Abstract
Spinocerebellar ataxia type 3 (SCA3)/Machado Joseph disease results from expansion of the polyglutamine domain in ataxin-3 (Atx3). Atx3 is a transcriptional co-repressor, as well as a deubiquitinating enzyme that appears to function in cellular pathways involved in protein homeostasis. In this study, we show that interactions of Atx3 with valosin-containing protein and hHR23B are dynamic and modulated by proteotoxic stresses. Heat shock, a general proteotoxic stress, also induced wild-type and pathogenic Atx3 to accumulate in the nucleus. Mapping studies showed that two regions of Atx3, the Josephin domain and the C-terminus, regulated heat shock-induced nuclear localization. Heat shock-induced nuclear localization of Atx3 was not affected by a casein kinase-2 inhibitor or by mutating a predicted nuclear localization signal. However, serine-111 of Atx3 was required for nuclear localization of the Josephin domain and regulated nuclear localization of full-length Atx3. Atx3 null cells were more sensitive to toxic effects of heat shock suggesting that Atx3 had a protective function in the cellular response to heat shock. Importantly, we found that oxidative stress also induced nuclear localization of Atx3; both wild-type and pathogenic Atx3 accumulated in the nucleus of SCA3 patient fibroblasts following oxidative stress. Heat shock and oxidative stress are the first processes identified that increase nuclear localization of Atx3. Observations in this study provide new and important insights for understanding SCA3 pathology as the nucleus is likely a key site for early pathogenesis.
INTRODUCTION
The polyglutamine neurodegenerative disease family consists of nine members including Huntington's disease, spinal and bulbar muscular atrophy, dentatorubral pallidoluysian atrophy and spinocerebellar ataxias (SCA) types 1, 2, 3, 6, 7 and 17 (1). Expansion of the polyglutamine domain destabilizes the mutant protein, leading to increased misfolding, altered protein–protein interactions and neuronal toxicity and death. A hallmark of polyglutamine diseases is the formation of neuronal inclusions, most often in the nucleus; inclusions consist of misfolded disease protein, ubiquitin, proteasomes, molecular chaperones and an array of other proteins (2–7). Spinocerebellar ataxia type-3 (SCA3) is caused by the expansion of the polyglutamine domain in ataxin-3 (Atx3) (8). Atx3 is widely expressed in cells and tissues and is present in both the cytoplasm and nucleus (9–12). Studies with cell models (5,7,13–17) and transgenic mice (18–20) show that pathogenic Atx3 as well as other polyglutamine disease proteins are more toxic when in the nucleus than in the cytoplasm. These and other studies are consistent with the nucleus being an important site of pathology in polyglutamine diseases including SCA3. Although the nucleus appears to be a primary site of pathogenesis in SCA3, the cellular processes that regulate Atx3 nuclear localization are not fully understood. Therefore, it is important to characterize cellular interactions and processes that regulate Atx3 nuclear localization.
Atx3 is a transcriptional co-repressor (21–24) as well as a deubiquitinating enzyme (DUB) (25,26) that functions in cellular pathways that regulate protein homeostasis (25,27–37). Atx3 preferentially cleaves K63-linked ubiquitin and mixed linkage ubiquitin chains (37) and its DUB activity is regulated by select proteotoxic stresses (38). Ubiquitin interacting motifs (UIMs) in Atx3 bind ubiquitin (25,28,29,39) and both its ubiquitin binding and DUB activities are important in suppressing polyglutamine-mediated neurodegeneration in Drosophila (31). Recently, our lab and others linked Atx3 to pathways that regulate protein homeostasis. Atx3 regulates ER-associated degradation through its interaction with valosin-containing protein (VCP/p97) (34,35) and its DUB activity, and UIMs are required for aggresome formation (30). Atx3 also interacts with the human homolog of Rad23, hHR23A/B (27,28). HHR23 appears to function in the ubiquitin proteasome pathway by binding ubiquitinated proteins and shuttling them to the proteasome for degradation (40–43). These properties of Atx3 suggest that it functions in cellular pathways that regulate protein homeostasis.
To investigate potential functions of Atx3 in cellular pathways that respond to altered protein homeostasis, we initiated studies to determine if proteotoxic stressors alter Atx3 interactions with VCP and hHR23B. In the early phase of this study, we found that heat shock, a general proteotoxic stress, resulted in nuclear accumulation of Atx3. Given the potential importance of this observation relative to SCA3 pathogenesis, we focused experiments on characterizing heat shock-induced nuclear localization of Atx3. Mapping studies showed that two regions of Atx3 as well as serine-111 regulate heat shock-induced nuclear localization of Atx3. However, DUB activity, UIMs and a predicted NLS were not required for Atx3 heat shock-induced nuclear localization. Atx3 knockout fibroblasts were more sensitive to toxic effects of heat shock compared with wild-type cells, suggesting that Atx3 has a protective function in the cellular response to heat shock. Importantly, we determined that oxidative stress, a stress that increases during aging and is linked to neurodegenerative diseases, resulted in nuclear accumulation of wild-type and pathogenic Atx3 in SCA3 patient fibroblasts. This raises the possibility that recurring oxidative stress during aging may be linked to SCA3 pathogenesis.
RESULTS
Cellular localization and interactions of Atx3 are altered following proteotoxic stress
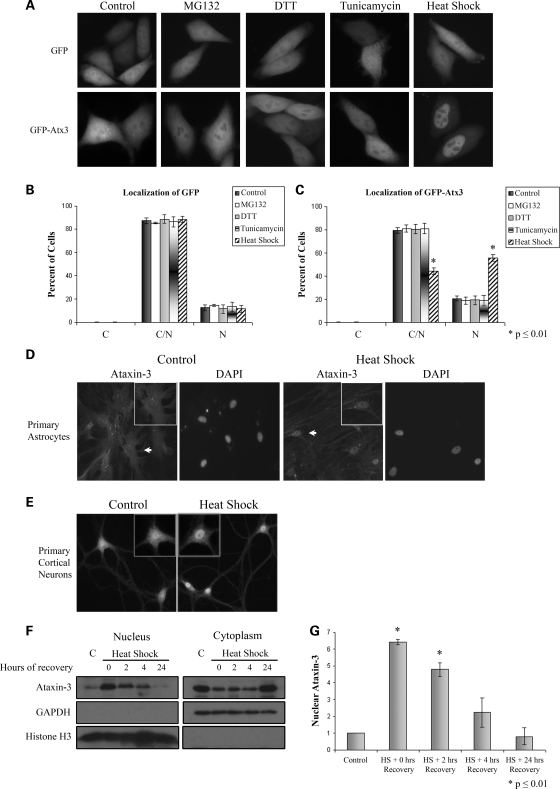
Several studies are consistent with Atx3 functioning in protein degradation pathways through interactions with effectors such as VCP and hHR23B (27,28,34,35). To determine if proteotoxic stresses regulated interactions of Atx3 with VCP and hHR23B, cells were treated with MG132 (inhibits proteasomes), DTT (activates the unfolded protein response), tunicamycin (activates the unfolded protein response) or heat shock (a general proteotoxic stress) and time courses of interactions were characterized. Most stressors resulted in dynamic changes in the interactions of Atx3 with VCP and hHR23B with each interaction and stressor showing a unique pattern (Supplementary Material, Fig. S1). These data are consistent with Atx3 functioning in cellular pathways that respond to altered protein homeostasis by altering its interactions with key effectors in these pathways. To determine if changes in cellular localization were associated with altered interactions, HeLa cells were transfected with GFP, GFP-Atx3, HA-VCP or HA-hHR23B. HA-VCP and HA-hHR23B did not alter their distribution following proteotoxic stress (Supplementary Material, Fig. S2). GFP-Atx3 was present in both the cytoplasm and the nucleus in control cells and its localization was not altered by treatment with MG132, DTT or tunicamycin (Fig. 1A). When cells were heat-shocked, however, GFP-Atx3 accumulated in the nucleus while the GFP control remained distributed evenly throughout the cell (Fig. 1A). Accumulation of GFP-Atx3 in the nucleus was rapid in HeLa cells with an increase in nuclear Atx3 beginning within the first 15 min of heat shock (data not shown). Data were quantified by counting the number of cells displaying GFP fluorescence that was primarily cytoplasmic, nuclear or distributed evenly between the cytoplasm and nucleus (Fig. 1B and C). Compared with control cells heat shock was the only stressor that resulted in a significant increase in the number of cells with GFP-Atx3 primarily localized to the nucleus (20% control versus 56% heat shock, P ≤ 0.01) (Fig. 1C). Heat shock is the first cellular process identified that induces nuclear localization of Atx3; because the nucleus is likely an early site of pathogenesis in SCA3 we initiated studies to further characterize heat shock-induced nuclear localization of Atx3.
Figure 1.
