Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by progressive weakness from loss of motor neurons. The fundamental pathogenic mechanisms are unknown and recent evidence is implicating a significant role for abnormal exon splicing and RNA processing. Using new comprehensive genomic technologies, we studied exon splicing directly in 12 sporadic ALS and 10 control lumbar spinal cords acquired by a rapid autopsy system that processed nervous systems specifically for genomic studies. ALS patients had rostral onset and caudally advancing disease and abundant residual motor neurons in this region. We created two RNA pools, one from motor neurons collected by laser capture microdissection and one from the surrounding anterior horns. From each, we isolated RNA, amplified mRNA, profiled whole-genome exon splicing, and applied advanced bioinformatics. We employed rigorous quality control measures at all steps and validated findings by qPCR. In the motor neuron enriched mRNA pool, we found two distinct cohorts of mRNA signals, most of which were up-regulated: 148 differentially expressed genes (P ≤ 10−3) and 411 aberrantly spliced genes (P ≤ 10−5). The aberrantly spliced genes were highly enriched in cell adhesion (P ≤ 10−57), especially cell–matrix as opposed to cell–cell adhesion. Most of the enriching genes encode transmembrane or secreted as opposed to nuclear or cytoplasmic proteins. The differentially expressed genes were not biologically enriched. In the anterior horn enriched mRNA pool, we could not clearly identify mRNA signals or biological enrichment. These findings, perturbed and up-regulated cell–matrix adhesion, suggest possible mechanisms for the contiguously progressive nature of motor neuron degeneration. Data deposition: GeneChip raw data (CEL-files) have been deposited for public access in the Gene Expression Omnibus (GEO), www.ncbi.nlm.nih.gov/geo, accession number GSE18920.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is an adult-onset idiopathic fatal neurodegenerative disease caused by highly selective loss of motor neurons (1). The main clinical features are insidious onset of upper and lower motor neuron degeneration and relatively linear progression over time. The main neuropathological features are loss of motor neurons. The age at onset is usually over 25 years and incidence increases slightly with age. The overall incidence of ALS is 2–3 per 100 000 and the prevalence is 6–10 per 100 000. Ninety to 95% of cases are sporadic (SALS) and 5–10% of cases are familial (FALS), for which ∼30% of the mutations have been identified to date. There are no effective treatments and death occurs in 90% of patients within 3–5 years. The mechanisms for the highly selective degeneration are unknown (2) and it is increasingly thought that non-neuronal cells are involved in the process (3,4). It is now recognized that close associations between ALS and frontotemporal lobar dementia with ubiquitin and TDP-43 positive immunoreactivity exist at the clinical, pathological, and molecular levels (5).
Alternative exon splicing is now believed to be one of the major factors determining the complexity and functionality of the eukaryotic genome (6–8). It is one of the main mechanisms of gene and cell regulation, modulates the properties of encoded proteins by regulating expression and adding and deleting functional domains and likely involves most genes. The control of exon splicing involves regulatory proteins binding to a variety of sites on transcripts such as exon splicing enhancers that bind SR proteins, polypyrimidine tracts, and splice motifs. The primary mediator of splicing is the spliceosome, composed of over 100 splicing factors including proteins, RNA and small nuclear ribonucleoproteins, which associate with pre-mRNA to mediate intron excision and exon ligation (9). Abnormal splicing can result in disease and the list of diseases attributed to this continues to grow (10). Appreciation of the significance of exon splicing in the genome has grown recently due to better analytic techniques (11) and is now entering an even more rapid growth phase with technology to perform deep sequencing (12).
There is increasing evidence that implicates abnormalities of RNA processing and exon splicing in ALS pathogenesis (13). TDP-43, a protein that has known functions in RNA processing and regulation of exon splicing, mislocalizes from the nucleus to the cytoplasm in SALS, suggesting its nuclear pre-mRNA functions are fundamentally compromised (14–16). Mutations in the TARDBP gene encoding the TDP-43 protein are found in both FALS and SALS, indicating its role can be primary (17–19). There is strong evidence that the toxic property resides in the C-terminal fragments, an architecture that would likely compromise nuclear functions, including exon splicing. Recently, the FUS/TLS gene has been identified in FALS by virtue of its function in RNA processing (20,21). Among the other genes identified to date in ALS as well other motor neuron degenerative disorders, there is over-representation of genes encoding proteins that are involved in RNA processing; these include: SETX, SMN, GARS, ATXN7, IGHMBP2 and ANG (22). A newly reported association has been identified between ALS and variants of a component of RNA polymerase II, elongator protein 3 (23). And spinal muscular atrophy (SMA), the most common motor neuron disease in childhood, is caused by homozygous loss of the SMN1 gene, which plays key roles in exon splicing (24).
Motor phenotypes of ALS indicate that underlying motor neuron degeneration is a focal process that progresses contiguously along neuronal anatomy (25). Neuropathological stages can be defined in relation to the site of onset, advanced in the region of onset and progressively less radially away (26). This creates opportunity for functional genomics analysis, as genomic signals can be isolated from regions in early to intermediate stages of degeneration and profiled using newly available laser and microchip technologies (27,28). To pursue this, we created a tissue repository specifically for simultaneous genomic and structural studies, used histopathology to identify suitable neuron-rich regions, validated RNA quality and profiled exon and gene expression in motor neurons and its anterior horn microenvironment. Using powerful bioinformatics approaches, we identified biological signals in motor neuron enriched mRNA pools that appear to be perturbed in SALS, the strongest of which is abnormal exon splicing, especially in cell-matrix adhesion genes. The pathogenesis of SALS remains elusive and mainly pursued by way of laboratory models of genetic disease—our findings identify important new aspects directly in human sporadic disease.
RESULTS
Special processing of CNS coupled with laser capture microdissection permits collection and selective enrichment of relevant cellular compartments for genomic analysis
Major challenges facing genomic analysis of human degenerative disorders of the CNS are precise localization and isolation of the degenerating compartment and the proper acquisition of materials. As SALS disease typically exhibits a focal onset and subsequent spread (25,26), patients whose disease began in bulbar and arm regions have caudally advancing disease and often have lumbar regions in relatively early stages of degeneration at the time of death. To exploit this, we developed an on-call autopsy system and rapidly harvested their nervous systems, usually completed within 6 h of death, and specifically processed them for downstream genomic and structural studies. We used histopathology to confirm that the lumbar regions distal to the site of onset had early or intermediate stage of disease pathology with abundant residual motor neurons and that they contained TDP-43 cytoplasmic deposits characteristic of SALS. Based upon this analysis and upon analysis of RNA quality as discussed below, we then chose nervous systems from the frozen inventory and collected from them motor neurons by laser capture microdissection in SALS (n = 12) and controls (n = 10) (Table 1 and Fig. 1A). After we had completed collection of motor neurons by microdissection, we collected the remaining anterior horn region to create a second sample set, reasoning that this might be important for understanding disease pathogenesis given the strong evidence for non-autonomous degeneration in ALS (3,4). For standardization, we studied lumbar regions in all cases, disease and control.
Table 1.
Demographic details
| Primary diagnosis | CNS ID number | Age | Gender | Site of onset | Disease course (years) | PMI (h) |
|---|---|---|---|---|---|---|
| SALS | 14 | 73 | Female | Bulbar | 1.5 | 6 |
| SALS | 16 | 61 | Male | Arm | 2.5 | 3.5 |
| SALS | 17 | 55 | Male | Arm | 2 | 3 |
| SALS | 18 | 80 | Female | Bulbar | 2 | 4 |
| SALS | 27 | 74 | Male | Bulbar | 3.25 | 4 |
| SALS | 33 | 54 | Male | Arm | 6.5 | 5 |
| SALS | 34 | 81 | Female | Bulbar | 1 | 3.5 |
| SALS | 35 | 74 | Female | Bulbar | 5.75 | 5 |
| SALS | 60 | 58 | Female | Bulbar | 3 | 3 |
| SALS | 63 | 68 | Male | Arm | 2.5 | 5 |
| SALS | 64 | 47 | Male | Arm | 3 | 6.5 |
| SALS | 68 | 72 | Female | Leg | 1.5 | 4 |
| Control | 10 | 78 | Male | NA | NA | 2.5 |
| Control | 19 | 80 | Female | NA | NA | 2.5 |
| Control | 26 | 49 | Male | NA | NA | 4 |
| Control | 39 | 77 | Male | NA | NA | 2 |
| Control | 42 | 61 | Male | NA | NA | 6 |
| Control | 44 | 80 | Female | NA | NA | 5 |
| Control | 55 | 71 | Male | NA | NA | 13 |
| Control | 59 | 73 | Male | NA | NA | 8 |
| Control | 65 | 82 | Male | NA | NA | 4 |
| Control | 67 | 77 | Male | NA | NA | 4 |
Figure 1.
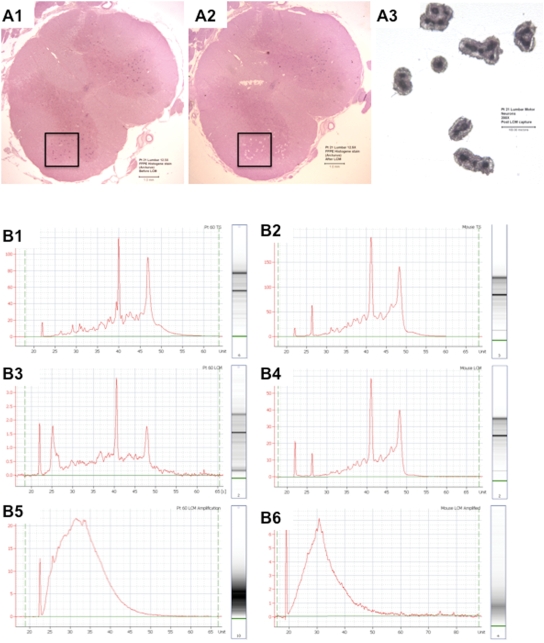
Laser capture microdissection (LCM) permits collection of high quality RNA from SALS motor neurons. (A): (A1) This is a low-power transverse view of SALS lumbar spinal cord stained with H&E before LCM. The rectangle indicates the anterior horn. Note the relative abundance of residual lumbar motor neurons (purple spots) in this nervous system where disease onset was in arm and respiratory muscles. (A2) Same, after LCM—the small white blanks are where motor neurons were microdissected. Note the specificity that is achieved within the complex cytoarchitecture of the spinal cord. (A3) This is a mid-power view of motor neurons captured and adhering to a thermoplastic polymer film on an LCM cap. The cap fits into a microtube for RNA isolation. [Scale bars are 1 mm in (A1) and (A2) and 100 µm in (A3)]. (B) These are electropherograms (left side of each panel) and digital gels (right side of each panel) generated by digital micro-electrophoresis that is used to assess RNA quality. The column on the left (B1, B3 and B5) is from a SALS nervous system and the column on the right (B2, B4 and B6) is from a SOD1 G93A ALS1 transgenic mouse for comparison. The top row (B1 and B2) shows total RNA before processing by laser capture microdissection (LCM). The middle row (B3 and B4) shows total RNA of motor neurons after isolation by LCM. The bottom row (B5 and B6) shows messenger RNA from the laser captured motor neurons after amplification. The similarities in these tracings illustrate the high quality of RNAs generated from SALS patient materials.
