Abstract
The lipid peroxidation product 4-hydroxynonenal (4-HNE) forms as a consequence of oxidative stress. By electrophilic attack on biological macromolecules, 4-HNE mediates signaling or may cause toxicity. A major route of 4-HNE disposal is via glutathione conjugation, in the mouse catalyzed primarily by glutathione transferase mGSTA4-4. Unexpectedly, mGsta4-null mice, in which 4-HNE detoxification is impaired, have an extended life span. This finding could be explained by the observed activation of the transcription factor Nrf2 in the knockout mice, which in turn leads to an induction of antioxidant and antielectrophilic defenses. Especially, the latter could provide a detoxification mechanism that contributes to enhanced longevity. We propose that disruption of 4-HNE conjugation elicits a hormetic response in which an initially increased supply of 4-HNE is translated into activation of Nrf2, leading to a new steady state in which the rise of 4-HNE concentrations is dampened, but life-extending detoxification mechanisms are concomitantly induced.
Keywords: Aging, Life span, Mouse, 4-hydroxynonenal, Glutathione transferase
TWO broad types of interventions have the capacity to modulate life span. The first type relies on modifications of high-level signaling pathways that affect multiple targets. The second type is aimed at altering these targets directly, thus bypassing the cellular- or organism-level signaling circuits. The most striking examples of the first category of life span–modulating changes are provided by hypomorphic or null mutations in the insulin/insulin-like signaling pathway of Caenorhabditis elegans (1), which can lead to up to a 10-fold extension of life span (2). In the mouse, several spontaneous and introduced mutations that directly affect components of the insulin growth factor (IGF) signaling pathway or that decrease IGF secretion, including the mutations present in dwarf mice, also extend life span (3), establishing IGF signaling as an important aspect of mammalian aging. The targets of IGF signaling include stress response, repair, and detoxification-related genes, consistent with the finding that stress resistance and longevity are often highly correlated (2,4–7) and that Ames dwarf mice show an increased resistance to oxidative stress (8). Another near-universal life-prolonging intervention is caloric restriction (9,10). The mechanism of its effect on aging remains unknown, in spite of multiple theories that have been advanced. In fact, different types of caloric/dietary restriction act via distinct mechanisms (11). Nonetheless, at least some dietary restriction regimens appear to involve elevated stress resistance (12). TOR (Target of Rapamycin)signaling may explain certain aspects of caloric restriction (13), and it is at the center of the hypertrophy/metabolic hyperactivity theory of aging (14).
The second broad class of experimental interventions able to modulate life span directly targets the metabolic processes that, in normal physiology, are regulated by the high-level signaling pathways discussed in the preceding paragraph. These processes, and their associated enzymes, include antioxidant defenses (15), other detoxification reactions (16), DNA (17,18), and, to a lesser extent, protein repair mechanisms (19), regulation of protein synthesis (20), degradation (21), and organellar autophagy (22), heat shock factors (23), modulation of membrane lipid unsaturation (24,25), and others. Experimental alteration of any of these metabolic reactions is expected to exert a smaller effect on life span than changes to high-level signaling but may also carry a lighter burden of side effects. For this reason, we have previously investigated in C. elegans the longevity effects of a particular type of detoxification enzymes that act on lipid-derived electrophiles (6,26,27). In the present communication, we sought to extend these studies to a mammalian system. We expected that disruption of mGsta4, a murine gene encoding a major antielectrophilic enzyme, will parallel the effect of a similar intervention in C. elegans and curtail the life span of the knockout mice. Surprisingly, the opposite was true. In the present article, we report this observation and provide a possible explanation for this unexpected effect.
METHODS
Animals
The methods used to disrupt the mGsta4 gene were described previously (28). The resulting mGsta4-null mice in the 129/sv genetic background were outcrossed to C57BL/6 animals for 10 generations. The resulting mGsta4-null mice in the C57BL/6 background were used for all experiments described in this article, unless otherwise indicated. The work was performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee.
Life Span Determination
Progeny resulting from matings of mice heterozygous with respect to the mGsta4 gene (+/− animals) was genotyped as described in the following section. Females of the −/− and +/+ genotypes were identified and were entered into the life span study until each group reached 50 animals. The study was fully populated with littermates over a span of 4 months. Mice were initially maintained at four animals per cage and were not combined as their numbers decreased during the course of the study. The animals had free access to water and to Harlan Teklad LM-485 Mouse/Rat Sterilizable Diet and were maintained on a 12-hour dark/12-hour light cycle. Mice were examined, and deaths were recorded daily. Because of this frequency of recording, and because no animals were removed from the study or lost to obvious infections or other accidents, neither left censoring nor right censoring was necessary.
Genotyping for the mGsta4 Gene
Multiplex polymerase chain reaction (PCR) genotyping on tail biopsy samples was carried out using the Extract-N-Amp Tissue PCR Kit (Sigma, St Louis, MO). The sense-strand primer was 5′-tccaatacacaaaaatgcatga, common to both amplicons, and the antisense primers were 5′-gatggccctggtctgtgtcagc (specific for the wild-type mGsta4 allele) and 5′-ctgtccatctgcacgagactagtg (derived from the neo cassette and thus specific for the disrupted mGsta4 allele). PCR was continued for 33 cycles using an annealing temperature of 56°C. A product of 284 bp was diagnostic for the wild-type allele and a product of 543 bp for the disrupted mGsta4 allele.
Determination of Transcript Levels by Reverse Transcription Real-Time Polymerase Chain Reaction
Total RNA was isolated from mouse tissues by the guanidinium thiocyanate method (TRI Reagent; Molecular Research Center, Cincinnati, OH) (29). Complementary DNA was prepared with the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA) according to the manufacturer's directions using the supplied mixture of oligo-dT and random primers. Reverse transcription real-time polymerase chain reaction (RT-qPCR) amplification reactions were performed on a DNA Engine Opticon 2 Detection System (MJ Research, Waltham, MA) with the FastStart SYBR Green Master mix (Roche Diagnostics, Indianapolis, IN) in a total volume of 20 μL containing 0.3 μM gene-specific primers. The primers are listed in Table 1. The cycling protocol was initial denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 61°C for 30 seconds, and 72°C for 30 seconds. The ribosomal protein S3 (Rps3) transcript was used as a reference for normalization.
Table 1.
