Abstract
Mutations in the mitochondrial DNA (mtDNA) encoded subunit 6 of ATPase (ATP6) are associated with variable disease expression, ranging from adult onset neuropathy, ataxia and retinitis pigmentosa (NARP) to fatal childhood maternally inherited Leigh's syndrome (MILS). Phenotypical variations have largely been attributed to mtDNA heteroplasmy. However, there is often a discrepancy between the levels of mutant mtDNA and disease severity. Therefore, the correlation among genetic defect, bioenergetic impairment and clinical outcome in NARP/MILS remains to be elucidated. We investigated the bioenergetics of cybrids from five patients carrying different ATP6 mutations: three harboring the T8993G, one with the T8993C and one with the T9176G mutation. The bioenergetic defects varied dramatically, not only among different ATP6 mutants, but also among lines carrying the same T8993G mutation. Mutants with the most severe ATP synthesis impairment showed defective respiration and disassembly of respiratory chain complexes. This indicates that respiratory chain defects modulate the bioenergetic impairment in NARP/MILS cells. Sequencing of the entire mtDNA from the different mutant cell lines identified variations in structural genes, resulting in amino acid changes that destabilize the respiratory chain. Taken together, these results indicate that the mtDNA background plays an important role in modulating the biochemical defects and clinical outcome in NARP/MILS.
INTRODUCTION
The F1F0-ATPase (ATP synthase or complex V) catalyzes the final step of oxidative phosphorylation (OXPHOS) by coupling proton translocation from the mitochondrial intermembrane space to the matrix to the synthesis of ATP. The mammalian F1F0-ATPase consists of the F1 soluble portion, where the sites catalyzing ATP synthesis are located, and the F0 portion embedded in the mitochondrial inner membrane, which functions as a proton channel. The passage of protons in the F0, at the interface between the ATP6 subunit and the c-ring, is responsible for conformational modifications transmitted to the F1 portion through the rotation of the stalk, providing energy for ATP synthesis (1–3).
In humans, mutations in the mitochondrial DNA (mtDNA) encoded ATP6 subunit cause complex disorders with heterogeneous expression and severity, ranging from adult onset neurogenic muscle weakness, ataxia and retinitis pigmentosa (NARP) to a fatal infantile subacute necrotizing encephalomyelopathy, maternally inherited form of Leigh syndrome (MILS).
The first ATP6 mutation reported, a T8993G resulting in the substitution of a highly conserved leucine to arginine (L156R) (4), is the most frequent mutation associated with NARP/MILS. The T8993C and T9176G mutations, which replace conserved leucines of ATP6 with proline (L156P) and arginine (L217R), respectively, have also been described in NARP/MILS pedigrees (5,6).
Since MTATP6 mutations are generally heteroplasmic (i.e. a mixture of mutant and wild type mtDNA) with uniform tissue distribution and lack age-related variation (7), disease severity and age of onset have been correlated with the proportion of mtDNA mutation in blood (8). However, the genotype/phenotype correlation, emphasized in previous reports, does not apply to all pedigrees. In T8993G mutants, both NARP and MILS can be found within the same family (9,10). Moreover, in the same families, oligosymptomatic children share the same mutation load of symptomatic siblings (11), and in some cases high T8993G mutation loads are not associated with symptoms of Leigh or NARP (12). Overall, the T8993C mutation is associated with a milder phenotype than the T8993G, often with late onset and slow progression (13), and the mutation load required to trigger neurological symptoms is very high (8).
The mechanisms whereby ATP6 mutations cause ATP synthesis defects remain to be fully elucidated. Mutant ATP6 subunit could impede proton translocation in the F0 and prevent the c-ring rotation by modifying charge distribution in the proton channel (14). Alternatively, the mutations may cause structural changes at the interface with the c-ring, resulting in inefficient coupling between proton transport and ATP synthesis (15). Both hypotheses are supported by findings of increased membrane potential (Δψ) and matrix pH in T8993G mutant cybrids (16) and lymphocytes (17). It is unclear whether the mutations cause ATP6 misfolding and hinder the assembly of complex V. Detached F1 sub-complexes were detected by Blue native gel electrophoresis (BN-PAGE) in post-mortem tissue from MILS patients (18), in NARP/MILS cybrids (19,20) and skeletal-muscle (21); on the other hand, ATPase disassembly was not found in different subsets of cybrids (22) and patients’ fibroblasts (23). Finally, ATP synthesis impairment cannot be the only pathogenic mechanism in ATP6 mutants, since the T8993C mutation does not result in overt ATP synthesis defects (24–26).
Taken together, epidemiological and biochemical evidence show that significant differences exist in the penetrance of ATP6 mutations, even among individuals harboring similar proportions of the same mutation. We hypothesized that these differences could be the consequence of modifying genetic factors, which modulate the disease phenotype.
Here, we have characterized the genetic, bioenergetic and molecular properties of cells derived from NARP/MILS patients. Our results demonstrate that mtDNA variations can explain the phenotypic differences among individuals harboring ATP6 mutations.