RNA and microarray quality are high
The success of genomic profiling depends upon both RNA and microarray quality. To ensure RNA quality, we used digital micro-electrophoresis and performed extensive validation of RNA quality (Supplementary Material S1). We found that our human RNA quality was high, RNA quality was sufficiently preserved through the steps of microdissection, and mRNA signals amplified to yield quantities and qualities necessary for microarray analysis (Fig. 1B). When RNA was optimal, we proceeded to label it, hybridize it to exon arrays, and process the arrays to CEL files. To ensure microarray quality, we examined CEL files in multiple ways (see Materials and Methods) and we found they were uniformly high (Supplementary Material S1). We always processed equal numbers of SALS and controls together to minimize batch effects and upon completion of the studies, we measured batch effects and they were minimal (data not shown), indicating evenness of acquired data between disease and control. We compared both RNA and microarray quality with our analogous experiments in ALS1 G93A transgenic mice using 3′ gene expression arrays, which were performed under ideal laboratory-controlled conditions, and found they were comparable (RNA quality is shown in Figure 1B and CEL file data are not shown). Thus, we were reassured of the high quality and reliability of the signals we acquired with our approach.
Genomic analysis identifies differentially expressed gene signals in the motor neuron enriched mRNA pool
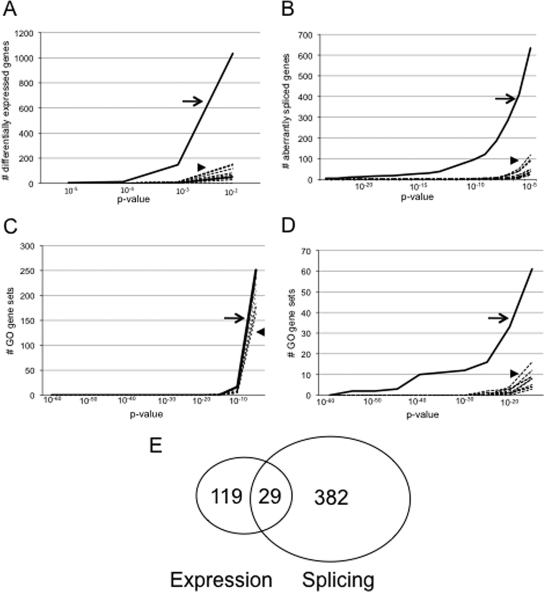
We next looked at differential gene expression. One of the main challenges of doing this is ascertaining significance when there are multiple tests and uncertain levels of noise. To determine noise levels in our data, we created sham test groups by permuting CEL file test categories and computing differential gene expression 10 times and then compared differential gene expression in our true data with the sham data (see Materials and Methods). In the motor neuron enriched mRNA pool, we found robust differential expression and readily definable noise levels: we estimate a data-specific 5% noise threshold of P≈ 10−3 and at this level of significance, there are 148 differentially expressed genes in the true data set and an average of six genes (range 2–12) in the permuted data sets (Fig. 2A, Table 2 and Supplementary Material S2). Most of the differentially expressed genes are up-regulated rather than down-regulated—for a fold change ≥1.5, 71 genes are up-regulated and 14 genes are down-regulated—suggesting increasing rather than decreasing molecular activity in the motor neuron enriched mRNA pool.
Figure 2.
Microarray analysis robustly identifies disease-associated signals in the SALS motor neuron enriched mRNA pool. (A) This graph shows the cumulative numbers of differentially expressed genes as a function of P-values. In this and in the other graphs, the solid line (arrow) shows the true comparison between SALS and control and the dashed lines (arrowhead) show sham comparisons of randomly permuted groups and indicate the noise levels in the data. Highly distinct differential gene expression is identified. (B) This graph shows the cumulative numbers of aberrantly spliced genes as a function of P-values. Note not only that significant aberrant gene splicing is identified, but also that the degree of abnormality is greater than seen with differential gene expression. (C) This graph shows cumulative numbers of Gene Ontology gene sets that enrich the differentially expressed genes identified in (A) as a function of P-value. There is no apparent biological enrichment of the differentially expressed genes. (D) This graph shows cumulative numbers of Gene Ontology gene sets that enrich the aberrantly spliced genes identified in (C) as a function of P-value. By comparison to differential gene expression, the aberrantly spliced genes are robustly enriched biologically. (E) Venn diagram comparing differentially expressed and aberrantly spliced genes in the SALS motor neuron enriched RNA pools. Note that the two cohorts of genes are distinctive and have only slight overlap.
Table 2.
Top 25 aberrantly spliced and top 25 differentially expressed genes
| Gene symbol | Gene name | RefSeq | Alt. splice P-value | Differential expression P-value | Fold-change (A versus C) | # Markers (exons) |
|---|---|---|---|---|---|---|
| ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 | NM_007168 | 2.84E–17 | 0.0147702 | 2.07853 | 37 |
| ACADVL | Acyl-Coenzyme A dehydrogenase, very long chain | NM_000018 | 0.07572 | 9.62E-05 | −1.36709 | 20 |
| AGTRL1 | Angiotensin II receptor-like 1 | NM_005161 | 0.00280309 | 0.000100449 | 2.80667 | 6 |
| AQP4 | Aquaporin 4 | NM_001650 | 1.35E-27 | 1.21E−06 | 3.17926 | 13 |
| ASPA | Aspartoacylase (Canavan disease) | NM_000049 | 0.199691 | 0.000120487 | 2.94572 | 6 |
| C1orf198 | Chromosome 1 open reading frame 198 | AK096166 | 2.66E−16 | 0.00111187 | 1.7294 | 17 |
| C21orf33 | Chromosome 21 open reading frame 33 | NM_004649 | 0.0218155 | 0.00010392 | −1.30992 | 10 |
| C3 | Complement component 3 | NM_000064 | 1.89E−23 | 0.00107972 | 2.39112 | 43 |
| C4A | Complement component 4A (Rodgers blood group) | NM_007293 | 1.88E−31 | 0.000487554 | 2.3395 | 48 |
| CENTD1 | Centaurin, delta 1 | NM_015230 | 9.18E−08 | 8.68E−05 | 1.86511 | 37 |
| COG5 | Component of oligomeric golgi complex 5 | NM_006348 | 1.79E−05 | 0.000102532 | 1.63965 | 28 |
| COL1A2 | Collagen, type I, alpha 2 | NM_000089 | 2.02E−24 | 0.0319974 | 1.44001 | 47 |
| COL6A3 | ollagen, type VI, Calpha 3 | NM_004369 | 3.34E−20 | 0.0945555 | 1.36456 | 45 |
| CPVL | Carboxypeptidase, vitellogenic-like | NM_019029 | 3.89E−11 | 6.99E−06 | 2.06165 | 17 |
| CXCL16 | Chemokine (C-X-C motif) ligand 16 | NM_022059 | 0.00011424 | 2.01E−05 | 1.99739 | 10 |
| DST | Dystonin | NM_183380 | 9.73E−24 | 0.323111 | 1.0925 | 125 |
| EDNRB | Endothelin receptor type B | NM_000115 | 2.64E−19 | 1.64E−05 | 3.01733 | 17 |
| ELAVL3 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like | NM_032281 | 1.12E−05 | 0.000109817 | −2.14778 | 18 |
| EXOSC6 | Exosome component 6 | NM_058219 | 0.804989 | 0.000105887 | −1.21483 | 7 |
| FAM46C | Family with sequence similarity 46, member C | NM_017709 | 6.43E−08 | 0.000117789 | 2.4746 | 6 |
| FLJ21963 | FLJ21963 protein | NM_024560 | 2.64E−12 | 2.35E−05 | 3.01776 | 17 |
| FLJ44874 | FLJ44874 protein | AK126822 | 0.597731 | 1.44E−07 | −1.46746 | 3 |
| FLNC | Filamin C, gamma (actin-binding protein 280) | NM_001458 | 2.80E−16 | 0.289023 | 1.0877 | 48 |
| FN1 | Fibronectin 1 | NM_212482 | 2.24E−21 | 0.0133788 | 2.50828 | 63 |
| HIF3A | Hypoxia inducible factor 3, alpha subunit | NM_022462 | 6.33E−16 | 0.0968477 | 1.24509 | 27 |
| HLA-B | Major histocompatibility complex, class I, B | NM_005514 | 0.011742 | 0.000145132 | 2.07808 | 3 |
| HLA-DMB | Major histocompatibility complex, class II, DM beta | NM_002118 | 0.00334092 | 0.000122339 | 2.64162 | 3 |
| HLA-DRA | Major histocompatibility complex, class II, DR alpha | NM_019111 | 0.0176529 | 9.96E−06 | 3.75813 | 9 |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145 kDa | NM_005541 | 2.28E−15 | 0.00485491 | 1.54023 | 25 |
| KIAA0644 | KIAA0644 gene product | NM_014817 | 5.30E−18 | 0.0764209 | 1.16279 | 15 |
| KRT73 | Keratin 73 | NM_175068 | 0.156092 | 6.78E−05 | −1.35392 | 9 |
| LRP1 | Low-density lipoprotein-related protein 1 (alpha-2-macroglo | NM_002332 | 2.68E−19 | 0.0219457 | 1.28675 | 105 |
| MACF1 | Microtubule-actin cross-linking factor 1 | NM_012090 | 7.85E−22 | 0.00408213 | 1.24486 | 115 |
| MYBPC1 | Myosin binding protein C, slow type | NM_002465 | 1.90E−21 | 0.00967833 | 1.31138 | 31 |
| OR2W3 | Olfactory receptor, family 2, subfamily W, member 3 | NM_001001957 | 0.602669 | 0.000100305 | −1.45266 | 13 |
| OR5AS1 | Olfactory receptor, family 5, subfamily AS, member 1 | NM_001001921 | 0.548751 | 0.000141626 | −1.3982 | 3 |
| PADI2 | Peptidyl arginine deiminase, type II | NM_007365 | 9.72E−14 | 0.000114651 | 1.6916 | 18 |
| PARP9 | Poly (ADP-ribose) polymerase family, member 9 | NM_031458 | 0.606104 | 2.15E−05 | 3.48026 | 3 |
| PCDHGC5 | Protocadherin gamma subfamily C, 5 | NM_018929 | 2.24E−21 | 0.00182203 | 1.23154 | 100 |
| PYGM | Phosphorylase, glycogen; muscle (McArdle syndrome, glycogen | NM_005609 | 7.14E−19 | 0.000661684 | 1.64136 | 20 |
| SEMA5B | Sema domain, seven thrombospondin Repeats (type 1 and | NM_001031702 | 6.12E−25 | 0.746505 | 1.02067 | 31 |
