Abstract
Escherichia coli pseudouridine synthase RluF is dedicated to modifying U2604 in a stem–loop of 23S RNA, while a homologue, RluB, modifies the adjacent base, U2605. Both uridines are in the same RNA stem, separated by ~4 Å. The 3.0 Å X-ray crystal structure of RluF bound to the isolated stem–loop, in which U2604 is substituted by 5-fluorouridine to prevent catalytic turnover, shows RluF distinguishes closely spaced bases in similar environments by a selectivity mechanism based on a frameshift in base pairing. The RNA stem–loop is bound to a conserved binding groove in the catalytic domain. A base from a bulge in the stem, A2602, has folded into the stem, forcing one strand of the RNA stem to translate by one position and thus positioning U2604 to flip into the active site. RluF does not modify U2604 in mutant stem–loops that lack the A2602 bulge and shows dramatically higher activity for a stem–loop with a mutation designed to facilitate A2602 refolding into the stem with concomitant RNA strand translation. Residues whose side chains contact rearranged bases in the bound stem–loop, while conserved among RluFs, are not conserved between RluFs and RluBs, suggesting that RluB does not bind to the rearranged stem loop.
Keywords: pseudouridine synthase, ribosomal RNA, RNA modification, S4 domain, induced fit
Introduction
All stable cellular RNA is posttranscriptionally modified.1,2 The modifications affect the structure, conformational stability, and chemical properties of the RNA3 and tend to cluster in functionally important regions of RNA, such as the peptidyl-transferase center of the ribosome or the anticodon stem–loop of tRNA.4–6 The role of RNA modification in stabilizing active conformations of tRNA and ribosomal RNA (rRNA) is well documented.7 Modifications in both tRNA and rRNA correlate with translational efficiency and accuracy.8,9
RNA modification can be critical for processes involving molecular recognition. For example, modifications of yeast , and possibly of other RNA transcripts, prevent them from being degraded by the exosome.10,11 RNA modifications of the steroid receptor RNA activator regulate transcriptional activation by hormone-responsive nuclear receptors.12
Pseudouridine (Ψ), the C5-glycosyl isomer of uridine, is the most common RNA modification. In eukaryotes and archaea, rRNA pseudouridylation is accomplished by ribonucleoprotein particles composed of a conserved protein assembly and a target-specific guide snoRNA (small nucleolar RNA) that can base-pair to rRNA sequences on either side of the target.13 However, bacteria use separate, single-chain Ψ synthases to catalyze pseudouridylation at one or a few sites in tRNAs and rRNAs. The Ψ synthase sequences are not highly homologous, but they have several conserved sequence motifs,14,15 and the catalytic domains of Ψ synthases have a conserved structural core.16,17 The bacterial Ψ synthases have been grouped into five families based on sequence similarity, and the families are named for their first discovered member, RsuA, RluA, TruA, TruB, and TruD,14,15,18 where the name of the representative enzyme indicates its substrate (uridine of ribosomal large or small subunit or of tRNA). A sixth Ψ synthase family, represented by Pus10, is conserved in archaea and eukaryotes, but not in bacteria.19
A challenge in structure–function correlation for the single-chain Ψ synthases is to determine how these homologous enzymes select their targets and why different enzymes, even within a single family, show different levels of specificity. For example, TruA and RluD are region specific: they modify more than one site within a single loop of rRNA or tRNA.20,21 Other Ψ synthases modify two or more sites that lie in different secondary structures of rRNA, or even in different substrates.22 TruB modifies the same site on multiple substrates (all elongator tRNAs). Structures and RNA-binding mechanisms for all of these types of Ψ synthases have recently been characterized.23–27 Here we present the structure of an RNA complex of RluF, a Ψ synthase that primarily targets a single site on rRNA.
Escherichia coli RluF is a Ψ synthase from the RsuA family that modifies U2604 in the Watson–Crick base-paired stem of a 23S RNA stem–loop at the peptidyltransferase center of the ribosome.22 Another Ψ synthase from the RsuA family, RluB, specifically modifies only U2605, the nucleotide adjacent to the U2604 in the stem.22 Thus RluF and RluB are able to select target bases that reside in the same RNA stem and that are less than 4 Å apart. While RluB is monospecific to U2605, RluF can modify this base in addition to its primary target, the adjacent U2604, although U2605 is modified at a much-reduced rate. The structural features of RluF and RluB that are responsible for their different selectivities must be highly specialized, since the two enzymes are homologues with the same three conserved domains and an overall sequence identity of 31%. Except at their N-and C-termini, they align with each other with no inserts or deletions larger than five residues.
For some RNA-modifying enzymes, elaborate rearrangements of RNA secondary structure are key to discrimination of the cognate substrate;23,28–31 however, the target bases in these cases lie in flexible loops. Extensive substrate rearrangement seemed less likely to be a mechanism for selectivity for RluF or RluB, since these enzymes modify bases in a stable RNA stem. A 22-base rRNA fragment containing U2604 and U2605 is predicted to form the same stem–loop structure it has in the ribosome, with a predicted free energy of approximately −10 kcal,32 and the rearrangement of this RNA structure upon binding to the enzyme would be energetically costly. Initially, we thought the more likely mechanism for extruding the target base from the stem into the active site would be simple displacement of the base by a side chain from the enzyme, which would leave most of the base pairing in the stem intact. Such a mechanism is used, for example, by many DNA (cytosine C5) methyltransferases and DNA glycosylases.33,34
To discover how RluF binds to its target and to shed light on how RluF and RluB distinguish between stacked uridines separated by less than 4 Å, we determined the crystal structure of full-length RluF bound to the 22-nt RNA stem–loop spanning the RluF substrate site. Contrary to expectations, we found that a major reorganization of the base pairing in the stem of the 22-mer occurs on RluF binding. Based on this result, we engineered a new stem–loop designed to minimize the energetic cost of the reorganization and showed that this novel substrate is modified at a 37-fold faster initial rate than the wild-type hairpin.
Results and Discussion
Co-crystallization of RluF–RNA
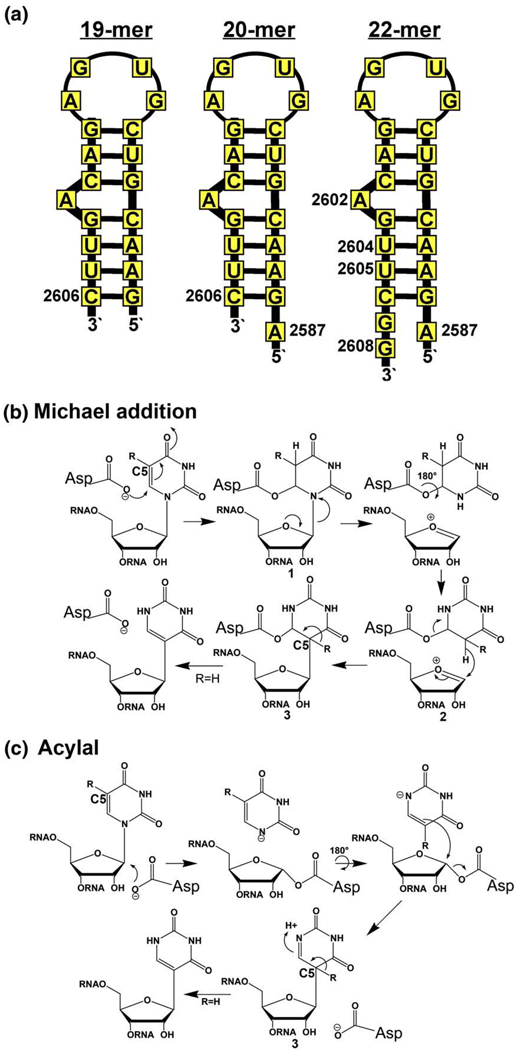
In an attempt to identify a minimal substrate for RluF–RNA crystallization, we synthesized three 23S RNA fragments that included the target-containing stem–loop (Fig. 1a) and analyzed the ability of the enzyme to pseudouridylate these substrates. RluF showed low activity for an isolated 19-mer stem–loop, nucleotides 2588–2606, which represents the basic stem–loop in the ribosome structure. However, a 20-nt fragment, which contained one extra nucleotide at the 5′ end of the stem–loop, and a 22-nt fragment, which contained one extra nucleotide at the 5′ end and two extra nucleotides at the 3′ end of the stem–loop, were much better substrates: after 5 min, RluF modified 0.15 pmol uridine per picomole enzyme for the 19-mer, 0.68 pmol uridine per picomole enzyme for the 20-mer, and 0.89 pmol uridine per picomole enzyme for the 22-mer.