RESULTS
Bioenergetic differences among ATP6 mutants
The cybrid system is a well-established cell culture model, where patients’ mtDNA is transferred to mtDNA-less human osteosarcoma cells to study the effects of mtDNA mutations on mitochondrial function independent of the nuclear background. In this way, cybrid cell lines harboring homoplasmic (i.e. 100%) mutant or wild-type (WT) mtDNA can be obtained from the same heteroplasmic individual.
Homoplasmic cybrids were generated from five patients harboring mutations in the ATP6 gene: three with the T8993G mutation (JC, AT, TU), one with the T9176G mutation (LR) and one with the T8993C mutation (DM).
Parental osteosarcoma cells (line 143B) were used as a normal reference in this study. To ensure that 143B cells are a representative control for bioenergetic assays, we compared them to cybrid lines from subjects with no history of mitochondrial diseases. We found variability among these control cybrids, but no statistically significant differences with 143B cells were detected in ATP synthesis, respiration, respiratory chain activities (Supplementary Material, Table S1) and assembly, assessed by BN-PAGE (data not shown).
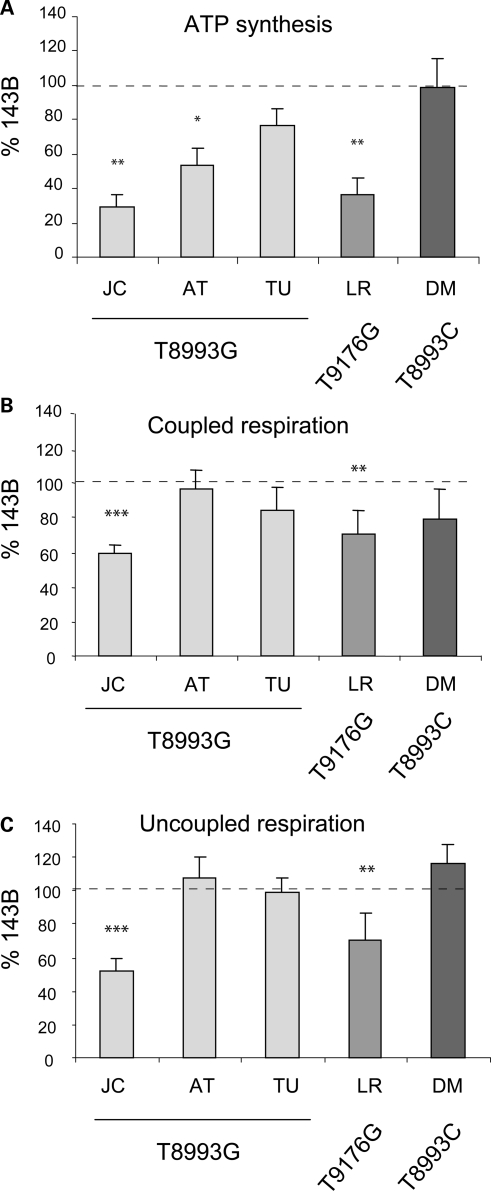
Mitochondrial ATP synthesis was measured in digitonin permeabilized ATP6 mutant cybrids using pyruvate and malate as substrates (27). For each cybrid line, digitonin was titrated to obtain the maximal ATP synthesis rate. Despite all being homoplasmic for the ATP6 mutations, the various cybrids had variable degrees of mitochondrial ATP synthesis defects (Fig. 1A). Surprisingly, cell lines containing the same T8993G homoplasmic mutation displayed widely different ATP synthesis defects relative to 143B, ranging from the most affected JC (29% residual activity) to the least affected TU (77% residual activity). ATP synthesis was severely impaired in the T9176G mutant LR (37% residual activity), but normal in the T8993C mutant DM (98% residual activity).
Figure 1.
Bioenergetic variations among the different homoplasmic ATP6 mutant cell lines. (A) ATP synthesis activities using pyruvate and malate as substrates. (B and C) Measurements of oxygen consumption in intact cells using pyruvate as substrate in the absence (coupled respiration) or in the presence of FCCP (uncoupled respiration). The values are average of at least three independent measurements and are expressed as a percentage of 143B cells. Statistically significant differences between 143B and mutant cell lines are indicated: *P < 0.05, **P < 0.005, *** < 0.0005 versus 143B.
Since ATPase activity is coupled to the mitochondrial electron transfer and proton translocation, it directly reflects the efficiency of the respiratory chain (RC). Thus, variations in ATP synthesis may depend upon differences in RC function upstream of the ATPase. Mitochondrial respiration measured in intact cells using pyruvate as substrate correlated with ATP synthesis: cell lines with severe ATP synthesis impairment (JC and LR) showed significantly decreased mitochondrial respiration (Fig. 1B). Uncoupled respiration (with 1 µM FCCP, Fig. 1C), an index of maximum respiratory capacity independent of ATPase activity, confirmed the RC electron transfer impairment. This data suggest that in the JC and LR lines, RC defects contribute to the severe ATP synthesis impairment.