| SF1 | Splicing factor 1 | NM_004630 | 3.25E−18 | 0.0117229 | 1.26463 | 32 |
| SLC1A3 | Solute carrier family 1 (glial high affinity glutamate transporter) | NM_004172 | 3.81E−05 | 0.000106293 | 2.50395 | 15 |
| SLC35A4 | Solute carrier family 35, member A4 | NM_080670 | 0.793629 | 7.95E−05 | −1.49356 | 8 |
| SLC4A4 | Solute carrier family 4, sodium bicarbonate co-transporter | NM_001098484 | 1.64E−16 | 0.000145173 | 2.17261 | 31 |
| TJP2 | Tight junction protein 2 (zona occludens 2) | NM_004817 | 9.14E−16 | 0.000452437 | 2.01894 | 31 |
| TRBV19 | T-cell receptor beta variable 19 | BC073930 | 1.48E−15 | 0.695085 | 1.02684 | 30 |
| UTRN | Utrophin | NM_007124 | 1.56E−20 | 0.00473928 | 1.78074 | 74 |
Exon splicing is significantly aberrant in the motor neuron enriched mRNA pool and is distinct from and more pronounced than differential gene expression
We next looked at exon splicing. To determine aberrant exon splicing, we used an ANOVA-based analysis that generates P-values indicating the probability of aberrant splicing occurring in a particular gene. The ANOVA-based analysis separates out many variables, most importantly overall gene expression levels. As with differential gene expression, one of the main challenges is ascertaining significance with multiple tests and uncertain levels of noise and we employed the same strategy of permutation analysis that we used in analyzing differential gene expression. In the motor neuron enriched mRNA pool, we again found robust signals and readily definable noise levels: we estimate a data-specific 5% noise threshold of P≈10−5 and at this level of significance, there are 411 aberrantly spliced genes in the true data set and an average of 21 genes (range 5–51) in the permuted data sets (Fig. 2B, Table 2 and Supplementary Material S2). Comparing aberrantly spliced genes to differentially expressed genes, we see a nearly 3-fold greater number (411 versus 148) using our data-specific noise levels to define the respective cohorts. Analysis of these cohorts indicates only 29 genes overlap—7% of the 411 aberrantly spliced genes and 21% of the 148 differentially expressed genes (Fig. 2E)—thus showing that the cohorts of genes are distinct from each other. The greater numbers of aberrantly spliced genes than differentially expressed genes cannot be attributed to the greater degree of multiple testing since this and other methods of analysis we used correct for this. As with the differential gene expression, most of the aberrantly spliced genes were up-regulated rather than down-regulated. To examine the architecture of aberrant splicing in these genes, we classified splicing abnormalities in the top 100 genes and found that 56% had changes in internal exons, 51% in 5′ regions, 36% in 3′ regions and 42% in multiple locations (both internal and terminal) (data not shown).
The aberrantly spliced genes but not the differentially expressed genes in the motor neuron enriched mRNA pool are biologically enriched
One of the main challenges of microarray analysis is ascertaining biological importance of identified genes—statistical significance does not equate to biological significance. Multiple approaches have been designed to meet these challenges (29,30) and one approach is by scoring enrichment from the Gene Ontology (GO) (www.geneontology.org) by way of a χ2 test (see Materials and Methods). Accordingly, we calculated this for both differentially expressed genes and for aberrantly spliced genes. Enrichment analyses often can be difficult to interpret and P-values deceptively low and thus in order to understand our results, we also calculated enrichment in each of the 10 permuted data sets using a comparable number of genes. Analysis of differentially expressed genes in the motor neuron enriched mRNA pool did not reveal clearly definable biological enrichment above noise levels (Fig. 2C). But enrichment analysis of the aberrantly spliced gene cohort, in contrast, revealed robust enrichment above noise levels: we estimate a data-specific 5% noise level threshold of P≈10−20 and at this level of significance, there are 33 pathways in our true data and an average of 1.6 pathways (range 0–4) in our permuted data (Fig. 2D and Supplementary Material S2). Some gene sets have enrichment P-values <10−57.
The specific enrichments that we identified in the motor neuron enriched mRNA pool are predominately related to cell adhesion and extracellular matrix (ECM) biology (Table 3 and Supplementary Material S2). Transmembrane receptor protein tyrosine kinase activity, vascular endothelial growth factor receptor activity and regulation of cell motility were also identified (Table 3 and Supplementary Material S2). While the 33 gene sets with P ≤ 10−20 contained a total number of 355 genes, many genes appeared redundantly in the gene sets and only 111 of these were unique. Of these, 57 specifically relate to cell adhesion and related functions (Fig. 3). In the sub-categories of cell adhesion, genes associated with cell–matrix adhesion and cell–substrate adhesion were far more abundant than those associated with cell–cell adhesion (enrichment P = 10−31 versus P = 10−12, respectively). When examining the 57 genes identified by cell adhesion, genes encoding transmembrane (n = 24) and secreted (n = 21) proteins were more highly represented than genes encoding nuclear (n = 2) or cytoplasmic (n = 10) proteins (Table 4). Most of these 57 genes (n = 52) were up-regulated and only a few (n = 5) were down-regulated, although the up- and down-regulation did not always meet statistical significance (Fig. 3 and Table 4). A large number of genes were associated with integrin signaling, representing both ligands (collagens, laminins, fibronectin) and receptors.
Table 3.
Biological enrichment of aberrantly spliced genes
| Enrichment | Enrichment P-value | % genes in group that are present | Number of genes present | Number of genes in group | GO ID | GO category |
|---|---|---|---|---|---|---|
| Biological adhesion | 10−58 | 8.6758 | 57 | 657 | 22610 | BP |
| Cell adhesion | 10−58 | 8.6758 | 57 | 657 | 7155 | BP |
| Basement membrane | 10−48 | 28.9474 | 11 | 38 | 5604 | CC |
| Transmembrane receptor protein tyrosine kinase activity | 10−43 | 20.5882 | 14 | 68 | 4714 | MF |
| Collagen | 10−43 | 28.5714 | 10 | 35 | 5581 | CC |
| Extracellular matrix structural constituent | 10−42 | 18.75 | 15 | 80 | 5201 | MF |
| Vascular endothelial growth factor receptor activity | 10−42 | 50 | 6 | 12 | 5021 | MF |
| Laminin-1 complex | 10−41 | 62.5 | 5 | 8 | 5606 | CC |
| Laminin complex | 10−41 | 62.5 | 5 | 8 | 43256 | CC |
| Transmembrane receptor protein kinase activity | 10−41 | 18.2927 | 15 | 82 | 19199 | MF |
| Regulation of cell migration | 10−39 | 29.0323 | 9 | 31 | 30334 | BP |
| Cell–substrate adhesion | 10−31 | 17.3913 | 12 | 69 | 31589 | BP |
| Cell–matrix adhesion | 10−28 | 17.1875 | 11 | 64 | 7160 | BP |
| Extracellular region part | 10−27 | 6.38003 | 46 | 721 | 44421 | CC |
| Protein binding | 10−27 | 3.32847 | 228 | 6850 | 5515 | MF |
| Plasma membrane part | 10−25 | 4.74517 | 81 | 1707 | 44459 | CC |
| Regulation of cell motility | 10−25 | 14.2857 | 12 | 84 | 51270 | BP |
| Cell–substrate junction assembly | 10−25 | 60 | 3 | 5 | 7044 | BP |
| Cell junction assembly | 10−25 | 60 | 3 | 5 | 34329 | BP |
| Extracellular matrix organization and biogenesis | 10−24 | 19.5122 | 8 | 41 | 30198 | BP |
| Collagen binding | 10−23 | 29.4118 | 5 | 17 | 5518 | MF |
| Phosphate transport | 10−23 | 13.1868 | 12 | 91 | 6817 | BP |
| Hemidesmosome | 10−23 | 100 | 2 | 2 | 30056 | CC |
| Integrin-mediated signaling pathway | 10−23 | 16.3636 | 9 | 55 | 7229 | BP |
| Proteinaceous extracellular matrix | 10−22 | 9.00901 | 20 | 222 | 5578 | CC |
| Regulation of embryonic development | 10−22 | 50 | 3 | 6 | 45995 | BP |
| Stereocilium | 10−22 | 50 | 3 | 6 | 32420 | CC |
| Fibroblast growth factor receptor activity | 10−22 | 50 | 3 | 6 | 5007 | MF |
| Extracellular matrix | 10−21 | 8.77193 | 20 | 228 | 31012 | CC |
| Anatomical structure development | 10−21 | 5.15464 | 55 | 1067 | 48856 | BP |
| Structural molecule activity | 10−21 | 5.91716 | 40 | 676 | 5198 | MF |
| Integrin complex | 10−20 | 21.4286 | 6 | 28 | 8305 | CC |
| Developmental process | 10−20 | 4.04485 | 101 | 2497 | 32502 | BP |
BP, biological process; MF, molecular function; CC, cellular component.