Heat shock increases nuclear localization of ataxin-3. (A) HeLa cells were transfected with GFP vector or GFP-Atx3 and 24 h later exposed to either 10 μm MG132 for 8 h, 1 mm DTT for 4 h, 10 μg/ml tunicamycin for 8 h, heat-shocked at 42°C for 1 h or maintained as untreated controls. Following treatment, GFP proteins were visualized in live cells. MG132, tunicamycin or DTT treatment did not alter localization of GFP-Atx3 or GFP. Heat shock did not affect localization of GFP; however, it increased nuclear localization of GFP-Atx3. Immunofluorescence data were quantified by counting GFP (B) or GFP-Atx3 (C) transfected cells with fluorescence primarily localized to the cytoplasm (C), nucleus (N) or evenly distributed (C/N). Data represent the mean ± SD of the fraction of total cells counted in each group with a particular localization pattern in three independent experiments with treated cells compared with controls (*P ≤ 0.01). Primary rat astrocytes (D) or primary rat cortical neurons (E) were either heat-shocked at 42°C for 30 min or left at 37°C. Following heat shock, cells were fixed and immunostained with Atx3 polyclonal antibody. In control cells, endogenous Atx3 localized throughout the cell, but after heat shock endogenous Atx3 redistributed to a primarily nuclear localization. Astrocytes were stained with DAPI to confirm the location of the nucleus. Cells indicated by white arrows are shown in insets. (F) Mouse fibroblasts were maintained at 37°C (C) or heat-shocked at 42°C for 3 h and collected over the following 24 h for cellular fractionation. Endogenous Atx3 localized to the nucleus following a 3 h heat shock and over the next 24 h returned to the cytoplasm. Distribution of GAPDH and histone H3 indicates purity of cytoplasmic and nuclear fractions. (G) Heat shock-induced nuclear accumulation of endogenous Atx3 was quantitated using densitometry. Data represent the mean ± SD of three independent experiments with data normalized to nuclear Atx3 levels in control cells. Nuclear levels of endogenous Atx3 increased 6.5-fold immediately following heat shock (P ≤ 0.01) and remained elevated following 2 h of recovery (P ≤ 0.01).
Heat shock-induced nuclear accumulation of Atx3 was not restricted to HeLa cells. Atx3 responded to heat shock similarly in other cell types tested including HEK293T, mouse fibroblasts, primary astrocytes and primary neurons. In untreated astrocytes and neurons, endogenous Atx3 localized to both the cytoplasm and nucleus while heat shock at 42°C for 30 min induced endogenous Atx3 to accumulate in the nucleus (Fig. 1D and E).
To support the fluorescent microscopy data, cell fractionation studies were carried out using mouse fibroblasts. Fibroblasts were heat-shocked for 3 h at 42°C and collected 0, 2, 4 or 24 h after heat shock and used for cellular fractionation. Western blots showed that endogenous Atx3 localized to the nuclear fraction after heat shock and over the following 24 h returned to the cytoplasmic fraction (Fig. 1F). The distribution of GAPDH and histone H3 indicated the purity of the cytoplasmic and nuclear fractions, respectively. Quantitation of the data indicated that heat shock induced a 6.5-fold increase in nuclear Atx3 levels (P ≤ 0.01) immediately following heat shock (Fig. 1G). Levels remained significantly higher following 2 h of recovery and the trend continued out to 4 h of recovery although the difference at this time point was not significant.
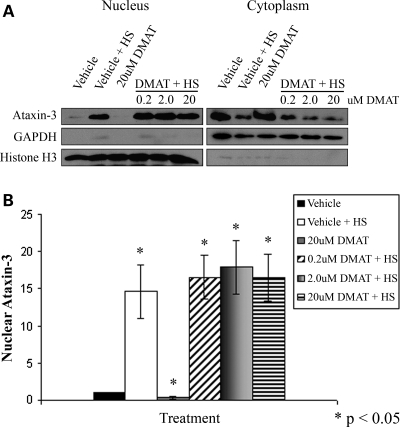
A recent publication indicated that CK2 phosphorylation of transfected Atx3 regulated its nuclear localization in control and heat-shocked HEK293 cells (44). To determine whether CK2 activity regulated heat shock-induced nuclear localization of endogenous Atx3, HEK293T cells were pre-treated with the CK2 inhibitor, DMAT, for 3 h, heat-shocked at 42°C for 1 h and cellular fractions prepared (Fig. 2A). Quantitation indicated that the CK2 inhibitor DMAT did not reduce heat shock-induced nuclear accumulation of endogenous Atx3 (Fig. 2B); however, it reduced nuclear Atx3 levels in cells maintained at 37°C by ∼60% (P < 0.05). To ensure that DMAT exposed to 42°C for 1 h was still active, 20 μm DMAT in DMEM was heated to 42°C for 1 h, returned to 37°C, and then tested for its ability to inhibit nuclear localization of Atx3 under basal conditions. DMAT heated at 42°C for 1 h still inhibited nuclear localization of Atx3 under basal conditions (Supplementary Material, Fig. S3). These data suggest that CK2 does not regulate heat shock-induced nuclear localization of Atx3 but appears to regulate nuclear localization of Atx3 under basal conditions.
Figure 2.
CK2 phosphorylation of ataxin-3 is not required for heat shock-induced nuclear localization. (A) HEK293T cells were treated with the CK2 inhibitor DMAT (0.2, 2.0 or 20 μm) or vehicle for 3 h and then heat-shocked for 1 h at 42°C and fractionated. Western blots indicated that following DMAT treatment, endogenous Atx3 still accumulated in the nucleus in response to heat shock similar to vehicle-treated cells. GAPDH and histone H3 were fractionation controls. (B) Heat shock-induced nuclear accumulation of endogenous Atx3 following vehicle or DMAT treatment was quantitated using densitometry. Data represent the mean ± SD of four independent experiments with all data normalized to vehicle control (*P < 0.05). No difference was detected between the DMAT + HS-treated groups and vehicle + HS group. DMAT treatment decreased the low level of nuclear Atx3 in cells maintained at 37°C by ∼60% (P < 0.05).
Effect of polyglutamine expansion and mutations affecting UIMs, DUB activity and a putative NLS on heat shock-induced nuclear accumulation of Atx3
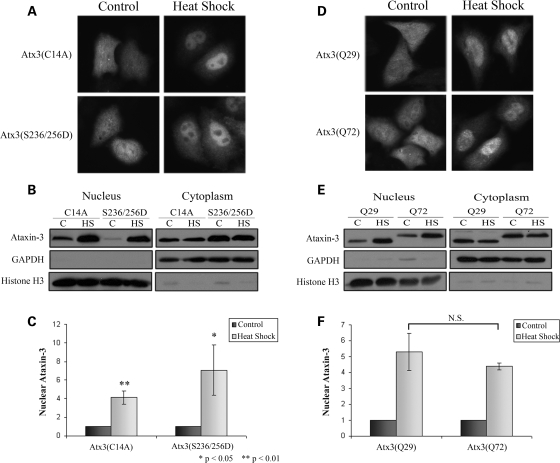
To determine whether Atx3 DUB activity or functional UIMs were required for nuclear localization, and whether the pathogenic protein responded to heat shock in a similar manner to the wild-type protein, myc-tagged pathogenic Atx3(Q72), Atx3(Q29)C14A (DUB mutant) and Atx3(Q29)S236/256D (UIM mutant) were transfected into cells, and myc immunostaining and cellular fractionations were performed on control and heat-shocked cells. Immunostaining of transfected HeLa cells showed that neither C14A nor S236/256D mutations blocked heat shock-induced nuclear localization of Atx3 (Fig. 3A). This was confirmed with cellular fractionations of transfected HEK293T cells (Fig. 3B). Densitometric analysis of these fractionations confirmed that Atx3 with either C14A or S236/256D mutations increased in the nucleus of heat-shocked cells compared with unstressed cells (Fig. 3C), indicating that DUB activity and functional UIMs were not required for heat shock-induced nuclear localization. Immunofluorescence (Fig. 3D) and cellular fractionation (Fig. 3E) data also indicated that pathogenic Atx3(Q72) accumulated in the nucleus similar to wild-type Atx3(Q29). Densitometric analysis of fractionations confirmed that Atx3(Q29) and Atx3(Q72) showed a similar increase in the nucleus following heat shock (5- and 4.5-fold increases, respectively) (Fig. 3F). These data raise the interesting possibility that pathogenic Atx3 may accumulate in the nucleus, a key site of pathogenesis in SCA3, as part of a normal cellular response of Atx3 to increased temperature or other stress.
Figure 3.