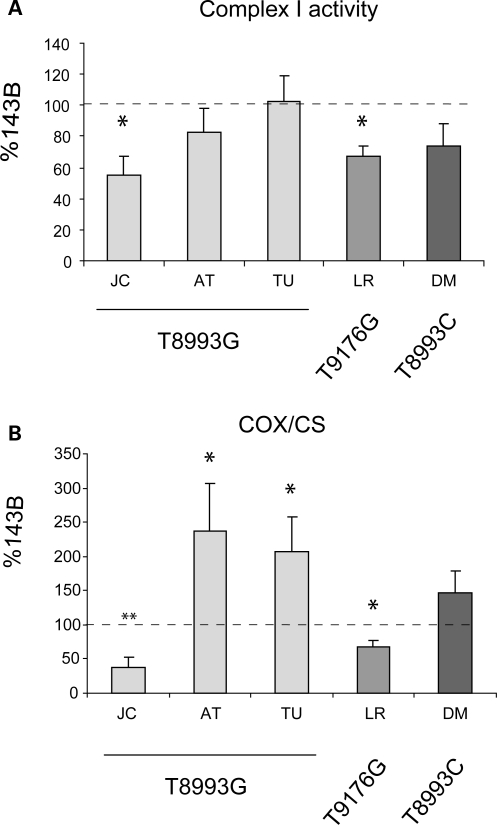
To determine which RC complexes are defective in JC and LR cells, we measured specific enzymatic activities. Complexes I (Fig. 2A) and IV (Fig. 2B) were significantly reduced in JC and LR, indicating that specific RC defects can worsen ATP synthesis defects in ATP6 mutant cells. Interestingly, AT and TU cells showed an increase in complex IV activity relative to 143B. Although this up-regulation may be interpreted as a compensatory mechanism, it did not increase respiration (Fig. 1B and C), suggesting that the other RC complexes are rate limiting for respiration in these cells.
Figure 2.
RC enzymatic activities. Complex I activity in isolated mitochondria (A) and complex IV (COX) activity normalized by citrate synthase (CS) activity in cell lysates (B). The values are average of at least three independent measurements and are expressed as a percentage of 143B cells. *P < 0.05, **P < 0.005, ***P < 0.0005 versus 143B.
OXPHOS complexes assembly is disrupted in RC defective ATP6 mutants
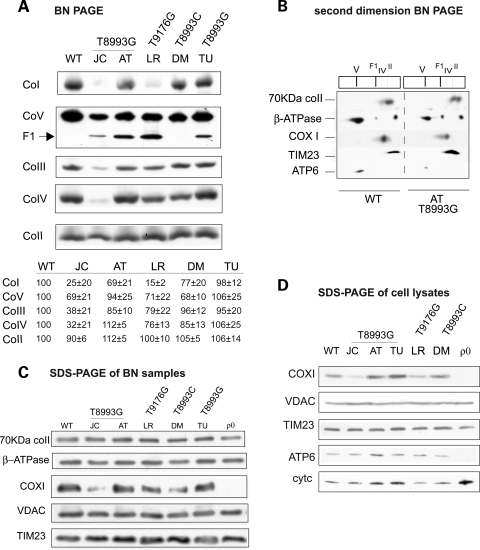
We investigated by BN-PAGE whether the decrease in OXPHOS function correlated with a decrease in the amounts of assembled complexes. JC and LR cells showed a loss of complexes I and IV (Fig. 3A). Complex III was also reduced in JC cells, but not in LR cells. BN-PAGE of the other mutant cell lines, which had normal respiration (AT, TU and DM), displayed some inter-experimental variability in RC assembly. AT had a trend for modest complex I and III defects (on average approximately 30 and 15%, respectively, n = 3), while DM had modest complex I and IV defects (on average approximately 23 and 15%, respectively, n = 3).
Figure 3.
RC complexes assembly and mitochondrial protein levels. (A) BN-PAGE of RC complexes. CoI, complex I; CoII, complex II; CoIII, dimer of complex III; CoIV, complex IV; CoV, complex V; F1, F1 portion of complex V. The amount of RC complexes expressed as percentage of WT, estimated by densitometry of western blot bands from at least three independent experiments, are indicated below each lane. (B) RC complexes subunits from 143B cell line (left) and AT mutant cybrid (right) resolved by first dimension BN-PAGE (as in A), followed by separation by second dimension denaturing SDS–PAGE probed with antibodies against the 70 kDa complex II subunit, β subunit of ATPase, subunit I of COX (COX I), the inner membrane translocator TIM23 and subunit 6 of ATPase (ATP6). (C) Western blot of mitochondrial proteins solubilized as in (A) resolved by denaturing SDS–PAGE, blotted and detected with specific antibodies. VDAC is the voltage-dependent anion channel (porin). (D) Western blot of whole cell lysates separated by denaturing SDS–PAGE, blotted, and detected with specific antibodies. Cytc, cytochrome c.