Figure 3.
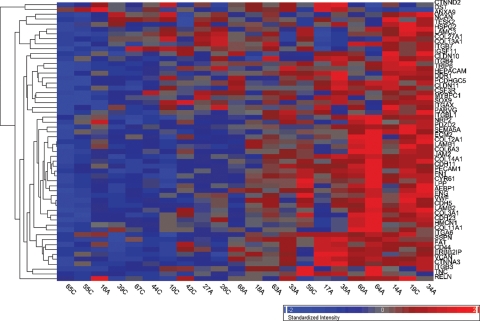
Heat map of the gene expression levels of the 57 cell adhesion genes identified by their aberrant splicing in the motor neuron enriched mRNA pool in our study. These 57 genes comprise the cell adhesion biological pathway as defined in the Gene Ontology. They were identified in our study by virtue of their marked over-representation in the 411 aberrantly spliced genes identified in the motor neuron enriched mRNA pool in SALS (P < 10−57). The colored bar indicates the range of intensity values for each gene and the heat maps display their overall gene expression levels, not their aberrant splicing. Note the relatively clean separation of SALS and control groups and the preponderant up-regulation or over-expression of these genes, which is seen in addition to their aberrant splicing. (C, control and A, SALS).
Table 4.
Aberrantly spliced cell adhesion genes
| Gene symbol | Gene name | RefSeq | Alt. splice P-value | Differential expression P-value | Fold change (A versus C) | Number of markers | Protein location |
|---|---|---|---|---|---|---|---|
| AEBP1 | AE binding protein 1 | NM_001129 | 2.44E−10 | 0.0186655 | 1.69315 | 27 | sec |
| ANXA9 | Annexin A9 | NM_003568 | 8.62E−07 | 0.971328 | −1.00249 | 14 | cyt |
| CDH11 | Cadherin 11, type 2, OB-cadherin (osteoblast) | NM_001797 | 2.94E−06 | 0.00458728 | 1.65623 | 21 | tm |
| CDH5 | Cadherin 5, type 2, VE-cadherin (vascular epithelium) | NM_001795 | 2.36E−06 | 0.102509 | 1.27449 | 19 | tm |
| CDH23 | Cadherin-like 23 | NM_022124 | 1.61E−07 | 0.156179 | 1.12818 | 71 | tm |
| CTNNA3 | Catenin (cadherin-associated protein), alpha 3 | NM_013266 | 6.53E−06 | 0.00648134 | 1.79781 | 25 | cyt |
| CTNND2 | Catenin (cadherin-associated protein), delta 2 | NM_001332 | 1.59E−06 | 0.0867763 | 1.1616 | 32 | cyt |
| CD44 | CD44 molecule (Indian blood group) | NM_000610 | 5.13E−12 | 0.000479249 | 1.89535 | 21 | tm |
| CLDN10 | Claudin 10 | NM_182848 | 6.03E−06 | 0.162268 | 1.20723 | 12 | tm |
| CLDN11 | Claudin 11 (oligodendrocyte transmembrane protein) | NM_005602 | 1.94E−06 | 0.00435624 | 1.53442 | 6 | tm |
| COL3A1 | Collagen, type III, alpha 1 (Ehlers-Danlos syndrome type) | NM_000090 | 3.84E−09 | 0.221719 | 1.17435 | 49 | sec |
| COL6A3 | Collagen, type VI, alpha 3 | NM_004369 | 3.34E−20 | 0.0945555 | 1.36456 | 45 | sec |
| COL11A1 | Collagen, type XI, alpha 1 | NM_001854 | 2.33E−09 | 0.178053 | 1.20705 | 46 | sec |
| COL12A1 | Collagen, type XII, alpha 1 | NM_004370 | 1.69E−10 | 0.0105983 | 1.79078 | 69 | sec |
| COL13A1 | Collagen, type XIII, alpha 1 | NM_005203 | 6.71E−09 | 0.169929 | 1.15626 | 25 | sec |
| COL14A1 | Collagen, type XIV, alpha 1 (undulin) | NM_021110 | 1.06E−08 | 0.00638787 | 1.58704 | 24 | sec |
| COL27A1 | Collagen, type XXVII, alpha 1 | NM_032888 | 1.43E−06 | 0.243964 | −1.10262 | 46 | sec |
| CSF3R | Colony stimulating factor 3 receptor (granulocyte) | NM_156039 | 2.04E−06 | 0.126098 | 1.10957 | 24 | tm |
| CYR61 | Cysteine-rich, angiogenic inducer, 61 | NM_001554 | 1.17E−07 | 0.0315499 | 1.55961 | 8 | sec |
| DDR1 | Discoidin domain receptor family, member 1 | NM_013993 | 4.24E−08 | 0.103136 | 1.15821 | 31 | tm |
| DST | Dystonin | NM_183380 | 9.73E−24 | 0.323111 | 1.0925 | 125 | cyt |
| ENG | Endoglin (Osler-Rendu-Weber syndrome 1) | NM_000118 | 5.65E−06 | 0.342529 | 1.19248 | 20 | tm |
| ERBB2IP | Erbb2 interacting protein | NM_018695 | 6.83E−07 | 0.000240051 | 2.08535 | 30 | cyt |
| ECM2 | Extracellular matrix protein 2, female organ and adipocyte | NM_001393 | 4.60E−06 | 0.0461937 | 1.59777 | 20 | sec |
| FAT | FAT // FAT tumor suppressor homolog 1 (Drosophila) | NM_005245 | 1.45E−08 | 0.00159728 | 1.86066 | 31 | tm |
| FN1 | Fibronectin 1 | NM_212482 | 2.24E−21 | 0.0133788 | 2.50828 | 63 | sec |
| HMCN1 | Hemicentin 1 | NM_031935 | 7.81E−07 | 0.209353 | 1.1854 | 116 | sec |
| HSPG2 | Heparan sulfate proteoglycan 2 | NM_005529 | 2.11E−09 | 0.938373 | −1.00658 | 101 | sec |
| HEPACAM | Hepatocyte cell adhesion molecule | NM_152722 | 2.96E−06 | 0.0223096 | 1.54885 | 13 | tm |
| IGSF11 | Immunoglobulin superfamily, member 11 | NM_001015887 | 5.57E−08 | 0.0724414 | 1.24835 | 13 | tm |
| ITGA6 | Integrin, alpha 6 | NM_000210 | 9.56E−08 | 0.0229754 | 2.06743 | 29 | tm |
| ITGAX | Integrin, alpha X (complement component 3 receptor 4 subunit) | NM_000887 | 7.10E−08 | 0.305373 | 1.09177 | 29 | tm |
| ITGB3 | Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | NM_000212 | 1.12E−06 | 0.394661 | 1.16482 | 20 | tm |
| ITGB4 | Integrin, beta 4 | NM_000213 | 4.07E−15 | 0.183707 | 1.14822 | 44 | tm |
| ITGB7 | integrin, beta 7 | NM_000889 | 1.28E−12 | 0.345837 | −1.07065 | 17 | tm |
| ITGBL1 | ITGBL1 // integrin, beta-like 1 (with EGF-like repeat domains) | NM_004791 | 2.24E−14 | 0.00392462 | 1.97861 | 16 | tm |
| JAM2 | Junctional adhesion molecule 2 | NM_021219 | 3.87E−08 | 0.00548407 | 2.02731 | 13 | tm |
| LAMB1 | Laminin, beta 1 | NM_002291 | 2.61E−13 | 0.0290629 | 1.61794 | 36 | sec |
| LAMB2 | Laminin, beta 2 (laminin S) | NM_002292 | 3.16E−06 | 0.155571 | 1.21405 | 40 | sec |
| LAMC3 | Laminin, gamma 3 | NM_006059 | 8.59E−06 | 0.370666 | −1.06083 | 35 | sec |
| LPP | LIM domain containing preferred translocation partner in lip | NM_005578 | 1.40E−06 | 0.147014 | 1.51955 | 13 | cyt |
| MYBPC1 | Myosin binding protein C, slow type | NM_002465 | 1.90E−21 | 0.00967833 | 1.31138 | 31 | cyt |
| NCAN | Neurocan | NM_004386 | 4.77E−10 | 0.206516 | 1.13639 | 25 | sec |
| NRP2 | Neuropilin 2 | NM_003872 | 5.65E−07 | 0.0447044 | 1.29315 | 36 | tm |
| PARVG | Parvin, gamma | NM_022141 | 2.03E−07 | 0.015984 | 1.25824 | 14 | cyt |
| PDZD2 | PDZ domain containing 2 | NM_178140 | 2.86E−07 | 0.0529539 | 1.34362 | 44 | cyt |
| PECAM1 | Platelet/endothelial cell adhesion molecule (CD31 antigen) | NM_000442 | 3.50E−06 | 0.023662 | 2.42764 | 17 | tm |
| PCDHGC5 | Protocadherin gamma subfamily C, 5 | NM_018929 | 2.24E−21 | 0.00182203 | 1.23154 | 100 | tm |
| RELN | Reelin | NM_173054 | 3.03E−06 | 0.0459772 | 1.59369 | 73 | sec |
| SSPN | Sarcospan (Kras oncogene-associated gene) | NM_005086 | 1.23E−07 | 0.00153469 | 1.87677 | 8 | tm |
| SEMA5A | Sema domain, seven thrombospondin Repeats | NM_003966 | 6.24E−06 | 0.132723 | 1.27857 | 29 | tm |
| SOX9 | SRY (sex determining region Y)-box 9 (campomelic dysplasia) | NM_000346 | 3.07E−07 | 0.584737 | 1.05446 | 9 | nuc |
| TNC | Tenascin C (hexabrachion) | NM_002160 | 2.55E−08 | 0.125224 | 1.66206 | 36 | sec |
| TESK2 | Testis-specific kinase 2 | NM_007170 | 6.75E−07 | 0.0632463 | 1.36536 | 14 | nuc |
| TRIP6 | Thyroid hormone receptor interactor 6 | NM_003302 | 2.74E−07 | 0.194651 | 1.16145 | 13 | cyt |
| VCAN | Versican | NM_004385 | 8.36E−11 | 0.000148832 | 2.61415 | 26 | sec |
| VWF | von Willebrand factor | NM_000552 | 5.40E−12 | 0.085195 | 1.51082 | 46 | sec |
cyt, cytoplasmic; nuc, nuclear; sec, secreted; tm, transmembrane.