Effect of UIM and DUB mutations and polyglutamine expansion on heat shock-induced nuclear accumulation of ataxin-3. (A) HeLa cells transfected with myc-Atx3(Q29)C14A (DUB mutant) or myc-Atx3(Q29)S236/256D (UIM mutant) were either kept at 37°C or heat-shocked at 42°C for 1 h and immunostained with an myc antibody. (B) HEK293T cells transfected with myc-Atx3(Q29)C14A, or myc-Atx3(Q29)S236/256D, were either kept at 37°C or heat-shocked at 42°C for 1 h and nuclear and cytoplasmic fractions prepared. GAPDH and histone H3 were fractionation controls. (C) Heat shock-induced nuclear accumulation of Atx3(Q29)C14A and myc-Atx3(Q29)S236/256D were quantitated using densitometry. Data represent the mean ± SD of three independent experiments with each heat-shock group normalized to their 37°C control (*P < 0.05; **P < 0.01). Western blots and immunofluorescence data indicated that neither DUB activity nor functional UIMs were required for heat shock-induced nuclear localization of Atx3. (D) Pathogenic myc-Atx3(Q72) and WT myc-Atx3(Q29) were transfected into HeLa cells and either heat-shocked at 42°C for 1 h or kept at 37°C and immunostained with an myc antibody. (E) HEK293T cells transfected with myc-Atx3(Q29) or myc-Atx3(Q72) were heat-shocked at 42°C for 1 h or kept at 37°C and nuclear and cytoplasmic fractions prepared. In response to heat shock both wild-type and pathogenic Atx3 increased similarly in the nucleus. (F) Heat shock-induced nuclear accumulation of myc-Atx3(Q29) and myc-Atx3(Q72) were quantitated using densitometry. Data represent the mean ± SD of three independent experiments with data normalized to their 37°C control and displayed as fold increase in response to heat shock.
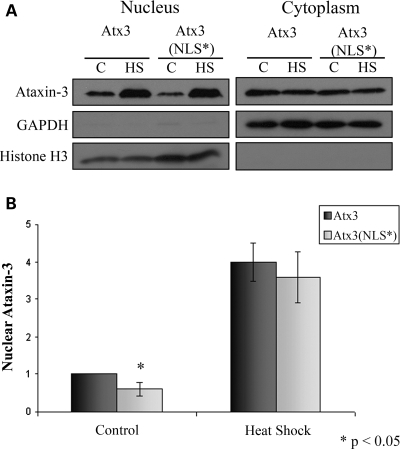
To determine whether the predicted nuclear localization signal (NLS) 282RKRR285 in Atx3 (10,45,46) regulated heat shock-induced nuclear localization, HEK293T cells were transfected with either myc-tagged wild-type Atx3 or the NLS mutant 282ANAA285 [Atx3(NLS*)] and 24 h later cells were heat-shocked and fractionated. Fractionation (Fig. 4A) and densitometry data (Fig. 4B) indicated that nuclear levels of Atx3(NLS*) were reduced by ∼40% (P < 0.05) under basal conditions, suggesting that this sequence modulates nuclear localization. Although the NLS regulated Atx3 nuclear localization in unstressed cells mutating the NLS did not alter accumulation of Atx3 in the nucleus in response to heat shock (Fig. 4A and B).
Figure 4.
Effect of NLS mutation on heat shock-induced nuclear accumulation of ataxin-3. (A) HEK293T cells transfected with myc-Atx3 or the NLS mutant myc-Atx3(NLS*) were heat-shocked at 42°C for 1 h or kept at 37°C and nuclear and cytoplasmic fractions prepared. In response to heat shock Atx3 and Atx3(NLS*) increased similarly in the nucleus. (B) Heat shock-induced nuclear accumulation of myc-Atx3 and myc-Atx3(NLS*) were quantitated using densitometry. Data represent the mean ± SD of three independent experiments with data normalized to Atx3 control (*P < 0.05).
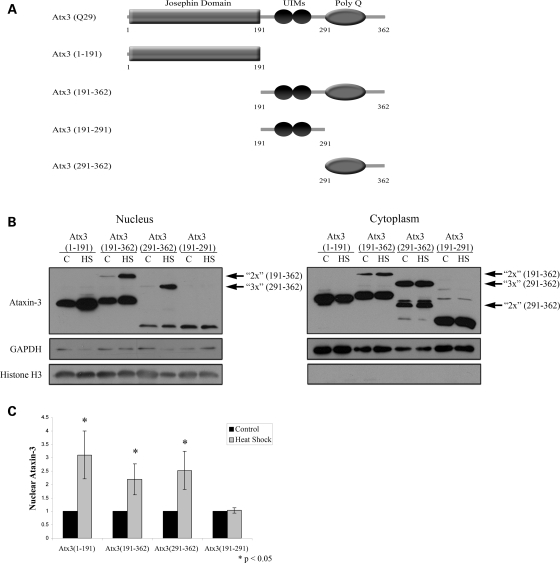
Atx3 contains two regions that regulate heat shock-induced nuclear localization
Under basal conditions, nuclear localization of Atx3 appears to be regulated by CK2 as well as a putative NLS adjacent to the polyglutamine domain. However, inhibiting CK2 or mutating the NLS sequence did not alter the nuclear accumulation of Atx3 following heat shock. To determine the region of Atx3 necessary for nuclear localization following heat shock, cells were transfected with myc-tagged Atx3 truncations (Fig. 5A) and cellular fractionations of control and heat-shocked cells were used to monitor the localization of these proteins (Fig. 5B). Truncations used were: Atx3(1–191) (N-terminal Josephin domain containing DUB activity), Atx3(191–362) (2 UIMs + PolyQ domain + C-terminus), Atx3(191–291) (contains two UIMs) and Atx3(291–362) (PolyQ + C-terminus) (Fig. 5A). Atx3 truncation constructs containing the polyQ domain [Atx3(191–362) and Atx3(291–362)] typically migrate in SDS PAGE gels at multiple discrete molecular weights possibly representing monomers, dimers and trimers (arrows denote potential dimers and trimers). Fractionation data (Fig. 5B) and quantitation of the western blots (Fig. 5C) indicated that Atx3 contained two areas that regulated heat shock-induced nuclear localization, one was present in the Josephin domain [Atx3(1–191)] and one in the polyQ + C-terminus [Atx3(291–362)] (Fig. 5B and C). In response to heat shock, Atx3(1–191) and Atx3(291–362) increased in the nucleus by 3- and 2.5-fold (P < 0.05), respectively (Fig. 5C). In contrast, Atx3(191–291) containing two UIMs did not accumulate in the nucleus following heat shock (Fig. 5B and C). Attempts to determine a region within Atx3(1–191) responsible for nuclear localization were unsuccessful due to the rapid degradation of truncated Josephin subdomains (data not shown). Interestingly, higher molecular weight multimers of Atx3(191–362) and Atx3(291–362) accumulated in the nucleus more than ‘monomeric’ forms (see Discussion).
Figure 5.
Mapping regions of ataxin-3 important for heat shock-induced nuclear localization. (A) Schematic of constructs used to map the regions of Atx3 involved in nuclear localization: full-length Atx3(Q29) (WT); Atx3(1–191) (N-terminal Josephin domain containing DUB activity); Atx3(191–362) (2 UIMs + PolyQ domain + C-terminus); Atx3(191–291) (2 UIMs) and Atx3(291–362) (PolyQ domain + C-terminus). (B) HEK293T cells were transfected with various myc-tagged Atx3 truncations to determine domains of Atx3 important for heat shock-induced nuclear localization. Both the N-terminal Josephin domain, Atx3(1–191), and the C-terminal half of the protein, Atx3(191–362), accumulated in the nucleus in response to heat shock, indicating that there were at least two sequences within Atx3 sufficient for nuclear localization. Atx3(291–362) also accumulated in the nucleus after heat shock, but Atx3(191–291) did not, suggesting that the second sequence important for regulating Atx3 heat shock-induced nuclear localization was located within amino acids 291–362. Atx3 truncations containing the polyQ domain typically migrate in SDS PAGE gels at multiple molecular weights that possibly represent multimers (these are identified with arrows and the number of potential ‘monomeric units’). (C) Heat shock-induced nuclear accumulation of each Atx3 truncation was quantitated using densitometry. Data represent the mean ± SD of three independent experiments with each heat-shock group normalized to their 37°C control (*P < 0.05).