The total amount of fully assembled complex V, detected either using an antibody against β-ATPase (Fig. 3A) or against ATP6 (Supplementary Material, Fig. S1) varied slightly among cell lines, and the results were similar with both antibodies. However, the differences in complex V assembly could not account for the variations in ATP synthesis, because LR and JC, which had severely reduced ATP synthesis, showed similar amounts of assembled complex V as DM, which had normal ATP synthesis (Fig. 1A).
All cybrids carrying the T8993G and the T9176G mutations (JC, AT, TU, LR), but not the T8993C mutation (DM), had detectable levels of complex V sub-complexes (F1). The F1 sub-complexes lacked ATP6, as shown by a second dimension BN-PAGE (Fig. 3B). To exclude that these sub-complexes derive from degradation, because of increased sensitivity of mutant complex V to protein solubilization, we decreased the amount of detergents to prevent degradation (Supplementary Material, Fig. S2A and B), or increased it to favor the accumulation of the F1 sub-complex (Supplementary Material, Fig. S2C). Detergent modification did not produce changes, suggesting that the F1 sub-complexes were not artifacts of solubilization. Interestingly, the formation of the F1 sub-complex was not specific to ATP6 mutants, because it occurred in a detergent-independent manner also in cybrids carrying mutations in MTCYTB (28) and in MTCOX1 (29) (Supplementary Material, Fig. S2D and E), resulting in complexes III and IV depletion, respectively.
To prove that comparable amounts of mitochondrial protein had been loaded in the BN-PAGE, samples were also separated by denaturing SDS–PAGE (Fig. 3C). No differences were found in the content of several mitochondrial membrane proteins (subunit 70 kDa of complex II, VDAC and TIM23) among the various cell lines (Fig. 3C). Furthermore, the amount of subunit β of complex V was unchanged in all cell lines. Western blots of total cells lysates under denaturing conditions showed that the steady-state levels of ATP6 (Fig. 3D) were similar in the various mutant cell lines and 143B control. COXI was decreased in JC and LR cells, whereas AT and TU cells showed an up-regulation of COXI and cytochrome c, in agreement with increased complex IV activity (Fig. 2B).
RC defects are transferable through re-cybridization of ATP6 mutants
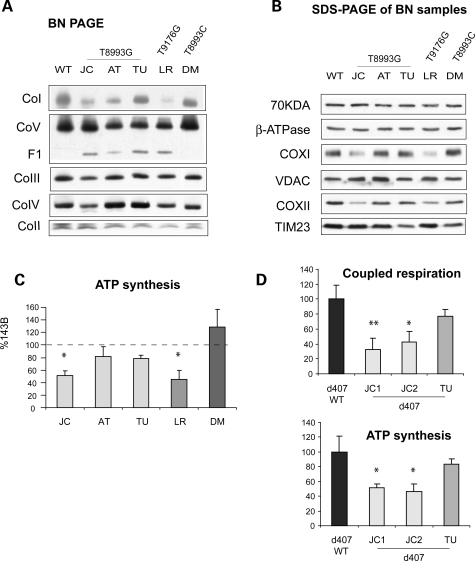
One caveat in the interpretation of results obtained from individual clones of cybrids is related to potential differences in nuclear gene expression due to chromosomal aneuplody. Therefore, the bioenergenic variations among T8993G mutants were confirmed in additional JC and TU homoplasmic cybrid clones (data not shown). Most importantly, we performed a second round of cybridization, where mtDNAs from enucleated cybrids were transferred into osteosarcoma ρ0 cells. The homoplasmic ATP6 mutations were confirmed by PCR-RFLP analysis (data not shown). Mass cultures from the second cybridization confirmed the biochemical differences among ATP6 mutants. The assembly of complexes I and IV were decreased in JC and LR cell lines; while complex I was moderately decreased in AT (Fig. 4A). F1 sub-complexes were detected in all mutants except DM (Fig. 4A). SDS–PAGE confirmed a decrease of COXI and COXII subunits of complex IV in JC and LR cell lines, with unchanged amounts of subunit 70 kDa of complex II, VDAC or TIM23 (Fig. 4B). ATP synthesis assays confirmed the defects observed in the original clones, where JC and LR were the most affected ones (Fig. 4C). To exclude the possibility that the observed biochemical differences among mutants were cell type specific, we transferred JC and TU mtDNAs in immortalized ρ0 retinal pigmentary epithelium cells (RPE d407) (30). The homoplasmic T8993G mtDNA genotype was confirmed in the RPE cybrids (data not shown). Two independent JC RPE clones showed decreased mitochondrial respiration and ATP synthesis when compared with parental RPE cells, whereas TU RPE cells had almost normal ATP synthesis and respiration (Fig. 4D).
Figure 4.
Bioenergetic variations were preserved after ‘re-cybridization’. (A) BN-PAGE of RC complexes from cybrids obtained by ‘re-cybridization’. (B) Western blot of mitochondrial proteins from cybrids obtained by ‘re-cybridization’ solubilized as in (A), resolved by denaturing SDS–PAGE, and detected with specific antibodies. (C) ATP synthesis in cybrids obtained by ‘re-cybridization’ expressed as a percentage of 143B cells. (D) Coupled respiration (top) and ATP synthesis (bottom) of ATP6 mutant cybrids obtained by ‘re-cybridization’ in a d407 retinal pigmentary epithelium background expressed as a percentage of d407 wild-type control cells. In (C) and (D), values are average of at least three independent measurements. *P < 0.05, **P < 0.005 versus wild-type cells.
