Abstract
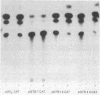
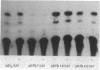
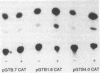
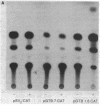
The function of the 5'-flanking region of a rat glutathione S-transferase Ya subunit structural gene has been examined in homologous and heterologous cells. By using the 5'-flanking region of the Ya subunit gene fused to the structural gene encoding chloramphenicol acetyltransferase, we have identified two cis-acting regulatory elements in the upstream region of the Ya gene. One element is required for maximum basal level expression in homologous cells, whereas maximum basal level expression in homologous cells, whereas the second element is required for inducible expression of the Ya gene by planar aromatic compounds such as beta-naphthoflavone. The cis-acting element required for inducible expression of the Ya gene by beta-naphthoflavone is functional only in cells with normal dioxin receptors.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alin P., Mannervik B., Jörnvall H. Cytosolic rat liver glutathione transferase 4-4. Primary structure of the protein reveals extensive differences between homologous glutathione transferases of classes alpha and mu. Eur J Biochem. 1986 Apr 15;156(2):343–350. doi: 10.1111/j.1432-1033.1986.tb09588.x. [DOI] [PubMed] [Google Scholar]
- Bass N. M., Kirsch R. E., Tuff S. A., Marks I., Saunders S. J. Ligandin heterogeneity : evidence that the two non-identical subunits are the monomers of two distinct proteins. Biochim Biophys Acta. 1977 May 27;492(1):163–175. doi: 10.1016/0005-2795(77)90223-9. [DOI] [PubMed] [Google Scholar]
- Bhargava M. M., Ohmi N., Listowsky I., Arias I. M. Subunit composition, organic anion binding, catalytic and immunological properties of ligandin from rat testis. J Biol Chem. 1980 Jan 25;255(2):724–727. [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Ding G. J., Ding V. D., Rodkey J. A., Bennett C. D., Lu A. Y., Pickett C. B. Rat liver glutathione S-transferases. DNA sequence analysis of a Yb2 cDNA clone and regulation of the Yb1 and Yb2 mRNAs by phenobarbital. J Biol Chem. 1986 Jun 15;261(17):7952–7957. [PubMed] [Google Scholar]
- Ding G. J., Lu A. Y., Pickett C. B. Rat liver glutathione S-transferases. Nucleotide sequence analysis of a Yb1 cDNA clone and prediction of the complete amino acid sequence of the Yb1 subunit. J Biol Chem. 1985 Oct 25;260(24):13268–13271. [PubMed] [Google Scholar]
- Ding V. D., Pickett C. B. Transcriptional regulation of rat liver glutathione S-transferase genes by phenobarbital and 3-methylcholanthrene. Arch Biochem Biophys. 1985 Aug 1;240(2):553–559. doi: 10.1016/0003-9861(85)90062-1. [DOI] [PubMed] [Google Scholar]
- Fujisawa-Sehara A., Sogawa K., Nishi C., Fujii-Kuriyama Y. Regulatory DNA elements localized remotely upstream from the drug-metabolizing cytochrome P-450c gene. Nucleic Acids Res. 1986 Feb 11;14(3):1465–1477. doi: 10.1093/nar/14.3.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferase AA from rat liver. Arch Biochem Biophys. 1976 Aug;175(2):710–716. doi: 10.1016/0003-9861(76)90563-4. [DOI] [PubMed] [Google Scholar]
- Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974 Nov 25;249(22):7130–7139. [PubMed] [Google Scholar]
- Homma H., Listowsky I. Identification of Yb-glutathione-S-transferase as a major rat liver protein labeled with dexamethasone 21-methanesulfonate. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7165–7169. doi: 10.1073/pnas.82.21.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel D. I., Whitlock J. P., Jr Induction of mRNA specific for cytochrome P1-450 in wild type and variant mouse hepatoma cells. J Biol Chem. 1983 Sep 10;258(17):10390–10394. [PubMed] [Google Scholar]
- Israel D. I., Whitlock J. P., Jr Regulation of cytochrome P1-450 gene transcription by 2,3,7, 8-tetrachlorodibenzo-p-dioxin in wild type and variant mouse hepatoma cells. J Biol Chem. 1984 May 10;259(9):5400–5402. [PubMed] [Google Scholar]
- Jakoby W. B., Ketterer B., Mannervik B. Glutathione transferases: nomenclature. Biochem Pharmacol. 1984 Aug 15;33(16):2539–2540. doi: 10.1016/0006-2952(84)90621-x. [DOI] [PubMed] [Google Scholar]
- Jones P. B., Durrin L. K., Fisher J. M., Whitlock J. P., Jr Control of gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Multiple dioxin-responsive domains 5'-ward of the cytochrome P1-450 gene. J Biol Chem. 1986 May 25;261(15):6647–6650. [PubMed] [Google Scholar]