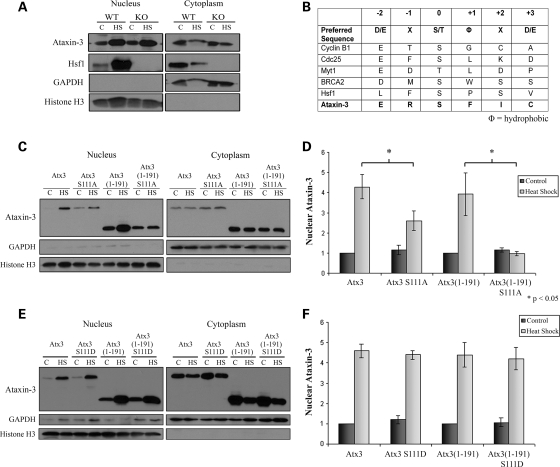
Effects of Hsf1 on heat shock-induced nuclear localization of Atx3
Hsf1 is a key regulator of the heat shock response and following heat shock rapidly undergoes homotrimerization, hyperphosphorylation and nuclear translocation (47–50). To determine if Hsf1 regulated Atx3 heat shock-induced nuclear localization, we first looked for an interaction between endogenous Atx3 and Hsf1 by immunoprecipitating Atx3 from control and heat-shocked mouse fibroblasts. Western blots indicated that Hsf1 did not co-immunoprecipitate with Atx3 in control or heat-shocked cells (Supplementary Material, Fig. S4). To further investigate whether Hsf1 regulated heat shock-induced Atx3 nuclear localization, Hsf1 WT and KO mouse fibroblasts were heat-shocked at 42°C for 1 h or kept at 37°C as a control and used for cellular fractionation. Immunoblots indicated that nuclear Atx3 increased in both Hsf1 WT and KO mouse fibroblasts in response to heat shock (Fig. 6A), indicating that Atx3 localized to the nucleus independent of Hsf1- and Hsf1-dependent processes.
Figure 6.
Ataxin-3 nuclear localization is regulated by serine-111 and independent of Hsf1. (A) Hsf1 WT and KO fibroblasts were either kept at 37°C or heat-shocked for 1 h at 42°C and cellular fractions prepared. Atx3 primarily localized to the nucleus after heat shock in both the Hsf1 WT and KO cells indicating that Hsf1 was not necessary for heat shock-induced nuclear localization of Atx3. GAPDH and histone H3 blots were controls for cell fractions. (B) Table showing the preferred sequence for Plk1 phosphorylation (52,53), several known substrates of Plk1 (Cyclin B1, Cdc25, Myt1, BRCA2 and Hsf1) and a potential phosphorylation site in Atx3 at serine-111. The sequence surrounding the Plk1 phosphorylation sites in known Plk1 substrates loosely resemble a predicted ‘preferred’ sequence. (C) HEK293T cells transfected with either Atx3, Atx3 S111A, Atx3(1–191) or Atx3(1–191) S111A were heat-shocked at 42°C for 1 h or kept at 37°C and then used for cellular fractionation. (D) Heat shock-induced nuclear accumulation of Atx3, Atx3 S111A, Atx3(1–191) and Atx3(1–191) S111A were quantitated using densitometry. Data represent the mean ± SD of three independent experiments comparing Atx3 with Atx3 S111A and Atx3(1–191) with Atx3(1–191) S111A (*P ≤ 0.05). Atx3 S111A and Atx3(1–191) S111A data were normalized to Atx3 and Atx3(1–191) data, respectively. Atx3 S111A showed a modest increase in the nucleus in response to heat shock but did not accumulate in the nucleus as much as wild-type Atx3. Nuclear localization of Atx3(1–191) increased following heat shock but Atx3(1–191) S111A did not accumulate in the nucleus in response to heat shock. (E) HEK293T cells transfected with either Atx3, Atx3 S111D, Atx3(1–191) or Atx3(1–191) S111D were heat-shocked at 42°C for 1 h or kept at 37°C and then used for cellular fractionation. Nuclear Atx3 S111D and Atx3(1–191) S111D increased in the nucleus in response to heat shock similar to Atx3 and Atx3(1–191). (F) Heat shock-induced nuclear accumulation of Atx3, Atx3 S111D, Atx3(1–191) and Atx3(1–191) S111D were quantitated using densitometry. Data represent the mean ± SD of three independent experiments. Atx3 S111D and Atx3(1–191) S111D data were normalized to Atx3 and Atx3(1–191) data, respectively. Atx3 S111D and Atx3(1–191) S111D increased in the nucleus similar to Atx3 and Atx3(1–191).
Serine-111 in Atx3 regulates heat shock-induced nuclear localization
Heat shock-induced nuclear localization of Atx3 was independent of Hsf1, but it remained possible that the pathway responsible for activating and inducing Hsf1 nuclear translocation was also responsible for Atx3 accumulating in the nucleus following heat shock. Phosphorylation of Hsf1 at serine-419 by Polo-like kinase 1 (Plk1) is important for heat shock-induced Hsf1 nuclear translocation (51). The preferred sequence for Plk1 phosphorylation is not tightly conserved, and phosphorylation sites of known Plk1 substrates often deviate from the predicted ‘preferred’ sequence (52,53) (Fig. 6B). The amino acid sequence flanking serine-111 in the Josephin domain of Atx3 resembled the ‘preferred’ sequence for Plk1 phosphorylation (Fig. 6B); therefore, we hypothesized that heat shock-activated Plk1 may phosphorylate Atx3 and induce nuclear localization. As a first step to test this hypothesis, S111A mutants were generated in full-length Atx3 and Atx3(1–191) and transfected into HEK293T cells. Cells were heat-shocked at 42°C or kept as controls at 37°C and cellular fractions prepared. The S111A mutation blocked nuclear accumulation of Atx3(1–191) which was consistent with serine-111 regulating nuclear localization of the Josephin domain following heat shock (Fig. 6C). The S111A mutation decreased nuclear accumulation of full-length Atx3; although, some Atx3 S111A still accumulated in the nucleus (Fig. 6C). These data were quantified confirming that the S111A mutation in Atx3(1–191) blocked heat shock-induced nuclear accumulation of the Josephin domain and resulted in a 39% reduction (P < 0.05) in the heat shock-induced nuclear accumulation of full-length Atx3 (Fig. 6D). These data were consistent with data in Figure 5B indicating that there was a second region of Atx3, Atx3(291–362), that regulated nuclear localization in response to heat shock.
Mutating serine-111 to alanine could inhibit nuclear accumulation by several mechanisms; an obvious mechanism would be blocking phosphorylation of serine-111. Serine phosphorylation can often be mimicked by mutating serine to aspartic acid; therefore, we mutated serine-111 to an aspartic acid in full-length Atx3 (Atx3 S111D) and the Josephin domain [Atx3(1–191) S111D]. These constructs were transfected into HEK293T cells and 24 h later heat-shocked and fractionated. Cellular fractionation (Fig. 6E) and densitometry data (Fig. 6F) indicated that the S111D mutations in Atx3(Q29) and Atx3(1–191) did not increase nuclear localization under basal conditions; however, in contrast to the alanine mutation constructs, the S111D mutation accumulated in the nucleus following heat shock. This is consistent with the S111D mutation mimicking phosphorylation and suggests that serine-111 phosphorylation is required for nuclear localization of Atx3 following heat shock but is not sufficient by itself to induce nuclear localization. Future studies will be needed to determine how serine-111 regulates nuclear localization and whether it is phosphorylated by Plk1 (or other kinase) or if serine-111 regulates nuclear localization independent of phosphorylation.
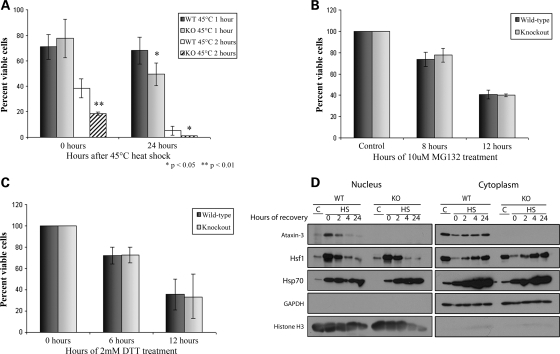
Atx3 knockout fibroblasts are sensitive to heat shock
To determine if Atx3 modulated acute heat toxicity or recovery from heat shock Atx3 WT and KO mouse primary fibroblasts were heat-shocked at 45°C for 1 or 2 h (preliminary studies indicated that heat shock at 42°C was not toxic to fibroblasts). The number of viable cells was determined immediately following heat shock or 24 h later. Immediately following a 1 h heat shock, there was a similar decrease in survival in both WT and KO cells with ∼75% of the cells alive compared with control cells (Fig. 7A). Twenty-four hours following the 1 h heat shock 68.3% of WT cells were viable while only 49.6% of KO cells were viable (P < 0.01; WT versus KO) compared with control cells. A greater number of Atx3 KO cells died acutely following the 2 h heat shock with 18.7% of KO cells and 38.4% of WT cells surviving immediately following heat shock (P < 0.01; WT versus KO). Twenty-four hours after the 2 h heat shock, 5.2% of WT cells and 1.2% of KO cells were viable (P < 0.05; WT versus KO). In contrast to heat shock, Atx3 WT and KO fibroblasts were equally sensitive to treatment with MG132 or DTT (Fig. 7B and C). These data were consistent with Atx3 selectively protecting cells from acute heat shock as well as helping cells recover from heat shock.
Figure 7.
