Abstract
Mutations in PTEN-induced putative kinase 1 (PINK1) or parkin cause autosomal recessive forms of Parkinson disease (PD), but how these mutations trigger neurodegeneration is poorly understood and the exact functional relationship between PINK1 and parkin remains unclear. Here, we report that PINK1 regulates the E3 ubiquitin-protein ligase function of parkin through direct phosphorylation. We find that phosphorylation of parkin by PINK1 activates parkin E3 ligase function for catalyzing K63-linked polyubiquitination and enhances parkin-mediated ubiquitin signaling through the IκB kinase/nuclear factor κB (NF-κB) pathway. Furthermore, the ability of PINK1 to promote parkin phosphorylation and activate parkin-mediated ubiquitin signaling is impaired by PD-linked pathogenic PINK1 mutations. Our findings support a direct link between PINK1-mediated phosphorylation and parkin-mediated ubiquitin signaling and implicate the deregulation of the PINK1/parkin/NF-κB neuroprotective signaling pathway in the pathogenesis of PD.
INTRODUCTION
Parkinson disease (PD) is the most common neurodegenerative movement disorder, yet the pathogenic mechanisms underlying the disease remain unclear. Although the majority of PD cases are idiopathic in nature, a small percentage of PD is associated with the inheritance of genetic mutations and is referred to as familial PD (1,2). Approximately 50% of all recessively transmitted early-onset familial PD cases are caused by homozygous mutations in parkin (3,4). Homozygous mutations in PTEN-induced putative kinase 1 (PINK1) are the second most common cause of early-onset recessive PD (5,6). In addition, heterozygous mutations in parkin or PINK1 have been implicated as significant risk factors in the development of late-onset PD (2,6). Despite compelling genetic evidence linking parkin and PINK1 mutations to PD, the molecular mechanisms by which the pathogenic mutations trigger neurodegeneration are poorly understood.
The parkin gene encodes a 465 amino acid cytosolic E3 ubiquitin-protein ligase that is expressed in many tissues and cell types (3,4). Parkin has been reported to regulate K48-linked polyubiquitination and proteasomal degradation of several putative substrate proteins (4,7), although it remains controversial regarding the validity of these proteins as physiological parkin substrates (8–10). We and others have recently shown that parkin can catalyze monoubiquitination and K63-linked polyubiquitination and function in a proteasome-independent manner for regulating endocytosis (11), NF-κB signaling (12) and aggresome formation (13). The importance of parkin E3 ligase activity to neuronal survival is highlighted by the finding that most PD-linked pathogenic parkin mutations cause severe impairment of parkin E3 ligase function (2,4,7). At present, very little is known about how parkin E3 ligase function is regulated in cells.
The PINK1 gene encodes a 581 amino acid putative mitochondrial serine/threonine kinase that is expressed in many tissues and cell types, including dopaminergic neurons (5,14,15). We and others have shown that recombinant PINK1 protein exhibits kinase activity in vitro (16–19) and that the PINK1 kinase activity is required for its cytoprotective function (20). Consistent with the presence of a mitochondrial targeting sequence at its N-terminus, PINK1 is localized to the mitochondria, where it is mainly found in the mitochondrial inner membrane and intermembrane space (14,17,19,21), although a fraction of PINK1 may exist in the mitochondrial outer membrane with the kinase domain facing the cytosol (22). There is also a cytosolic pool of PINK1 (16,23,24), as expected because PINK1 is encoded by the nuclear genome. Together, current data suggest that PINK1 could exert cytoprotective action through phosphorylation of mitochondrial as well as cytosolic substrate proteins.
The fact that homozygous mutations in parkin or PINK1 cause autosomal recessive PD indicates that the normal function of these proteins is required for cell survival, particularly the survival of dopaminergic neurons. Recent genetic studies in Drosophila reveal that loss of parkin or PINK1 function leads to a similar phenotype that includes reduced lifespan, motor deficits, mitochondrial abnormalities and degeneration of muscle and dopaminergic neurons (25–30). The Drosophila PINK1 mutant phenotype can be rescued by overexpression of parkin, suggesting that PINK1 acts upstream of parkin in a common pathway (27–30). Recent studies in mammalian cells further support a functional link between PINK1 and parkin (31) and suggest that PINK1 is capable of interacting with parkin (32–34) and phosphorylating parkin (35). However, the cellular role of PINK1–parkin interaction is unclear and whether PINK1-mediated parkin phosphorylation regulates the E3 ligase function of parkin remains to be determined.
In this study, we investigated the interaction of parkin with and the phosphorylation of parkin by wild-type and PD-linked mutant PINK1 proteins and their functional consequences. Our results reveal that PINK1-mediated phosphorylation activates parkin E3 ligase function for catalyzing K63-linked polyubiquitination and enhances parkin-mediated ubiquitin signaling through the NF-κB pathway. Moreover, our findings provide evidence supporting a link between the deregulation of the PINK1/parkin/NF-κB neuroprotective pathway and PD pathogenesis.
RESULTS
PINK1 interacts with parkin and this interaction is not affected by PINK1 pathogenic mutations or kinase-dead mutation
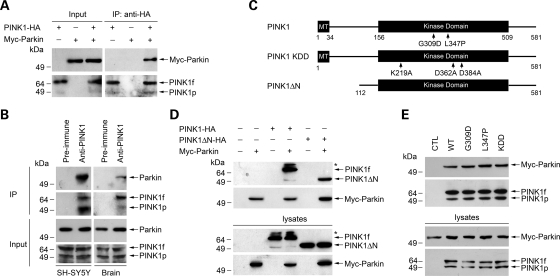
To determine whether PINK1 interacts with parkin, we first performed co-immunoprecipitation analysis using lysates from transfected human SH-SY5Y dopaminergic cells expressing N-terminal Myc-tagged parkin (Myc-parkin) and/or C-terminally HA-tagged PINK1 (PINK1-HA). Immunoprecipitation of the lysates with an anti-HA antibody revealed that Myc-parkin was co-immunoprecipitated with PINK1-HA (Fig. 1A). Furthermore, in the reciprocal co-immunoprecipitation experiments, anti-Myc antibody was able to co-immunoprecipitate PINK1-HA with Myc-parkin (Fig. 1D), providing evidence for an interaction between PINK1 and parkin in transfected SH-SY5Y cells. We then examined the association of endogenous PINK1 and Parkin in untransfected SH-SY5Y cells and human brain (Fig. 1B). The anti-PINK1 antibody, but not the pre-immune serum, co-immunoprecipitated endogenous PINK1 and parkin from SH-SY5Y cell lysates (Fig. 1B, left panel) and human brain homogenates (Fig. 1B, right panel), indicating an specific interaction between endogenous PINK1 and parkin in vivo.
Figure 1.