Primers Used for Reverse Transcription Real-Time Polymerase Chain Reaction Determination of Transcript Levels
| Transcript | Forward Primer (5′ → 3′) | Reverse Primer (5′ → 3′) | Amplicon (bp) | PrimerBank ID |
| Cat | ttgccgttcggttctccac | gaaaataggggtgttgtttccca | 134 | 6753272a3 |
| Sod1 | aaccagttgtgttgtcaggac | ccaccatgtttcttagagtgagg | 139 | 12805215a1 |
| Sod2 | acaaacctgagccctaagggt | gaaccttggactcccacagac | 128 | 31980762a3 |
| Prdx6 | cgccagagtttgccaagag | tccgtgggtgtttcaccattg | 115 | 6671549a1 |
| Gpx1 | ccgtgcaatcagttcggaca | tcacttcgcacttctcaaacaat | 124 | 6680075a3 |
| Gpx4 | tctgtgtaaatggggacgatg | acgcagccgttcttatcaat | 130 | |
| Txnrd1 | gcagtactgagtggcgtt | tgtgagggagcctcgtg | 107 | |
| Gclc | cgatgtctgagttcaacactgt | ggaatgaagtgatggtgcagag | 105 | 33468897a3 |
| Gclm | aggagcttcgggactgtatcc | gggacatggtgcattccaaaa | 105 | 6680019a1 |
| Gsta1+2 | tattatgtcccccagaccaaaga | cctgttgcccacaaggtagtc | 128 | 7110611a3 |
| Gsta3 | aaaccaggaaccgttacttccc | ttcaaccagggcaatatcagc | 107 | 31981724a3 |
| Gstm1 | agcaccacctggatggag | agtcagggttgtaacagagcat | 111 | |
| Aor | aggagggcgacaggatgat | ccaacctaataaagccgctaca | 101 | 13385466a2 |
| Akr1b3 | tgcggtgaaccagatcgag | ccaaggggactatatgctgtca | 102 | 31981909a3 |
| Akr1b7 | ccaatactgtcaatccaagggc | aatctccattactacggggtctt | 103 | 6753148a3 |
| Akr1b8 | agggcatctctgtcactgc | gctgaggttttctcgtgcttg | 130 | 6679791a2 |
| Rps3 | ttacaccaaccaggacagaaatc | tggacaactgcggtcaactc | 100 | 6755372a2 |
Notes: Primer sequences were from PrimerBank (30) (http://pga.mgh.harvard.edu/primerbank) except for primer sets for Gstm1, Txnrd1 and Gpx4, which were designed by us. The gene names are Cat = catalase; Sod1 = superoxide dismutase 1; Sod2 = superoxide dismutase 2; Prdx2 = peroxiredoxin 6; Gpx1 = glutathione peroxidase 1; Gpx4 = glutathione peroxidase 4 (mitochondrial phospholipid hydroperoxide glutathione peroxidase Phgpx); Txnrd1 = thioredoxin reductase 1 (the primer set recognizes the four known splice variants NM_001042523, NM_001042513, NM_015762, and NM_001042514); Gclc = glutamate–cysteine ligase, catalytic subunit; Gclm = glutamate–cysteine ligase, modifier subunit; Gsta1 and Gsta2 = glutathione transferase A1 and A2, respectively (complementary DNA derived from both genes is recognized by the primer set); Gsta3 = glutathione transferase A3; Gstm1 = glutathione transferase M1; Aor = alkenal/one oxidoreductase (also known as prostaglandin reductase Ptgr1 or leukotriene B4 12-hydroxydehydrogenase Ltb4dh) (31–33); Akr1b3 = aldo-keto reductase family 1, member B3; Akr1b7 = aldo-keto reductase family 1, member B7; Akr1b8 = aldo-keto reductase family 1, member B8; Rps3 = ribosomal protein S3.
Quantitation of Nrf2
Livers and skeletal muscle, harvested from 16-week-old female wild-type and mGsta4-null mice, were quick-frozen to liquid nitrogen temperature using Wollenberger tongs (34) and stored at −75°C until use. The level of active nuclear Nrf2 protein was determined by ELISA using the TransAM Nrf2 Kit (Active Motif, Carlsbad, CA) according to the manufacturer’s recommendations. Briefly, nuclei were prepared and tested for contamination with cytosolic proteins by Western blotting for actin using a monoclonal antibody against the C-terminus of actin (catalog number sc-8432; Santa Cruz Biotechnology, Santa Cruz, CA). Within the detection limits of the method, no cytosolic contamination was found in nuclei prepared from liver or skeletal muscle (data not shown). Nuclear extracts (10 μg protein) were added to wells coated with an oligonucleotide containing the antioxidant response element (ARE) consensus sequence. DNA-bound Nrf2 was recognized by subsequently added anti-Nrf2 primary antibody. Detection and quantitation was achieved using a secondary antibody conjugated with horseradish peroxidase. Specificity of the reaction was ascertained by competition with ARE-containing or mutated oligonucleotides supplied with the TransAM Nrf2 Kit.
Statistics
Survival curves were compared by the log-rank and the Gehan–Wilcoxon tests, as implemented in the NCSS software package (Number Cruncher Statistical Systems, Kaysville, UT, www.ncss.com). Both the chi-square and the randomization variants of the tests were employed; the randomization variant used 100,000 Monte Carlo samples of the data. Maximum likelihood estimates of Gompertz survival parameters (35) were obtained using WinModest software (36,37). Significance of differences in transcript levels was evaluated by the t test carried out on Ct (threshold cycle number) values that were obtained by RT-qPCR on groups of 3–5 mice per group and were normalized to the level of ribosomal protein S3 (Rps3) transcript.
RESULTS AND DISCUSSION
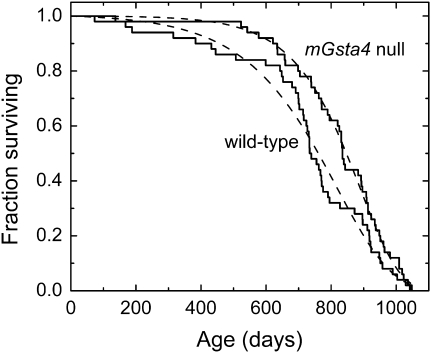
We have previously demonstrated that disruption of 4-hydroxynonenal (4-HNE)–conjugating enzymes in C. elegans curtailed life span, whereas overexpression of enzymes capable of 4-HNE disposal extended life (6,26,27). By analogy, we expected that mGsta4-null mice will have a shorter life span than wild-type mice. Survival analysis was carried out on mice in the C57BL/6 genetic background because this strain is relatively long lived and has variable causes of death but a low tumor incidence (38–40), characteristics that are attractive for aging studies. Against our expectations, the median life span of C57BL/6 mGsta4-null animals was longer by approximately 100 days than that of matched wild-type controls (Figure 1; summary statistics are provided in Table 2). The difference in survival was even greater initially (i.e., at 5%–15% mortality) but declined thereafter; the two groups were very similar or identical in maximal life span (Figure 1 and Table 2).
Figure 1.