There are no clearly identifiable gene signals in the anterior horn enriched mRNA pool
As previously stated, involvement of the microenvironment (‘non-neuronal neighbors’) is an important aspect of ALS pathobiology in transgenic mice expressing mutant SOD1 (3) and thus to study it in SALS, we collected and profiled the entire anterior horn after removal of motor neurons. We had comparable quality of data as the motor neuron enriched mRNA pool (Supplementary Material S1) but when applying the same bioinformatics analyses that we used for the motor neuron enriched pool, we could not identify any mRNA gene signals above noise levels either by way of differential gene expression, aberrant splicing, or biological enrichment (Supplementary Material S3), thus demonstrating the compartment-specific nature of our findings. To better understand this lack of signal in the anterior horn, we examined our analogous G93A mouse data that used 3′ gene expression arrays and determined signal strength in the anterior horn and in motor neuron enriched mRNA pools isolated by microdissection using a similar bioinformatics analysis. At both 20 and 60 post-natal days, we identified significant and comparable numbers of differentially expressed genes above experiment-specific noise levels in both pools (data not shown), thus demonstrating different strengths of genomic expression in the relevant histological compartments between mouse SOD1 and human SALS samples.
The biological enrichments of aberrant exon splicing in SALS and in an SMA mouse model of motor neuron degeneration are highly concordant
Recently, exon splicing in a mouse model of SMA motor neuron degeneration using whole genome exon arrays demonstrated widespread tissue-specific aberrant splicing, supporting the hypothesis that functional loss of the SMN1 protein in spliceosomes and aberrant splicing are fundamental in disease pathogenesis (24). To compare our findings to those in this study in uniform manner, we downloaded the CEL files of the spinal cord tissues from this study and processed them the same way we processed our own. With the bioinformatics approach reported in the study, 259 genes were identified as being aberrantly spliced in the spinal cord. Using our bioinformatics approach estimating data-specific noise thresholds, 74 genes were identified (data not shown). By either approach, there was significant aberrant splicing in degenerating tissues. There was a greater degree of aberrant splicing in our human SALS tissues (n = 411) than in the SMA mouse model tissues (n = 74), a difference likely due to greater compartment specificity in our study using microdissection. While this supports emerging ideas about a role of exon splicing in motor neuron diseases, in fact the ability to detect global splicing alterations is relatively recent with the development of exon arrays and few studies have been reported. To better understand this, we next compared the SALS and SMA data with others recently reported in the literature in non-motor neuron disease conditions (31,32). We downloaded CEL files from these studies and processed them the same way we processed our own data. We found even greater degrees of aberrant gene splicing occurring in these other conditions (data not shown), thus reinforcing that important biologic functions relate to exon splicing.
This functional specificity of exon splicing is clearly supported by enrichment analyses. The SMA mouse model study also identified enrichment of ECM biology in the cohort of aberrantly spliced genes, although quantitative analysis was not presented and the finding was not emphasized. To compare our findings to the ones reported in this study in uniform manner, we sought enrichment the same way we sought it in ours and we noted comparably robust enrichment: in the SMA study, we identified 26 gene sets with enrichment P ≤ 10−20, the cutoff we calculated for their noise. Some gene sets had enrichment P-values <10−78. The specific enrichments were dominantly related to ECM, collagen, cell adhesion, and related functions (Supplementary Material S2). While the 26 gene sets with enrichment P ≤ 10−20 contained a total number of 185 genes, many genes appeared redundantly in the gene sets and only 48 were unique. Of these, 32 specifically relate to ECM, cell adhesion, and related functions. Of the 48 unique genes from the SMA study and the 111 unique genes from our SALS study identified by the respective enrichment analyses, only 5 overlapped (col12a1, col14a1, fn1, hspg2 and itgb4) (Supplementary Material S2).
TARDBP/TDP-43, TDP-43 targets and other genes of ALS interest are mostly normal
We interrogated our data for currently known genes of interest in SALS pathobiology. A 1.5-fold change of TARDBP has been previously reported in brains of frontotemporal lobar dementia with ubiquitinated pathology using microarray analysis, although the replicates were small and tissue was not cell-specific (33). In our array data, we also found that this might be true in the motor neuron enriched mRNA pool (1.3-fold change with P = 0.008) but not the anterior horn enriched mRNA pool, but subsequent qPCR of all 12 SALS and 10 controls samples did not validate this (data not shown). TARDBP was normally spliced and we therefore conclude TARDBP mRNA transcription is normal in SALS. We also interrogated our data for known targets of TDP-43—CFTR, APO A-II, and CDK6—and we found they were neither differentially expressed nor aberrantly spliced. The representations for SMN1 and SMN2 are sparse on the array and what little could be assayed did not demonstrate differential gene expression (aberrant splicing could not be determined). We also interrogated our data for currently known genes of interest in FALS and motor neuron disease biology other than TARDBP—SOD1, FUS/TLS, SETX, ATXN7, IGHMBP2, ELP3 and ANG—and they were neither differentially expressed nor aberrantly spliced. As the targets of these genes are largely unknown, we cannot comment further on their possible aberrant splicing. Glycyl-tRNA synthetase (GARS), which is of putative interest in ALS (22), appeared to demonstrate significant down-regulation (−1.4-fold change with P = 0.0006) by microarray but this was not validated by subsequent qPCR (data not shown). In addition, we looked at genes identified by the Gene Ontology as involved in RNA processing and identified 39 genes as being either aberrantly spliced or differentially expressed, although this biological category was not abnormally enriched by enrichment analysis. We also found that the glutamate transporter EAAT2 (SLC1A2), which is potentially aberrantly spliced in ALS (34) was not abnormal, consistent with another report (35); however, we did observe that two other glutamate transporters, EAAT1 (SLC1A3) and EAAT4 (SLC1A6) might be differentially expressed (2.5-fold change with P = 0.0001 and −1.2-fold change with P = 0.007, respectively) and both validated by qPCR (P = 0.025) (Supplementary Material, S2).
qPCR validates microarray predictions of aberrantly spliced gene candidates
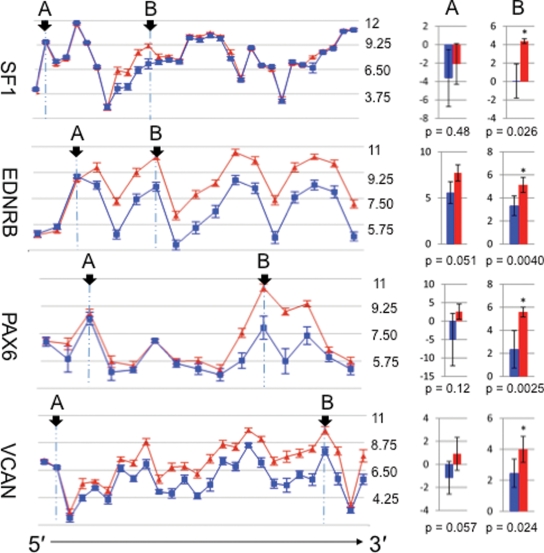
In order to validate aberrant exon splicing between the SALS and control as predicted by the exon arrays, 12 candidate genes were selected and exons within these genes predicted to exhibit differences and lack of difference were both assayed by TaqMan qPCR (see Materials and Methods). These genes were chosen from a variety of biological pathways to assay for splicing differences in various biological processes (Supplementary Material S4). Eight out of the 12 genes or 67% (EDNRB, PAX6, SF1, TMBIM1, TRBV19, VCAN, ARHGAP15 and AQP4) as illustrated in Figure 4 showed both of the changes predicted by the exon array in the same gene (Supplementary Material S4). This level of PCR validation is consistent with or above those in similar exon array studies (36–38) and we are thus reassured of the validity of the exon array predictions.
Figure 4.
Aberrant splicing predictions of the exon arrays validate by qPCR. Four examples of validation of aberrant splicing are demonstrated in this figure. The gene views on the left show the expression of exons as determined by exon array; the expression levels are shown on a log2 scale; the error bars show standard errors of means; SALS, red triangles; control, blue squares. Exons showing similar (A) and different (B) expression between SALS and control (arrows) were selected for validation by qPCR. The bar plots on the right show the results of qPCR: expression of the selected exons are normalized to GAPDH and shown on a log2 scale; the error bars show the 95% confidence intervals; SALS, red; control, blue. Asterisks indicate significant difference by t-test (α = 0.05) in expression as predicted by the microarray. In these examples, exons predicted to have differential expression and exons predicted not to have differential expression were confirmed by qPCR.
DISCUSSION
We made three key observations, the foundation of which is summarized in Table 5. The first key observation is that genomic abnormalities in SALS are highly localized to the motor neuron enriched mRNA pool (the degenerating anatomic compartment) and not apparent in the anterior horn enriched mRNA pool (the surrounding microenvironment). The strength of signal in the former and the lack of signal in the latter (which is itself enriched by removal of nearly 75% of the surrounding spinal cord) show this and the critical importance of anatomic and technical precision in identifying pathologic changes in vivo. The exact identity of the motor neuron enriched mRNA pool is not entirely clear. In favor of its mRNA deriving mostly from motor neurons is the anatomic specificity of microdissection and our preliminary histological studies of these same tissues in FFPE that do not show obvious surrounding hypercellularity. In favor of its containing mRNA also from non-motor cells is the difficulty of resolving cell morphology in the frozen cryosections on which microdissection is performed, the identification of biological enrichment that may be external to the cell itself (cell–adhesion, cell–matrix and ECM), and the identification of a number of genes that appear to be primarily glial. Thus, while enriched in microdissected motor neurons, it is possible that the mRNA signals are also enriched in other proximate cells or their mRNA-rich processes.
Table 5.