Taken together, these experiments demonstrated that the RC defects observed in a subset of ATP6 mutants were attributable exclusively to mtDNA.
MtDNA content is not responsible for RC defects in ATP6 mutants
To exclude that RC impairement in JC and LR cells resulted from mtDNA depletion incurred during cybridization, we measured mtDNA content in all cybrids cell lines. Real-time PCR analysis, using the MTCOX1 gene and the nuclear 18S as a reference, revealed no significant differences among JC, TU and LR mutant cybrids, WT cybrids and parental 143B cells (Table 1). AT and DM cybrids had significantly lower mtDNA content, which however did not correlate with the severity of the RC defects, since these lines had normal respiration and relatively modest changes in complexes assembly (Figs 1 and 3). In addition, the re-cybridized mutant cells did not show significant differences in mtDNA content when compared with WT cells (Supplementary Material, Table S2). Therefore, the RC defect observed in a subset of ATP6 mutants was not dependent on quantitative changes in mtDNA.
Table 1.
mtDNA content in cybrid and 143B cell lines expressed as mtDNA/nDNA
| COX1/18S | |
|---|---|
| 143B TK-206 | 1.13 ± 0.20 |
| WT | 1.01 ± 0.05 |
| JC (T8993G) | 1.03 ± 0.15 |
| AT (T8993G) | 0.52 ± 0.04 |
| TU (T8993G) | 1.14 ± 0.34 |
| LR (T9176G) | 1.01 ± 0.20 |
| DM (T8993C) | 0.70 ± 0.14 |
The mtDNA background determines the RC defects in ATP6 mutants
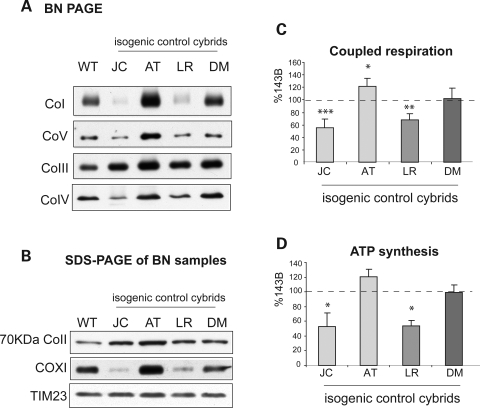
To test the hypothesis that the mtDNA that contains the ATP6 mutation modulates the RC phenotype, we studied isogenic homoplasmic control cybrids (i.e. cybrids from the same individual) devoid of ATP6 mutations from JC, AT, LR and DM. The lack of ATP6 mutations was confirmed by PCR-RFLP analysis (data not shown). Biochemical and structural studies reveled that JC and LR isogenic control cybrids had reduced amounts of assembled complexes I and IV (Fig. 5A), reduced levels of COXI (Fig. 5B) and decreased respiration (Fig. 5C), when compared with 143B cells. The isogenic controls did not show significant differences in mtDNA content when compared with WT cells (Supplementary Material, Table S2). These results indicate that JC and LR mtDNAs caused RC defects, independent of the ATP6 mutation. The ATP synthesis impairment in JC and LR isogenic controls (Fig. 5D) was milder (53 and 54% of 143B, respectively) than in the JC and LR ATP6 mutants (29 and 37% of 143B, respectively, Fig. 1A). Therefore, the ATP6 mutations and the RC defects associated with the mtDNA background act synergistically to impair OXPHOS.
Figure 5.
Bioenergetic variations in isogenic cybrids. (A) BN-PAGE of RC complexes from isogenic cybrids. (B) Western blot of mitochondrial proteins from isogenic control cybrid solubilized as in (A), resolved by denaturing SDS–PAGE and detected with specific antibodies. (C) ATP synthesis and (D) coupled respiration of isogenic cybrids. The values are average of at least three independent measurements and are expressed as percentage of 143B. *P < 0.05, **P < 0.005, ***P < 0.0005 versus 143B.
Taken together, these observations suggest that the mtDNA background can explain the bioenergetic variations observed, not only among different ATP6 mutants, but also among cybrids harboring the same T8993G mutation.
RC defects are associated with mtDNA sequence variations in ATP6 mutants
To investigate potential sequence variations that may affect mtDNA-encoded respiratory chain components and cause RC dysfunction, we sequenced the entire mtDNA of the ATP6 mutants. We identified numerous variations relative to the revised Cambridge reference sequence (31) (Table 2). Some of these variations were associated with specific mtDNA haplogroups, as determined by the phylogenetic tree (Supplementary Material, Fig. S3) (32–34). Patients JC, AT and DM belonged to different subgroups within haplogroup U, and TU belonged to haplogroup H, all descendents of the European lineage. Patient LR belonged to the N1b2 haplogroup, a West Asia lineage often associated with Ashkenazi population (35).