Interaction of parkin with wild-type and mutant PINK1. (A) Co-immunoprecipitation of parkin with PINK1. Lysates from SH-SY5Y cells expressing Myc-tagged parkin and/or C-terminal HA-tagged PINK1 (Input) were immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-Myc and anti-HA antibodies. PINK1f, PINK1 full-length form; PINK1p, a processed form of PINK1. (B) Endogenous PINK1 interacts with parkin in SH-SY5Y cells and human brain. SH-SY5Y cell lysates (left panel) or human brain homogenates (right panel) were subjected to immunoprecipitation with anti-PINK1 antibody or pre-immune serum followed by immunoblotting with anti-PINK1 and anti-parkin antibodies. (C) Schematic representation of PINK1 and its mutants encoded by C-terminal HA-tagged expression vectors. MT, mitochondrial targeting sequence. The locations of PINK1 point mutations are indicated on the domain structure. (D) N-terminal deletion has no effect on the interaction of PINK1 with parkin. Lysates from SH-SY5Y cells transfected with indicated constructs were immunoprecipitated with anti-Myc antibody followed by immunoblotting with anti-Myc and anti-HA antibodies. The asterisk indicates a band that probably represents a post-translationally modified form of PINK1. (E) Effects of PINK1 pathogenic mutations and kinase-dead mutation on the PINK1–parkin interaction. Lysates from SH-SY5Y cells co-transfected with Myc-tagged parkin and the empty HA vector (CTL) or indicated wild-type (WT) or mutant PINK1 constructs were immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-Myc and anti-HA antibodies.
To further define the interaction of PINK1 with parkin, we generated a PINK1 deletion construct, PINK1ΔN, which encodes a PINK1 mutant protein that lacks the N-terminal 111 residues including the mitochondrial targeting sequence (Fig. 1C) and is incapable of translocating from the cytosol to the mitochondria (24). Co-immunoprecipitation analysis revealed that the PINK1ΔN protein retained the ability to interact with parkin (Fig. 1D), suggesting that the site of parkin–PINK1 interaction is in the cytosol. Next, we assessed the interaction of parkin with PD-linked PINK1 mutants G309D and L347P and with the synthetic ‘kinase-dead’ PINK1 triple mutant KDD (19). We found that none of these PINK1 mutations had any significant effect on the ability of PINK1 to interact with parkin (Fig. 1E). This result, together with the previous finding that PINK1 kinase function is disrupted by the G309D, L347P and KDD mutations (16,17,19), indicates that the kinase activity of PINK1 is not required for the interaction of PINK1 with parkin.
The RING-finger 1 domain, but not parkin E3 ligase activity, is required for the interaction of parkin with PINK1
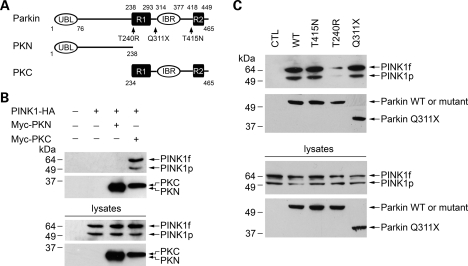
Parkin contains an ubiquitin-like (UBL) domain at the N terminus and two RING-finger motifs (R1 and R2) and an in-between RING-finger (IBR) domain at the C terminus (Fig. 2A). To define the specific region of parkin responsible for the interaction with PINK1, we made several parkin deletion and site-specific mutants and examined their ability to interact with PINK1 by co-immunoprecipitation analysis (Fig. 2). We found that the parkin C-terminal region (PKC) that encompasses the R1, IBR and R2 domains, but not the parkin N-terminal region (PKN) that contains the UBL domain and the linker region (Fig. 2A), was able to interact with PINK1 (Fig. 2B). The ability of parkin to interact with PINK1 was not affected by the PD-linked parkin truncation mutation Q311X (X is the stop codon) that deletes the IBR and R2 domains (Fig. 2C), indicating that the IBR and R2 domains are dispensable for the parkin–PINK1 interaction. Together, these data suggest that the PINK1-interacting region is between amino acids 234–311 where the R1 domain is located. In support of this notion, parkin T240R, a pathogenic mutation at the R1 domain, significantly reduced the interaction of parkin with PINK1, whereas parkin T415N, a pathogenic mutation near the R2 domain, had no effect on the parkin–PINK1 interaction (Fig. 2C). Since previous studies have shown that parkin Q311X and T415N mutations completely abolish the E3 ligase activity of parkin (36,37), the observation that parkin Q311X and T415N mutants retained the ability to interact with PINK1 (Fig. 2C) indicates that the E3 ligase activity of parkin is not required for the interaction of parkin with PINK1.
Figure 2.
Interaction of PINK1 with wild-type and mutant parkin. (A) Schematic representation of parkin and its mutants encoded by N-terminal Myc-tagged expression vectors. UBL, ubiquitin-like domain; R1 and R2, RING-finger 1 and RING-finger 2 motifs; IBR, in-between RING-finger domain. The locations of parkin pathogenic mutations are indicated on the domain structure. (B) PINK1 binds to the C-terminal region of parkin. Lysates from SH-SY5Y cells transfected with the indicated constructs were immunoprecipitated with anti-Myc antibody followed by immunoblotting with anti-HA and anti-Myc antibodies. (C) Effects of parkin pathogenic mutations on the PINK1–parkin interaction. Lysates from SH-SY5Y cells co-transfected with PINK1-HA and the empty Myc vector (CTL) or indicated wild-type (WT) or mutant parkin constructs were immunoprecipitated with anti-Myc antibody followed by immunoblotting with anti-PINK1 and anti-Myc antibodies.
PINK1 phosphorylates parkin and this phosphorylation is abrogated by PD-linked mutations
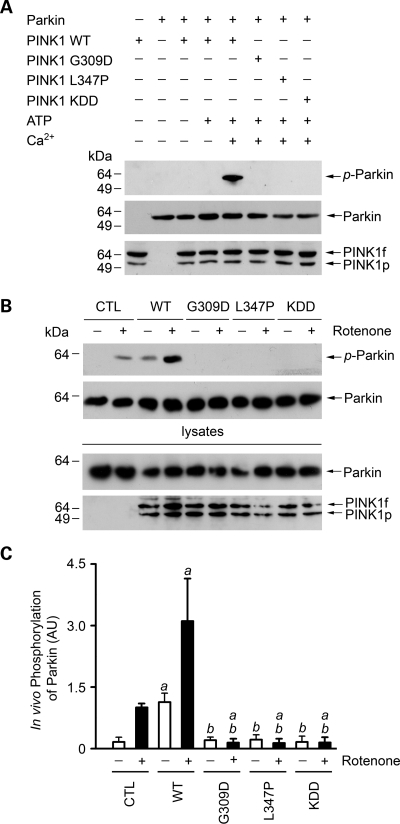
The observed interaction of PINK1 with parkin in cells (Figs 1 and 2) raises the possibility that parkin may be a substrate of PINK1 kinase. To examine this possibility, we performed in vitro phosphorylation assays to investigate the ability of wild-type and mutant PINK1 to phosphorylate parkin. We found that wild-type PINK1 was able to phosphorylate parkin in the presence of Ca2+, but not in the absence of Ca2+ (Fig. 3A). The Ca2+-dependent phosphorylation of parkin required the PINK1 kinase activity, as the phosphorylation was abolished by the kinase-dead PINK1 KDD mutation (Fig. 3A). Furthermore, the in vitro phosphorylation of parkin by PINK1 was abrogated by PD-linked PINK1 pathogenic mutations G309D and L347P (Fig. 3A), consistent with the previous reports that these pathogenic mutations disrupted PINK1 kinase function (16,17,19).
Figure 3.