Kaplan–Meier survival plots of female wild-type C57BL/6 mice (solid line labeled “wild-type”) and female mGsta4-null mice in C57BL/6 genetic background (solid line labeled “mGsta4 null”). The dashed lines correspond to the Gompertz models of the data, plotted using Gompertz parameters estimated by the maximum likelihood method (see Table 3). Fifty animals per group were used.
Table 2.
Descriptive Survival Statistics of Wild-Type and mGsta4-Null Mice
| Wild Type (d) | mGsta4 Null (d) | Change in mGsta4 Null vs Wild Type (%) | |
| Mean survival time | |||
| Experimental data (M ± SE) | 727 ± 31 | 820 ± 23 | 13 |
| Gompertz fitting | 730 | 822 | 13 |
| Median survival time | |||
| Experimental data | 735 | 836 | 14 |
| Gompertz fitting | 761 | 847 | 11 |
| 10% mortality | |||
| Experimental data | 384 | 622 | 62 |
| Gompertz fitting | 456 | 621 | 36 |
| 90% mortality | |||
| Experimental data | 952 | 996 | 5 |
| Gompertz fitting | 959 | 990 | 3 |
| Maximal life span | 1049 | 1043 | −1 |
Notes: Life span measurements were carried out on groups of 50 mice per genotype. Survival statistics are shown both for the original experimental data and for a fitted Gompertz curve using Gompertz parameters from Table 3. “Maximal life span” denotes the age at death of the last animal in the cohort.
Several statistical tests are available for comparison of survival curves. The log-rank test is used most frequently. This test has maximum power when the ratio of hazards of the two groups to be compared is constant over time (proportional hazards assumption). The log-rank test can tolerate moderate but not severe violation of the proportional hazards assumption (41,42). Inspection of the survival curves of Figure 1 indicates that the proportional hazards assumption may not hold as the mGsta4-null animals have low initial mortality followed by a steeper drop later in life, resulting in an early separation of the survival curves but an identical maximal life span, as noted previously. When the proportional hazards assumption does not apply, Gehan's generalized Wilcoxon test has been found to be more robust (41). Compared to the log-rank test, the Gehan–Wilcoxon test is more sensitive to differences between groups that occur at early time points of the life span (43). Even though the log-rank test may not be appropriate in our case because of the violation of the proportional hazards assumptions, its results are shown in Table 3 because of the familiarity of this test and its extensive use in the literature. The log-rank p value does not reach the customary threshold of .05; the relatively high p value reflects the convergence of the two survival curves toward the end of the life span. However, the observed p = .1 means that a separation of the survival curves equal to or greater than that shown in Figure 1 would occur by chance in only 1 in 10 life span determinations if there were no real longevity differences between the two populations of mice. Moreover, when tested by the Gehan–Wilcoxon procedure that is more appropriate for the data of Figure 1, the survival curves of wild-type and mGsta4-null mice were different at p = .018.
Table 3.
Analysis of Survival Curves of Wild-Type and mGsta4-Null Mice
| Wild Type | mGsta4 Null | |
| Gehan–Wilcoxon test for survival curves | ||
| Chi-square | p = .01 | |
| Randomization | p = .018 | |
| Log-rank test for survival curves | ||
| Chi-square | p = .096 | |
| Randomization | p = .102 | |
| Gompertz parameter A (95% confidence interval) | 4.36 × 10−5 (1.47–12.99) × 10−5 | 0.49 × 10−5 (0.10–2.41) × 10−5 |
| Likelihood ratio test for parameter A | p = .020 | |
| Gompertz parameter G (95% confidence interval) | 6.01 × 10−3 (4.72–7.65) × 10−3 | 8.35 × 10−3 (6.64–10.50) × 10−3 |
| Likelihood ratio test for parameter G | p = .047 | |
Notes: Survival curves (n = 50 mice per group; same animals as used for experiment summarized in Table 2) were compared by the Gehan–Wilcoxon and the log-rank tests, as implemented in the NCSS statistical software package. The p values are listed for both the chi-square approximation and the permutation (105 Monte Carlo samples) variants of each test. Maximum likelihood estimates of Gompertz parameters were obtained using the WinModest program (36,37). The values of these parameters for wild-type and mGsta4-null mice were compared by the likelihood ratio test of WinModest. In this test, the log likelihood estimate of the full model is subtracted from that of a “null hypothesis model” in which one of the parameters has been constrained to be identical for the two groups of mice. The resulting difference, multiplied by 2, is distributed as a chi-square random variable with 1 df.
As determined by a maximum likelihood test (implemented in WinModest software; 36,37), of the models tested (Gompertz, Gompertz–Makeham, logistic, and logistic-Makeham), the Gompertz model best describes the data shown in Figure 1. Estimates of Gompertz parameters and their 95% confidence limits are listed in Table 3. Both parameters of the Gompertz equation, A and G, were significantly different between wild-type and mGsta4-null mice (Table 3). It is, however, noteworthy that mGsta4-null mice have a lower value of parameter A but a higher value of parameter G than wild-type animals. Although the Gompertz function is a mathematical formalism without a priori biological meaning, parameter A is typically interpreted as the intrinsic or baseline mortality and parameter G as the rate of exponential increase in mortality with age. A low value of parameter A indicates low initial mortality, with a relatively prolonged “plateau” phase prior to substantial attrition. A high value of G has no significant effect at young age but leads to a steep decline in survivorship later in life. Accordingly, the survival curves of wild-type and mGsta4-null mice are most divergent early in life. Assuming the Gompertz models (broken lines in Figure 1) to be optimal representations of average survival curves that would have been obtained on multiple repetitions of the life span study, mGsta4-null mice live 36% longer at the 10th percentile of mortality. The median (i.e., at the 50th percentile of mortality) life span extension is 11%. At the 90th percentile of mortality, the two groups of mice have an approximately equal life span (3% extension for mGsta4-null animals; Table 2).
mGSTA4-4 has high catalytic efficiency for glutathione conjugation of the lipid peroxidation product 4-HNE (44,45), a toxicant (46,47) that causes electrophilic stress. Therefore, mGSTA4-4 is considered to have a protective detoxification role, and an extension of life span in animals lacking mGSTA4-4 appears counterintuitive. This apparent contradiction may be resolved by our earlier finding that in mice of the C57BL/6 genetic background that were used for the life span study, the steady-state level of 4-HNE in several tissues (liver, skeletal muscle, and white adipose tissue) was only marginally higher in mGsta4-null than in wild-type animals (48). In fact, in contrast to mice in the 129/sv background, for C57BL/6 animals, this difference of 4-HNE levels did not reach statistical significance in most experiments (48).