Fig. 1.
Design of a small RluF substrate for co-crystallization. (a) Three 23S RNA stem–loop fragments containing the RluF target base that were tested as RluF substrates (see Materials and Methods). RluF showed highest activity for the 22-mer. The Michael addition (b) and acylal (c) chemical mechanisms have been proposed for Ψ synthase catalysis.35,36 In both mechanisms, the final chemical step is proton abstraction from C5 of intermediate 3. When uridine is substituted with 5-fluorouridine (R=F), the reaction stalls at this intermediate; thus, the 22-mer small substrate in which the target uridine was replaced with 5-fluorouridine was used for co-crystallization with RluF.
For crystallization we synthesized a mechanism-based substrate analog inhibitor in which the target uridine of the 22-mer was replaced with its 5-fluoro derivative. The reaction catalyzed by Ψ synthase requires dual reactions of N-glycoside bond cleavage and C–C bond formation. An invariant Asp in the active site is required for catalysis, but its role is still debated. Either Michael addition of the Asp to C-6 of the pyrimidine ring (Michael addition mechanism; Fig. 1b)35 or nucleophilic displacement of the uracil by attack of the Asp at C1′ of the ribose (acylal mechanism, Fig. 1c)36 cleaves the glycosidic bond. Proton abstraction from C5 of the isomerized uridine is the final step in both mechanisms, and when the hydrogen is substituted with fluorine, this step cannot occur.
The 22-mer substrate analog inhibitor formed a covalent complex with the enzyme that could be isolated on SDS gels. Presumably, the covalent complex was intermediate 3 of Fig. 1b or a covalent adduct of the catalytic Asp with intermediate 3 of Fig. 1c (depending on whether the enzyme uses the Michael addition or acylal mechanism) where in either case R=F. This substrate analog was subsequently co-crystallized with RluF.
Crystals of the RluF–22-mer complex are of space group P21 with a=89.5 Å, b=84.1 Å, c=91.3 Å, and β=94.9° and contain four protein–RNA complexes per asymmetric unit. The four independent complexes have essentially identical structures at 3 Å resolution and form a pair of dimers related by approximate 2-fold symmetry. The dimers are not physiologic since RluF is a monomer in solution;37 however, the dimeric arrangement provides an important crystal contact. Within one dimer (A and B or C and D in Fig. 2a), the stem–loops of the two adjacent RNA molecules have swapped bases such that G2588 of one stem base pairs with C2606 of the other and vice versa (Fig. 2b). The dimer interface buries 587 Å2 surface area.
Fig. 2.
Asymmetric unit of the RluF–RNA crystals. (a) Stereo ribbon drawing of the four equivalent RluF–RNA complexes in the asymmetric unit of the crystal structure. Complexes A and B are related to complexes C and D by a pseudo 2-fold axis that is approximately perpendicular to the plane of the paper. The interface between A and B comprises only nucleotides from the RNA substrates; the A and B stem–loops have swapped base G2588 with each other. The same is true of complexes C and D. (b) Close-up stereo plot of the A–B dimer interface showing base swapping between adjacent RNAs. The figure was made with PyMOL (DeLano Scientific).
Overall structure of the full-length RluF–RNA complex
Limited proteolysis followed by mass spectrometry and N-terminal sequencing shows that RluF is composed of three domains separated by flexible linkers.37 Only residues corresponding to the N-terminal and central domains are visible in our RluF–RNA electron density maps. The C-terminal domain, shown by limited proteolysis to comprise residues Thr253–Arg290,37 is disordered. The final structure consists of the following protein residues: chain A, 4–243; chain B, 3–243; chain C, 4–242; chain D, 4–242. All 22 nucleotides of the stem-loop are present in all four complexes.
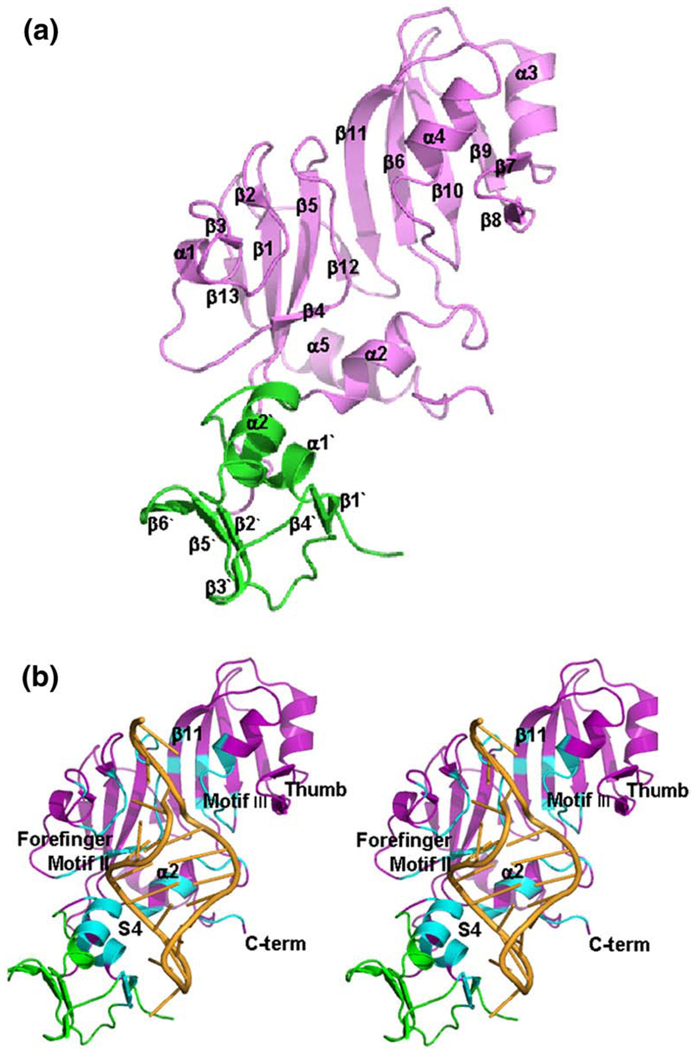
The N-terminal domain composed of amino acids Pro3–Pro61 (Fig. 3a, green ribbons), has the same fold as the RNA-binding domain in ribosomal protein S4.38 The S4-like fold in RluF had been predicted by sequence analysis,37,39 and RsuA from the same family has a very similar N-terminal S4-like domain.39 The rmsd for 51 Cαs that compose the structurally conserved S4 fold in the N-terminal domains of RsuA and RluF after superposition is 1.28 Å.
Fig. 3.
Conserved RluF–RNA interface. (a) Ribbon drawing of one RluF molecule, with the catalytic domain shown in lavender and the S4-like domain in green. Secondary-structure elements in the catalytic domain are labeled according to the structure of the apo ΔRluF catalytic domain.37 Labels for secondary-structure elements of the S4-like domain are distinguished by a prime. (b) Stereo ribbon drawing of the RluF–22-mer complex, with the RNA shown in orange. Residues that are in contact with the RNA are in cyan. Conserved structural features that have roles in substrate binding in other Ψ synthases are labeled. The figure was made with PyMOL (DeLano Scientific).
The central, catalytic domain of RluF, residues Gly66–Ser240, has an α–β fold that is conserved in other Ψ synthases (Fig. 3a, lavender ribbons). The previously reported crystal structure of apo ΔRluF, an N-terminal 65-amino-acid truncation of RluF, had revealed the conserved fold of the RluF catalytic domain,37 but the C-terminal domain was disordered, as it is in our full-length RluF–RNA crystals. Thus, while the crystallized apoprotein contained residues 66–290, the structure comprised only residues 66–240.
The RluF C-terminal domain residues, Thr253–Arg290, show significant sequence homology to the N-terminal 48 amino acids of ribosomal protein L2 (42% identical residues). The N-terminal region of L2 is intrinsically unstructured, allowing it to form an extensive interface with ribosomal RNA.40 Eighteen of the 48 disordered C-terminal residues in RluF are Lys or Arg, suggesting that the C-terminal domain may similarly associate with rRNA.