Phosphorylation of parkin by wild-type and mutant PINK1. (A) In vitro phosphorylation assays were performed by incubation of purified Myc-tagged parkin and FLAG-tagged wild-type (WT) or mutant PINK1 proteins as indicated in the absence or presence of 0.1 mm ATP and 0.5 mm CaCl2. Phosphorylation of parkin was detected by immunoblotting with anti-phosphoserine antibody (top panel), and the amounts of PINK1 and parkin proteins used in the phosphorylation assays were shown by immunoblotting with anti-Myc (middle panel) and anti-FLAG antibodies (bottom panel). (B) SH-SY5Y cells expressing Myc-tagged parkin and wild-type or mutant PINK1 or the vector-transfected control (CTL) were treated with 20 nm rotenone for 8 h as indicated. In vivo parkin phosphorylation was determined by immunoprecipitated with anti-Myc antibody followed by immunoblotting using anti-phosphoserine and anti-parkin antibodies. (C) Normalized levels of in vivo parkin phosphorylation by PINK1. Data represent mean ± SEM from three independent experiments. aSignificantly different from the corresponding vector-treated controls (P < 0.05). bSignificantly different from the corresponding wild-type PINK1-transfected controls (P < 0.05). AU, arbitrary units.
Next, we investigated the effects of overexpressing wild-type or mutant PINK1 on the in vivo phosphorylation of parkin in SH-SY5Y cells. We found that, in the vector-transfected control cells, while there was little phosphorylation of parkin under the basal condition, parkin was phosphorylated in response to treatment with rotenone (Fig. 3B and C). Overexpression of wild-type PINK1 significantly increased both basal and rotenone-induced phosphorylation of parkin (Fig. 3B and C). The ability of PINK1 to promote in vivo parkin phosphorylation was abolished by PD-linked PINK1 G309D and L347P mutations and the kinase-dead PINK1 KDD mutation (Fig. 3B and C). These data, together with the result that these mutations did not alter the interaction of PINK1 with parkin (Fig. 1), suggest that the PINK1 G309D, L347P and KDD mutants have a dominant-negative effect on parkin phosphorylation in vivo by competing with endogenous PINK1 for binding its substrate parkin.
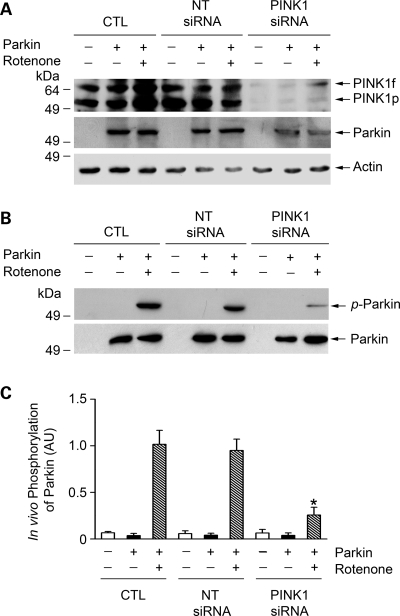
To provide further evidence supporting the role of PINK1 in regulating parkin phosphorylation, we examined the effect of small interfering RNA (siRNA)-mediated PINK1 depletion on parkin phosphorylation in SH-SY5Y cells. As shown in Fig. 4A, PINK1 siRNA specifically inhibited the expression of endogenous PINK1 in SH-SY5Y cells. We found that rotenone-induced phosphorylation of parkin was significantly reduced in PINK1-depleted cells compared with the vehicle or non-targeting (NT)-siRNA transfected control cells (Fig. 4B and C), further confirming that parkin is indeed a cellular substrate of PINK1 kinase.
Figure 4.
Depletion of endogenous PINK1 reduces parkin phosphorylation in dopaminergic cells. SH-SY5Y cells were transfected with Myc-tagged parkin and vehicle (CTL), non-targeting (NT) siRNA or PINK1 siRNA and treated with 20 nm rotenone for 8 h as indicated. (A) The levels of PINK1, parkin and actin in the cell lysates were analyzed by immunoblotting with anti-PINK1, anti-Myc and anti-actin antibodies. (B) In vivo parkin phosphorylation was determined by immunoprecipitated with anti-Myc antibody followed by immunoblotting using anti-phosphoserine and anti-parkin antibodies. (C) Normalized levels of in vivo parkin phosphorylation by PINK1. Data represent mean ± SEM from three independent experiments. *Significantly different from the corresponding rotenone-treated controls (P < 0.05). AU, arbitrary units.
PINK1-mediated phosphorylation enhances the E3 ligase activity of parkin for catalyzing K63-linked polyubiquitination in cooperation with the UbcH13/Uev1a E2 enzyme
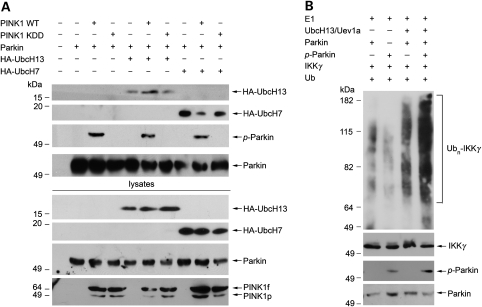
We and others have previously shown that parkin is a dual-function E3 ligase that can catalyze K48-linked polyubiquitination in cooperation with the E2 enzyme UbcH7 and K63-linked polyubiquitination with the heterodimeric E2 enzyme UbcH13/Uev1a (4,7,12,13). The molecular mechanism that regulates the ability of parkin to recruit distinct E2 enzymes for catalyzing different types of ubiquitination remains unknown. To determine whether PINK1 has a role in regulating the E2 recruitment by parkin, we investigated the effects of co-expressing wild-type or mutant PINK1 on the interaction of parkin with UbcH7 and UbcH13 in SH-SY5Y cells. We found that co-expression of wild-type PINK1 significantly enhanced the interaction of parkin with UbcH13 but not UbcH7 (Fig. 5A). The ability of PINK1 to promote the UbcH13 binding by parkin was abolished by the kinase-dead PINK1 KDD mutation (Fig. 5A), suggesting that the PINK1 kinase activity is required for facilitating the UbcH13 recruitment by parkin.
Figure 5.
Phosphorylation of parkin by PINK1 enhances the ability of parkin to bind UbcH13 and catalyze K63-linked polyubiquitination. (A) Lysates from SH-SY5Y cells expressing Myc-tagged parkin, HA-tagged UbcH7 or UbcH13 and wild-type or mutant PINK1 constructs were immunoprecipitated with anti-Myc antibody followed by immunoblotting with anti-HA, anti-phosphoserine and anti-parkin antibodies. (B) In vitro ubiquitination assays were performed by incubation of purified FLAG-tagged IKKγ, non-phosphorylated parkin or PINK1-phosphorylated parkin in the presence of E1, E2 (UbcH13/Uev1a) and ubiquitin as indicated. Ubiquitination of IKKγ was detected by immunoprecipitation with anti-FLAG antibody followed by immunoblotting using anti-ubiquitin antibody. The amounts of IKKγ, non-phosphorylated parkin and PINK1-phosphorylated parkin used in the ubiquitination assays (Input) were shown by immunoblotting with anti-FLAG, anti-parkin and anti-phosphoserine antibodies. Ubn-IKKγ, polyubiquitinated IKKγ.