Table 2.
mtDNA haplogroup associations and non-synonymous nucleotide changes relative to the revised Cambridge Sequence
| Cybrid cell line | GenBank ID | NARP mutation | Haplogroup | Non-synonymous base change | Amino acid change |
|---|---|---|---|---|---|
| JC | GQ891609 | T8993G | U5b | G6300A (COX1)a | Ala > Thr |
| C12092T (ND4) | Leu > Phe | ||||
| G13630A (ND5)a | Thr > Ala | ||||
| A13637G (ND5) | Gln > Arg | ||||
| A15326G (CYTB) | Thr > Ala | ||||
| T15372C (CYTB)a | Leu > Pro | ||||
| G15497A (CYTB) | Gly > Ser | ||||
| G15803A (CYTB) | Val > Met | ||||
| AT | GQ891610 | T8993G | U1 | G6081A (COX1) | Ala > Thr |
| A11928G (ND4) | Asn > Ser | ||||
| A15326G (CYTB) | Thr > Ala | ||||
| TU | GQ891611 | T8993G | H13 | A15218G (CYTB) | Thr > Ala |
| LR | GQ891612 | T9176G | N1b2 | C4735A (ND2) | Thr > Asn |
| A4917G (ND2) | Asn > Asp | ||||
| C4960T (ND2) | Ala > Val | ||||
| C8472T (ATP8) | Pro > Leu | ||||
| C12092T (ND4) | Leu > Phe | ||||
| A15326G (CYTB) | Thr > Ala | ||||
| DM | GQ891613 | T8993C | U2 | A15326G (CYTB) | Thr > Ala |
| C14766T (CYTB) | Thr > Ile |
In bold are substitutions of highly conserved amino acids in conserved stretches of proteins.
aSubstitutions never reported before and confirmed by RFLP analysis.
In JC mtDNA, the ND5/13637 has been previously reported as a secondary mutation, exacerbating Leber's hereditary optic atrophy (LHON) (36,37). The CYTB/15497 results in the substitution of an evolutionary conserved glycine to serine (G251S), and has been associated with obesity and exercise intolerance (38). Interestingly, hystocitoid cardiomyopathy has been reported in association with a G251D substitution (CYTB/G15498A mutation) (39). The ND4/12092 variation substitutes a conserved leucine with phenylalanine (L445F) (Supplementary Material, Fig. S4). The COX1/6300, ND5/13630 and CYTB/15372 have not been reported earlier. Protein alignments analysis (Supplementary Material, Fig. S4) showed that the COX1/6300 is located in a conserved region of the protein and results in the substitution of a highly conserved alanine with threonine (A133T). Although the other two mutations are located in conserved stretches of the proteins, the mutated amino acids are not conserved in different species (Supplementary Material, Fig. S4).
In AT mtDNA, the COX1/6081 alanine to threonine substitution (A60T) has been associated with prostate cancer (40). The ND4/11928 is in a highly conserved protein region, but a serine is present instead of an asparagine at position 390 (N390S) in some species (Supplementary Material, Fig. S4).
In LR mtDNA, several variations were found in ND2 and ND4 subunits, mostly polymorphisms specific to the rare N1b2 haplogroup (35). The ND2/4735 (T89N) is not in an evolutionary conserved region. The ND2/4917 is in a highly conserved stretch of the protein and the asparagine substituted with aspartic acid (N150D) is an evolutionary conserved amino acid (41) (Supplementary Material, Fig. S4). ND2/4917 is also a haplogroup T marker. This variation has been proposed to act synergistically with the ND4/11778 LHON mutation, and increase the probability of optic atrophy (36). The ND4/12092 variation substitutes a conserved leucine with phenylalanine (L445F) (Supplementary Material, Fig. S4).
Abnormal assembly kinetics of RC complexes in ATP6 mutant cells
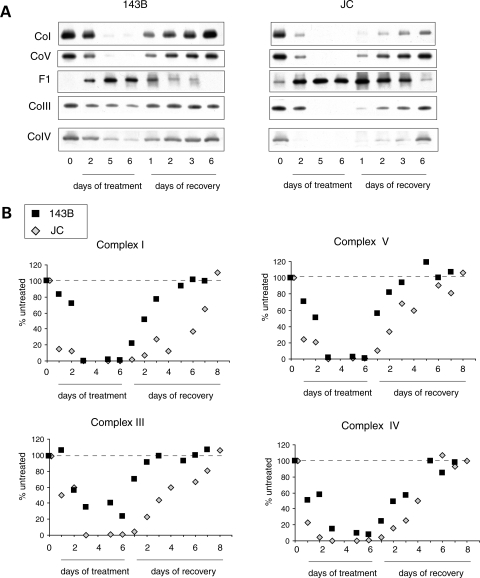
In order to study the kinetics of respiratory chain complexes assembly, we depleted the cells of mtDNA-encoded subunits by reversibly blocking mitochondrial translation with doxycycline (42). After 6 days in doxycycline, the inhibitor was removed to restore mitochondrial protein translation. Samples were collected at different time points during and after doxycycline treatment to follow complexes turnover and reconstitution rates. At each time point, complexes were quantified relative to untreated cells (day 0). Six days of doxycycline completely depleted all complexes in JC cells, whereas in 143B cells there were variable proportions of residual assembled complexes III and IV (Fig. 6A and B). The turnover rates of complexes I, III and IV were faster and the reconstitution rates slower in JC cells when compared with 143B controls. The results suggest an overall instability of the respiratory chain in JC mutant cells.