Summary of key findings
| Motor neuron enriched mRNA pool | Anterior horn enriched mRNA pool | |
|---|---|---|
| Cellularity | Homogeneous (single-cell enriched) | Heterogeneous (glia, endothelium, small neurons, degenerating neurons) |
| Anatomic size | Highly discrete | Comparatively vast |
| Differential gene expression | 148 genes | Not detectable above noise |
| Biological enrichment of differentially expressed genes | Not detectable above noise | Not detectable above noise |
| Aberrant exon splicing | 411 genes | Not detectable above noise |
| Biological enrichment of aberrantly spliced genes | Cell–matrix adhesion and extracellular matrix biology; also transmembrane receptor protein tyrosine kinase activity, vascular endothelial growth factor receptor activity, and regulation of cell motility | Not detectable above noise |
A striking finding is lack of signal and biologic enrichment in the anterior horn enriched mRNA pool. While this lack of signal validates our strategic approach of separating the two compartments, it is surprising given the strong evidence indicating that motor neuron degeneration is non-autonomous in mice expressing mutant SOD1 (3,4) and the strong evidence supporting the role of glia in magnifying the motor neuron pathology associated with ALS (39–41). In our comparable data from mutant SOD1 transgenic mice, we did in fact show equal signal strengths between the motor neuron and the anterior horn enriched mRNA pools. There are a number of potential explanations for this difference in human disease and the mutant SOD1 transgenic mouse model. In humans, the anterior horn is much larger anatomically than in mice and the critical interactions may reside in very close apposition to the neurons and thus detectable in the motor neuron enriched mRNA pool but diluted in the anterior horn enriched mRNA pool. In humans, the larger-sized anterior horn compared with mice may allow greater cellular heterogeneity and greater noise. In humans, the changes are sequential and summating, whereas in mice they are temporally synchronized. In humans, the effects in the anterior horn may be subtler than in mice and not detectable with our bioinformatics-based approach. Or, of course, non-autonomous cell effects may not be as significant in SALS pathognesis as in the SOD1 mouse model, upon which most of the non-autonomous cell degeneration literature is based.
The second key observation of our study is that exon splicing is more significant and relevant than differential gene expression in SALS—signals are more clearly identified and their biological enrichment more robustly defined by profiling aberrant exon splicing than differential gene expression. This supports the critical biological (6–8) and pathologic (10) importance of exon splicing in general and its interest to ALS (14–24) in particular. TDP-43 is the leading candidate in SALS pathogenesis and its exact role in pathogenesis is fundamental but remains unclear (14). Since one of its main known functions is exon splicing and in SALS it mislocalizes from the nucleus to the cytoplasm, it is important to note that we could not detect expression or splicing changes in its encoding gene TARDBP or any of its known targets including CFTR, APO A-II, CDK6 and SMN1. Together, these suggest that its role in SALS pathobiology is not likely at its own mRNA level and that it likely involves some other yet-to-be-identified targets or mechanisms—to date, a comprehensive list of TDP-43 targets and actions has not been defined. Alternatively, TDP-43 may involve functions other than exon splicing and the importance of the aberrant splicing that was detected by us is separate. The important role exon splicing may play in motor neuron degeneration is highlighted by SMA (42). SMA is caused by homozygous loss of the SMN gene, which is essential for biogenesis of small nuclear ribonucleoprotein particles that are critical components of the RNA splicing apparatus (43). Thus, splicing abnormalities are expected to play major roles in motor neuron degeneration and have been demonstrated in a mouse model to be both widespread and tissue-specific, differing within different neuronal tissues and between neuronal and non-neuronal tissues (24). Together with our study, splicing abnormalities are shown to be cell-specific as well as tissue-specific.
The mechanistic bases of the aberrant splicing in ALS are unclear. A number of RNA processing factors are themselves aberrantly spliced or differentially expressed in our SALS samples (Supplementary Material S2), which may provide an explanation for at least part of the splicing changes in SALS. We do not know if the genes encoding these RNA processing factors are mutated in SALS, similar to FUS/TLS or TDP-43, or if they are abnormally regulated downstream of primary disease-causing genes, but they represent interesting candidates for further analysis. For example, SF1 and SFRS2 have been shown to associate with FUS/TLS (44,45) and SFRS2 also alters the splicing of the TDP-43 target APO A-II (46), while SFRS5 alters the splicing of two TDP-43 targets (APO A-II and CFTR) (46,47). In addition, ELAVL3 (HuC) has a role in paraneoplastic neurologic disorders (48) and neuronal development (49). Interestingly, mutant SOD1 competes with HuC (and HuR) for binding to the 3′-UTR of vascular endothelial growth factor (VEGF) (50). RNASET2 is of interest due to the potential role of angiogenin (also an RNase) in ALS (22), as well as the recently demonstrated effects of RNASET2 loss on neuronal development (51). PTPBP1 represents a key regulator of neuronal development through its effects on repressing neural PTP (also called PTBP2) (52), as loss of PTPBP1 results in neural differentiation associated with PTPBP2 activation (53). There is also recent evidence of PTPBP1 involvement in the proliferation and migration of gliomas (54).
The third key observation of our study is the specific identification in SALS of perturbation of cell adhesion and ECM biology, especially cell–matrix as opposed to cell–cell adhesion genes. Other identified pathways included transmembrane receptor protein tyrosine kinase activity, vascular endothelial growth factor receptor activity [already of great interest in ALS pathobiology (55)] and regulation of cell motility (Tables 3 and 5). Since these were only identified in the motor neuron enriched mRNA pool and not in the larger surroundings of the anterior horn, their gene action is at or very proximate to neurons. Most of the cell adhesion genes we identified encode proteins that are either secreted or transmembrane as opposed to cytosolic or nuclear. The mechanisms by which these genes are likely to exert their effects are on the structure of the encoded proteins because the key to their identification was the aberrant splicing of the exons in the genes encoding them. Their abnormal action is likely to be part of an active rather than passive process since they are also mostly over-expressed or up-regulated. Where in the cascade of motor neuron degeneration their perturbations occurs or whether their perturbations are primary or secondary are unknown—their identification depended on pooling motor neurons within each nervous system and analysis of signals among multiple nervous systems. But the perturbations arguably are relatively upstream: (i) studies were performed in regions to which the degenerative process was newly advancing and therefore neuron-rich (in fact, these studies are only feasible in such regions) (26); (ii) the neurons in these regions showed relatively inconspicuous changes histologically when examined in companion FFPE tissues; and (iii) microdissection has an intrinsic bias for selectively collecting larger and healthier motor neurons. The studies on the mouse model of SMA highlight the functional and biological importance of the specific perturbations of cell–matrix and ECM biology: what is most concordant between SALS and SMA motor neuron degeneration is not aberrant exon splicing per se—there are only five genes that overlap—but the nearly identical and highly robust biological enrichments of the two distinct aberrant spliced gene cohorts. The source and coordination of the aberrant splicing is unknown and would only be identified by our method if altered in either expression or splicing. While we did observe alterations of RNA processing factors (mentioned above and Supplementary Material S2), there are many other possible alterations such as point mutations or post-translational changes that we are not able to observe. It is even possible, as demonstrated by SMA, that alterations in only one factor could cause multiple splicing abnormalities, but for the same reasons we might not be able to detect such.
Motor neurons are surrounded by a rich structural as well as cellular matrix and there are a number of potential scenarios where altered cell–matrix interactions could significantly affect neuron function. Disruption of cell–matrix molecules could adversely affect glia as well as neurons, thus resulting in or resulting from dysfunction of both cell categories (56). A few possibilities are outlined. (i) Alterations in the ECM could aberrantly modify the diffusion kinetics of paracrine factors such as growth factors, purines, etc., resulting in too much or too few of these factors reaching adjacent cells (57). (ii) Alterations in ECM could adversely affect neurons via integrin adhesion and signaling (58). (iii) Cell–matrix interactions are critical for neuronal migration, which occurs primarily during development as neural progenitor cells migrate to their proper positions before differentiating (59). Abnormal cell–matrix interactions could alter the normal formation and wiring of the nervous system, thus priming a delayed failure (60). (iv) Alterations in the ECM might cause or reflect derangements to the release of or diffusion of chemoattractants that activate neural progenitor cells and promote their migration toward diseased structures to try to ‘rescue’ degenerating motor neurons (61); alterations in the ECM might allow propagation of a maladapted process. (v) The basal lamina in the CNS, which is comprised predominately of ECM proteins, serves an important role in modulating the permeability of the blood brain barrier (62). Cleavage of proteins in the basal lamina by matrix metalloproteinases produced by invading inflammatory cells is required to facilitate their access to the CNS (63), thus alterations in ECM protein isoforms could alter blood brain barrier permeability. In addition, we have found that a protein crucial for determining the water permeability of the blood brain barrier, aquaporin-4 (62), demonstrates increased expression of a specific isoform in SALS (see Results). Overexpression of aquaporin-4 has been reported in ALS mouse (64) and rat models (65). It is interesting to note evidence from mouse ALS models indicating blood brain barrier changes early in the disease process (66). And (vi), ECM proteins can directly affect cells or alter the interactions of other ECM proteins with adjacent cells. For example, the ECM protein versican, which demonstrates altered splicing in SALS (see Results), is thought to restrict neuronal plasticity in adults as a component of perineuronal nets (67,68) and is upregulated in a rat model of ALS (66). Additionally, versican promotes the assembly of hyaluronan (69), which can activate microglia though binding to the CD44 receptor (70) (which also undergoes altered splicing in SALS).
These three key observations of our study are especially interesting in light of our recent postulations that motor neuron degeneration in SALS is an actively spreading and propagating process (71). That the aberrantly spliced genes are up-regulated is consistent with the process being active rather than passive. That the aberrantly spliced genes have perturbed cell–matrix and ECM adhesion biology is consistent with the importance of interactions in the microenvironment that could advance in space and over time. In this regard, it is important to note that there are two general types of inter-neuronal communication: one is synaptic communication between neurons that are in series (such as between upper and lower motor neurons or between neurons and interneurons) and the other is local side-to-side communication between neurons that are in parallel and involves local paracrine signaling and the neuron microenvironment. The recent identification that one or more soluble factors are selectively toxic to motor neurons is especially intriguing (72). One of many scenarios that can be imagined is that perturbed cell adhesion induces local changes and release of toxic soluble factors recruiting nearby cells, thus propagating a non-accelerating pathologic process. In the final analysis, our findings do not distinguish primary from secondary changes in pathobiology and add more pieces to the complex puzzle. Many challenges and questions remain, an immediate one being biological validation of our largely bioinformatics-based findings.