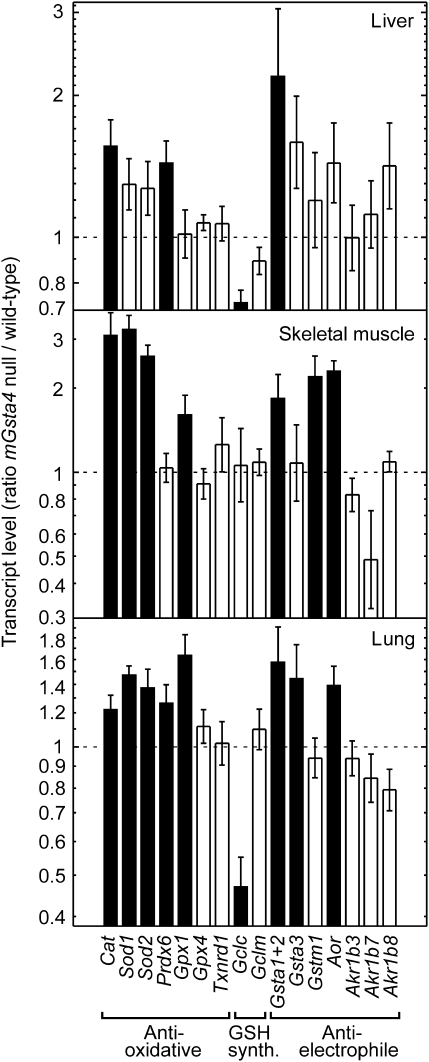
To determine why the steady-state tissue concentration of 4-HNE is not significantly elevated in C57BL/6 mGsta4-null mice in spite of disruption of a major gene involved in 4-HNE metabolism, we examined the expression of other genes that may be relevant to 4-HNE metabolism. The selected genes fall into three functionally distinct, albeit partially overlapping, classes: antioxidant, antielectrophile, and genes necessary for glutathione synthesis (Figure 2). Products of antioxidant genes could prevent lipid peroxidation and thus limit the formation of 4-HNE. Proteins encoded by antielectrophilic genes, such as glutathione transferases, alkenal/one oxidoreductase, and aldo-keto reductases, may act directly on 4-HNE (by conjugation, reduction of the double bond between carbon atoms 2 and 3, and reduction of the aldehyde moiety, respectively). Some of the enzymes may carry out both functions; for example, certain glutathione transferases have glutathione peroxidase activity against phospholipid hydroperoxides (antioxidant role) as well as the ability to conjugate 4-HNE with glutathione (antielectrophilic activity) (49). Likewise, enzymes necessary for glutathione biosynthesis contribute to subsequent antioxidant and antielectrophilic reactions because of the dual metabolic role of glutathione as a reductant and a nucleophile (50). The increased transcript levels of a number of antioxidant and antielectrophilic genes in several tissues of mGsta4-null mice (Figure 2) are consistent with a lower rate of formation, and higher rate of disposal, of 4-HNE in knockout animals. This shift in metabolism may explain why 4-HNE tissue concentrations in C57BL/6 mGsta4-null mice are almost identical to the wild-type level (48).
Figure 2.
Differences between mGsta4-null and wild-type C57BL/6 mice in transcript levels of selected genes involved in antioxidant and antielectrophilic defenses and in glutathione synthesis. Transcript levels were determined by reverse transcription real-time polymerase chain reaction in liver, skeletal muscle, and lung of 16-week-old female animals (n = 5 per group). The bars represent ratios (± SD of the ratio) of mean transcript levels (normalized to the level of ribosomal protein S3 [Rps3] transcript) of mGsta4-null and wild-type animals. Filled bars indicate that the difference between mGsta4-null and wild-type mice is statistically significant (p < .05 by t test).
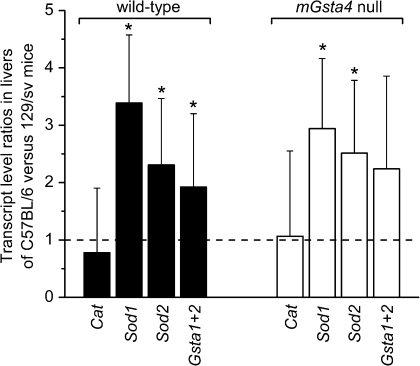
In contrast to C57BL/6 mice, the concentration of 4-HNE is significantly increased in tissues of mGsta4 knockouts in the 129/sv genetic background (48). This strain dependence of the phenotype is of interest, even though 129/sv animals were not used for life span studies in the present work. A direct comparison of transcript levels for catalase, Sod1, Sod2, and glutathione transferases A1 and A2 in livers of C57BL/6 and of 129/sv mice shows that, with the exception of catalase, these genes are more highly expressed in C57BL/6 animals (Figure 3). The strain comparison was carried out separately for wild-type mice (i.e., wild-type C57BL/6 were compared with wild-type 129/sv) and for their mGsta4-null counterparts (i.e., knockout C57BL/6 were compared with knockout 129/sv). The transcript ratios were similar for wild-type and knockout mice, that is, with or without disruption of the mGsta4 gene, the Sod1 transcript level was approximately threefold higher in C57BL/6 than in 129/sv, Sod2 transcripts were 2.5-fold higher, and glutathione transferase A1+A2 transcripts were approximately twofold higher (Figure 3). The resulting lower level of antioxidant and antielectrophilic defense mechanisms in 129/sv versus C57BL/6 animals, both wild type and mGsta4 null, could contribute to the initial higher 4-HNE concentration, at least in the liver and skeletal muscle of 129/sv mice, and to the further increase in 4-HNE upon mGsta4 disruption (48). In contrast, in C57BL/6 animals, lipid peroxidation and 4-HNE generation would be limited, and any 4-HNE that formed would be more efficiently removed, leading to lower and relatively invariant tissue levels of 4-HNE.
Figure 3.
Comparison of transcript levels of catalase (Cat), superoxide dismutase 1 and 2 (Sod1 and Sod2, respectively), and glutathione transferases 1 plus 2 (Gsta1+2) in livers of C57BL/6 and 129/sv mouse strains. Transcript levels were determined by reverse transcription real-time polymerase chain reaction. The bars represent ratios (± SD of the ratio) of mean transcript levels (normalized to the level of ribosomal protein S3 [Rps3] transcript) of three animals per group. Statistically significant differences between the C57BL/6 and 129/sv strains (i.e., transcript ratios of C57BL/6 vs 129/sv animals differing from 1; p < .05 by t test) are indicated by asterisks. The comparison was carried out separately for wild-type (filled bars) and for mGsta4-null (open bars) mice in the C57BL/6 and 129/sv genetic backgrounds.