Ataxin-3 knockout fibroblasts are more sensitive to toxic heat shock. Atx3 WT and KO primary mouse fibroblasts were either (A) heat-shocked or treated with (B) MG132 or (C) DTT. Cell viability at each time point is expressed relative to the number of viable cells in parallel sets of control cultures. Data represent mean ± SD of three independent experiments; statistical comparisons are for WT versus KO cells at the same time point (*P < 0.05; **P < 0.01). (A) Atx3 WT and KO primary fibroblasts were heat-shocked at 45°C for 1 or 2 h or kept at 37°C as a control. Cell viability was measured immediately following heat shock and again 24 h later and represented as a percentage of control cells. Atx3 WT and KO primary mouse fibroblasts were treated with (B) 10μm MG132 for 8 or 12 h or (C) 2 mm DTT for 6 or 12 h and the number of viable cells were measured and represented as a percentage of control cells. Atx3 WT and KO fibroblasts responded similarly to MG132 and DTT treatment. (D) Atx3 WT and KO mouse fibroblasts were either kept at 37°C or heat-shocked at 42°C for 3 h and collected over the following 24 h for preparation of nuclear and cytoplasmic fractions. As expected no Atx3 was detected in Atx3 KO cells. In Atx3 WT cells endogenous Atx3 accumulated in the nucleus following heat shock and then returned to the cytoplasm between 4 and 24 h following heat shock. Both WT and KO fibroblasts initiated early events in the heat shock response resulting in Hsf1 translocating to the nucleus following heat shock as well as Hsf1 returning to the cytoplasm following induction of hsp70 expression.
To ensure that the basic response to heat shock was intact in Atx3 KO cells, Atx3 WT and KO cells were heat-shocked and fractionated. In both Atx3 WT and KO cells, Hsf1 translocated to the nucleus after heat shock and Hsp70 levels increased similarly in the cytoplasmic and nuclear compartments of both WT and KO cells (Fig. 7D). Taken together, this indicated that early Hsf1 functions appeared normal in Atx3 KO cells following heat shock and suggested that Atx3 may be functioning independent of Hsf1 during heat shock or in a subset of Hsf1-dependent processes.
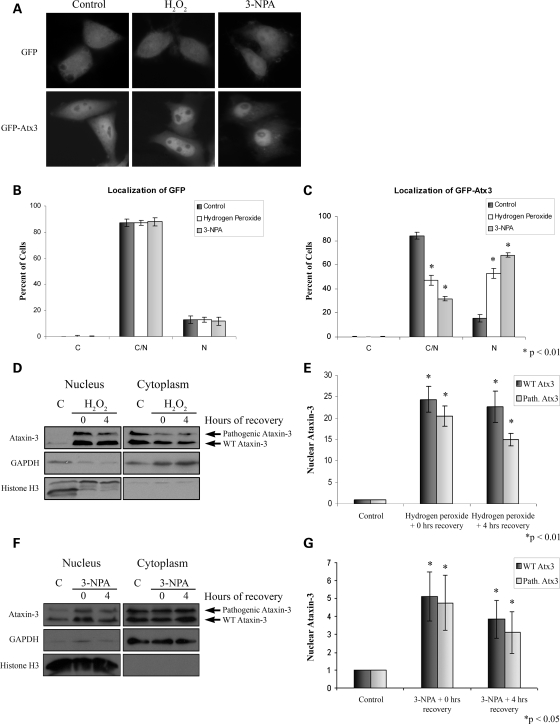
Oxidative stress induces nuclear localization of Atx3
Heat shock caused a striking accumulation of Atx3 in the nucleus; although, it seems unlikely that nuclear accumulation of pathogenic Atx3 in SCA3 patients results from recurring heat or temperature stress in the brain. However, a proteotoxic stress that is more commonly present in the brain such as oxidative stress might be responsible for nuclear accumulation of pathogenic Atx3. Oxidative stress occurs with increasing frequency in the aging brain and is linked to many neurodegenerative diseases, including polyglutamine diseases (54–59). Therefore, we investigated whether oxidative stress increased nuclear localization of Atx3. HeLa cells were transfected with GFP-Atx3(Q29) or GFP vector as a control. Twenty-four hours later, cells were treated with either 1 mm H2O2 for 5 h or 10 mm 3-nitropropionic acid (3-NPA) for 12 h and GFP fluorescence was visualized. GFP-Atx3(Q29) accumulated in the nucleus in response to both oxidative stressors while the distribution of GFP was not altered by H2O2 or 3-NPA (Fig. 8A). Data were quantified by counting the number of GFP or GFP-Atx3 transfected cells displaying primarily cytoplasmic, nuclear, or evenly distributed fluorescence. H2O2 and 3-NPA treatment did not alter the distribution of GFP (Fig. 8B) but increased the number of cells with a nuclear distribution of GFP-Atx3 3–4-fold (Fig. 8C; P < 0.01). To support this observation and determine whether endogenous Atx3 responded similar to transfected Atx3, cellular fractionations were performed on fibroblasts from an SCA3 patient. Cells were exposed to 2.5 mm H2O2 for 4 h and allowed to recover for 0 or 4 h. Both WT and pathogenic Atx3 accumulated in the nucleus following H2O2 treatment (Fig. 8D). Quantitation of these fractionations indicated that immediately following 4 h of H2O2 treatment nuclear levels of wild-type and pathogenic Atx3 were 20–25-fold greater than untreated control cells (Fig. 8E; P < 0.01), and remained 15–20-fold higher following 4 h of recovery (Fig. 8E; P < 0.01). SCA3 patient fibroblasts exposed to 10 mm 3-NPA for 24 h and allowed to recover for 0 or 4 h were fractionated and showed increased nuclear levels of WT and pathogenic Atx3 (Fig. 8F). Quantitation of these data indicated that following 24 h of 3-NPA treatment nuclear levels of WT and pathogenic Atx3 were 4–5-fold greater than untreated control cells (Fig. 8G; P < 0.05), and remained 3–4-fold higher following 4 h of recovery (Fig. 8G; P < 0.05). These data indicated that oxidative stress induced nuclear accumulation of endogenous WT and pathogenic Atx3.
Figure 8.
Wild-type and pathogenic ataxin-3 accumulate in the nucleus following oxidative stress. (A) HeLa cells were transfected with either GFP or GFP-Atx3 and exposed to 1 mm H2O2 for 5 h or 10 mm 3-NPA for 12 h. Following treatment GFP-Atx3 accumulated in the nucleus. Immunofluorescence data were quantitated by counting GFP (B) or GFP-Atx3 (C) transfected cells with fluorescence primarily localized to the cytoplasm (C), nucleus (N), or evenly distributed (C/N). Data represent the mean ± SD of the fraction of total cells counted in each group with a particular localization pattern in three independent experiments; treated cells were compared with control cells (*P < 0.01). (D) SCA3 patient fibroblasts expressing both WT and pathogenic Atx3 were exposed to 2.5 mm H2O2 for 4 h and cellular fractionations were performed following 0 or 4 h of recovery. Endogenous wild-type and pathogenic Atx3 accumulated in the nucleus of SCA3 patient fibroblasts following treatment with H2O2. (E) H2O2-induced nuclear accumulation of endogenous wild-type and pathogenic Atx3 were quantitated using densitometry. Data represent the mean ± SD of three independent experiments; for statistics H2O2- treated cells were compared with control cells (*P < 0.01). (F) SCA3 patient fibroblasts were exposed to 10 mm 3-NPA for 24 h and subcellular fractionations prepared following 0 or 4 h of recovery. Endogenous wild-type and pathogenic Atx3 accumulated in the nucleus of SCA3 patient fibroblasts following treatment with 3-NPA. (G) 3-NPA-induced nuclear accumulation of endogenous wild-type and pathogenic Atx3 was quantitated using densitometry. Data represent the mean ± SD of three independent experiments; 3-NPA-treated cells were compared with control cells (*P < 0.05).
DISCUSSION
Cells are exposed constantly to intrinsic and environmental stresses (60). These stresses often cause accumulation of damaged and misfolded proteins that alter protein homeostasis. To restore homeostasis stress, response pathways such as the heat shock response are activated in cells to reduce the number of aberrant proteins (60–62). In the present study, we show that two proteotoxic stresses, heat shock and oxidative stress, which increase damaged and misfolded proteins and activate the heat shock response, also induce nuclear accumulation of Atx3. Atx3 is a deubiquitinating enzyme present in the cytoplasm and nucleus that appears to function in cellular pathways regulating protein homeostasis as well as transcription (21–37). Nuclear accumulation of Atx3 following heat and oxidative stress suggests it has a nuclear function such as transcriptional regulation. Interestingly, two other cellular stresses that increase misfolded and aberrant proteins, proteasome inhibition and ER stress, did not induce nuclear accumulation of Atx3; this may reflect a primarily cytoplasmic function for Atx3 following these proteotoxic stresses. It seems likely that most proteotoxic stresses activate a set of common pathways like the heat shock response as well as activating select stress pathways that reflect the specific cellular stress. Data in Supplementary Material, Figure S1 support this idea and indicate that different proteotoxic stresses differentially alter interactions of Atx3 with protein quality control effectors, VCP and hHR23B.