Given that UbcH13 is the catalytic subunit of the UbcH13/Uev1a E2 enzyme and the UbcH13–parkin interaction has been shown to recruit the UbcH13/Uev1a heterodimer to parkin (13,38), the result of Fig. 5A prompted us to test whether PINK1-mediated phosphorylation regulates the E3 ligase activity of parkin for catalyzing K63-linked polyubiquitination. To address this question, we perform in vitro ubiquitination assays using the IκB kinase (IKK) subunit IKKγ, a known substrate for parkin-mediated K63-linked polyubiquitination (12). Consistent with the previous reports (12,13), we found that parkin was able to facilitate in vitro polyubiquitination of IKKγ in cooperation with the UbcH13/Uev1a E2 enzyme (Fig. 5B). Phosphorylation of parkin by PINK1 significantly increased the E3 ligase activity of parkin for catalyzing IKKγ polyubiquitination (Fig. 5B), supporting a direct role for PINK1-mediated phosphorylation in activating parkin E3 ligase function for catalyzing K63-linked polyubiquitination of IKKγ.
PINK1 is a major kinase for regulating the E3 ligase function of parkin in mediating K63-linked polyubiquitination of IKKγ in dopaminergic cells
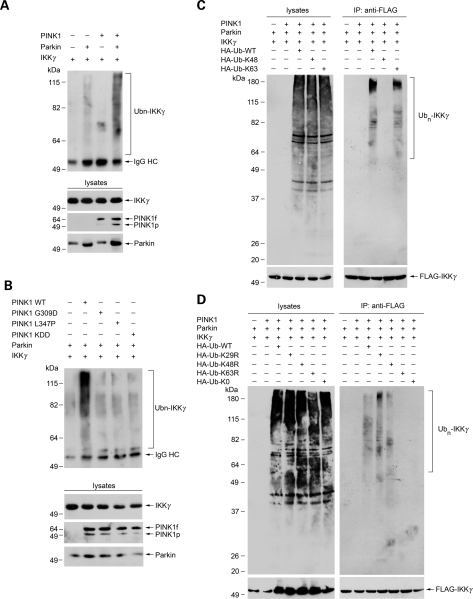
To characterize the cellular role of PINK1 in regulating parkin E3 ligase activity for catalyzing K63-linked polyubiquitination, we performed in vivo ubiquitination assays to assess the effects of wild-type and mutant PINK1 on parkin-mediated ubiquitination of IKKγ in SH-SY5Y cells. In agreement with the previous report (12), we found that IKKγ was polyubiquitinated by parkin in SH-SY5Y cells (Fig. 6A). The parkin-mediated IKKγ polyubiquitination was significantly increased by co-expression of wild-type PINK1 (Fig. 6A). This facilitatory effect was virtually abolished by the kinase-dead PINK1 KDD mutation and PD-linked PINK1 pathogenic mutations G309D and L347P (Fig. 6B), indicating that the PINK1 kinase activity is required for the ability of PINK1 to promote parkin-mediated IKKγ polyubiquitination in dopaminergic cells.
Figure 6.
Wild-type but not mutant PINK1 promotes parkin-mediated K63-linked polyubiquitination of IKKγ. (A and B) PINK1 promotes parkin-mediated IKKγ polyubiquitination in a kinase-dependent manner. SH-SY5Y cells were transfected with FLAG-tagged IKKγ, Myc-tagged parkin and C-terminally HA-tagged wild-type or mutant PINK1 as indicated. In vivo ubiquitination of IKKγ was determined by immunoprecipitation with anti-FLAG antibody followed by immunoblotting using anti-ubiquitin antibody. The expression of IKKγ, parkin and PINK1 in the cell lysates were confirmed by immunoblotting with anti-FLAG, anti-HA and anti-parkin antibodies. Ubn-IKKγ, polyubiquitinated IKKγ. IgG HC, immunoglobulin heavy chain. (C and D) Parkin-mediated IKKγ polyubiquitination occurs via the K63 linkage. Lysates from SH-SY5Y cells transfected with the indicated constructs were subjected to immunoprecipitations with anti-FLAG antibody followed by immunoblotting with anti-FLAG and anti-HA antibodies.
We then determined the linkage of parkin-mediated IKKγ polyubiquitination by using ubiquitin mutants Ub-K48 and Ub-K63, which contain arginine substitutions of all of its lysine residues except the one at position 48 or 63 and thus only allow the formation of K48-and K63-linked polyubiquitin chains, respectively (13). As shown in Fig. 6C, parkin promoted polyubiquitination of IKKγ in cells expressing wild-type ubiquitin (Ub-WT) or Ub-K63, but not in cells expressing Ub-K48, suggesting that parkin-mediated IKKγ polyubiquitination occurs via the K63-linkage. To further confirm this linkage, we used another set of ubiquitin mutants, Ub-K29R, Ub-K48R, Ub-K63R, which contain a single lysine-to-arginine mutation at position 29, 48 and 63, respectively. The Ub-K29R, Ub-K48R and Ub-K63R mutants are expected to solely disrupt the assembly of K29-, K48- and K63-linked polyubiquitin chains, respectively. As a control, we also included a polymerization-defective mutant of ubiquitin (Ub-K0), in which all lysine residues of ubiquitin were changed to arginines, and therefore is unable to form polyubiquitin chains. We found that parkin-mediated IKKγ polyubiquitination was greatly reduced by replacement of Ub-WT with Ub-K63R and Ub-K0, but not by the replacement with Ub-K29R and Ub-K48R (Fig. 6D). The in vivo ubiquitination data (Fig. 6) agree well with the in vitro ubiquitination result (Fig. 5B) and provide further support for the notion that PINK1-mediated phosphorylation enhances the E3 ligase function for catalyzing K63-linked polyubiquitination of IKKγ.
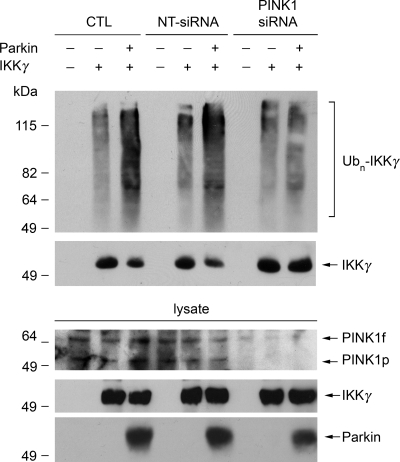
To determine the role of endogenous PINK1 in regulating the E3 ligase function of parkin for catalyzing K63-linked polyubiquitination, we examined the effect of siRNA-mediated PINK1 depletion on parkin-mediated K63-linked polyubiquitination of IKKγ in SH-SY5Y cells. We found that the polyubiquitination of IKKγ by parkin was significantly reduced in PINK1-depleted cells compared with the vehicle or NT-siRNA transfected control cells (Fig. 7), indicating that PINK1 plays an essential role in regulating parkin E3 ligase activity for catalyzing K63-linked polyubiquitination of IKKγ in vivo.
Figure 7.