- Jones P. B., Durrin L. K., Galeazzi D. R., Whitlock J. P., Jr Control of cytochrome P1-450 gene expression: analysis of a dioxin-responsive enhancer system. Proc Natl Acad Sci U S A. 1986 May;83(9):2802–2806. doi: 10.1073/pnas.83.9.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. B., Galeazzi D. R., Fisher J. M., Whitlock J. P., Jr Control of cytochrome P1-450 gene expression by dioxin. Science. 1985 Mar 22;227(4693):1499–1502. doi: 10.1126/science.3856321. [DOI] [PubMed] [Google Scholar]
- Jones P. B., Miller A. G., Israel D. I., Galeazzi D. R., Whitlock J. P., Jr Biochemical and genetic analysis of variant mouse hepatoma cells which overtranscribe the cytochrome P1-450 gene in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1984 Oct 25;259(20):12357–12363. [PubMed] [Google Scholar]
- Lai H. C., Grove G., Tu C. P. Cloning and sequence analysis of a cDNA for a rat liver glutathione S-transferase Yb subunit. Nucleic Acids Res. 1986 Aug 11;14(15):6101–6114. doi: 10.1093/nar/14.15.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H. C., Li N., Weiss M. J., Reddy C. C., Tu C. P. The nucleotide sequence of a rat liver glutathione S-transferase subunit cDNA clone. J Biol Chem. 1984 May 10;259(9):5536–5542. [PubMed] [Google Scholar]
- Lai H. C., Tu C. P. Rat glutathione S-transferases supergene family. Characterization of an anionic Yb subunit cDNA clone. J Biol Chem. 1986 Oct 15;261(29):13793–13799. [PubMed] [Google Scholar]
- Litwack G., Ketterer B., Arias I. M. Ligandin: a hepatic protein which binds steroids, bilirubin, carcinogens and a number of exogenous organic anions. Nature. 1971 Dec 24;234(5330):466–467. doi: 10.1038/234466a0. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Jensson H. Binary combinations of four protein subunits with different catalytic specificities explain the relationship between six basic glutathione S-transferases in rat liver cytosol. J Biol Chem. 1982 Sep 10;257(17):9909–9912. [PubMed] [Google Scholar]
- Miller A. G., Israel D., Whitlock J. P., Jr Biochemical and genetic analysis of variant mouse hepatoma cells defective in the induction of benzo(a)pyrene-metabolizing enzyme activity. J Biol Chem. 1983 Mar 25;258(6):3523–3527. [PubMed] [Google Scholar]
- Okey A. B., Bondy G. P., Mason M. E., Nebert D. W., Forster-Gibson C. J., Muncan J., Dufresne M. J. Temperature-dependent cytosol-to-nucleus translocation of the Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin in continuous cell culture lines. J Biol Chem. 1980 Dec 10;255(23):11415–11422. [PubMed] [Google Scholar]
- Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977 Dec;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Pickett C. B., Telakowski-Hopkins C. A., Ding G. J., Argenbright L., Lu A. Y. Rat liver glutathione S-transferases. Complete nucleotide sequence of a glutathione S-transferase mRNA and the regulation of the Ya, Yb, and Yc mRNAs by 3-methylcholanthrene and phenobarbital. J Biol Chem. 1984 Apr 25;259(8):5182–5188. [PubMed] [Google Scholar]
- Prochaska H. J., De Long M. J., Talalay P. On the mechanisms of induction of cancer-protective enzymes: a unifying proposal. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8232–8236. doi: 10.1073/pnas.82.23.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rothkopf G. S., Telakowski-Hopkins C. A., Stotish R. L., Pickett C. B. Multiplicity of glutathione S-transferase genes in the rat and association with a type 2 Alu repetitive element. Biochemistry. 1986 Mar 11;25(5):993–1002. doi: 10.1021/bi00353a007. [DOI] [PubMed] [Google Scholar]
- Sogawa K., Fujisawa-Sehara A., Yamane M., Fujii-Kuriyama Y. Location of regulatory elements responsible for drug induction in the rat cytochrome P-450c gene. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8044–8048. doi: 10.1073/pnas.83.21.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telakowski-Hopkins C. A., Rodkey J. A., Bennett C. D., Lu A. Y., Pickett C. B. Rat liver glutathione S-transferases. Construction of a cDNA clone complementary to a Yc mRNA and prediction of the complete amino acid sequence of a Yc subunit. J Biol Chem. 1985 May 10;260(9):5820–5825. [PubMed] [Google Scholar]
- Telakowski-Hopkins C. A., Rothkopf G. S., Pickett C. B. Structural analysis of a rat liver glutathione S-transferase Ya gene. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9393–9397. doi: 10.1073/pnas.83.24.9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C. P., Lai H. C., Li N. Q., Weiss M. J., Reddy C. C. The Yc and Ya subunits of rat liver glutathione S-transferases are the products of separate genes. J Biol Chem. 1984 Aug 10;259(15):9434–9439. [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Galeazzi D. R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin receptors in wild type and variant mouse hepatoma cells. Nuclear location and strength of nuclear binding. J Biol Chem. 1984 Jan 25;259(2):980–985. [PubMed] [Google Scholar]