The 22-mer stem–loop binds to an RNA-binding groove whose floor is formed by β11 and whose walls are formed by residues of the loop regions from two of the characteristic Ψ synthase sequence motifs, motifs II and III14,15 (Fig. 3b). The motif III loop binds to the major groove of the stem, while the motif II loop binds to the minor groove of the stem and contains the catalytic Asp107 and the highly conserved Arg105. The guanidinium group of Arg105 is intercalated between the bases of G2603 and U2605, taking the place of the target base, U2604, which has flipped out of the stem and into the catalytic site.
Besides motifs II and III, several other regions of the protein bind to the RNA stem–loop (Fig. 3b). Together the RNA–protein interactions bury over 3090 Å2 surface area, approximately 30% of the stem–loop surface. A loop connecting β2 and β3 binds to the minor groove of the stem (labeled “forefinger loop” in Fig. 3b). Helix α2, which belongs to the conserved core of the catalytic domain, also binds to the minor groove, as do residues at the C-terminal end of the catalytic domain. Approximately one-third of the RNA–protein interface involves the N-terminal, S4-like domain. The helix–turn–helix motif of this domain binds in the major groove of the stem, while residues in β4′ interact with the RNA loop (Fig. 3b).
The common elements of RNA binding: motifs II and III, α2, thumb and forefinger loops, and R105
The RNA-binding surface of RluF has many similarities to the RNA-binding surfaces of other Ψ synthases. The loop regions of motif II and motif III, β11, and helix α2 are all conserved segments of the catalytic core whose counterparts in TruB, TruA, and RluA also bind to RNA.23–26 Arg105 of RluF that displaces the target base from a base stack has a conserved equivalent in RluA. In RluA, this Arg has been shown through mutagenesis to be essential for activity.23
The RluF β2–β3 forefinger loop binds to the substrate minor groove (Fig. 3b). It is one of two variable loops that bind to RNA in some, but not all, Ψ synthases. Its counterpart in RluA is proposed to have a key role in protein-induced substrate reorganization.23 The second variable loop, named the thumb loop, is a key binding determinant in both RluA and TruB,23,24 but it is too short in RluF to make contact with the RNA stem–loop (Fig. 3b).
The crystal structures of RluF–, RluA–, and TruB– RNA complexes show that the RNA substrates adopt similar conformations in the binding grooves of the catalytic cores. After superimposition of the conserved protein cores of the RluF, TruB, and RluA complexes, the target uridines and the nucleotides that are 5′ to the target uridine (A2602, G2603 in RluF; G30, A31 in RluA; and G53, U54 in TruB) also closely align (Fig. 4a).
Fig. 4.
Conserved active-site structure. (a) Stereo plot of the superimposed substrate complexes of RluF (lavender protein, orange RNA with blue highlighting the RNA in the binding groove), TruB (dark red protein, yellow RNA) and RluA (green protein, cyan RNA) showing conservation of RNA conformation in the groove. The Cαs of the conserved protein cores were used for alignment. (b) Stereo stick rendition of the RluF active site (green carbons indicate protein; yellow carbons indicate RNA) overlaid with a 2Fo-Fc map. Red dotted lines indicate putative hydrogen bonds. The figure was made with PyMOL (DeLano Scientific).
Conserved structure of the catalytic site implies a common catalytic mechanism
The crystal structure shows that 5-fluoro-U2604, which was substituted for U2604 to prevent catalytic turnover, has isomerized to (5S, 6R)-5-fluoro-6-hydroxy-Ψ (Fig. 4b), the hydration product of 3 in Fig. 1b, where R=F. This result verifies that U2604 is pseudouridylated in the 22-mer, and in this respect the hairpin behaves analogously to the physiologic substrate. In crystal structures of TruB24,26,27 and RluA23 complexed with substrates in which the target uridine is replaced with 5-fluorouridine, the 5-fluorouridine is also isomerized and hydrated to 5-fluoro-6-hydroxy-Ψ.
The 5-fluoro-6-hydroxy-Ψ binds in a highly conserved site adjacent to the catalytic Asp107 (Fig. 4b). The carboxyl group of the catalytic Asp is positioned to accept hydrogen bonds from the pyrimidine N3 and from the ribose O2′ of the 5-fluoro-6-hydroxy-Ψ, as it is in TruB and RluA RNA complexes. Conserved Tyr137 lies on the opposite side of the pyrimidine ring with its hydroxyl group pointing towards the 5-fluoro substituent. The equivalent tyrosine in TruB has been shown to be essential for catalysis and has been proposed to act as a base for proton abstraction in the final step of catalysis (Fig. 1b and c).41 Sitting above the target base is Leu203. A Leu or Ile is present in other Ψ synthases at this position and imparts a shape to the active-site cavity that is much more compatible with the “L-shaped” intermediate 3 of Fig. 1b or c (or the hydration product of 3 present in our structure) than with substrate or product, in which the pyrimidine and ribose rings are approximately coplanar. For example, mutation of the catalytic Asp of the Ψ synthase TruB to Asn allowed crystallization of an analog of the Michaelis complex, in which the target base bound in the active site but did not undergo isomerization.42 The target base in this complex makes unfavorably close contacts of ~ 3 Å with the conserved Leu. Thus, Leu203 may help drive isomerization or product release by destabilizing the Michaelis or product complexes, respectively. Conserved Arg190 makes a hydrogen bond or salt bridge with the catalytic Asp and is adjacent to Arg187; Ψ synthases in the RsuA and RluA families typically have a positively charged residue at the position equivalent to Arg187.
A. 1-nt frameshift of base pairs at the end of the substrate stem–loop
In ribosomal 23S RNA, the stem–loop containing the RluF target, U2604, contains seven Watson–Crick base pairs, interrupted by a bulge at A2602 [Protein Data Bank (PDB) code 2I2T,43 Fig. 5a and c]. This secondary structure is also predicted for the isolated stem–loop.32 Binding to RluF, however, induces a radically different structure, particularly in the region of the stem distal to the loop (Fig. 5b and d). The base pairing of the stem adjacent to the loop (Watson–Crick base pairs G2592–C2601, U2593–A2600, C2594–G2599) is preserved: the loop and these three base pairs form a large number of hydrogen bonds or ionic interactions with the S4-like domain. These multiple interactions serve to align the site of the adjacent A2602 stem bulge with the bottom of the RNA binding groove of the catalytic domain. A2602 no longer bulges; instead it has refolded into the stem, while the remaining Watson–Crick base pairs in the stem have separated and the substrate stand has translated one position in the 3′ direction. The effect of this strand translation is to align U2604 with the active site cavity, allowing it to flip into the active site, and so allowing RluF to induce the initial steps of isomerization.
Fig. 5.
Stem–loop base pairing and interactions with RluF. (a) Cartoon rendition of the region of 23S RNA on either side of the RluF target base (PDB code 2I2T).43 Putative hydrogen bonds are shown with dashed lines. (b) Cartoon rendition of the same RNA region as it appears in the structure of RluF–RNA complex. (c) Schematic of the 22-mer RNA stem–loop in the ribosome. (d) Schematic of the 22-mer RNA stem–loop in the crystal structure with likely protein–RNA hydrogen bonds or salt bridges in the RluF–RNA structure shown. (a) and (b) were made with PyMOL (DeLano Scientific).
With the caveat that hydrogen bonding in the rearranged region of the stem–loop cannot be conclusively assigned at 3 Å resolution, we have tentatively identified the following base pairs (Fig. 5b). The bulge nucleotide A2602 forms a distorted (single hydrogen bond) A–C reverse Hoogsteen base pair with C2591. G2603 forms a G–A N1–N1 carbonyl–amino base pair with A2590. Arg105 intercalates between bases G2603 and U2605 such that its guanidinium group is in plane with and across from the A2589 base. U2605 forms a reverse Hoogsteen base pair with A2587. G2588 is flipped out of the stem and forms a Watson–Crick base pair with C2606′ from a neighboring complex in the crystal structure, and G2588′ from the neighboring molecule in turn forms a Watson–Crick base pair with C2606 (Fig. 2b).