Figure 6.
Assembly kinetics of RC complexes. (A) BN-PAGE of 143B (left) and JC (right) cells grown for 6 days in the presence of doxycicline followed by 6 days without the drug. Mitochondrial RC complexes were studied at different time points during the treatment and the recovery. Note that the JC blots are subjected to a longer exposure than 143B ones in order to obtain similar band intensities in the two lines. (B) Quantification of complexes I, III, IV and V band intensities in doxycline treated cells expressed as a percentage of the respective complexes from untreated cells (day 0). Each value is the average of two independent experiments.
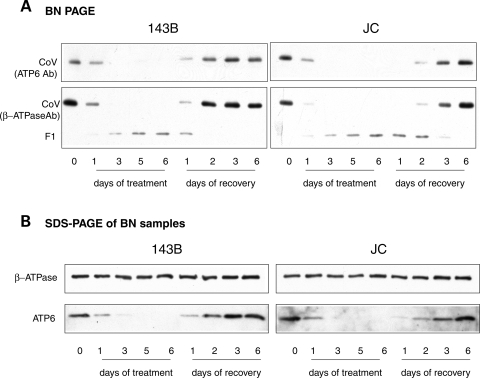
Complex V progressively decreased during doxycycline treatment, while the amount of F1 sub-complex increased (Fig. 6A and B). During the recovery phase, complex V increased, while the F1 decreased, until only traces of F1 remained in JC cells. The experiment repeated using antibodies against ATP6 reproduced the same pattern of complex V assembly kinetics (Fig. 7A). However, a denaturing western blot of the samples, which detects the total amount of proteins (i.e. assembled in complex V plus unassembled), showed a delay in the recovery of ATP6 levels in JC cells when compared with WT (Fig. 7B). These results clearly demonstrate that the F1 sub-complex is the result of incomplete assembly rather than the effect of complex V degradation. The availability of the mtDNA-encoded ATP6 subunit appears to be a limiting factor for complex V assembly. Since no differences were found in the steady state levels of ATP6 subunit between ATP6 mutant and WT cells (Fig. 3D), it is likely that ATP6 mutations delay the assembly of F1 with F0 by slowing ATP6 synthesis or increasing ATP6 instability and degradation.
Figure 7.
Assembly kinetics of complex V. (A) BN-PAGE of complex V from 143B (left) and JC (right) cells grown and treated as in Figure 6. Samples were separated in parallel on two different gels, blotted and probed with antibodies against ATP6 subunit (top panels) and β-ATPase subunit (bottom panels). (B) Western blot of mitochondrial proteins solubilized as in (A), resolved by denaturing SDS–PAGE and detected with specific antibodies.
Growth in galactose reveals a correlation between RC impairment and bioenergetic defects in ATP6 mutants
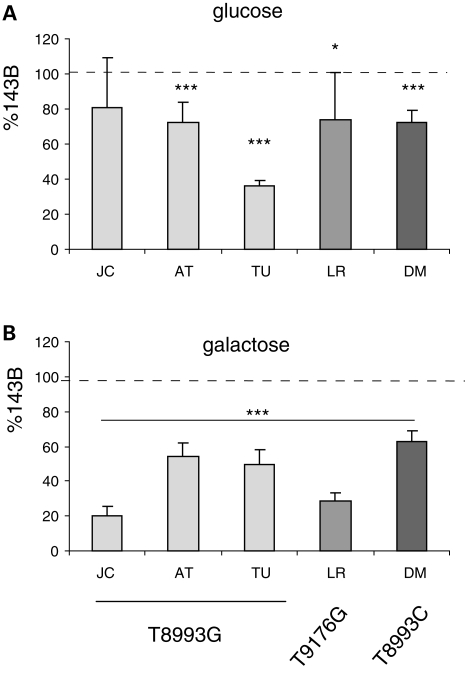
We investigated the physiological significance of the bioenergetic differences in the ATP6 mutants by growth in medium containing either glucose or galactose as energetic substrates. Cell replication after 3 days in glucose medium was only decreased by 20–30% in JC, AT, LR and DM cells (Fig. 8A). Surprisingly, TU showed a delayed growth in glucose medium, for which we do not have an explanation at the moment. In galactose medium, where cells have to rely primarily on OXPHOS for ATP production (43), the survival of JC and LR was severely compromised after 3 days (20 and 28% of WT, respectively, Fig. 8B). Importantly, AT, TU, DM cell lines, with milder ATP synthesis and respiration defects, showed a much less severe cell growth defect (50–60% of control, Fig. 8B).