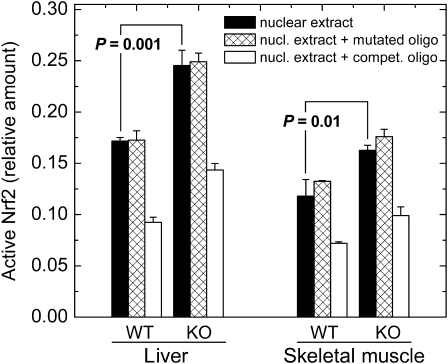
The mechanism by which multiple antioxidant and antielectrophilic genes are induced in mGsta4-null mice is of considerable interest. The constitutive as well as induced expression of many such genes is regulated by the transcription factor Nrf2 (51,52). Although other signaling pathways also modulate the expression of genes involved in tissue protection, the hypothesis that Nrf2 plays a role in the induction of gene expression shown in Figure 2 is particularly attractive because of the documented ability of 4-HNE to activate Nrf2 (53,54). To test this hypothesis directly, we determined the amount of active nuclear Nrf2 in C57BL/6 wild-type and mGsta4-null mice. In liver and skeletal muscle, the two tissues that were evaluated, active Nrf2 was increased by 43% and 38%, respectively, by knocking out mGsta4 (Figure 4). The assay was specific, as demonstrated by competition with an ARE-containing, but lack of competition with a mutated, oligonucleotide (Figure 4).
Figure 4.
Quantitation of active nuclear Nrf2 by ELISA. Nrf2 was measured in nuclear extracts from liver and skeletal muscle of wild-type and mGsta4-null 16-week-old female mice in the C57BL/6 genetic background. Filled bars represent M ± SD of three mice per group; statistical significance was evaluated by the t test and is shown in the Figure. Crosshatched bars represent competition with a mutated specific oligonucleotide, and open bars represent competition with an antioxidant response element–containing specific oligonucleotide. KO = mGsta4-null mice; WT = wild-type.
Whereas the increase in active Nrf2 was statistically significant for both liver and skeletal muscle, its magnitude, approximately 40% (Figure 4), was less than the severalfold increase typically reported for cells or animals treated with a variety of activators (e.g., 55). However, several considerations indicate that a 40% activation of Nrf2 is likely to be biologically relevant. First, there are published precedents for biological effects of a moderate increase of active Nrf2. For example, an approximately 30% increase of active Nrf2, as determined by electrophoretic mobility shift assays (EMSA), was sufficient to induce the expression of ARE-containing genes in cells treated with a polyphenol (56; EMSA was quantitated by densitometry of the published figure). Similarly, a less than twofold nuclear accumulation of Nrf2 was functionally significant, although a greater accumulation resulted in a more pronounced effect (55). Second, activation of Nrf2 is typically reported in terms of an increase of nuclear Nrf2 protein, determined by Western blotting or by immunofluorescence. However, not all nuclear Nrf2 may be active, suggesting that antibody-based assays could overestimate Nrf2 activation. In fact, in a recent article in which both assays were presented, a sevenfold increase in nuclear Nrf2 protein corresponded to a twofold increase in ARE-binding activity (quantitated by densitometry of figures from 56). Our assay (Figure 4) is based on Nrf2 activity rather than amount. Finally, it is worth noting that the majority of published accounts of Nrf2 activation involve a short-term acute treatment with an inducer. This is in contrast with the mGsta4-null mice in which active Nrf2 is likely to be chronically elevated. A lifelong increase in Nrf2 activity, even if moderate compared with the effects of acute treatments, is expected to cause a sustained augmentation of multiple downstream defense and repair mechanisms and thus have a cumulative effect on life span extension.
Interestingly, the transcript levels for Gclc and Gclm, subunits of glutamate–cysteine ligase that catalyzes the rate-limiting step of glutathione synthesis, were not increased in mGsta4-null mice (Figure 2), even though 4-HNE treatment induces the expression of these genes in cultured cells (57); in particular, submicromolar 4-HNE exerts its inducing effect on Gcl via Nrf2 signaling (58). The discrepancy between our results and those obtained in cultured cells could be due to differences in parameters such as prevailing partial pressure of oxygen and the effective concentration of 4-HNE, to an adaptation of the knockout mouse to a chronic physiological shift, or to a more complex regulatory network in the context of a multi-tissue intact organism. The regulation of glutathione synthesis involves several distinct signaling pathways (59), and the effects of 4-HNE on such signaling systems may depend on the concentration of the compound, on the cell type, and on the physiological context (reviewed in 60). Of particular interest is the recent finding that inhibition of Jun N-terminal kinase (JNK) prevented the induction by 4-HNE of Gclc and Gclm expression but not that of other Nrf2-dependent genes (57,61). Further work will be needed to elucidate whether the lack of Gclc and Gclm induction in mGsta4-null mice is linked to differential regulation of these genes by JNK or determined by other signaling events.
Taken together, our results are most readily interpreted as follows. Impaired 4-HNE conjugation in mGsta4-null mice results in an elevated tissue level of 4-HNE. This modulates several signaling cascades (62–65) including, but not limited to, the activation of Nrf2 (61). Enhanced Nrf2 signaling elicits a defense response. In particular, upregulation of antioxidant enzymes limits lipid peroxidation and 4-HNE formation, whereas antielectrophilic detoxification enzymes dispose of 4-HNE. Both types of reactions lower the concentration of 4-HNE and should be viewed as part of a negative feedback loop that stabilizes 4-HNE levels. In 129/sv mice, this feedback loop is only partially effective because of a relatively low expression of the defense mechanisms, resulting in a steady-state concentration of 4-HNE that is significantly higher in mGsta4-null than in wild-type animals. In contrast, the more robust detoxification systems of C57BL/6 mice reduce the steady-state concentration of 4-HNE in the knockout to almost wild-type levels. However, in both strains, there is a persistent elevation of antioxidant and antielectrophilic enzymes when mGsta4 is disrupted.
How do the biochemical and physiological changes precipitated by the disruption of the mGsta4 gene lead to an extended life span in the knockout mice (Figure 1)? Several possibilities could be considered. The primary effect of the loss of mGSTA4-4, the product of the mGsta4 gene, is an impairment in the glutathione conjugation of its substrates, mainly α,β-unsaturated carbonyl compounds such as 4-HNE (44,45) and, at least in the case of rat and human GSTA4-4, certain isoprostanes (66). 4-HNE is probably the physiologically most relevant substrate of mGSTA4-4, and experimental modulation of 4-HNE conjugation has been shown to affect life span in C. elegans (6,26,27). However, disruption of 4-HNE conjugation in C. elegans caused a shortening of life span, unlike the extension observed in the mGsta4-null mouse (Figure 1). The key to understanding this seemingly paradoxical result is the fact that overexpression or silencing of 4-HNE–conjugating enzymes in C. elegans resulted in decreased or elevated 4-HNE levels, respectively. This is in contrast to mice in which the upregulation of a defense response virtually abrogates any possible increase in 4-HNE concentrations but, via the negative feedback loop discussed previously, translates it into a steady-state increase of a variety of detoxification enzymes. In other words, the absence of a significant change in 4-HNE levels effectively rules out a direct effect of 4-HNE on the life span of mGsta4-null mice. It is, however, likely that longevity is affected by the secondary response, that is, the elevated antioxidant and antielectrophilic activities.