The swapping of bases G2588 and G2588′ from neighboring RNAs (Fig. 2b) is unlikely to occur in solution since RluF is active as a monomer. G2588 may still flip out of the stem, but if it does not flip out, it would be able to form a noncanonical base pair with U2605. A2587 would then be likely to form a noncanonical base pair with C2606. Whether G2588 flips out of the stem–loop has little bearing on the salient features of the rearrangement, which are the folding of A2602 into the stem and the strand translation of the nucleotides 3′ to A2602. Since A2587 and G2588 are not in the protein interface, the protein can accommodate either the observed or the hypothetical base-pairing arrangement.
It is unlikely that the mechanism of RluF–substrate complex formation involves only a local rearrangement of A2602 and the adjacent base pairs, G2603–C2591 and U2604–A2590. Translation of G2603 and U2604 without disruption of the U2605–A2589 base pair would produce a distorted RNA structure not compatible with the RluF binding groove. Moreover, as described below, our activity data are consistent with the more extensive base-pair rearrangement seen in the crystal structure.
In the ribosome, A2587 forms hydrogen bonds with G2608; these interactions would be expected to help maintain the hairpin structure of the stem–loop. In the crystal structure, A2587 has formed a new base pair with U2605, thus stabilizing the refolded RNA, while G2608 does not interact with protein or RNA. We find that adding A2587 alone (without G2608) to the 19-nt stem–loop, which is a very poor RluF substrate, increases the rate of pseudouridylation by RluF at least ninefold, to about one-fourth the rate for the 22-mer (data not shown). Thus, A2587 appears to have an important role in stabilizing the rearranged RNA during RluF pseudouridylation of isolated stem–loops. On the other hand, deletion of G2608 from the 22-mer increases the rate of pseudouridylation (data not shown); thus, its putative role in stabilizing the hairpin structure of the unbound RNA stem–loop is unimportant or even detrimental to activity. In other words, the most active of the tested hairpins is the 21-mer that contains A2587, which stabilizes the rearranged stem–loop seen in the crystal structure, and excludes a G2608, which could pair with A2587 to stabilize the unrearranged stem–loop. These results argue that the base-pair frameshift seen in the crystal structure is not an artifact of crystallization, but rather a part of the catalytic mechanism.
S4 binding to RNA induces a change in helix α2 that drives the base-pair frameshift
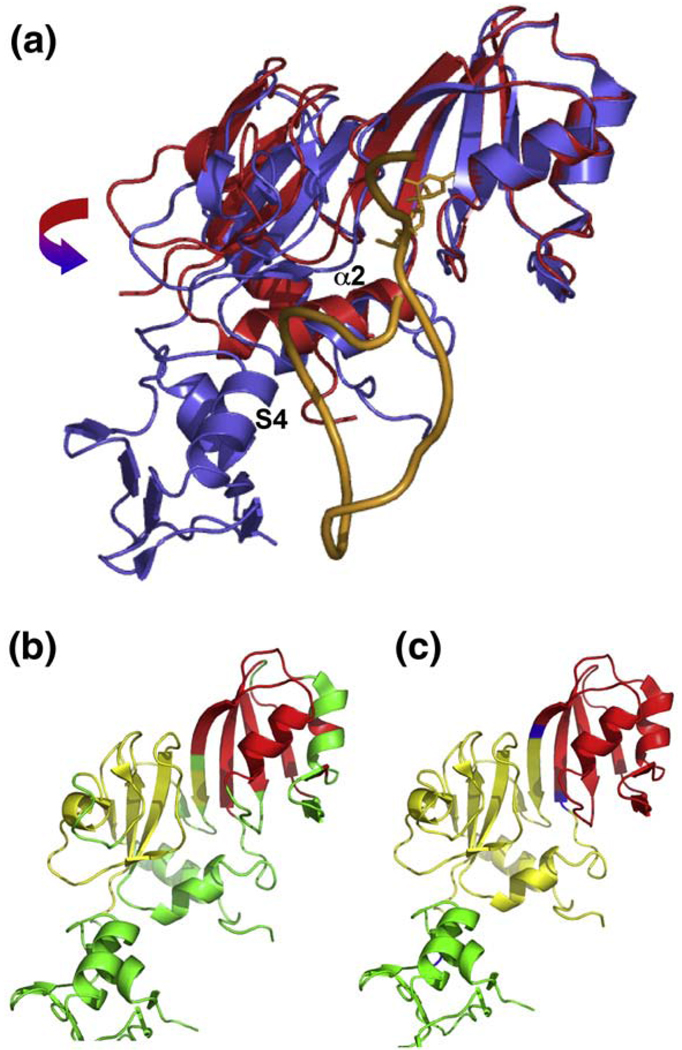
To interpret the adaptation and induced fit that occurs on RNA binding we compared our structure of full-length RNA-bound RluF to the structure of apo ΔRluF.37 The apo ΔRluF structure is truncated, missing the N-terminal S4-like domain and the flexible linker connecting it to the catalytic domain.37 However, there is evidence that truncation of the S4-like domain has no effect on the conformation of the catalytic domain. Crystal structures of other Ψ synthases that have S4-like domains tethered to their catalytic domains show that in the absence of substrate, the S4-like domains can adopt several different orientations, even within a single-crystal structure, and are frequently not visible in electron density maps.39,44,45 The crystal structures of apo-RluD with and without the N-terminal domain have been solved and the catalytic domains of these two structures show no significant conformational differences.46 Thus, it is highly likely that the conformational differences between the catalytic domains of RluF–RNA and apo ΔRluF are due solely to complex formation rather than to the absence of the S4-like domain from the apo enzyme.
The comparison of the apo ΔRluF and our RluF–RNA structures shows that the catalytic domain of RluF undergoes large changes upon complex formation; after superposition of residues 66–240, the rmsd in Cα positions is 2.2 Å. The most extensive protein conformational change that occurs when RluF binds to its substrate is a hinge motion that encloses the RNA stem (Fig. 6a). The hinge axis lies at the back of the RNA binding groove, approximately parallel to the β-strands in the core β-sheet. An algorithm for locating protein domains47,48 based on difference-distance analysis49 identifies subdomains of ~ 40–50 amino acids that remain relatively rigid during closure of the binding groove (Fig. 6b, yellow and red). TruB also optimizes its contacts with its RNA substrate through hinged bending of its central β-sheet.26,27 A crystal structure of apo TruB with two crystallographically independent protein molecules shows the catalytic domains in distinct conformations that indicate a hinged bending of the central β-sheet, which becomes ordered in the presence of substrate.50 Hinged bending of the central β-sheet is also revealed by crystallographically independent structures of apo RluE51 and is predicted to occur in RluE and RluF based on elastic network model analysis52,53 (Fig. 6c). Thus, flexibility of the core β-sheet appears to be a general feature of Ψ synthases that is exploited for RNA complex formation.
Fig. 6.
RNA-induced protein conformational changes. (a) Superposition of the apo ΔRluF (red) and RluF–RNA (blue) structures, showing the hinge movement and α2-shift that take place on substrate binding. The RNA is the orange ribbon and the modified target base is shown with sticks. The proteins were aligned by superimposing the Cαs of the red subdomain in (b). (b) Ribbon plot of RluF in which the two subdomains from the catalytic core that move as rigid bodies during substrate binding are highlighted in yellow and red. (c) Ribbon of RluF that highlights in yellow and red the subdomains that would move as rigid bodies in a hinge motion predicted by HingeProt for apo RluF. Predicted hinge residues are located on the floor of the RNA binding groove and are colored blue. The figure was made with PyMOL (DeLano Scientific).
Other conformational changes occur upon RNA-binding: the C-terminal residues of the catalytic domain move toward the binding groove and helix α2 shifts and its C-terminus unwinds (Fig. 6a). Helix α2 must shift its position in order for the S4-like domain to bind in the major groove of the stem–loop. The shift and unwinding of the helix also allow helix α2 residues to form hydrogen bonds to the stem–loop, the S4-like domain, and the C-terminal residues of the catalytic domain. Helix α2 is a major part of the protein–RNA interface, and the unwound C-terminal region of α2 contains Arg128, which donates hydrogen bonds to the RNA backbone at C2601 and A2602.