Depletion of endogenous PINK1 reduces K63-linked polyubiquitination of IKKγ by parkin. SH-SY5Y cells were transfected with FLAG-tagged IKKγ, Myc-tagged parkin and vehicle (CTL), NT-siRNA or PINK1 siRNA as indicated. In vivo ubiquitination of IKKγ was detected by immunoprecipitation with anti-FLAG antibody followed by immunoblotting using anti-ubiquitin antibody. The levels of PINK1, IKKγ and parkin in the cell lysates were determined by immunoblotting with anti-PINK1, anti-FLAG and anti-Myc antibodies. All experiments were replicated three times with similar results. Ubn-IKKγ, polyubiquitinated IKKγ.
PINK1 functions upstream of parkin in NF-κB activation and cytoprotection against rotenone-induced apoptosis
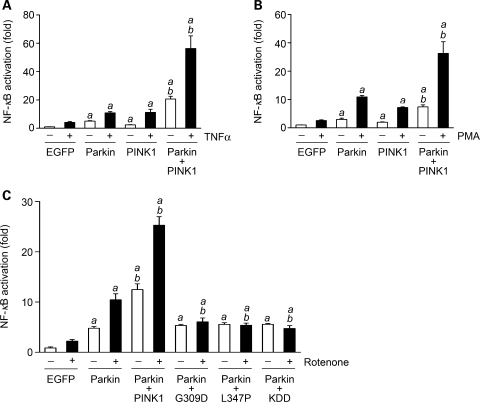
The K63-linked polyubiquitination of IKKγ is a critical step in the cytoprotective signaling pathway that activates NF-κB, a ubiquitously expressed transcription factor which mediates transcription of a number of pro-survival genes (39–41). Our finding that PINK1-mediated phosphorylation enhances the E3 ligase activity of parkin for catalyzing K63-linked polyubiquitination of IKKγ (Figs 5–7) raises the possibility that PINK1 may function together with parkin in the activation of the NF-κB signaling pathway. To test this possibility, we assessed the role of PINK1 and parkin in the activation of the NF-κB by using a well-established NF-κB-responsive luciferase reporter assay (12). We found that expression of exogenous parkin or PINK1 in SH-SY5Y cells not only stimulated NF-κB-dependent transcription under the basal condition (Fig. 8), but also enhanced NF-κB activation in response to treatment with TNFα (Fig. 8A), PMA/ionomycin (Fig. 8B) or rotenone (Fig. 8C). The NF-κB activation was further increased in SH-SY5Y cells co-expressing exogenous parkin and PINK1 compared with the cells expressing exogenous parkin or PINK1 alone (Fig. 8). The effect of PINK1 to increase parkin-stimulated NF-κB activation was abrogated by the kinase-dead PINK1 KDD mutation and PD-linked PINK1 pathogenic mutations G309D and L347P (Fig. 8C), suggesting that the ability of PINK1 to enhance the NF-κB-activating potential of parkin is dependent on PINK1 kinase activity.
Figure 8.
Wild-type but not mutant PINK1 enhances parkin-stimulated NF-κB activation. SH-SY5Y cells transfected with NF-κB-Luc reporter plasmid, EGFP or wild-type parkin, and indicated wild-type or mutant PINK1 were treated with 25 ng/ml TNFα (A), 10 ng/ml PMA and 160 ng/ml ionomycin (B) or 20 nm rotenone (C) for 3 h and harvested after additional 8 h. NF-κB activation was determined by NF-κB reporter assay and is expressed as the fold of NF-κB activation relative to the vehicle-treated EGFP control. Data represent mean ± SEM from three independent experiments. aSignificantly different from the corresponding EGFP controls (P < 0.05). bSignificantly different from the corresponding parkin-transfected cells (P < 0.05).
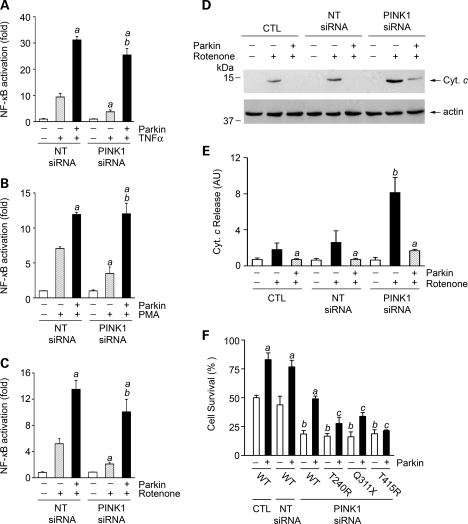
To provide evidence for the involvement of endogenous PINK1 in regulating the NF-κB signaling pathway, we examined the effect of siRNA-mediated PINK1 depletion on NF-κB activation in SH-SY5Y cells. We found that depletion of endogenous PINK1 resulted in a significant decrease in the NF-κB activation in response to treatment with TNFα (Fig. 9A), PMA/ionomycin (Fig. 9B) or rotenone (Fig. 9C), supporting an essential role for PINK1 in regulating the NF-κB signaling pathway. The phenotype of impaired NF-κB activation in PINK1-depleted cells was rescued by expression of exogenous parkin (Fig. 9A–C), suggesting that PINK1 acts upstream of parkin in activation of the NF-κB signaling pathway.
Figure 9.
PINK1 depletion impairs NF-κB activation and cell survival and this phenotype can be rescued by wild-type but not mutant parkin. (A–C) SH-SY5Y cells transfected with NF-κB-Luc reporter plasmid, indicated siRNAs and Myc-tagged parkin were treated with 25 ng/ml TNFα (A), 10 ng/ml PMA and 160 ng/ml ionomycin (B) or 20 nm rotenone (C) for 3 h and harvested after additional 8 h. NF-κB activation was determined by NF-κB reporter assay and is expressed as the fold of NF-κB activation relative to the vehicle-treated NT-siRNA transfected control. Data represent mean ± SEM from three independent experiments. aSignificantly different from the drug-treated NT-siRNA transfected control (P < 0.05). bSignificantly different from the drug-treated PINK1 siRNA-transfected cells in the absence of parkin overexpression (P < 0.05). (D–F) SH-SY5Y cells transfected with vehicle (CTL), NT-siRNA or PINK1 siRNA and indicated wild-type or mutant parkin construct were treated with 40 nm rotenone for 24 h. (D) The levels of cytochrome c (Cyt. c) and actin in the cytosolic fractions were determined by immunoblotting with anti-cytochrome c and anti-actin antibodies. (E) The level of cytochrome c released to the cytosol is normalized to the level of actin in each cell sample. Data represent mean ± SEM from three independent experiments. aSignificantly different from the corresponding rotenone-treated cells in the absence of parkin overexpression (P < 0.05). bSignificantly different from the rotenone-treated control (CTL) cells (P < 0.05). AU, arbitrary units. (F) The extent of cell survival was assessed by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Data represent mean ± SEM from three independent experiments. aSignificantly different from the same vehicle or siRNA-transfected cells in the absence of parkin overexpression (P < 0.05). bSignificantly different from the CTL cells in the absence of parkin overexpression (P < 0.05). cSignificantly different from the PINK1 siRNA-transfected cells with overexpression of wild-type parkin (P < 0.05).
