proteomics Clin Appl. 2007;1:983–1003. doi: 10.1002/prca.200700086. [DOI] [PubMed] [Google Scholar]
- 21.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–46. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 22.Norin M, Sundstrom M. Structural proteomics: Developments in structure-to-function predictions. Trends Biotechnol. 2002;20:79–84. doi: 10.1016/s0167-7799(01)01884-4. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto H, Komori N. Protein identification on two-dimensional gels archived nearly two decades ago by in-gel digestion and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Anal Biochem. 1999;270:176–9. doi: 10.1006/abio.1999.4054. [DOI] [PubMed] [Google Scholar]
- 24.Weston AD, Hood L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–96. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 25.van der Greef J, Stroobant P, van der Heijden R. The role of analytical sciences in medical systems biology. Curr Opin Chem Biol. 2004;8:559–65. doi: 10.1016/j.cbpa.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Ideker I. Systems biology 101—what you need to know. Nat Biotechnol. 2004;22:473–5. doi: 10.1038/nbt0404-473. [DOI] [PubMed] [Google Scholar]
- 27.Marko-Varga G, Fehniger TE. Proteomics and disease—the challenges for technology and discovery. J Proteome Res. 2004;3:167–78. doi: 10.1021/pr049958+. [DOI] [PubMed] [Google Scholar]
- 28.Domon B, Broder S. Implications of new proteomics strategies for biology and medicine. J Proteome Res. 2004;3:253–60. doi: 10.1021/pr034082c. [DOI] [PubMed] [Google Scholar]
- 29.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–9. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 30.Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665–85. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 31.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–21. [PMC free article] [PubMed] [Google Scholar]
- 32.Carrette O, Burkhard PR, Sanchez JC, Hochstrasser DF. State-of-the-art two-dimensional gel electrophoresis: a key tool of proteomics research. Nat Protoc. 2006;1:812–23. doi: 10.1038/nprot.2006.104. [DOI] [PubMed] [Google Scholar]
- 33.Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–9. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 34.Barderas MG, Wigdorovitz A, Merelo F, et al. Serodiagnosis of African swine fever using the recombinant protein p30 expressed in insect larvae. Journal Virological Methods. 2000;89:129–36. doi: 10.1016/s0166-0934(00)00209-3. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez-Barderas M, Gallego-Delgado J, Mas S, et al. Isolation of circulating human monocytes with high purity for proteomic analysis. Proteomics. 2004;4:432–7. doi: 10.1002/pmic.200300629. [DOI] [PubMed] [Google Scholar]
- 36.Durán MC, Mas S, Martin-Ventura JL, et al. Proteomic analysis of human vessels: application to atherosclerotic plaques. Proteomics. 2003;3:973–8. doi: 10.1002/pmic.200300389. [DOI] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 40.Barderas MG, Tunon J, Darde VM, et al. Circulating human monocytes in the acute coronary syndrome express a characteristic proteomic profile. J Proteome Res. 2007;6(2):876–6. doi: 10.1021/pr0601990. [DOI] [PubMed] [Google Scholar]
- 41.Nieto-Sampedro M. Neurite outgrowth inhibitors in gliotic tissue. Adv Exp Med Biol. 1999;468:207–24. doi: 10.1007/978-1-4615-4685-6_17. [DOI] [PubMed] [Google Scholar]
- 42.Koda M, Nishio Y, Hashimoto M, et al. Up-regulation of macrophage migration-inhibitory factor expression after compression-induced spinal cord injury in rats. Acta Neuropathol. 2004;108:31–6. doi: 10.1007/s00401-004-0853-z. [DOI] [PubMed] [Google Scholar]
- 43.Giger RJ, Venkatesh K, Chivatakarn O, et al. Mechanisms of CNS myelin inhibition: evidence for distinct and neuronal cell type specific receptor systems. Restor Neurol Neurosci. 2008;26:97–115. [PMC free article] [PubMed] [Google Scholar]
- 44.Jacobsson G, Piehl F, Meister B. VAMP-1 and VAMP-2 gene expression in rat spinal motoneurones: differential regulation after neuronal injury. Eur J Neurosci. 1998;10:301–16. doi: 10.1046/j.1460-9568.1998.00050.x. [DOI] [PubMed] [Google Scholar]
- 45.Wu GJ, Chen WF, Sung CS, et al. Preventive effects of intrathecal methylprednisolone administration on spinal cord ischemia in rats: the role of excitatory amins acid metabolizing systems. Neuroscience. 2007;147:294–303. doi: 10.1016/j.neuroscience.2007.04.040. [DOI] [PubMed] [Google Scholar]
- 46.Cao F, Yang XF, Liu WG, et al. Elevation of neuron-specific enolase and S-100beta protein level in experimental acute spinal cord injury. J Clin Neurosci. 2008;15:541–4. doi: 10.1016/j.jocn.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Wu X, Yoo S, Wrathall JR. Real-time quantitative PCR analysis of temporal-spatial alterations in gene expression after spinal cord contusion. J Neurochem. 2005;93:943–52. doi: 10.1111/j.1471-4159.2005.03078.x. [DOI] [PubMed] [Google Scholar]
- 48.Kang SK, So HH, Moon YS, Kim CH. Proteomic analysis of injured spinal cord tissue proteins using 2-DE and MALDI-TOF MS. Proteomics. 2006;6:2797–812. doi: 10.1002/pmic.200500621. [DOI] [PubMed] [Google Scholar]
- 49.Shen A, Liu Y, Zhao J, et al. Temporal-spatial expressions of p27kip1 and its phosphorylation on Serine-10 after acute spinal cord injury in adult rat: Implications for post-traumatic glial proliferation. Neurochem Int. 2008;52:1266–75. doi: 10.1016/j.neuint.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Li GL, Farooque M, Holtz A, Olsson Y. Expression of the ubiquitin carboxyl-terminal hydrolase PGP 9.5 in axons following spinal cord compression trauma. An immunohistochemical study in the rat. APMIS. 1997;105:384–90. doi: 10.1111/j.1699-0463.1997.tb00585.x. [DOI] [PubMed] [Google Scholar]
- 51.Willis D, Li KW, Zheng JQ, et al. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J Neurosci. 2005;25:778–91. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li JY, Henning Jensen P, Dahlström A. Differential localization of alpha-, beta- and gamma-synucleins in the rat CNS. Neuroscience. 2002;113:463–78. doi: 10.1016/s0306-4522(02)00143-4. [DOI] [PubMed] [Google Scholar]