Helix α2 is part of the catalytic core of Ψ synthases. In apo-ΔRluF, it makes only nonspecific van der Waals contacts with the rest of the protein; thus, it is relatively free to move independently of the rigid subdomains on either side of the binding groove (Fig. 6b). The structures of α2 and the loop between α2 and β6 are not highly conserved among the Ψ synthases. The loop has been seen in a variety of conformations even among different structures of the same apoenzyme,51 suggesting that like the central β-sheet, it has an intrinsic flexibility that allows it to adapt to substrate binding. We propose that in RluF, a shift in helix α2 is induced by binding of the S4-like domain to the major groove of the substrate stem-loop, and that the shift in α2 triggers repositioning of A2602 into the stem with concomitant strand translation of the 3′ nucleotides.
Protein–RNA interactions
The energetic cost of RNA rearrangement is presumably paid for by favorable RNA–protein interactions. Figure 5d shows the hydrogen bonds or salt bridges between the protein and the RNA stem–loop, with the caveat that at 3 Å resolution, side-chain positions are clearly resolved in electron density but hydrogen-bond lengths are not. Eighteen residues in RluF have side chains that potentially form a total of 32 hydrogen bonds to the RNA. Ten of the residues are Lys or Arg, some of which provide counterions for phosphates at nine positions, and six are Asn, Asp, or Glu.
Seven of these residues are part of the N-terminal S4-like domain and interact primarily with the loop end and adjacent three base pairs of the substrate. Several residues from the substrate-binding groove form hydrogen bonds with the phosphate backbone or ribose moieties of nucleotides surrounding the target base, while catalytic Asp107 forms hydrogen bonds to the target base itself, as shown in Fig. 4b. Finally, Asn124 and Arg128 from helix α2 and Glu242 are in contact with the RNA backbone of the two unrearranged base pairs adjacent to the rearranged region of the stem. Arg128 also is positioned to form a hydrogen bond to the ribose O2′ of A2602, the bulge nucleotide that is incorporated into the base stack when the stem–loop binds to the protein.
The S4-like domain anchors the loop, thus determining which stem bases can access the active site
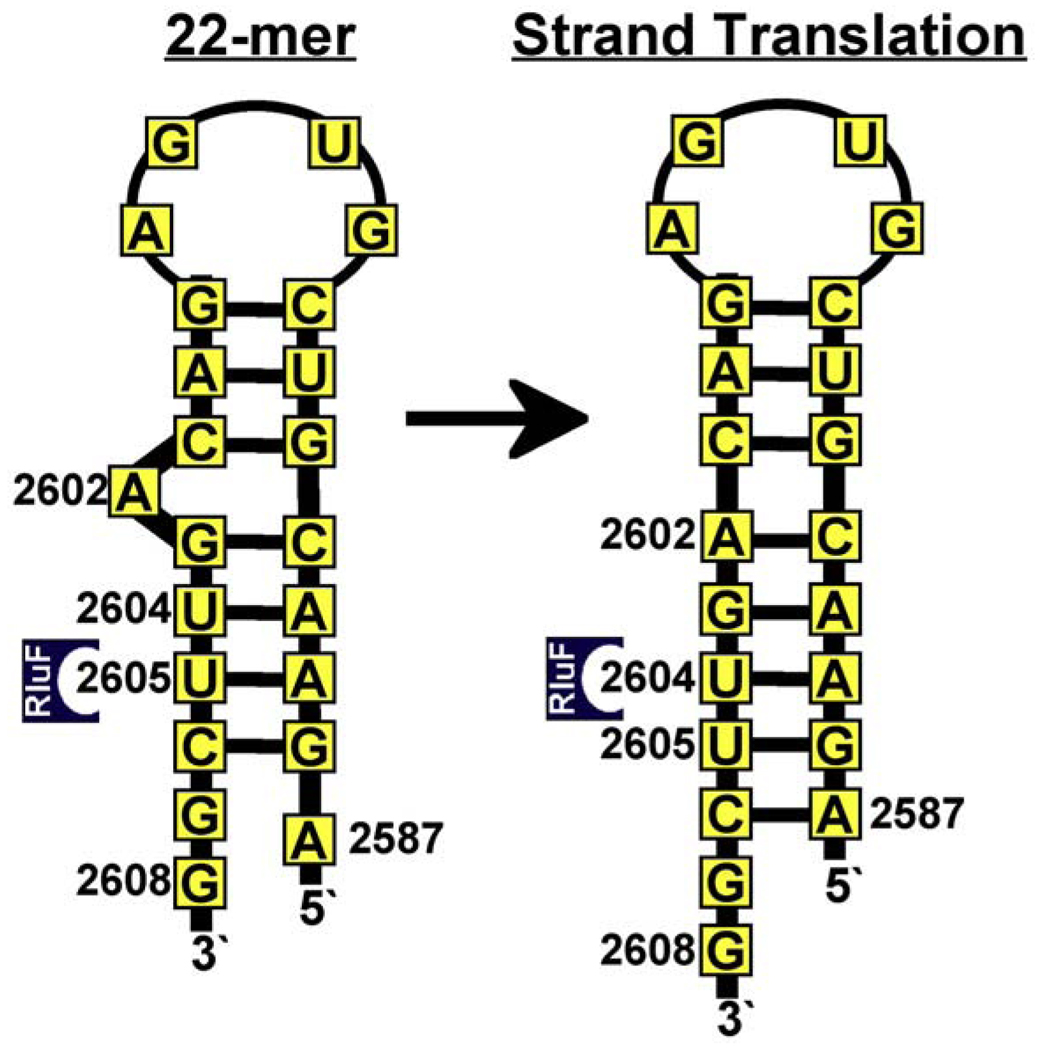
The hydrogen bonds and ionic interactions that anchor the loop and adjacent three base pairs of the substrate to the protein dictate how the stem–loop is aligned in the RluF RNA-binding groove and restrict the nucleotides that can enter its active site to those at a specific distance from the bound loop. This mechanism is reminiscent of the so-called ruler mechanisms for target selectivity described for the Ψ synthase TruA25 as well as for the endonuclease Dicer54 and the eukaryl splicing endonucleases.55 However, the RluF–RNA structure indicates that RluF uses a unique variation of the ruler mechanism in which the distances of stem nucleotides from the bound loop are altered by a protein-induced rearrangement of the RNA base pairing, wherein A2602 becomes base-paired and translates all succeeding nucleotides by one (Fig. 7).
Fig. 7.
Proposed mechanism for RluF target selectivity. Cartoon illustrating the ruler mechanism for target selection. The loop and adjacent three Watson–Crick base pairs of the 22-mer stem–loop are anchored to the S4-like domain via numerous ionic and hydrogen-bond interactions. Tethering of the distal end of the stem–loop determines which bases can flip into the active site and be pseudouridylated. Prior to stem–loop rearrangement, U2605 is positioned for modification. After A2602 refolding and RNA strand translation, U2604 is positioned for modification.
If the stem–loop had maintained its predicted solution secondary structure (with optimized base pairing, as in the ribosome) upon binding, the protein interactions with the distal (loop) end of the stem–loop would have aligned U2605 against the RluF active site. However, as A2602 refolded into the stem, U2604 became aligned, flipped into the site, and underwent the initial steps of catalysis. Since the primary biological target of RluF is U2604, we propose that repositioning of A2602 into the stem and concomitant translation of the stem nucleotides 3′ to A2602 is an essential step in target selection.
A similar mechanism is used by the restriction endonuclease Ecl18kI, which flips the central nucleotides of its recognition sequence in substrate DNA out of the DNA duplex.56 The base flipping kinks the DNA and shifts the DNA register by 1 bp, thereby altering the distance between the scissile bonds to match the distance between active sites in the Ecl18kI dimer.56
Test of the frameshift mechanism
In order to test the hypothesis that RluF can modify U2604 only if the stem–loop base pairs rearrange, but can modify the alternate nucleotide U2605 when stem–loop rearrangement is hindered or prevented, we measured RluF activity against a series of stem–loop mutants designed to either prevent the rearrangement by elimination of the bulge A2602, or facilitate the rearrangement by replacing a Watson–Crick base pair (U–A) by a noncanonical base pair that should reduce the stability to the frameshift (Fig. 8). To predict whether a mutation would facilitate rearrangement, we calculated its secondary structure and associated free energy using the Vienna RNAfold server.32 The RNAfold server predicts secondary structure by free-energy minimization using empirical thermodynamic parameters.57,58
Fig. 8.