Given the critical role of the NF-κB signaling pathway in promoting cell survival (39,40), we next investigated the functional relationship between parkin and PINK1 in cytoprotection against rotenone-induced apoptosis. Analyses of cell viability and cytochrome c release revealed that siRNA-mediated depletion of endogenous PINK1 significantly enhanced the susceptibility of SH-SY5Y cells to rotenone-induced cytochrome c release (Fig. 9D and E) and cell death (Fig. 9F). The pro-apoptotic phenotype of PINK1 depletion could be rescued by wild-type parkin (Fig. 9), suggesting that parkin acts downstream of PINK1 in cytoprotection against rotenone-induced apoptosis. The ability of parkin to rescue PINK1 depletion phenotype was abolished by PD-linked parkin T240R, Q311X and T415N mutations (Fig. 9F), providing further support for a loss-of-function pathogenic mechanism for these mutations.
DISCUSSION
Despite compelling genetic evidence indicating that PINK1 functions upstream of parkin in a common pathway (27–30), whether and how PINK1 regulates parkin function remain unclear. The present study reveals that PINK1 regulates the E3 ligase function of parkin for catalyzing K63-linked polyubiquitination through direct phosphorylation. Our data indicate that PINK1-mediated phosphorylation activates parkin-mediated ubiquitin signaling through the IKK/NF-κB pathway and support a link between the impairment of the PINK1/parkin/NF-κB signaling pathway and neurodegeneration in PD.
Our finding that PINK1 interacts with parkin in SH-SY5Y dopaminergic cells and human brain is consistent with recent reports (32–35). However, in contrast to the previous studies (32,33), our biochemical analysis shows that the interaction of PINK1 with parkin is not dependent on the kinase activity of PINK1, as neither PINK1 kinase-dead mutation KDD nor the PD-linked PINK1 pathogenic mutations (G309D and L347P) that are known to disrupt PINK1 kinase function (16,17,19) had any apparent effect on the ability of PINK1 to interact with parkin. Furthermore, our deletion and mutagenesis studies reveal that the PINK1–parkin interaction requires the RING-finger 1 domain of parkin, but not the E3 ligase activity of parkin.
Recent studies from our group and others have shown that PINK1 is localized in both the mitochondria and the cytosol (14,16,17,19,21–24), suggesting that PINK1 could exert cytoprotective action through phosphorylation of mitochondrial as well as cytosolic substrates. Our previous work has identified molecular chaperone protein TNF receptor-associated protein 1 as a mitochondrial PINK1 substrate (19). The present study shows that parkin is a cytosolic substrate for PINK1 kinase, as PINK1 not only binds parkin, but also phosphorylates parkin both in vitro and in vivo. The observed reduction in parkin phosphorylation by siRNA-mediated depletion of endogenous PINK1 in SH-SY5Y cells further supports that PINK1 is a major kinase for phosphorylating parkin in the dopaminergic cells.
Our findings indicate that there is direct connection between PINK1-mediated phosphorylation and parkin-mediated ubiquitination. Protein ubiquitination is a dynamic post-translational modification that serves diverse cellular roles (42,43). Whereas K48-linked polyubiquitination acts as the canonical signal for targeting the substrate to the proteasome for degradation, K63-linked polyubiquitination function in an proteasome-independent manner for regulating multiple cellular processes, including signal transduction and endocytic trafficking (42–44). Recent evidence indicates that parkin has dual E3 ligase activities for catalyzing K48-linked polyubiquitination and K63-linked polyubiquitination (4,7,12,13), but how these two distinct E3 ligase activities are regulated is not understood. Our result that phosphorylation of parkin by PINK1 increases the ability of parkin to bind UbcH13 (the catalytic subunit of the UbcH13/Uev1a E2 enzyme for mediating K63-linked polyubiquitination) but not UbcH7 (the E2 enzyme for mediating K48-linked polyubiquitination) provides a mechanism for preferentially enhancing the E3 ligase activity of parkin for catalyzing K63-linked polyubiquitination. In support of this notion, our result of in vitro ubiquitination analysis reveals that PINK1-mediated phosphorylation up-regulates parkin E3 ligase activity for catalyzing K63-linked polyubiquitination in cooperation with the UbcH13/Uev1a E2 enzyme.
Ample evidence indicates that parkin has the capacity to protect cells against cell death induced by a wide variety of stress, including oxidative stress, proteasome inhibition, mitochondrial dysfunction and proteotoxic stress caused by overexpression of mutant α-synuclein or tau (7,45). Although the molecular mechanism underlying the cytoprotective action of parkin remains incompletely understood, a recent study has shown that the cytoprotective action of parkin is linked to its E3 ligase activity for catalyzing K63-linked polyubiquitination of IKKγ and thereby activating the cytoprotective NF-κB stress response pathway (12). Our in vivo ubiquitination analysis reveals that parkin-mediated K63-linked polyubiquitination of IKKγ in SH-SY5Y cells is reduced by siRNA-mediated depletion of endogenous PINK1 and is significantly enhanced by co-expression of wild-type PINK1 but not the kinase-dead mutant PINK1. These results, together with the in vitro ubiquitination data, provide strong evidence supporting that PINK1 is a major kinase for activating the E3 ligase function of parkin for catalyzing K63-linked polyubiquitination of IKKγ in the dopaminergic cells. Our works further shows that, correlating with reduced parkin-mediated K63-linked polyubiquitination of IKKγ, PINK1 depletion impairs NF-κB activation and sensitizes the dopaminergic cells to rotenone-induced apoptosis. The impaired NF-κB activation and pro-apoptotic phenotype of PINK1 depletion can be rescued by parkin overexpression. Collectively, these findings point to a specific pathway in which PINK1 exerts its cytoprotective function by phosphorylating downstream effector parkin to enhance K63-linked polyubiquitination of IKKγ and subsequent NF-κB activation and cell survival.
Our work indicates that the PINK1/parkin/NF-κB cytoprotective signaling pathway is disrupted by PD-linked PINK1 pathogenic mutations G309D and L347P. We find that PINK1 G309D and L347P mutations abolish the kinase activity of PINK1 to phosphorylate its physiological substrate parkin and impair the ability of PINK1 to promote parkin-mediated K63-linked polyubiquitination of IKKγ and NF-κB activation. We and others have previously shown that PINK1 G309D and L347P mutations inhibit the cytoprotective action of PINK1 against cell death induced by oxidative stress and mitochondrial toxins (5,19,20). In addition, Henn et al. (12) have shown that parkin-mediated K63-linked polyubiquitination of IKKγ and NF-κB activation are reduced by PD-linked parkin pathogenic mutations R42P and G430D that have impaired cytoprotective capacity. Together, these results suggest that deregulation of the PINK1/parkin/NF-κB cytoprotective pathway, which could be caused by PINK1 or parkin mutations, is a common pathogenic mechanism leading to neurodegeneration in early-onset familial PD.