Typically, wild-type and pathogenic Atx3 localize diffusely throughout the cell; however, following heat shock they transiently accumulated in the nucleus. This occurred in all cells tested ranging from tumor cell lines to primary neurons and astrocytes. Nuclear localization of Atx3 had a similar time course as the major heat shock transcriptional regulator, Hsf1 (Fig. 7D); although, Hsf1 was not required for Atx3 nuclear localization (Fig. 6A) and Atx3 was not required for Hsf1 nuclear localization (Fig. 7D). Mapping domains of Atx3 responsible for nuclear localization indicated that there were two regions that regulated heat shock-induced nuclear localization; one region was the Josephin domain and the other was the C-terminal domain containing the polyglutamine sequence. Interestingly, high molecular weight multimers of truncated Atx3 containing the polyglutamine domain, Atx3(191–362) and Atx3(291–362), accumulated in the nucleus more than ‘monomeric’ forms. A possible explanation for this is that monomers of Atx3 subregions should be small enough to diffuse in and out of the nucleus while dimers and trimers may be restricted to nucleocytoplasmic shuttling due to their size and/or conformation. Alternatively, dimerization/trimerization could mask a nuclear export signal (NES) and trap these proteins in the nucleus. A similar mechanism is responsible for nuclear accumulation of p53 in response to DNA damage; p53 forms tetramers that mask an NES thereby trapping p53 in the nucleus (63). Atx3 contains two NES sequences in the N-terminal Josephin domain (46) but to date no NES has been identified in the region of Atx3 comprising these C-terminal truncations.
Mutating the UIMs and the DUB active site of Atx3 did not alter nuclear localization indicating that DUB activity and functional UIMs were not necessary for heat shock-induced nuclear localization. It was notable that Atx3(191–291) did not accumulate in the nucleus following heat shock; this domain contains two UIMs and a site predicted to be an NLS, 282RKRR285 (10,45,46). Mutating 282RKRR285 to 282ANAA285 in full-length Atx3 did not inhibit Atx3 heat shock-induced nuclear localization but significantly decreased nuclear levels of Atx3 under basal conditions (Fig. 4A and B). These data were consistent with published reports suggesting that the sequence is a functional NLS (45,46); however, our data show that this ‘basal’ NLS does not regulate heat shock-induced nuclear localization. Recently, CK2 phosphorylation of Atx3 was suggested to regulate nuclear localization of Atx3 (44). Our data indicated that the CK2 inhibitor, DMAT, decreased nuclear Atx3 in cells maintained under basal conditions at 37°C but did not alter heat shock-induced nuclear localization of endogenous Atx3. This suggests that CK2 regulates the basal level of Atx3 in the nucleus but does not regulate nuclear localization of Atx3 following heat shock.
Mutating serine-111 to an alanine in the Josephin domain blocked heat shock-induced nuclear localization of Atx3(1–191) and decreased nuclear localization of full-length Atx3 (Fig. 6C and D). Serine-111 was mutated and tested because it is a potential Plk phosphorylation site and Plk1 phosphorylation of Hsf1 regulates its nuclear translocation following heat shock (51). To begin testing whether phosphorylation at serine-111 might regulate heat shock-induced nuclear localization of Atx3, serine-111 was mutated to an aspartic acid to mimic phosphorylation (Fig. 6E and F). The S111D mutation did not increase nuclear localization of full-length Atx3 or Atx3(1–191) under basal conditions; however, in response to heat shock both proteins accumulated in the nucleus. Therefore, mutating serine-111 to alanine decreased nuclear localization following heat shock while mutating serine-111 to aspartic acid supported nuclear localization of Atx3. This suggests that phosphorylation of serine-111 is required but not sufficient for nuclear localization of Atx3 following heat shock. Therefore, a possible scenario is that in response to heat shock Plk phosphorylates Atx3 at serine-111 and in combination with unknown post-translational modifications or protein interactions regulates nuclear localization of Atx3. Ultimately, to determine whether Plk phosphorylation of Atx3 regulates heat shock-induced nuclear localization knockdowns of Plk1,2, 3 and 4 will need to be tested in cells. Plk1 phosphorylation of Hsf1 at serine-419 is responsible for nuclear localization of Hsf1 following heat shock in HEK293 cells (51); however, Plk1 is not present in the brain (64) and therefore cannot be responsible for Atx3 heat shock-induced nuclear localization in neurons and astrocytes (Fig. 1C and D). Neurons contain Plk2 and Plk3 (64) and these kinases are obvious candidates for phosphorylating Atx3 and increasing its nuclear localization in neurons.
In response to heat or oxidative stress both wild-type and pathogenic Atx3 accumulated in the nucleus. Although the function of nuclear Atx3 following these stresses is unknown, one possibility is that Atx3 modulates transcription associated with cellular stress. Several studies support the role of Atx3 as a co-repressor and transcriptional regulator (21–23). Preliminary microarray data with Atx3 wild-type and null cells are consistent with Atx3 modulating expression of a subset of genes in response to heat shock (unpublished data). Atx3-dependent changes in gene expression following heat shock may be responsible for the enhanced survival of wild-type fibroblasts compared with Atx3 null fibroblasts. Alternatively, or in addition to transcriptional effects, Atx3 appears to function in cellular stress pathways (25,27–37). For instance, data in Supplementary Material, Figure S1 show stress-dependent dynamic interactions of wild-type Atx3 with two important regulators of protein degradation pathways, VCP and hHR23B. These interactions or similar ones also may be important in both the acute response to stress as well as recovery from stress.
Several studies are consistent with the nucleus being a primary site of pathogenesis in many polyglutamine diseases (5,7,13–20); therefore, identifying processes that regulate nuclear localization of polyglutamine disease proteins is important for understanding pathogenesis and developing potential therapeutics. In a recent study, Truant and colleagues (65) showed that ER stress decreased targeting of huntingtin to the ER and increased levels in the nucleus. ER stress-induced nuclear localization of pathogenic huntingtin resulted in cellular toxicity (65). Nuclear localization of polyglutamine expanded Atx3 also is thought to be critical for SCA3 pathogenesis (5,7,15,17,20); therefore, an important observation in the present study is that select cellular stresses cause Atx3 to accumulate in the nucleus. Importantly, oxidative stress resulted in nuclear localization of wild-type and pathogenic Atx3. Oxidative stress and reactive oxygen species (ROS) increase in the brain during aging and are linked to many neurodegenerative diseases including polyglutamine diseases (54–59). Over its lifetime a neuron may respond thousands of times to increasing ROS or other stresses by transiently increasing nuclear localization of Atx3. In the case of pathogenic Atx3, each transient buildup in the nucleus may cause incremental damage eventually resulting in dysfunction of key neural functions and in time, neuronal death. Alternatively, dysfunction/toxicity may be a statistically rare event where the probability of a lethal/toxic occurrence increases with the frequency of high nuclear levels of pathogenic Atx3 which may be directly related to its response to oxidative/cellular stress.
In the current study a number of important observations were made including heat and oxidative stress-dependent nuclear localization of wild-type and pathogenic Atx3, serine-111 in the Josephin domain regulating nuclear accumulation of Atx3, and a functional protective role of Atx3 in the cellular response to heat shock. These observations raise a large number of questions that need to be addressed including: does stress-induced nuclear localization of pathogenic Atx3 compromise and/or protect cellular function and viability, do Plk or other kinases regulate Atx3 nuclear localization following stress, which amino acid(s) in the C-terminus of Atx3 regulate nuclear localization and how can this information be used to identify agents/therapeutics that disrupt pathogenic properties of Atx3 while maintaining its protective effects.
MATERIALS AND METHODS
Plasmids and antibodies
The following plasmids were previously described (5,30): myc-Atx3(Q29), myc-Atx3(Q72), myc-Atx3(Q29)C14A, myc-Atx3(Q29)S236/256D, myc-Atx3(Q29)282RKRR/ANAA285 and myc-Atx3(1–191). Myc-Atx3(191–362), myc-Atx3(291–362) and myc-Atx3(191–291) were subcloned into pCDNA3. Myc-Atx3(Q29) was subcloned into the GFP N1 vector. S111A and S111D mutations in myc-Atx3(Q29) and myc-Atx3(1–191) were generated using the QuickChange Site-Directed Mutagenesis kit (Stratagene) as per the manufacturer's instructions using the following primers: S111A forward: 5′-C GAT CCT ATA AAT GAA AGA GCA TTT ATA TGC AAT TAT AAG G-3′ and reverse: 5′-C CTT ATA ATT GCA TAT AAA TGC TCT TTC ATT TAT AGG ATC G-3′; S111D forward: 5′-C GAT CC T ATA AAT GAA AGA GAC TTT ATA TGC AAT TAT AAG G-3′ and reverse: 5′-C CTT ATA ATT GCA TAT AAA GTC TCT TTC ATT TAT AGG ATC G-3′. HA-tagged VCP was provided by Akira Kakizuka and Gen Sobue (Kyoto University and Nagoya University, Japan). The following antibodies were commercially available or made in our laboratory: HA monoclonal (Sigma), myc monoclonal 9E10 (Developmental Studies Hybridoma Bank at the University of Iowa), Hsf1 and Hsp70 monoclonals (Assay Designs), GAPDH monoclonal (Advanced Immunochemical), histone H3 polyclonal (Upstate), anti-Atx3 monoclonal 1H9 (Chemicon) and anti-Machado Joseph disease (MJD) polyclonal (made in our laboratory).