RluF activity against stem–loop mutants. (a) Mutant stem–loops with U2604 as the potential target for RluF. C05G89=(U2605C, A2589G); C05G89Δ02=(U2605C, A2589G) with A2602 deleted, C05=U2605C; C05Δ02=U2605C with A2602 deleted. The predicted minimum free energies (kilocalories per mole) and initial rates of uridine modification (picomoles uridine per picomole enzyme per minute) are listed above the stem–loops. Initial rates were determined by least-squares fit to the linear region of the progress curves for reaction mixtures containing 50 nm RluF and 500 nm [5-3H] Ura-RNA in TNE buffer (see Materials and Methods). Each rate is the average of three measurements. (b) Stem–loops with U2605 as the potential target for RluF. C04G90=(U2604C, A2590G); C04G90Δ02=(U2604C, A2590G) with A2602 deleted; C04=U2604C; C04Δ02=U2604C with A2602 deleted. Free energies and initial rates are listed as in (a).
We used a tritium release assay in which tritiated uridine is incorporated at every uridine position in the synthetic loops and thus cannot distinguish between pseudouridylation at 2604 and 2605. Therefore, when U2604 was tested as a target, U2605 was mutated to C, with or without a compensating mutation that preserves Watson–Crick (C–G) base pairing. When U2605 was tested as a target, U2604 was mutated to C, with or without the compensating mutation.
In the mutant stem–loops containing the target base, U2604, there is a correlation between dramatically enhanced enzyme activity and the relative ability, as predicted based on base pair inventory, of the RNA substrate to undergo the rearrangement diagrammed in Fig. 7 (Fig. 8a). The substrate in which U2605 is mutated to C, but its base pair partner is not mutated (abbreviated as C05), has a non-Watson–Crick base pair in the stem, which is predicted to destabilize the hairpin structure, facilitating strand translation. In fact, the predicted most favored structure for this mutant in solution is the strand-translated stem–loop in which A2602 is folded into the stem and C2605 is base-paired with G2588 (Fig. 8a, second from the left).32 Thus, C05 would be preorganized to allow U2604 access to the RluF active site. Regarding the initial rate of modification for C05 as the standard for comparison of all mutant substrates, its rate is more than 60-fold higher than for any of the other mutant stem–loops, and incidentally more than 37-fold higher than for the wild-type 22-mer.
When both U2605 and its base-pair partner A2589 are mutated (to C and G, respectively) there is then no net loss in the number of Watson–Crick base pairs relative to the wild-type 22-mer (Fig. 8a, C05G89). The replacement of a U–A base pair with a C–G base pair predicts that the stem–loop will be even more stable than wild-type and therefore more difficult to rearrange. The initial rate for modification of C05G89 is 141-fold slower than for C05. These results are consistent with base pair rearrangement being a critical, slow step in the modification of U2604; the extra stabilization provided by the mutations diminishes U2604 access to the active site.
The importance of the A2602 bulge rearrangement is highlighted by two mutant substrates with A2602 deleted. The simple deletion alone (C05Δ02) is modified at a rate 636-fold lower than that of C05, which has similar base pairing within the stem. Although C05Δ02 is predicted to be less stable than C05G89 it is modified at a 4.5-fold slower initial rate. Thus, the specific RNA rearrangement involving A2602 repositioning and strand translation is important for U2604 modification, even in a stem–loop with a destabilized stem. The substrate, C05G89Δ02, which has Watson–Crick base pairs throughout the stem and no bulge, would be expected to be very stable and, without A2602, could not undergo strand translation. Modification of this substrate is not detectable. These results show that in RluF activity correlates with facility of the substrate to undergo the specific rearrangement of A2602 refolding and strand translation. This contrasts with the Ψ synthase TruA, for which substrate flexibility per se correlates with activity.25
The ruler mechanism predicts that U2605 would be prealigned with the RluF active site in the docked stem–loop prior to RNA rearrangement. Consistent with this mechanism, all mutant stem–loops that preserve U at 2605 (i.e., with U2604 mutated to C) are RluF substrates, regardless of whether A2602 is present to allow strand translation (Fig. 8b). The activity of RluF toward these mutants correlates with predicted stability of the hairpin structure, suggesting that U2605 modification requires only flipping of U2605 into the RluF active site and is facilitated by factors that maintain the position of U2605 in the structure of the stem.
In summary, when U2604 is the target (as in the wild-type ribosome) the lesser the stability of the stem–loop the greater the activity, presumably because flexibility or strand translation is required to access the active site. No rearrangement is required for U2605 access to the active site, and rate of activity on this noncognate base is increased when the stem is stabilized against any frameshift. Hence, the more stable is the stem, the harder it is for the enzyme to evoke the strand translation necessary for the normal function of RluF, namely, its ability to correctly select U2604 for pseudouridylation.
The assays of stem–loop mutants confirm that stem–loop rearrangement is critical for modification of U2604 by RluF. The isolated stem–loop is predicted to form a very stable hairpin secondary structure; thus, rearrangement of the stem–loop probably does not occur to an appreciable extent until the stem–loop binds to the protein. The driving force for the rearrangement could be unfavorable interactions between bulged-out A2602 and residues in the C-terminal end of helix α2, particularly Arg128. Binding of the S4-like domain to the major groove of the stem–loop is accompanied by a shift of helix α2 that would create these unfavorable interactions.
In vivo specificity of RluF
The ability of RluF to modify both series of mutant substrates, with U2604 or with U2605 mutated to C (Fig. 8), suggests that RluF might pseudouridylate both U2604 and U2605 in stem–loop fragments of the physiologic substrate. This is consistent with the fact that although its major target in vivo is U2604, RluF appears to also pseudouridylate U2605 to a small degree.22 rRNA fragments larger than a 22-mer may be required to fully recapitulate the RluF target selectivity seen in vivo, as is the case for the rRNA-modifying Ψ synthase RluD. RluD exhibits highest activity and target specificity with full or partially assembled ribosomes.59 RluF must modify U2604 before complete ribosome assembly, however, since the stem–loop that contains U2604 is not accessible in the fully assembled ribosome.43 Larger substrate fragments may interact with RluF partly through the disordered C-terminal domain of RluF. Thus C-terminal domain interactions with rRNA may decrease Km. They may also facilitate the rearrangement of the stem–loop required for U2604 modification, and so could further enhance the activity on U2604.
How RluF and RluB target adjacent uridines in the same stem–loop
Comparison of the RluF and RluB sequences in light of the unusual structure of the RluF–RNA complex provides clues to the reasons for different substrate specificities of the two enzymes. In the RluF–RNA complex, most of the residues that are hydrogen-bonded to the loop end of the stem–loop (distal to the active site) are conserved or have conservative substitutions in RluB, while residues that are hydrogen-bonded to the rearranged nucleotides of the RNA stem (with the exception of the target base) are not conserved, or even similar in RluB. This observation suggests that the RNA stem–loop may initially dock to RluF and RluB in a similar manner, but that the two enzymes trigger different rearrangements of the RNA after docking.
In particular, we propose that the steric clashes between the bulge nucleotide, A2602, and residues at the C-terminal end of helix α2, including Arg128, are the driving force for A2602 repositioning and concomitant frameshift of four base pairs that translates U2604 to the active-site cavity in RluF. Arg128 is conserved in RluF from different species but is a proline in RluB. Proline is much less bulky than arginine and might allow RluB to accommodate the stem–loop bulge rather than inducing A2602 refolding. The crystal structure of a stem–loop-bound RluB will assess this proposal.
Conclusion
RNA plasticity (its ability to adopt alternate secondary structures) allows riboswitches and some ribozymes to rearrange their structures in order to form binding sites for small-molecule ligands.60 The plasticity and flexibility of RNA are also frequently exploited for formation of tight and specific enzyme–RNA complexes.23,28–31 In the latter examples, another layer of complexity is added to molecular recognition in that both binding partners, RNA and protein, may change conformation during complex formation.
RluF is the latest example of an RNA-modifying enzyme whose substrate undergoes radical structural rearrangements upon binding.23,28–30 The striking difference between RluF and the previous examples is that the RluF target base lies in an RNA stem rather than in a flexible loop and the substrate conformational change requires disruption of multiple base pairs. This energetically costly process is driven by interactions of the RNA with the protein. The mechanism adds to the repertoire of strategies used by RNA-modifying enzymes for achieving their exquisite, often unique specificity for a single position in the RNA components of the cell.