MATERIALS AND METHODS
Expression constructs
Conventional molecular biological techniques were used to generate the following expression constructs: C-terminal HA- or FLAG-tagged human wild-type PINK1, pathogenic PINK1 G309D and L347P mutants, PINK1 catalytically inactive KDD triple mutant (K219A/D362A/D384A) and PINK1ΔN (residues 112–581); N-terminal Myc-tagged parkin T240R, T415N and Q311X mutants, and parkin deletion mutants PKN (residues 1–238) and PKC (residues 234–465). Myc-tagged wild-type parkin, FLAG-tagged IKKγ and NF-κB-Luc were provided by T. Suzuki (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan), K. Winklhofer (Adolf-Butenandt Institute, Munich, Germany) and Z. Chen (Southwestern Medical Center, Dallas, TX, USA), respectively. HA-tagged Ub-WT, Ub-K48, Ub-K63 and Ub-K0 were provided by T. Dawson (Johns Hopkins University, Baltimore, MD, USA); and Ub-K29R, Ub-K48R and Ub-K63R were provided by M. Wooten (Auburn University, Auburn, AL, USA). The pGL4.73-hRluc/SV40 plasmid was obtained from Promega. All expression constructs were sequenced to ensure that the fusion was in the correct reading frame and there were no unwanted changes in the codons.
Antibodies
The rabbit polyclonal anti-PINK1 antibody was generated and described in our previous study (19). Other antibodies used in this study include the following: anti-HA (12CA5), anti-Myc (9E10.3, Neomarkers), anti-parkin (Cell Signaling), anti-phosphoserine (anti-pS) and anti-phosphothreonine (anti-pT) (Chemicon), anti-actin (Chemicon), anti-cytochrome c (Santa Cruz), anti-FLAG (M2, Sigma), anti-ubiquitin FL76 (Santa Cruz Biotechnology, Inc.) and anti-ubiquitin P4G7 (Covance). All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.
Cell transfection and co-immunoprecipitation
Human SH-SY5Y cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine medium and 100 U/ml penicillin and streptomycin. Cells were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Cells lysates were prepared from transfected cells and immunoprecipitations performed as described previously (46) using the indicated antibodies. Immunocomplexes were resolved by SDS–PAGE and analyzed by immunoblotting analysis with the appropriate antibodies and horseradish peroxidase-conjugated secondary antibodies. Results were visualized using enhanced chemiluminescence.
siRNAs transfection
For depletion of PINK1 in SH-SY5Y cells, PINK1 siRNA (Dharmacon) was generated against the following human PINK1 mRNA sequences, 5′-GAAAUCCGACAACAUCCUUUU-3′. A control siRNA (NT-siRNA) with no known mammalian homology (siCONTROL Non-Targeting siRNA #1, Dharmacon) was used as a negative control. SH-SY5Y cells were transfected with the indicated siRNAs using the Lipofectamine 2000 (Invitrogen) as described previously (47). Experiments were performed 48 h after siRNA treatment.
In vitro and in vivo phosphorylation assays
In vitro kinase assays were performed as we described previously (19) by incubation of affinity-purified FLAG-tagged wild-type or mutant PINK1 with Myc-tagged parkin for 60 min at 30°C in 100 µl of reaction buffer containing 50 mm Tris–HCl (pH 7.5), 8 mm MgCl2, 2 mm MnCl2 and 0.1 mm ATP in the absence or presence of 0.5 mm CaCl2. The reaction mixtures were resolved by SDS–PAGE, and phosphorylation of parkin was determined by immunoblotting with anti-phosphoserine and anti-phosphothreonine antibodies. For detection of in vivo phosphorylation of parkin, Myc-tagged parkin protein was immunoprecipitated from cell lysates by using anti-Myc antibody followed by immunoblotting with anti-phosphoserine and anti-phosphothreonine antibodies. The relative level of in vivo parkin phosphorylation was determined by quantification of the intensity of the phosphorylated parkin band followed by normalization to the total level of parkin.
In vitro and in vivo ubiquitination assays
In vitro ubiquitination assays were performed as we described previously (13,48) by incubation of affinity-purified FLAG-tagged IKKγ and non-phosphorylated parkin or PINK1-phosphorylated parkin at 37°C for 2 h in 60 µl of reaction buffer (50 mm Tris–HCl, pH 7.5, 2.5 mm MgCl2, 2 mm DTT and 2 mm ATP) containing 200 ng of E1 (Boston Biochem), 400 ng of E2 ubiquitin-conjugating enzyme (UbcH13/Uev1a) (Boston Biochem), 10 µg ubiquitin (Boston Biochem). Ubiquitination of FLAG-IKKγ was detected by immunoprecipitation with anti-FLAG antibody followed by immunoblotting using anti-ubiquitin antibody. In vivo ubiquitination assays were performed as we described previously (48–50). Briefly, lysates from SH-SY5Y cells co-transfected with FLAG-tagged IKKγ, HA-tagged wild-type or mutant ubiquitin, and other indicated constructs were immunoprecipitated under denaturing conditions with anti-FLAG antibody. Ubiquitination of FLAG-IKKγ was detected by immunoblotting using anti-HA or anti-ubiquitin antibodies as indicated.
NF-κB reporter assays
SH-SY5Y cells were cotransfected with the NF-κB-Luc reporter plasmid encoding firefly luciferase under the control of an NF-κB-responsive promoter, pGL4.73-hRluc/SV40 internal control Renilla luciferase plasmid and other indicated constructs. At 24 or 48 h post-transfection, cells were treated with 25 ng/ml TNFα, 10 ng/ml PMA and 160 ng/ml ionomycin or 20 nm rotenone at 37°C for 3 h and harvested after additional 8 h. NF-κB luciferase activity in cell lysates was measured as described (51) using the Dual Luciferase Reporter Assay system (Promega) and normalized to the internal control Renilla luciferase activity.
Cell viability and cytochrome c release assays
Cell viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay as described (19). The biochemical cytochrome c release assay was performed as we described previously (19). Briefly, cells were homogenized and then fractionated into the mitochondria and cytosol fractions. The release of cytochrome c to the cytosol was determined by immunoblotting with anti-cytochrome c antibody.
Statistical analysis
All experiments were performed in triplicate and repeated at least three times. Data were subjected to statistical analysis by ANOVA and a P-value of less than 0.05 was considered statistically significant.
FUNDING
This work was supported by grants from National Institutes of Health (GM082828, GM082828-02S1, ES015813, AG021489, AG034126 and NS050650).