MATERIALS AND METHODS
Tissue acquisition and repository
All nervous systems were acquired by way of an Investigational Review Board and Health Insurance Portability and Accountability Act compliant process. The SALS nervous systems were from patients who had been followed during the clinical course of their illness and met El Escorial criteria for definite ALS (73). Upon death, autopsies were performed immediately by an on-call tissue acquisition team. Control nervous systems were from patients from the hospital's critical care unit when life support was withdrawn (7), patient on hospice (1) and from the Pennsylvania Tissue Repository (2). Tissue collections were completed within 6 h, usually within 4 h, of death and the entire motor system was dissected and elaborately archived for downstream applications by creating two parallel tissue sets from alternating adjacent regions. For molecular studies, segments were embedded in cutting media, frozen on blocks of dry ice and stored at −80°C. For structural studies, the adjacent segments were fixed in 70% neutral buffered formalin, embedded in paraffin (formalin-fixed paraffin-embedded or FFPE) and stored at room temperature.
Microdissection and total RNA isolation
Thirty to forty 8–10 µm cryocut frozen tissue sections stained with cresyl violet acetate were microdissected from each nervous system on a Pixcell IIe Laser Capture Microdissection (LCM) System (Arcturus Bioscience) in a 2-h long epoch for the motor neuron enriched RNA pool. Microdissected cells were captured on CapSure™ Macro LCM Caps (Arcturus Bioscience). After completion of LCM, the surrounding remaining anterior horn region was collected to create a second parallel RNA pool. RNA isolations were based on a guanadinium isothiocyanate and 2-mercaptoethanol extraction and column purification with RNeasy® Micro Kit (Qiagen).
mRNA amplification and exon array hybridization
For exon profiling, total RNA was amplified to cDNA probe using random priming with WT-Ovation™ Pico RNA Amplification System (NuGEN). The antisense–sense orientation was converted to sense transcript-cDNA (ST-cDNA) using WT-Ovation™ Exon Module (NuGEN). ST-cDNA was fragmented and biotin labeled using FL-Ovation™ cDNA Biotin Module v2 (NuGEN). Probe was hybridized to GeneChip® Human Exon 1.0 ST Arrays (Affymetrix). This array determines both exon and gene expression and are more resilient to RNA degradation changes than traditional 3′ gene arrays, which depend solely on integrity of the 3′ exon (74). Hybridized chips were stained and washed on a GeneChip 450 fluidics Station (Affymetrix) and scanned with a GeneChip 3000 scanner (Affymetrix). Initial data was processed to CEL files using GCOS Version 1.4 (Affymetrix). The microarray data presented herein and deposited in the GEO is MIAME compliant.
Quality control
RNA quality were assessed using digital microelectrophoresis with either a PicoChip™ or NanoChip™ (Agilent), which generated electropherograms, digital gels, and RNA Integrity numbers (RINs) (75). RNA quality was ascertained at each stage of processing—acquisition, microdissection, amplification, fragmentation—and studies were advanced only if optimal. Microarray quality was assessed by visualization of the microarray CEL files, percent positive calls on the microarray, area under the curve (AUC) values, distribution histograms of non-normalized expression scores, box plots of mean and 25th and 75th percentiles probe scores before and after quantile normalization, median normalized non-background corrected scores of probes designated as background, graphs of mean absolute difference between transformed background-corrected and normalized probe scores and frequency histograms of GC counts (76).
Bioinformatics
Primary bioinformatics analysis used Genomics Suite Version 6.4 (Partek). CEL files were imported into the software and their backgrounds underwent RMA and quantile normalizations. Statistical analysis was performed by creating from the individual CEL files two groups, control or SALS, and then comparing them to each other. The exon arrays have three types of probe sets: core, extended and full. The core probe set contains ∼17 800 transcript clusters and ∼284 000 probe sets to RefSeq and GenBank full-length mRNA's; the extended probe set contains ∼129 000 transcript clusters and ∼523 000 probe sets (plus core probes), with additional probes to expressed sequence tags (EST's) and partially annotated mRNA's; and the full probe set contains ∼262 000 transcript clusters and ∼580 000 probe sets (plus core and extended probes) with additional probes to all predicted exons in the genome. For our analysis, we have used the core probe set because of the depth of annotation available. The software uses a proprietary Exon Splice ANOVA algorithm to calculate both each gene's differential expression, which is determined by a composite expression of all its exons comparing between disease and control, and each gene's exon splice index, which is determined by exon-to-exon comparison between disease and control after correcting for other variables, including overall differential gene expression. Among the factors measured by the ANOVA are batch effects. Signal strengths and noise levels were measured in two ways. First, noise levels specific to our data were estimated by randomly permuting CEL file classifications to create comparisons between sham groups; 10 permutations were performed. Second, multiple test correction was performed by Bonferroni method; these data are reported in the Supplementary Material. Enrichment analysis was determined by generating an enrichment score equal to the P-value of a χ2 test comparing identified genes and their biological functionality as defined by Gene Ontology (www.geneontology.org). Heat maps were generated by first summarizing the exons of each gene using the ‘gene summary’ command, followed by the ‘cluster based on significant genes’ command. Additional bioinformatics analysis was performed using Expression Console™ Version 1.1 (Affymetrix) and XRAY Version 3.92 (Biotique Systems).
Validation of array results by qPCR
Genes identified as being aberrantly spliced were selected for validation by qPCR. Exons exhibiting significant differences as well as lack of differences of expression between SALS and control were analyzed. Target sequences to these exons were obtained from Affymetrix. To these, primers and probes were designed using Applied Biosystem's Primer Express software program (Applied Biosystems Inc.). In the case of TDP-43, GARS, EAAT1 (SLC1A3), EAAT4 (SLC1A6) and GAPDH, standard TaqMan® probes were used (Applied Biosystems, Inc). qPCR was performed using either a 7900HT Fast Real Time PCR System or a 7500 Real Time PCR system using either a 384 or 96-well format (Applied Biosystems, Inc). Four-fold standard dilution curves were generated on each plate for each probe in an RT step using RNeasy-purified HEK 293 RNA (Qiagen). Test samples consisted of amplified cDNA from the motor neuron enriched RNA pools and were run in triplicate. The resulting data were normalized to endogenous GAPDH run in parallel on each plate. Values from 3–12 SALS and 3–10 control were separately determined, averaged between groups, and compared by two-tailed Welch's t-test using logarithmic scaling. We called a gene valid only if two conditions were both met: an exon predicted to have differential expression and an exon predicted not to have differential expression were confirmed by qPCR.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from Microsoft Research, the National Institutes of Health (NS051738), the Department of Defense (USAMRAA W81XWH-07-0246), the Wyckoff family, the Moyer Foundation, Mrs Lois Caprile and the Benaroya Foundation.
Supplementary Material
REFERENCES
- 1.Rowland L.P., Shneider N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Bruijn L.I., Miller T.M., Cleveland D.W. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 3.Boillee S., Vande Velde C., Cleveland D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Neusch C., Bähr M., Schneider-Gold C. Glia cells in amyotrophic lateral sclerosis: new clues to understanding an old disease? Muscle Nerve. 2007;35:712–724. doi: 10.1002/mus.20768. [DOI] [PubMed] [Google Scholar]
- 5.Liscic R.M., Grinberg L.T., Zidar J., Gitcho M.A., Cairns N.J. ALS and FTLD: two faces of TDP-43 proteinopathy. Eur J. Neurol. 2008;15:772–780. doi: 10.1111/j.1468-1331.2008.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Kim E., Goren A., Ast G. Alternative splicing: current perspectives. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- 8.Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T.J., Sladek R., Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 9.Wahl M.C., Will C.L., Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 10.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 11.Clark T.A., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shendure J. The beginning of the end for microarrays? Nature Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 13.Lagier-Tourenne C., Cleveland D.W. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 15.Arai T., Hasegawa L.M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 16.Buratti E., Baralle F.E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 17.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 19.Van Deerlin V.M., Leverenz J.B., Bekris L.M., Bird T.D., Yuan W., Elman L.B., Clay D., Wood E.M., Chen-Plotkin A.S., Martinez-Lage M., et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwiatkowski T.J., Bosco D.A., LeClerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 21.Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beleza-Meireles A., Al-Chalabi A. Genetic studies of amyotrophic lateral sclerosis: controversies and perspectives. Amyotroph. Lateral Scler. 2008;26:1–14. doi: 10.1080/17482960802585469. [DOI] [PubMed] [Google Scholar]
- 23.Simpson C.L., Lemmens R., Miskiewicz K., Broom W.J., Hansen V.K., van Vught P.W., Landers J.E., Sapp P., Van Den Bosch L., Knight J., et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravits J., Paul P., Jorg C. Focality of upper and lower motor neuron degeneration at the clinical onset of ALS. Neurology. 2007;68:1571–1575. doi: 10.1212/01.wnl.0000260965.20021.47. [DOI] [PubMed] [Google Scholar]
- 26.Ravits J., Laurie P., Fan Y., Moore D.H. Implications of ALS focality: rostral-caudal distribution of lower motor neuron loss postmortem. Neurology. 2007;68:1576–1582. doi: 10.1212/01.wnl.0000261045.57095.56. [DOI] [PubMed] [Google Scholar]