Cell culture, transfections and immunoprecipitations
HEK293T and HeLa cells were cultured in DMEM containing 10% fetal bovine serum (FBS), 100 unit/ml penicillin and 100 μg/ml streptomycin. Hsf1 WT and KO mouse embryonic fibroblasts were generously provided by Ivor Benjamin (University of Texas Southwestern) and cultured in DMEM containing 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, MEM non-essential amino acids (Invitrogen), 1 mm sodium pyruvate, 1 mm glutamine and 0.1 mm 2-mercaptoethanol. Atx3 WT and KO mouse primary skin fibroblasts were harvested from day 0 postnatal Atx3 WT and KO mice and cultured in DMEM containing 10% FBS, 100 unit/ml penicillin and 100 μg/ml streptomycin. Primary cultures of cortical neurons and astrocytes were established from cortices of E16 rat pups. Neurons were grown on poly-lysine coated glass coverslips in Neurobasal medium containing B27 supplement, 24 μg/ml penicillin and 24 μg/ml streptomycin. Astrocytes were grown on coverslips in DMEM containing 10% FBS, 100 μg/ml penicillin and 100 μg/ml streptomycin. SCA3 patient fibroblasts were obtained from Coriell (Camden, NJ, USA) and grown on collagen coated plates in MEM containing 10% FBS, 100 μg/ml penicillin, 100 μg/ml streptomycin, and 1 mm glutamine. HEK293T cells were transfected using Lipofectamine 2000 (Invitrogen) and HeLa cells were transfected using Effectene (Qiagen) according to manufacturer's protocol. Twenty-four hours after transfection, cells were used for experiments as described.
For immunoprecipitations, cleared cell lysates in lysis buffer [30 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100 and protease inhibitor cocktail (Roche)] were incubated with anti-MJD rabbit polyclonal antibody and Protein A agarose (Invitrogen) for 3 h at 4°C. Beads were washed three times in 500 μl of lysis buffer and bound proteins were separated by SDS–PAGE and analyzed by immunoblotting.
Immunofluorescence
Cells were grown on glass bottom microwell dishes (MatTek) or glass coverslips and fixed in 4% paraformaldehyde for 15 min and permeabilized with 0.05% Triton X-100. Cells were blocked with 2% goat serum in PBS and incubated with one of the following primary antibodies: anti-MJD, anti-myc or anti-HA for 90 min at room temperature. Cells were washed with PBS and incubated with goat secondary antibodies conjugated to Alexa fluor 568 (Molecular Probes) for 1 h at room temperature. Fluorescence was visualized using a Nikon TE-2000U microscope.
Cell treatments
For typical heat shock experiments cell cultures were sealed with parafilm, heat-shocked in a 42°C water bath and then allowed to recover at 37°C in the incubator for the indicated time. Compounds used to induce proteotoxic stress included 10 μm MG132 (American Peptide) to inhibit the proteasome, 1 or 2 mm DTT (Sigma) or 10 μg/ml tunicamycin (Sigma) to induce the unfolded protein response and 1 or 2.5 mm H2O2 (Sigma) or 10 mm 3-NPA (Sigma) for oxidative stress. To inhibit CK2 HEK293T cells were treated with 0.2, 2.0 or 20 μm DMAT (Calbiochem) for 3 h. In all cases, cells were exposed to stressors in serum-containing medium, except for neurons that were in complete Neurobasal medium.
Cell fractionation and densitometry
Cell fractionations were performed using a modified protocol of the Cell Compartment Fractionation kit (Qiagen). Briefly, cell pellets were resuspended in cold buffer CE1 and incubated on ice for 10 min. The lysate was centrifuged at 1000g for 10 min and the supernatant containing cytosolic proteins was saved. The pellet was resuspended in cold buffer CE2 and incubated on ice for 30 min and then centrifuged at 1000g for 10 min to pellet the nuclear fraction. The supernatant containing the membrane fraction was transferred to a new tube and the nuclear pellet was washed in cold PBS and lysed with buffer [30 mm Tris, pH 7.4, 150 mm NaCl and 1% Triton X-100]. Nuclear lysate was incubated on ice for 10 min, sonicated and centrifuged at 21 000g for 10 min and the resulting supernatant contained the soluble nuclear proteins. Western blots were quantified using Image J software (NIH) and nuclear Atx3 bands were normalized to the histone H3 loading control. Quantitation of Atx3 truncations was performed on monomeric species.
Toxic stress paradigms
Atx3 WT and KO primary mouse fibroblasts were heat-shocked in a 45°C incubator for 1 or 2 h and viable cells counted immediately following heat shock or 24 h later using trypan blue staining and a hemocytometer. For measuring cell viability following activation of the unfolded protein response, Atx3 WT and KO primary fibroblasts were grown in 96-well plates and treated with 2 mm DTT for 0, 6, or 12 h and cell viability was measured using Calcein AM and a fluorescent plate reader. Cell viability of Atx3 WT and KO primary fibroblasts following 10 μm MG132 treatment was determined following 0, 8, or 12 h of MG132 treatment. Viable cells were counted by trypan blue exclusion using a hemocytometer. In all stress paradigms viability was determined at each time point for a parallel set of control cells.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (NS042625 to R.N.P., CA009677 to C.P.R.).
Supplementary Material
Acknowledgements
Conflict of Interest statement. None declared.
REFERENCES
- 1.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Paulson H.L., Perez M.K., Trottier Y., Trojanowski J.Q., Subramony S.H., Das S.S., Vig P., Mandel J.L., Fischbeck K.H., Pittman R.N. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 3.Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., Bates G.P. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 4.Cummings C.J., Mancini M.A., Antalffy B., DeFranco D.B., Orr H.T., Zoghbi H.Y. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 5.Perez M.K., Paulson H.L., Pendse S.J., Saionz S.J., Bonini N.M., Pittman R.N. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. US A. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai Y., Koppenhafer S.L., Shoesmith S.J., Perez M.K., Paulson H.L. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi Y., Okamoto T., Taniwaki M., Aizawa M., Inoue M., Katayama S., Kawakami H., Nakamura S., Nishimura M., Akiguchi I., et al. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 9.Paulson H.L., Das S.S., Crino P.B., Perez M.K., Patel S.C., Gotsdiner D., Fischbeck K.H., Pittman R.N. Machado-Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann. Neurol. 1997;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
- 10.Tait D., Riccio M., Sittler A., Scherzinger E., Santi S., Ognibene A., Maraldi N.M., Lehrach H., Wanker E.E. Ataxin-3 is transported into the nucleus and associates with the nuclear matrix. Hum. Mol. Genet. 1998;7:991–997. doi: 10.1093/hmg/7.6.991. [DOI] [PubMed] [Google Scholar]
- 11.Trottier Y., Cancel G., An-Gourfinkel I., Lutz Y., Weber C., Brice A., Hirsch E., Mandel J.L. Heterogeneous intracellular localization and expression of ataxin-3. Neurobiol. Dis. 1998;5:335–347. doi: 10.1006/nbdi.1998.0208. [DOI] [PubMed] [Google Scholar]
- 12.Perez M.K., Paulson H.L., Pittman R.N. Ataxin-3 with an altered conformation that exposes the polyglutamine domain is associated with the nuclear matrix. Hum. Mol. Genet. 1999;8:2377–2385. doi: 10.1093/hmg/8.13.2377. [DOI] [PubMed] [Google Scholar]
- 13.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 14.Peters M.F., Nucifora F.C., Jr, Kushi J., Seaman H.C., Cooper J.K., Herring W.J., Dawson V.L., Dawson T.M., Ross C.A. Nuclear targeting of mutant Huntingtin increases toxicity. Mol. Cell. Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- 15.Fujigasaki H., Uchihara T., Koyano S., Iwabuchi K., Yagishita S., Makifuchi T., Nakamura A., Ishida K., Toru S., Hirai S., et al. Ataxin-3 is translocated into the nucleus for the formation of intranuclear inclusions in normal and Machado-Joseph disease brains. Exp. Neurol. 2000;165:248–256. doi: 10.1006/exnr.2000.7479. [DOI] [PubMed] [Google Scholar]
- 16.Nucifora F.C., Jr, Ellerby L.M., Wellington C.L., Wood J.D., Herring W.J., Sawa A., Hayden M.R., Dawson V.L., Dawson T.M., Ross C.A. Nuclear localization of a non-caspase truncation product of atrophin-1, with an expanded polyglutamine repeat, increases cellular toxicity. J. Biol. Chem. 2003;278:13047–13055. doi: 10.1074/jbc.M211224200. [DOI] [PubMed] [Google Scholar]
- 17.Kretzschmar D., Tschape J., Bettencourt Da Cruz A., Asan E., Poeck B., Strauss R., Pflugfelder G.O. Glial and neuronal expression of polyglutamine proteins induce behavioral changes and aggregate formation in Drosophila. Glia. 2005;49:59–72. doi: 10.1002/glia.20098. [DOI] [PubMed] [Google Scholar]