As originally formulated, the oxidative stress theory of aging (67,68) postulates that antioxidant defenses prolong life. Within this conceptual framework, the induction of catalase, Sod, and several other antioxidant enzymes (Figure 2) could be relevant to the observed life span extension in mGsta4-null mice (Figure 1). However, the oxidative stress theory of aging has been recently subjected to increasing criticism (69–73), in part because of the disappointing results of attempts to extend life by application of antioxidants, whether chemical or enzymatic. It could be argued that both the original enthusiasm and the recent rejection of the oxidative stress theory are oversimplifications and that a more nuanced approach is called for (15,69). In some (but certainly not all) situations, oxidative damage appears to contribute to aging, and antioxidant processes or interventions may prolong life (e.g., 74–76). Thus, it appears premature to rule out a causative role of antioxidant enzymes in the extension of life span of mGsta4-null mice, although this hypothesis remains to be tested.
A stronger case can be made for antielectrophilic enzymes, also upregulated in mGsta4-null animals (Figure 2). Consistent with the latter finding is the activation of Nrf2 (Figure 4), as it has been recently shown that Nrf2 is more important for the detoxification of electrophiles than for removal of reactive oxygen species (77). Induction of several classes of detoxification enzymes, including those that act on electrophiles, correlated with extended life span in several systems. This gave rise to the recently formulated “green” theory of aging (16,78). The theory emphasizes the role in aging of a broad spectrum of detoxification, stress response, and repair mechanisms, of which antioxidants are but a subset. We have accumulated evidence in C. elegans that overexpression of antielectrophilic enzymes has the potential to extend life (6, reviewed in 79). The increased longevity of mGsta4-null mice (Figure 1) could be the result of elevated antielectrophilic enzymes, as shown in Figure 2, and/or of additional detoxification proteins whose expression is dependent on Nrf2.
In summary, we postulate that the impairment of 4-HNE conjugation in mGsta4-null mice tends to increase the tissue level of 4-HNE. This, however, leads to the establishment of a new steady state in which 4-HNE activates signaling pathways including Nrf2, which in turn induces the expression of a large set of response genes. Some of the now overexpressed gene products create a negative feedback loop through which they control the concentration of 4-HNE. The same and additional Nrf2-dependent genes contribute to the extension of life span. This physiological shift in the mGsta4-null animals could be visualized in terms of translating the chronic but moderate increase in 4-HNE, the primary effect of disrupting a glutathione transferase, into a secondary induction of several stress response mechanisms. Such translation of the loss of a specific detoxification reaction into a generalized resistance has been observed in other physiological contexts; for example, plants with a silenced glutathione transferase can become refractory to fungal infection (80). It has been shown by modeling (81,82) that, in certain situations, an initial insult can be actually overcompensated by the resulting steady-state response. In mGsta4-null mice, the increased capacity for detoxification could be viewed as a hormetic response (83–85) to the chronic oversupply of 4-HNE; this hormetic effect could be responsible for the observed extension of life span.
FUNDING
National Institutes of Health grants R01 ES07804 and R01 AG18845 (to P.Z.). P.Z. is a recipient of a VA Research Career Scientist Award.
References
- 1.Kenyon C. A conserved regulatory system for aging. Cell. 2001;105:165–168. doi: 10.1016/s0092-8674(01)00306-3. [DOI] [PubMed] [Google Scholar]
- 2.Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 3.Bartke A. Impact of reduced IGF-1/insulin signaling on aging in mammals: novel findings. Aging Cell. 2008;7:285–290. doi: 10.1111/j.1474-9726.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 4.Lithgow GJ, Walker GA. Stress resistance as a determinate of C. elegans lifespan. Mech Ageing Dev. 2002;123:765–771. doi: 10.1016/s0047-6374(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 5.Murakami S. Stress resistance in long-lived mouse models. Exp Gerontol. 2006;41:1014–1019. doi: 10.1016/j.exger.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 6.Ayyadevara S, Engle MR, Singh SP, et al. Lifespan and stress resistance of Caenorhabditis elegans are increased by expression of glutathione transferases capable of metabolizing the lipid peroxidation product 4-hydroxynonenal. Aging Cell. 2005;4:257–271. doi: 10.1111/j.1474-9726.2005.00168.x. [DOI] [PubMed] [Google Scholar]
- 7.Tazearslan C, Ayyadevara S, Bharill P, Shmookler Reis RJ. Positive feedback between transcriptional and kinase suppression in nematodes with extraordinary longevity and stress resistance. PLoS Genet. 2009;5:e1000452. doi: 10.1371/journal.pgen.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bokov AF, Lindsey ML, Khodr C, Sabia MR, Richardson A. Long-lived Ames dwarf mice are resistant to chemical stressors. J Gerontol A Biol Sci Med Sci. 2009;64A:819–827. doi: 10.1093/gerona/glp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masoro EJ. Caloric restriction-induced life extension of rats and mice: a critique of proposed mechanisms. Biochim Biophys Acta. 2009;1790:1040–1048. doi: 10.1016/j.bbagen.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 13.Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blagosklonny MV. Aging: ROS or TOR. Cell Cycle. 2008;7:3344–3354. doi: 10.4161/cc.7.21.6965. [DOI] [PubMed] [Google Scholar]
- 15.Gruber J, Schaffer S, Halliwell B. The mitochondrial free radical theory of ageing—where do we stand? Front Biosci. 2008;13:6554–6579. doi: 10.2741/3174. [DOI] [PubMed] [Google Scholar]
- 16.Gems D, McElwee JJ. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech Ageing Dev. 2005;126:381–387. doi: 10.1016/j.mad.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijg J. The role of DNA damage and repair in aging: new approaches to an old problem. Mech Ageing Dev. 2008;129:498–502. doi: 10.1016/j.mad.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Ruan H, Tang XD, Chen ML, et al. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18:228–235. doi: 10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Hipkiss AR. Error-protein metabolism and ageing. Biogerontology. 2009;10:523–529. doi: 10.1007/s10522-008-9188-9. [DOI] [PubMed] [Google Scholar]