Materials and Methods
RluF expression and purification
The rluF gene was cloned into pET-30b (Novagen) and the construct was transformed into E. coli BL21(DE3). Cells were grown in LB medium containing kanamycin (10 µg/mL) to an optical density of 0.6 at 600 nm, induced with isopropyl-1-thio-β-d-galactopyranoside (1 mM), and grown for an additional 4 h before harvesting. The cell pellet was suspended in buffer A containing 25 mM Tris, pH 7.4 (RT), 300 mM NaCl, and Complete EDTA-free protease inhibitor cocktail (Roche Diagnostics). After lysis using an Emulsiflex-C5 homogenizer (Avestin, Inc., Canada), the lysate was centrifuged at 30,000g for 20 min and loaded onto a 5-mL HiTrap Heparin HP column (GE Healthcare). A gradient of 0–100% buffer B (25 mM Tris, pH 7.4, 2 M NaCl) was used to elute RluF. Fractions were pooled, DTT was added to 3 mM, and the protein was concentrated to 10 mg/mL using an Amicon 10-kDa centrifugal concentration device. NaCl concentration was about 750 mM in the pool. Protein was aliquoted and stored at −80 °C.
RNA synthesis
The [5-3H]Ura-RNA substrates were synthesized by in vitro transcription with T7 RNA polymerase using the MEGAshortscript kit (Ambion) and using 20% (of total UTP) as [5-3H]-UTP (Moravek). The templates were produced by annealing (95 °C, 3 min followed by slow cool down) a T7 promoter sequence (5′-TAATACGACTCACTATAG) to a DNA template containing a sequence complementary to T7 and each of the following sequences from the 23S rRNA, one at a time: 22-mer: fragment 2587–2608 (5′-AGAACGUCGUGAGACAGUUCGG); 19-mer: fragment 2588–2606 (5′-GAACGUCGUGAGACAGUUC); 20-mer: fragment 2587–2606 (5′-AGAACGUCGUGAGACAGUUC). The mutated RNA substrates were synthesized in the same way based on the 22-mer sequence and including the following mutations: C04G90: U2604C and A2590G (5′-AGAGCGUCGUGAGACAGCUCGG); C04G90ΔA02: U2604C and A2590G with A2602 deleted (5′-AGAGCGUCGUGAGAC-GCUCGG); C05G89: U2605C and A2589G (5′-AGGACGUCGUGAGACA-GUCCGG); C05G89ΔA02: U2605C and A2589G with A2602 deleted (5′-AGGACGUCGUGAGAC-GUCCGG); C05: U2605C (5′-AGAACGUCGUGAGACAGUCCGG); C05ΔA02: U2605C with A2602 deleted (5′-AGAACGUCGUGAGAC-GUCCGG); C04: U2604C (5′-AGAACGUCGUGAGACAGCUCGG); C04ΔA02: U2604C with A2602 deleted (5′-AGAACGUCGUGAGAC-GCUCGG).
The [5-3H]Ura-RNAs were purified on DEAE–Sepharose (GE Healthcare) packed in disposable Poly-Prep columns (Bio-Rad). After washing with TE, free [5-3H]UTP and short transcripts eluted at 0.1–0.2 M NaCl, while RNA eluted at 0.4 M NaCl. RNA was then precipitated using ammonium acetate and resuspended in 50 µL TE.
Tritium release assay
The tritium release assays were performed as described previously36 with minor modifications. Reactions containing 50 nM RluF and 500 nM [5-3H]Ura-RNA (~2.0×104 dpm/pmol) in TNE buffer (20 mM Tris–Cl, pH 8.0, 0.1 M NH4Cl, and 2 mM DTT) were incubated at room temperature. Aliquots (20 µL) were removed at different time intervals and quenched with 1 mL of 5% Norit-A in 0.1 N HCl. Mixtures were centrifuged (5000g, 5 min) and the supernatants were again treated with 0.5 mL of 5% Norit-A in 0.1 N HCl. Mixtures were centrifuged again and supernatants were filtered using Ultrafree-MC filters (Amicon), and 0.5 mL of each filtrate was counted in 5 mL of Aquasol-2 (Perkin Elmer).
Crystallization of an RluF–RNA complex
For crystallization, the 23S rRNA fragment 2587–2608 with the [F]5U2604 modification (5′-AG AAC GUC GUG AGA CAG (U[5F])UC GG) was purchased from Dharmacon. RluF was thawed and the NaCl concentration was lowered to 184 mM by dilution with a no-salt buffer. The protein was then reconcentrated to 12.5 mg/mL with an Amicon Microcon 10-kDa unit. Enzyme–RNA complexes were formed by incubating 390 µM RluF with molar excess (1.2-fold) RNA on ice for 1 h., then used for crystallization.
Crystals were grown at room temperature using the hanging-drop vapor-diffusion method. The complex solution (1 µL) was mixed with an equal volume of well buffer: 200 mM ammonium acetate, and 25% PEG (polyethylene glycol) 3350. Crystals belonged to space group P21 with four complexes in the asymmetric unit.
Data collection and structure determination
Crystals were transferred to the well buffer solution containing 20% (v/v) ethylene glycol shortly before immersion in liquid nitrogen. Diffraction data from single crystals were collected to 3.0 Å at −170 °C on a Quantum 315r image plate at the Advanced Light Source (ALS, Lawrence Berkeley Lab, CA). Data were processed and scaled using the HKL-2000 program suite61 (Table 1).
Table 1.
Statistics for data collection and refinement
| Crystal data | |
| Space group | P21 |
| Cell constants | |
| a (Å) | 89.55 |
| b (Å) | 84.13 |
| c (Å) | 91.35 |
| β (°) | 94.87 |
| Complexes/ASU | 4 |
| Vm (Å3/Da), % solvent | 2.2, 44 |
| Data collection statistics | |
| Resolution (last shell) (Å) | 90.91–3.0 (3.08–3.0) |
| Wavelength (Å) | 1.11 |
| No. of reflections | 27, 252 |
| Redundancy (last shell) | 3.3 (2.9) |
| Completeness (last shell) (%) | 98.7 (87.64) |
| Rmerge (last shell) | 0.121 (0.48) |
| I/σ (last shell) | 9.64 (1.87) |
| Refinement statistics | |
| Resolution (Å) | 3.0 |
| Reflections in working set | 25,651 |
| Reflections in test set (5.0%) | 1355 |
| Rcrystal (last shell) (%) | 23.1 (38.4) |
| Rfree (last shell) (%) | 27.9 (43.0) |
| rmsd bonds (Å) | 0.014 |
| rmsd angles (°) | 1.85 |
| Average B-factor (Å2) | |
| Protein (each chain) | 53 |
| RNA (each chain) | 58 |
| Water atoms | 49 |
| Ramachandran distribution (%) | |
| Most favorable | 83.2 |
| Allowed | 15.2 |
| Generously allowed | 1.1 |
| Disallowed | 0.5 |
The structure was solved by molecular replacement using the catalytic domain of apo-ΔRluF as a search model (PDB code 2GML). The program Phaser62 was used to locate two of the protein molecules in the asymmetric unit. The N-terminal domains and RNAs were built into 2Fo-Fc and Fo-Fc maps using CHAIN63 and Coot.64 An RluF–RNA complex was then used as a molecular replacement search model to place all four complexes in the asymmetric unit using MOLREP.65
The structure was refined by manual building in Coot alternated with positional and isotropic B-factor refinement using CNS66 and REFMAC567 (Table 1). Tight noncrystallographic symmetry (NCS) restraints (σ=0.05 Å for NCS-related atoms) were used for all residues of the protein and medium restraints (σ=0.5 Å for NCS-related atoms) were used for the RNA. This combination of NCS restraints gave the lowest Rfree.
After refinement, the well-ordered residue Met209 was the only residue whose (ϕ,Ψ) angles were in a disallowed region of the Ramachandran plot (ϕ=45° and Ψ=−117°). Met209 unambiguously adopts the same conformation in the 2.6 Å apo ΔRluF structure (ϕ=56° and Ψ=−123°). The strained backbone conformation of this residue allows it to form a tight β-turn between β11 and β12 while burying its side chain in a hydrophobic pocket in the core of the enzyme.
Protein Data Bank accession numbers
Coordinates and structure factors for the X-ray crystal structure of the RluF-22mer complex have been deposited in the PDB with accession code 3DH3.