- 18.Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 19.Katsuno M., Adachi H., Kume A., Li M., Nakagomi Y., Niwa H., Sang C., Kobayashi Y., Doyu M., Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 20.Bichelmeier U., Schmidt T., Hubener J., Boy J., Ruttiger L., Habig K., Poths S., Bonin M., Knipper M., Schmidt W.J., et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J. Neurosci. 2007;27:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Macfarlan T., Pittman R.N., Chakravarti D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J. Biol. Chem. 2002;277:45004–45012. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- 22.Evert B.O., Vogt I.R., Vieira-Saecker A.M., Ozimek L., de Vos R.A., Brunt E.R., Klockgether T., Wullner U. Gene expression profiling in ataxin-3 expressing cell lines reveals distinct effects of normal and mutant ataxin-3. J. Neuropathol. Exp. Neurol. 2003;62:1006–1018. doi: 10.1093/jnen/62.10.1006. [DOI] [PubMed] [Google Scholar]
- 23.Evert B.O., Araujo J., Vieira-Saecker A.M., de Vos R.A., Harendza S., Klockgether T., Wullner U. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J. Neurosci. 2006;26:11474–11486. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou A.H., Yeh T.H., Ouyang P., Chen Y.L., Chen S.Y., Wang H.L. Polyglutamine-expanded ataxin-3 causes cerebellar dysfunction of SCA3 transgenic mice by inducing transcriptional dysregulation. Neurobiol. Dis. 2008;31:89–101. doi: 10.1016/j.nbd.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Burnett B., Li F., Pittman R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 26.Scheel H., Tomiuk S., Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum. Mol. Genet. 2003;12:2845–2852. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- 27.Wang G., Sawai N., Kotliarova S., Kanazawa I., Nukina N. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum. Mol. Genet. 2000;9:1795–1803. doi: 10.1093/hmg/9.12.1795. [DOI] [PubMed] [Google Scholar]
- 28.Doss-Pepe E.W., Stenroos E.S., Johnson W.G., Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell. Biol. 2003;23:6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai Y., Berke S.S., Cohen R.E., Paulson H.L. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J. Biol. Chem. 2004;279:3605–3611. doi: 10.1074/jbc.M310939200. [DOI] [PubMed] [Google Scholar]
- 30.Burnett B.G., Pittman R.N. The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. Proc. Natl Acad. Sci. U S A. 2005;102:4330–4335. doi: 10.1073/pnas.0407252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warrick J.M., Morabito L.M., Bilen J., Gordesky-Gold B., Faust L.Z., Paulson H.L., Bonini N.M. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 32.Nicastro G., Menon R.P., Masino L., Knowles P.P., McDonald N.Q., Pastore A. The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc. Natl Acad. Sci. U S A. 2005;102:10493–10498. doi: 10.1073/pnas.0501732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao Y., Senic-Matuglia F., Di Fiore P.P., Polo S., Hodsdon M.E., De Camilli P. Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain. Proc. Natl Acad. Sci. U S A. 2005;102:12700–12705. doi: 10.1073/pnas.0506344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong X., Pittman R.N. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q., Li L., Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J. Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrigues A.J., Coppola G., Santos C., Costa Mdo C., Ailion M., Sequeiros J., Geschwind D.H., Maciel P. Functional genomics and biochemical characterization of the C. elegans orthologue of the Machado-Joseph disease protein ataxin-3. FASEB J. 2007;21:1126–1136. doi: 10.1096/fj.06-7002com. [DOI] [PubMed] [Google Scholar]
- 37.Winborn B.J., Travis S.M., Todi S.V., Scaglione K.M., Xu P., Williams A.J., Cohen R.E., Peng J., Paulson H.L. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todi S.V., Winborn B.J., Scaglione K.M., Blount J.R., Travis S.M., Paulson H.L. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donaldson K.M., Li W., Ching K.A., Batalov S., Tsai C.C., Joazeiro C.A. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc. Natl Acad. Sci. USA. 2003;100:8892–8897. doi: 10.1073/pnas.1530212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertolaet B.L., Clarke D.J., Wolff M., Watson M.H., Henze M., Divita G., Reed S.I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson C.R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell. Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 42.Chen L., Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao H., Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 44.Mueller T., Breuer P., Schmitt I., Walter J., Evert B.O., Wullner U. CK2-dependent phosphorylation determines cellular localization and stability of ataxin-3. Hum. Mol. Genet. 2009;18:3334–3343. doi: 10.1093/hmg/ddp274. [DOI] [PubMed] [Google Scholar]
- 45.Macedo-Ribeiro S., Cortes L., Maciel P., Carvalho A.L. Nucleocytoplasmic shuttling activity of ataxin-3. PLoS One. 2009;4:e5834. doi: 10.1371/journal.pone.0005834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antony P.M., Mantele S., Mollenkopf P., Boy J., Kehlenbach R.H., Riess O., Schmidt T. Identification and functional dissection of localization signals within ataxin-3. Neurobiol. Dis. 2009;36:280–292. doi: 10.1016/j.nbd.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 47.Westwood J.T., Clos J., Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- 48.Baler R., Dahl G., Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol. Cell. Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarge K.D., Murphy S.P., Morimoto R.I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guettouche T., Boellmann F., Lane W.S., Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S.A., Yoon J.H., Lee S.H., Ahn S.G. Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. J. Biol. Chem. 2005;280:12653–12657. doi: 10.1074/jbc.M411908200. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima H., Toyoshima-Morimoto F., Taniguchi E., Nishida E. Identification of a consensus motif for Plk (Polo-like kinase) phosphorylation reveals Myt1 as a Plk1 substrate. J. Biol. Chem. 2003;278:25277–25280. doi: 10.1074/jbc.C300126200. [DOI] [PubMed] [Google Scholar]
- 53.Johnson E.F., Stewart K.D., Woods K.W., Giranda V.L., Luo Y. Pharmacological and functional comparison of the polo-like kinase family: insight into inhibitor and substrate specificity. Biochemistry. 2007;46:9551–9563. doi: 10.1021/bi7008745. [DOI] [PubMed] [Google Scholar]
- 54.Perez-Severiano F., Rios C., Segovia J. Striatal oxidative damage parallels the expression of a neurological phenotype in mice transgenic for the mutation of Huntington's disease. Brain Res. 2000;862:234–237. doi: 10.1016/s0006-8993(00)02082-5. [DOI] [PubMed] [Google Scholar]
- 55.Bogdanov M.B., Andreassen O.A., Dedeoglu A., Ferrante R.J., Beal M.F. Increased oxidative damage to DNA in a transgenic mouse model of Huntington's disease. J. Neurochem. 2001;79:1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- 56.Perluigi M., Poon H.F., Maragos W., Pierce W.M., Klein J.B., Calabrese V., Cini C., De Marco C., Butterfield D.A. Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol. Cell. Proteomics. 2005;4:1849–1861. doi: 10.1074/mcp.M500090-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Miyata R., Hayashi M., Tanuma N., Shioda K., Fukatsu R., Mizutani S. Oxidative stress in neurodegeneration in dentatorubral-pallidoluysian atrophy. J. Neurol. Sci. 2008;264:133–139. doi: 10.1016/j.jns.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 58.Yu Y.C., Kuo C.L., Cheng W.L., Liu C.S., Hsieh M. Decreased antioxidant enzyme activity and increased mitochondrial DNA damage in cellular models of Machado-Joseph disease. J. Neurosci. Res. 2009;87:1884–1891. doi: 10.1002/jnr.22011. [DOI] [PubMed] [Google Scholar]
- 59.Beal M.F. Mitochondrial dysfunction in neurodegenerative diseases. Biochim. Biophys. Acta. 1998;1366:211–223. doi: 10.1016/s0005-2728(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 60.Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 61.Morimoto R.I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes. Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hetz C., Glimcher L.H. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol. Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stommel J.M., Marchenko N.D., Jimenez G.S., Moll U.M., Hope T.J., Wahl G.M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seeburg D.P., Pak D., Sheng M. Polo-like kinases in the nervous system. Oncogene. 2005;24:292–298. doi: 10.1038/sj.onc.1208277. [DOI] [PubMed] [Google Scholar]
- 65.Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.