- 22.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy genes in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinkraus KA, Smith ED, Davis C, et al. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in C. elegans. Aging Cell. 2008;7:394–404. doi: 10.1111/j.1474-9726.2008.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulbert AJ, Pamplona R, Buffenstein R, Buttemer WA. Life and death: metabolic rate, membrane composition, and life span of animals. Physiol Rev. 2007;87:1175–1213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- 25.Pamplona R, Barja G. Highly resistant macromolecular components and low rate of generation of endogenous damage: two key traits of longevity. Ageing Res Rev. 2007;6:189–210. doi: 10.1016/j.arr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Ayyadevara S, Dandapat A, Singh SP, et al. Life span and stress resistance of Caenorhabditis elegans are differentially affected by glutathione transferases metabolizing 4-hydroxynon-2-enal. Mech Ageing Dev. 2007;128:196–205. doi: 10.1016/j.mad.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayyadevara S, Dandapat A, Singh SP, et al. Lifespan extension in hypomorphic daf-2 mutants of Caenorhabditis elegans is partially mediated by glutathione transferase CeGSTP2-2. Aging Cell. 2005;4:299–307. doi: 10.1111/j.1474-9726.2005.00172.x. [DOI] [PubMed] [Google Scholar]
- 28.Engle MR, Singh SP, Czernik PJ, et al. Physiological role of mGSTA4-4, a glutathione S-transferase metabolizing 4-hydroxynonenal: generation and analysis of mGsta4 null mouse. Toxicol Appl Pharmacol. 2004;194:296–308. doi: 10.1016/j.taap.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dick RA, Kensler TW. The catalytic and kinetic mechanisms of NADPH-dependent alkenal/one oxidoreductase. J Biol Chem. 2004;279:17269–17277. doi: 10.1074/jbc.M400427200. [DOI] [PubMed] [Google Scholar]
- 32.Dick RA, Kwak MK, Sutter TR, Kensler TW. Antioxidative function and substrate specificity of NAD(P)H-dependent alkenal/one oxidoreductase. A new role for leukotriene B4 12-hydroxydehydrogenase/15-oxoprostaglandin 13-reductase. J Biol Chem. 2001;276:40803–40810. doi: 10.1074/jbc.M105487200. [DOI] [PubMed] [Google Scholar]
- 33.Itoh K, Yamamoto K, Adachi M, Kosaka T, Tanaka Y. Leukotriene B412-hydroxydehydrogenase/15-ketoprostaglandinΔ13-reductase (LTB412-HD/PGR) responsible for the reduction of a double-bond of the α,β-unsaturated ketone of an aryl propionic acid non-steroidal anti-inflammatory agent CS-670. Xenobiotica. 2008;38:249–263. doi: 10.1080/00498250701767667. [DOI] [PubMed] [Google Scholar]
- 34.Wollenberger A, Ristau O, Schoffa G. Eine einfache Technik der sehr schnellen Abkühlung grösserer Gewebestücke. Pflugers Arch Gesamte Physiol Menschen Tiere. 1960;270:399–412. [PubMed] [Google Scholar]
- 35.Arking R. Measuring age-related changes in populations. In: Arking R, editor. Biology of Aging: Observations and Principles. 2nd ed. Sunderland, MA: Sinauer Associates; 1998. pp. 27–59. [Google Scholar]
- 36.Pletcher SD. Model fitting and hypothesis testing for age-specific mortality data. J Evol Biol. 1999;12:430–439. [Google Scholar]
- 37.Promislow DEL, Tatar M, Pletcher SD, Carey JR. Below-threshold mortality: implications for studies in evolution, ecology and demography. J Evol Biol. 1999;12:314–328. [Google Scholar]
- 38.Turturro A, Duffy P, Hass B, Kodell R, Hart R. Survival characteristics and age-adjusted disease Incidences in C57BL/6 mice fed a commonly used cereal-based diet modulated by dietary restriction. J Gerontol A Biol Sci Med Sci. 2002;57A:B379–B389. doi: 10.1093/gerona/57.11.b379. [DOI] [PubMed] [Google Scholar]
- 39.Treuting PM, Linford NJ, Knoblaugh SE, et al. Reduction of age-associated pathology in old mice by overexpression of catalase in mitochondria. J Gerontol A Biol Sci Med Sci. 2008;63:813–822. doi: 10.1093/gerona/63.8.813. [DOI] [PubMed] [Google Scholar]
- 40.Anisimov VN. Mutant and genetically modified mice as models for studying the relationship between aging and carcinogenesis. Mech Ageing Dev. 2001;122:1221–1255. doi: 10.1016/s0047-6374(01)00262-7. [DOI] [PubMed] [Google Scholar]
- 41.Lee ET, Desu MM, Gehan EA. A Monte Carlo study of the power of some two-sample tests. Biometrika. 1975;62:425–432. [Google Scholar]
- 42.Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care. 2004;8:389–394. doi: 10.1186/cc2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein JP, Moeschberger ML. Survival Analysis: Techniques for Censored and Truncated Data. 2nd ed. New York, NY: Springer Science+Business Media; 2003. [Google Scholar]
- 44.Zimniak P, Singhal SS, Srivastava SK, et al. Estimation of genomic complexity, heterologous expression, and enzymatic characterization of mouse glutathione S-transferase mGSTA4-4 (GST 5.7) J Biol Chem. 1994;269:992–1000. [PubMed] [Google Scholar]
- 45.Nanduri B, Hayden JB, Awasthi YC, Zimniak P. Amino acid residue 104 in an alpha-class glutathione S-transferase is essential for the high selectivity and specificity of the enzyme for 4-hydroxynonenal. Arch Biochem Biophys. 1996;335:305–310. doi: 10.1006/abbi.1996.0511. [DOI] [PubMed] [Google Scholar]
- 46.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 47.Eckl PM, Ortner A, Esterbauer H. Genotoxic properties of 4-hydroxyalkenals and analogous aldehydes. Mutat Res. 1993;290:183–192. doi: 10.1016/0027-5107(93)90158-c. [DOI] [PubMed] [Google Scholar]
- 48.Singh SP, Niemczyk M, Saini D, Awasthi YC, Zimniak L, Zimniak P. Role of the electrophilic lipid peroxidation product 4-hydroxynonenal in the development and maintenance of obesity in mice. Biochemistry. 2008;47:3900–3911. doi: 10.1021/bi702124u. [DOI] [PubMed] [Google Scholar]
- 49.Zimniak P. Substrates and reaction mechanisms of GSTs. In: Awasthi YC, editor. Toxicology of Glutathione Transferases. Boca Raton, FL: CRC Press; 2006. pp. 71–101. [Google Scholar]
- 50.Forman HJ, Zhang H, Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med. 2009;30:1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osburn WO, Kensler TW. Nrf2 signaling: An adaptive response pathway for protection against environmental toxic insults. Mutat Res. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen ZH, Saito Y, Yoshida Y, Sekine A, Noguchi N, Niki E. 4-Hydroxynonenal induces adaptive response and enhances PC12 cell tolerance primarily through induction of thioredoxin reductase 1 via activation of Nrf2. J Biol Chem. 2005;280:41921–41927. doi: 10.1074/jbc.M508556200. [DOI] [PubMed] [Google Scholar]