- 27.Mirnics K., Pevsner J. Progress in the use of microarray technology to study the neurobiology of disease. Nat. Neurosci. 2004;7:434–439. doi: 10.1038/nn1230. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y.M., Yamamoto M., Kobayashi Y., Yoshihara T., Liang Y., Terao S., Takeuchi H., Ishigaki S., Katsuno M., Adachi H., et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann. Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 29.Curtis R., Oresic M., Vidal-Puig A. Pathways to the analysis of microarray data. Trends in Biotechnology. 2005;23:429–435. doi: 10.1016/j.tibtech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Bussemaker H., Ward L., Boorsma A. Dissecting complex transcriptional responses using pathway-level scores based on prior information. BMC Bioinformatics. 2007;8(Suppl. 6):1–7. doi: 10.1186/1471-2105-8-S6-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKee A.E., Neretti N., Carvalho L.E., Meyer C.A., Fox E.A., Brodsky A.S., Silver P.A. Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome Biol. 2007;8:R159. doi: 10.1186/gb-2007-8-8-r159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hang X., Li P., Li Z., Qu W., Yu Y., Li H., Shen Z., Zheng H., Gao Y., Wu Y., et al. Transcription and splicing regulation in human umbilical vein endothelial cells under hypoxic stress conditions by exon array. BMC Genomics. 2009;10:126. doi: 10.1186/1471-2164-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra M., Paunesku T., Woloschak G.E., Siddique T., Zhu L.J., Lin S., Greco K., Bigio E.H. Gene expression analysis of frontotemporal lobar degeneration of the motor neuron disease type with ubiquitinated inclusions. Acta Neuropathol. 2007;114:81–94. doi: 10.1007/s00401-007-0240-7. [DOI] [PubMed] [Google Scholar]
- 34.Lin C.L., Bristol L.A., Jin L., Dykes-Hoberg M., Crawford T., Clawson L., Rothstein J.D. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- 35.Flowers J.M., Powell J.F., Leigh P.N., Andersen P., Shaw C.E. Intron 7 retention and exon 9 skipping EAAT2 mRNA variants are not associated with amyotrophic lateral sclerosis. Ann. Neurol. 2001;49:643–649. [PubMed] [Google Scholar]
- 36.Gardina P.J., Clark T.A., Shimada B., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwan T., Benovoy D., Dias C., Gurd S., Serre D., Zuzan H., Clark T.A., Schweitzer A., Staples M.K., Wang H., et al. Heritability of alternative splicing in the human genome. Genome Res. 2007;17:1210–1218. doi: 10.1101/gr.6281007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung L.H., Heiner M., Hui J., Schreiner S., Benes V., Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Giorgio F.P., Carrasco M.A., Siao M.C., Maniatis T., Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nature Neuroscience. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchetto M.C., Muotri A.R., Mu Y., Smith A.M., Cezar G.G., Gage F.H. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 41.King A.E., Dickson T.C., Blizzard C.A., Woodhouse A., Foster S.S., Chung R.S., Vickers J.C. Neuron-glia interactions underlie ALS-like axonal cytoskeletal pathology. Neurobiol. Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.04.004. doi:10.1016/j.neurobiolaging.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Talbot K., Davies K.E. Is good housekeeping the key to motor neuron survival? Cell. 2008;133:572–574. doi: 10.1016/j.cell.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Ule J. Ribonucleoprotein complexes in neurologic diseases. Curr. Opin. Neurobiol. 2008;18:516–523. doi: 10.1016/j.conb.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 44.Zhang D., Paley A.J., Childs G. The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J. Biol. Chem. 1998;273:18086–18091. doi: 10.1074/jbc.273.29.18086. [DOI] [PubMed] [Google Scholar]
- 45.Yang L., Embree L.J., Tsai S., Hickstein D.D. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J. Biol. Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- 46.Mercado P.A., Ayala Y.M., Romano M., Buratti E., Baralle F.E. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buratti E., Stuani C., De Prato G., Baralle F.E. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007;35:4359–4368. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirose G. Paraneoplastic neurologic syndromes. Intern. Med. 1996;35:925–929. doi: 10.2169/internalmedicine.35.925. [DOI] [PubMed] [Google Scholar]
- 49.Akamatsu W., Okano H.J., Osumi N., Inoue T., Nakamura S., Sakakibara S., Miura M., Matsuo N., Darnell R.B., Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc. Natl Acad. Sci. USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X., Lu L., Bush D.J., Zhang X., Zheng L., Suswam E.A., King P.H. Mutant copper-zinc superoxide dismutase associated with amyotrophic lateral sclerosis binds to adenine/uridine-rich stability elements in the vascular endothelial growth factor 3'-untranslated region. J. Neurochem. 2009;108:1032–1044. doi: 10.1111/j.1471-4159.2008.05856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henneke M., Diekmann S., Ohlenbusch A., Kaiser J., Engelbrecht V., Kohlschütter A., Krätzner R., Madruga-Garrido M., Mayer M., Opitz L., et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat. Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- 52.Boutz P.L., Stoilov P., Li Q., Lin C.H., Chawla G., Ostrow K., Shiue L., Ares M., Jr, Black D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makeyev E.V., Zhang J., Carrasco M.A., Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheung H.C., Hai T., Zhu W., Baggerly K.A., Tsavachidis S., Krahe R., Cote G.J. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009;132:2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zacchigna S., Lambrechts D., Carmeliet P. Neurovascular signaling defects in neurodegeneration. Nat. Rev. Neurosci. 2008;9:169–181. doi: 10.1038/nrn2336. [DOI] [PubMed] [Google Scholar]
- 56.Nave K.A., Trapp B.D. Axon-glial signaling and the glial support of axon function. Annu. Rev. Neurosci. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- 57.Stritt C., Stern S., Harting K., Manke T., Sinske D., Schwarz H., Vingron M., Nordheim A., Knöll B. Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat. Neurosci. 2009;12:418–427. doi: 10.1038/nn.2280. [DOI] [PubMed] [Google Scholar]
- 58.Gibson R.M., Craig S.E., Heenan L., Tournier C., Humphries M.J. Activation of integrin alpha5beta1 delays apoptosis of Ntera2 neuronal cells. Mol. Cell Neurosci. 2005;28:588–598. doi: 10.1016/j.mcn.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 59.Kosodo Y., Huttner W.B. Basal process and cell divisions of neural progenitors in the developing brain. Dev. Growth Differ. 2009;51:251–261. doi: 10.1111/j.1440-169X.2009.01101.x. [DOI] [PubMed] [Google Scholar]
- 60.Dityatev A., Fellin T. Extracellular matrix in plasticity and epileptogenesis . Neuron Glia Biol. 2009;5:1–13. doi: 10.1017/S1740925X09000118. [DOI] [PubMed] [Google Scholar]
- 61.Cayre M., Canoll P., Goldman J.E. Cell migration in the normal and pathological postnatal mammalian brain. Prog. Neurobiol. 2009;88:41–63. doi: 10.1016/j.pneurobio.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolburg H., Noell S., Wolburg-Buchholz K., Mack A., Fallier-Becker P. Agrin, aquaporin-4, and astrocyte polarity as an important feature of the blood-brain barrier. Neuroscientist. 2009;15:180–193. doi: 10.1177/1073858408329509. [DOI] [PubMed] [Google Scholar]
- 63.Agrawal S., Anderson P., Durbeej M., van Rooijen N., Ivars F., Opdenakker G., Sorokin L.M. Dystroglycan is selectively cleaved at the parenchymal basement membrane at sites of leukocyte extravasation in experimental autoimmune encephalomyelitis. J. Exp. Med. 2006;203:1007–1019. doi: 10.1084/jem.20051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaiser M., Maletzki I., Hülsmann S., Holtmann B., Schulz-Schaeffer W., Kirchhoff F., Bähr M., Neusch C. Progressive loss of a glial potassium channel (KCNJ10) in the spinal cord of the SOD1 (G93A) transgenic mouse model of amyotrophic lateral sclerosis. J. Neurochem. 2006;99:900–912. doi: 10.1111/j.1471-4159.2006.04131.x. [DOI] [PubMed] [Google Scholar]
- 65.Nicaise C., Soyfoo M.S., Authelet M., De Decker R., Bataveljic D., Delporte C., Pochet R. Aquaporin-4 overexpression in rat ALS model. Anat. Rec. (Hoboken) 2009;292:207–213. doi: 10.1002/ar.20838. [DOI] [PubMed] [Google Scholar]
- 66.Zhong Z., Deane R., Ali Z., Parisi M., Shapovalov Y., O'Banion M.K., Stojanovic K., Sagare A., Boillee S., Cleveland D.W., Zlokovic B.V. ALS-causing SOD1 mutants generate vascular changes prior to motor neuron degeneration. Nat. Neurosci. 2008;11:420–422. doi: 10.1038/nn2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mizuno H., Warita H., Aoki M., Itoyama Y. Accumulation of chondroitin sulfate proteoglycans in the microenvironment of spinal motor neurons in amyotrophic lateral sclerosis transgenic rats. J. Neurosci. Res. 2008;86:2512–2523. doi: 10.1002/jnr.21702. [DOI] [PubMed] [Google Scholar]
- 68.Vitellaro-Zuccarello L., Bosisio P., Mazzetti S., Monti C., De Biasi S. Differential expression of several molecules of the extracellular matrix in functionally and developmentally distinct regions of rat spinal cord. Cell Tissue Res. 2007;327:433–447. doi: 10.1007/s00441-006-0289-y. [DOI] [PubMed] [Google Scholar]
- 69.Suwan K., Choocheep K., Hatano S., Kongtawelert P., Kimata K., Watanabe H. Versican/PG-M assembles hyaluronan into extracellular matrix and inhibits CD44-mediated signaling toward premature senescence in embryonic fibroblasts. J. Biol. Chem. 2009;284:8596–8604. doi: 10.1074/jbc.M806927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang M.J., Kuo J.S., Lee W.W., Huang H.Y., Chen W.F., Lin S.Z. Translational event mediates differential production of tumor necrosis factor-alpha in hyaluronan-stimulated microglia and macrophages. J. Neurochem. 2006;97:857–871. doi: 10.1111/j.1471-4159.2006.03776.x. [DOI] [PubMed] [Google Scholar]
- 71.Ravits J., La Spada A. Motor phenotype heterogeneity, focality and spread: deconstructing motor neuron degeneration. Neurology. 2009;73:805–811. doi: 10.1212/WNL.0b013e3181b6bbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagai M., Re D.B., Nagata T., Chalazonitis A., Jessell T.M., Wichterle H., Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat. Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brooks B.R. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. J. Neurol. Sci. 1994;124(suppl.):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 74.Boelen M.J., te Meerman G.J., Gibcus J.H., Blokzijl T., Boezen H.M., Timens W., Postma D.S., Groen H.J., van den Berg A. Microarray amplification bias: loss of 30% differentially expressed genes due to long probe—poly(A)-tail distances. BMC Genomics. 2007;8:277–290. doi: 10.1186/1471-2164-8-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bolstad B.M., Collin F., Brettschneider J., Cope L., Simpson K., Irizarry R., Speed T.P. Quality assessment of Affymetrix GeneChip data. In: Gentleman R., editor. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. chapter 3. New York: Springer Science+Business Media, Inc.; 2005. pp. 33–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.