ACKNOWLEDGEMENTS
We thank Drs Zhijian Chen, Toshiaki Suzuki, Ted Dawson, Konstanze Winklhofer and Marie Wooten for providing plasmids.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dawson T.M., Dawson V.L. Rare genetic mutations shed light on the pathogenesis of Parkinson disease. J. Clin. Invest. 2003;111:145–151. doi: 10.1172/JCI17575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schapira A.H. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 3.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 4.Hattori N., Mizuno Y. Pathogenetic mechanisms of parkin in Parkinson's disease. Lancet. 2004;364:722–724. doi: 10.1016/S0140-6736(04)16901-8. [DOI] [PubMed] [Google Scholar]
- 5.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 6.Tan E.-K., Skipper L.M. Pathogenic mutations in Parkinson disease. Hum. Mutat. 2007;28:641–653. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- 7.Moore D.J. Parkin: a multifaceted ubiquitin ligase. Biochem. Soc. Trans. 2006;34:749–753. doi: 10.1042/BST0340749. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg M.S., Fleming S.M., Palacino J.J., Cepeda C., Lam H.A., Bhatnagar A., Meloni E.G., Wu N., Ackerson L.C., Klapstein G.J., et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 9.Ko H.S., von Coelln R., Sriram S.R., Kim S.W., Chung K.K.K., Pletnikova O., Troncoso J., Johnson B., Saffary R., Goh E.L., et al. Accumulation of the authentic Parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J. Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Periquet M., Corti O., Jacquier S., Brice A. Proteomic analysis of parkin knockout mice: alterations in energy metabolism, protein handling and synaptic function. J. Neurochem. 2005;95:1259–1276. doi: 10.1111/j.1471-4159.2005.03442.x. [DOI] [PubMed] [Google Scholar]
- 11.Fallon L., Belanger C.M.L., Corera A.T., Kontogiannea M., Regan-Klapisz E., Moreau F., Voortman J., Haber M., Rouleau G., Thorarinsdottir T., et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nat. Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- 12.Henn I.H., Bouman L., Schlehe J.S., Schlierf A., Schramm J.E., Wegener E., Nakaso K., Culmsee C., Berninger B., Krappmann D., et al. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J. Neurosci. 2007;27:1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olzmann J.A., Li L., Chudaev M.V., Chen J., Perez F.A., Palmiter R.D., Chin S. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandhi S., Muqit M.M., Stanyer L., Healy D.G., Abou-Sleiman P.M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A.J., et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 15.Hoepken H.-H., Gispert S., Morales B., Wingerter O., Del Turco D., Musch A., Nussbaum R.L., Muler K., Droe S., Brandt U., et al. Mitochondrial dysfunction, peroxidation damage and changes in glutathione metabolism in PARK6. Neurobio. Dis. 2007;25:401–411. doi: 10.1016/j.nbd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D.W., Petsko G.A., Cookson M.R. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl Acad. Sci. USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E.M., Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 18.Sim C.H., Lio D.S., Mok S.S., Masters C.L., Hill A.F., Culvenor J.G., Cheng H.C. C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum. Mol. Genet. 2006;15:3251–3262. doi: 10.1093/hmg/ddl398. [DOI] [PubMed] [Google Scholar]
- 19.Pridgeon J.W., Olzmann J.A., Chin L.S., Li L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petit A., Kawarai T., Paitel E., Sanjo N., Maj M., Scheid M., Chen F., Gu Y., Hasegawa H., Salehi-Rad S., et al. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J. Biol. Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 21.Muqit M.M., Abou-Sleiman P.M., Saurin A.T., Harvey K., Gandhi S., Deas E., Eaton S., Payne Smith M.D., Venner K., Matilla A., et al. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J. Neurochem. 2006;98:156–169. doi: 10.1111/j.1471-4159.2006.03845.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E.A., Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl Acad. Sci. USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weihofen A., Ostaszewski B., Minami Y., Selkoe D.J. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum. Mol. Genet. 2008;17:602–616. doi: 10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- 24.Haque M.E., Thomas K.J., D'Souza C., Callaghan S., Kitada T., Slack R.S., Fraser P., Cookson M.R., Tandon A., Park D.S. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc. Natl Acad. Sci. USA. 2008;8:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greene J.C., Whitworth A.J., Kuo I., Andrews L.A., Feany M.B., Pallanck L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pesah Y., Pham T., Burgess H., Middlebrooks B., Verstreken P., Zhou Y., Harding M., Bellen H., Mardon G. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131:2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 27.Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 28.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.-M., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 29.Wang D., Qian L., Xiong H., Liu J., Neckameyer W.S., Oldham S., Xia K., Wang J., Bodmer R., Zhang Z. Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila. Proc. Natl Acad. Sci. USA. 2006;103:13520–13525. doi: 10.1073/pnas.0604661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.-W., Yang L., Beal M.F., Vogel H., Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl Acad. Sci. USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Exner N., Treske B., Paquet D., Holmstrom K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H.-H., Gasser T., et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Um J.W., Stichel-Gunkel C., Lubert H., Lee G., Chung K.C. Molecular interaction between parkin and PINK1 in mammalian neuronal cells. Mol. Cell. Neurosci. 2009;40:421–432. doi: 10.1016/j.mcn.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 33.Shiba K., Arai T., Sato S., Kubo S.-i., Ohba Y., Mizuno Y., Hattori N. Parkin stabilizes PINK1 through direct interaction. Biochem. Biophy. Res. Commun. 2009;383:331–335. doi: 10.1016/j.bbrc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C., Xia K., Wei J., Ze'ev R., Zhuang X., et al. Parkin, PINK1 and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y., Park J., Kim S., Song S., Kwon S.-K., Lee S.-H., Kitada T., Kim J.-M., Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophy. Res. Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Gao J., Chung K.K.K., Huang H., Dawson V.L., Dawson T.M. Parkin functions as an E2-dependent ubiquitin-protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl Acad. Sci. USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sriram S.R., Li X., Ko H.S., Chung K.K.K., Wong E., Lim K.L., Dawson V.L., Dawson T.M. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum. Mol. Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- 38.Doss-Pepe E.W., Chen L., Madura K. Alpha-synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J. Biol. Chem. 2005;280:16619–16624. doi: 10.1074/jbc.M413591200. [DOI] [PubMed] [Google Scholar]
- 39.Krappmann D., Scheidereit C. A pervasive role of ubiquitin conjugation in activation and termination of IkappaB kinase pathways. EMBO Rep. 2005;6:321–326. doi: 10.1038/sj.embor.7400380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattson M.P., Meffert M.K. Roles for NF-κB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell. Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welchman R.L., Gordon C., Mayer R.J. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 43.Pickart C.M., Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opi. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Haglund K., Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson T.M. Parkin and defective ubiquitination in Parkinson's disease. J. Neural. Transm. Suppl. 2006;70:209–213. doi: 10.1007/978-3-211-45295-0_32. [DOI] [PubMed] [Google Scholar]
- 46.Olzmann J.A., Brown K., Wilkinson K.D., Rees H.D., Huai Q., Ke H., Levey A.I., Li L., Chin S. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 2004;279:8506–8515. doi: 10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- 47.Deng H., Jankovic J., Guo Y., Xie W., Le W. Small interfering RNA targeting the PINK1 induces apoptosis in dopaminergic cells SH-SY5Y. Biochem. Biophys. Res. Commun. 2005;337:1133–1138. doi: 10.1016/j.bbrc.2005.09.178. [DOI] [PubMed] [Google Scholar]
- 48.Kim B.Y., Olzmann J.A., Barsh G.S., Chin L.-S., Li L. Spongiform neurodegeneration-associated E3 ligase mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol. Biol. Cell. 2007;18:1129–1142. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wheeler T.C., Chin L.-S., Li Y., Roudabush F.L., Li L. Regulation of synaptophysin degradation by mammalian homologues of seven in absentia. J. Biol. Chem. 2002;277:10273–10282. doi: 10.1074/jbc.M107857200. [DOI] [PubMed] [Google Scholar]
- 50.Lee J.T., Wheeler T.C., Li L., Chin S. Ubiquitination of α-synuclein by Siah-1 promotes α-synuclein aggregation and apoptotic cell death. Hum. Mol. Genet. 2007;17:906–917. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 51.Chin L.-S., Fu Q., Kachinsky A.M., Jabren G., Niu Y., Li L. Neuron-specific and developmental regulation of the synapsin II gene expression in transgenic mice. Mol. Brain Res. 1999;67:239–246. doi: 10.1016/s0169-328x(99)00066-2. [DOI] [PubMed] [Google Scholar]