- 54.Levonen AL, Landar A, Ramachandran A, et al. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldring CEP, Kitteringham NR, Elsby R, et al. Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology. 2004;39:1267–1276. doi: 10.1002/hep.20183. [DOI] [PubMed] [Google Scholar]
- 56.Na H-K, Kim E-H, Jung J-H, Lee H-H, Hyun J-W, Surh Y- J. (-)-Epigallocatechin gallate induces Nrf2-mediated antioxidant enzyme expression via activation of PI3K and ERK in human mammary epithelial cells. Arch Biochem Biophys. 2008;476:171–177. doi: 10.1016/j.abb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Dickinson DA, Iles KE, Watanabe N, et al. 4-hydroxynonenal induces glutamate cysteine ligase through JNK in HBE1 cells. Free Radic Biol Med. 2002;33:974–987. doi: 10.1016/s0891-5849(02)00991-7. [DOI] [PubMed] [Google Scholar]
- 58.Zhang H, Court N, Forman HJ. Submicromolar concentrations of 4-hydroxynonenal induce glutamate cysteine ligase expression in HBE1 cells. Redox Rep. 2007;12:101–106. doi: 10.1179/135100007X162266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Awasthi YC, Sharma R, Sharma A, et al. Self-regulatory role of 4-hydroxynonenal in signaling for stress-induced programmed cell death. Free Radic Biol Med. 2008;45:111–118. doi: 10.1016/j.freeradbiomed.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Forman HJ. Signaling pathways involved in phase II gene induction by α,β-unsaturated aldehydes. Toxicol Ind Health. 2009;25:269–278. doi: 10.1177/0748233709102209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yun MR, Im DS, Lee SJ, et al. 4-Hydroxynonenal enhances CD36 expression on murine macrophages via p38 MAPK-mediated activation of 5-lipoxygenase. Free Radic Biol Med. 2009;46:692–698. doi: 10.1016/j.freeradbiomed.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 63.Rinna A, Forman HJ. SHP-1 Inhibition by 4-hydroxynonenal activates Jun N-terminal kinase and glutamate cysteine ligase. Am J Respir Cell Mol Biol. 2008;39:97–104. doi: 10.1165/rcmb.2007-0371OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kutuk O, Basaga H. Apoptosis signalling by 4-hydroxynonenal: a role for JNK-c-Jun/AP-1 pathway. Redox Rep. 2007;12:30–34. doi: 10.1179/135100007X162329. [DOI] [PubMed] [Google Scholar]
- 65.Sampey BP, Carbone DL, Doorn JA, Drechsel DA, Petersen DR. 4-Hydroxy-2-nonenal adduction of extracellular signal-regulated kinase and the inhibition of hepatocyte Erk-Elk-AP-1 signal transduction. Mol Pharmacol. 2006;71:871–883. doi: 10.1124/mol.106.029686. [DOI] [PubMed] [Google Scholar]
- 66.Hubatsch I, Mannervik B, Gao L, Roberts LJ, Chen Y, Morrow JD. The cyclopentenone product of lipid peroxidation, 15-A2t-isoprostane (8-isoprostaglandin A2), is efficiently conjugated with glutathione by human and rat glutathione transferase A4-4. Chem Res Toxicol. 2002;15:1114–1118. doi: 10.1021/tx020027r. [DOI] [PubMed] [Google Scholar]
- 67.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 68.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 69.Gems D, Doonan R. Antioxidant defense and aging in C. elegans: is the oxidative damage theory of aging wrong? Cell Cycle. 2009;8:1077–1083. doi: 10.4161/cc.8.11.8595. [DOI] [PubMed] [Google Scholar]
- 70.Perez VI, Van Remmen H, Bokov A, Epstein CJ, Vijg J, Richardson A. The overexpression of major antioxidant enzymes does not extend the lifespan of mice. Aging Cell. 2009;8:73–75. doi: 10.1111/j.1474-9726.2008.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lapointe J, Stepanyan Z, Bigras E, Hekimi S. Reversal of the mitochondrial phenotype and slow development of oxidative biomarkers of aging in long-lived Mclk1+/−mice. J Biol Chem. 2009;284:20364–20374. doi: 10.1074/jbc.M109.006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez VI, Bokov A, Van Remmen H, et al. Is the oxidative stress theory of aging dead? Biochim Biophys Acta. 2009;1790:1005–1014. doi: 10.1016/j.bbagen.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0138-8. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schriner SE, Linford NJ, Martin GM, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 75.Brown JC, McClelland GB, Faure PA, Klaiman JM, Staples JF. Examining the mechanisms responsible for lower ROS release rates in liver mitochondria from the long-lived house sparrow (Passer domesticus) and big brown bat (Eptesicus fuscus) compared to the short-lived mouse (Mus musculus) Mech Ageing Dev. 2009;130:467–476. doi: 10.1016/j.mad.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 76.Dai DF, Santana LF, Vermulst M, et al. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reisman SA, Yeager RL, Yamamoto M, Klaassen CD. Increased Nrf2 activation in livers from Keap1-knockdown mice increases expression of cytoprotective genes that detoxify electrophiles more than those that detoxify reactive oxygen species. Toxicol Sci. 2009;108:35–47. doi: 10.1093/toxsci/kfn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 79.Zimniak P. Detoxification reactions: relevance to aging. Ageing Res Rev. 2008;7:281–300. doi: 10.1016/j.arr.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hernandez I, Chacon O, Rodriguez R, et al. Black shank resistant tobacco by silencing of glutathione S-transferase. Biochem Biophys Res Commun. 2009;387:300–304. doi: 10.1016/j.bbrc.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Q, Pi J, Woods CG, Andersen ME. Phase I to II cross-induction of xenobiotic metabolizing enzymes: a feedforward control mechanism for potential hormetic responses. Toxicol Appl Pharmacol. 2009;237:345–356. doi: 10.1016/j.taap.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Q, Andersen ME. Dose response relationship in anti-stress gene regulatory networks. PLoS Comput Biol. 2007;3:e24. doi: 10.1371/journal.pcbi.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calabrese EJ, Baldwin LA. Hormesis: the dose-response revolution. Annu Rev Pharm Toxicol. 2003;43:175–197. doi: 10.1146/annurev.pharmtox.43.100901.140223. [DOI] [PubMed] [Google Scholar]
- 84.Gems D, Partridge L. Stress-response hormesis and aging: “that which does not kill us makes us stronger”. Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 85.Wu D, Cypser JR, Yashin AI, Johnson TE. The U-shaped response of initial mortality in Caenorhabditis elegans to mild heat shock: does it explain recent trends in human mortality? J Gerontol A Biol Sci Med Sci. 2008;63A:660–668. doi: 10.1093/gerona/63.7.660. [DOI] [PubMed] [Google Scholar]