Acknowledgements
We thank Sun Hur and Sanjay Agarwalla for assistance and advice on performing and interpreting the tritium release assays and Pat Greene for advice throughout the project and for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM51232 to J.F.M.
Abbreviations used
- Ψ
pseudouridine
- S4-like domain
domain with the same fold as the RNA-binding domain of ribosomal protein S4
- ΔRluF
N-terminal truncation of RluF
- C05
stem–loop mutant U2605C
- C04
stem–loop mutant U2604C
- C05G89
stem–loop mutant (U2605C, A2589G)
- C04G90
stem–loop mutant (U2604C, A2590G)
- Δ02
A2602 deletion
- rRNA
ribosomal RNA
- NCS
noncrystallographic symmetry
References
- 1.Bjork G. Stable RNA modification. In: Neidhardt FC, Curtiss R, editors. 2nd edit. Washington, DC: ASM Press; 1996. pp. 861–886. [Google Scholar]
- 2.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishitani R, Yokoyama S, Nureki O. Structure, dynamics, and function of RNA modification enzymes. Curr. Opin. Struct. Biol. 2008;18:330–339. doi: 10.1016/j.sbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 5.Ofengand J, Malhotra A, Remme J, Gutgsell NS, Del Campo M, Jean-Charles S, et al. Pseudouridines and pseudouridine synthases of the ribosome. Cold Spring Harb. Symp. Quant. Biol. 2001;66:147–159. doi: 10.1101/sqb.2001.66.147. [DOI] [PubMed] [Google Scholar]
- 6.Sundaram M, Durant PC, Davis DR. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry. 2000;39:12575–12584. doi: 10.1021/bi0014655. [DOI] [PubMed] [Google Scholar]
- 7.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejby M, Sorensen MA, Pedersen S. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc. Natl Acad. Sci. USA. 2007;104:19410–19415. doi: 10.1073/pnas.0706558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Patton JR, Ghosh SK, Fischel-Ghodsian N, Shen L, Spanjaard RA. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol. Endocrinol. 2007;21:686–699. doi: 10.1210/me.2006-0414. [DOI] [PubMed] [Google Scholar]
- 13.Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514:17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- 14.Gustafsson C, Reid R, Greene PJ, Santi DV. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res. 1996;24:3756–3762. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hur S, Stroud RM, Finer-Moore J. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: a structural perspective. J. Biol. Chem. 2006;281:38969–38973. doi: 10.1074/jbc.R600034200. [DOI] [PubMed] [Google Scholar]
- 17.Mueller EG. Chips off the old block. Nat. Struct. Biol. 2002;9:320–322. doi: 10.1038/nsb0502-320. [DOI] [PubMed] [Google Scholar]
- 18.Kaya Y, Ofengand J. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. Rna. 2003;9:711–721. doi: 10.1261/rna.5230603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol. 2007;373:1243–1254. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Ku J, Pookanjanatavip M, Gu X, Wang D, Greene PJ, Santi DV. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry. 1998;37:15951–15957. doi: 10.1021/bi981002n. [DOI] [PubMed] [Google Scholar]
- 21.Kammen HO, Marvel CC, Hardy L, Penhoet EE. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J. Biol. Chem. 1988;263:2255–2263. [PubMed] [Google Scholar]
- 22.Del Campo M, Kaya Y, Ofengand J. Identification and site of action of the remaining four putative pseudouridine synthases in Escherichia coli. RNA. 2001;7:1603–1615. [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D’Amare AR. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol. Cell. 2006;24:535–545. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Hoang C, Ferre-D’Amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 25.Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol. Cell. 2007;26:189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl Acad. Sci. USA. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phannachet K, Huang RH. Conformational change of pseudouridine 55 synthase upon its association with RNA substrate. Nucleic Acids Res. 2004;32:1422–1429. doi: 10.1093/nar/gkh287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: a consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc. Natl Acad. Sci. USA. 2008;105:6876–6881. doi: 10.1073/pnas.0802247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a post-transcriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 30.Lee TT, Agarwalla S, Stroud RM. A unique RNA fold in the RumA–RNA–cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 31.Zhou C, Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc. Natl Acad. Sci. USA. 2008;105:16142–16147. doi: 10.1073/pnas.0805680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 34.Stivers JT. Extrahelical damaged base recognition by DNA glycosylase enzymes. Chemistry. 2008;14:786–793. doi: 10.1002/chem.200701501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu X, Liu Y, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. USA. 1999;96:14270–14275. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–351. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- 37.Sunita S, Zhenxing H, Swaathi J, Cygler M, Matte A, Sivaraman J. Domain organization and crystal structure of the catalytic domain of E. coli RluF, a pseudouridine synthase that acts on 23S rRNA. J. Mol. Biol. 2006;359:998–1009. doi: 10.1016/j.jmb.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 38.Davies C, Gerstner RB, Draper DE, Ramakrishnan V, White SW. The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J. 1998;17:4545–4558. doi: 10.1093/emboj/17.16.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivaraman J, Sauve V, Larocque R, Stura EA, Schrag JD, Cygler M, Matte A. Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP. Nat. Struct. Biol. 2002;9:353–358. doi: 10.1038/nsb788. [DOI] [PubMed] [Google Scholar]
- 40.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 41.Phannachet K, Elias Y, Huang RH. Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase. Biochemistry. 2005;44:15488–15494. doi: 10.1021/bi050961w. [DOI] [PubMed] [Google Scholar]
- 42.Hoang C, Hamilton CS, Mueller EG, Ferre-D’Amare AR. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Sci. 2005;14:2201–2206. doi: 10.1110/ps.051493605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl Acad. Sci. USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Del Campo M, Ofengand J, Malhotra A. Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli. RNA. 2004;10:231–239. doi: 10.1261/rna.5187404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizutani K, Machida Y, Unzai S, Park SY, Tame JR. Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli. Biochemistry. 2004;43:4454–4463. doi: 10.1021/bi036079c. [DOI] [PubMed] [Google Scholar]
- 46.Sivaraman J, Iannuzzi P, Cygler M, Matte A. Crystal structure of the RluD pseudouridine synthase catalytic module, an enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli. J. Mol. Biol. 2004;335:87–101. doi: 10.1016/j.jmb.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Montfort WR, Perry KM, Fauman EB, Finer-Moore JS, Maley GF, Hardy L, et al. Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate. Biochemistry. 1990;29:6964–6977. doi: 10.1021/bi00482a004. [DOI] [PubMed] [Google Scholar]
- 48.Perry KM, Fauman EB, Finer-Moore JS, Montfort WR, Maley GF, Maley F, Stroud RM. Plastic adaptation toward mutations in proteins: structural comparison of thymidylate synthases. Proteins. 1990;8:315–333. doi: 10.1002/prot.340080406. [DOI] [PubMed] [Google Scholar]
- 49.Nishikawa K, Ooi T. Comparison of homologous tertiary structure of proteins. J. Theor. Biol. 1974;43:351–374. doi: 10.1016/s0022-5193(74)80066-4. [DOI] [PubMed] [Google Scholar]
- 50.Chaudhuri BN, Chan S, Perry LJ, Yeates TO. Crystal structure of the apo forms of psi 55 tRNA pseudouridine synthase from Mycobacterium tuberculosis: a hinge at the base of the catalytic cleft. J. Biol. Chem. 2004;279:24585–24591. doi: 10.1074/jbc.M401045200. [DOI] [PubMed] [Google Scholar]
- 51.Pan H, Ho JD, Stroud RM, Finer-Moore J. The crystal structure of E. coli rRNA pseudouridine synthase RluE. J. Mol. Biol. 2007;367:1459–1470. doi: 10.1016/j.jmb.2007.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 54.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 55.Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 56.Bochtler M, Szczepanowski RH, Tamulaitis G, Grazulis S, Czapinska H, Manakova E, Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermo-dynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 58.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaidyanathan PP, Deutscher MP, Malhotra A. RluD, a highly conserved pseudouridine synthase, modifies 50S subunits more specifically and efficiently than free 23S rRNA. RNA. 2007;13:1868–1876. doi: 10.1261/rna.711207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- 61.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 62.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sack JS. CHAIN—a crystallographic modelling program. J. Mol. Graphics. 1988;6:244–245. [Google Scholar]
- 64.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 65.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 66.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 67.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]