Figure 8.
Cell replication in glucose and galactose medium. Equal numbers of cells were plated and the total cell count was obtained after 3 days of growth either in glucose (A) or galactose (B) medium. Cell growth is expressed as percentage of 143B. *P < 0.05, ***P < 0.0005.
DISCUSSION
In a thorough study of the three ATP6 mutations most commonly associated with NARP/MILS, we found a wide variation of bioenergetic phenotypes, not only among different ATP6 mutants, but also among cell lines carrying the same mutation. Two cell lines, JC (T8993G) and LR (T9176G), were severely defective in ATP synthesis and mitochondrial respiration. We demonstrated compromised assembly and enzymatic activities of RC complexes I and IV, which are not directly affected by the ATP6 mutation. Decreased complex I, III and IV activities have also been reported in a subset of muscle biopsies from NARP/MILS subjects (20,44) with high proportions of the T8993G or the T8993C mutations, but not in other individuals with similar mutations loads, demonstrating a variable biochemical expression of the disease in patients.
MtDNA genetic analyses of our panel of ATP6 mutants revealed several point mutations resulting in amino acid substitutions in subunits of the RC complexes. Mutations involving evolutionary conserved amino acids were found in the most affected cell lines (Table 2), COX1/6300, ND4/12092 and CYTB/15497 in JC and ND2/4917 and ND4/12092 in LR. These mutations may directly affect the electron transfer efficiency or the assembly of RC complexes.
Detrimental effects on complex I have been previously proposed for the ND2/4735, ND2/4960, ND2/4917 and ND4/12092 variations (35), all of which were found in LR. The ND2/4917 is a polymorphism related to the T haplogroup, which is often associated as a secondary mutation with the ND4/11778 LHON mutation (36). Interestingly, the ND4/12092 substitution was common to JC and LR lines, which displayed the most severe RC defects among the ones tested, suggesting that this mutation may play a particularly detrimental role.
Further investigations will be required to determine if the other mutations of less conserved residues in JC and LR cells (Table 2) act synergistically with the substitutions of conserved residues. In addition, it cannot be excluded that polymorphisms in the D-loop control region or in non-protein coding genes, such rRNA and tRNAs, could contribute to the observed phenotypes, since polymorphic variations in these regions might affect protein translation efficiency and thus the expression levels of mitochondrial proteins.
Since no mtDNA variations were detected in complex IV, the cause of the partial complex IV defect in LR is unclear. In our previous studies, we showed that RC complexes I, III and IV are organized in functional supercomplexes and that a decrease of either complex III or IV result in a decrease of RC supercomplexes and of complex I assembly (45). Therefore, in LR, a decreased complex I may destabilize the supercomplex, and down-regulate complex IV.
Different groups have obtained conflicting results on the effects of mutant ATP6 on complex V assembly and stability. In some reports, the steady-state levels of mutant complex V were normal and no assembly defects were detected (22,23); while other groups showed defective complex V assembly (18,20,21). Various explanations have been proposed to account for such discrepancies, including different cell types, proportions of MTATP6 mutation, fresh versus postmortem tissue and methods of mitochondrial solubilization. In our panel of homoplasmic cell lines, we unequivocally demonstrated by complex V assembly kinetics that the F1 sub-complex is not a product of disassembly/degradation of complex V. We show that the F1 sub-complex is a stable assembly intermediate waiting to bind ATP6 subunit to complete complex V assembly.
The restrictive metabolic conditions in galactose medium revealed a growth defect in ATP6 mutant cybrids with otherwise normal respiration and ATP synthesis (AT, TU and DM). This suggests that OXPHOS measurements may underestimate bioenergetic defects. In biochemical assays, where the OXPHOS steps are extrapolated from the physiological context, without substrate limitations or product inhibition, high activity rates can be obtained, despite the presence of mutant ATP6. Instead, galactose medium mimics the metabolic challenges to which vulnerable organs are exposed in NARP/MILS patients and is a functional parameter indicative of total cell metabolism efficiency, in more physiological conditions.
Often, discrepancy between mutation loads and predicted clinical phenotypes (11,12), unexpected deterioration of symptoms leading to premature death (46,47), and inexplicable resolution of symptoms with favorable outcome (48), have been observed in NARP/MILS patients, suggesting that genetic modifiers are at play in determining the disease phenotype.
A clear example of interplay between primary pathogenic mtDNA mutations and mtDNA background is LHON. Haplogroup J has been associated with LHON (49,50), and the probability of visual loss is increased when the primary mutations 11778/ND4, 14484/ND6 and 3460/ND1 occur in the J2, J1 and K haplogroups, respectively (51,52). In a recent study, mtDNA haplotypes have been shown to play a role in the assembly kinetics of OXPHOS complexes in LHON cybrids (53), suggesting a mechanism whereby mtDNA variations can modulate the phenotype of LHON mutations.
Here, we showed that mtDNA background plays an important role in modulating the biochemical phenotype of NARP/MILS. We propose that mtDNA variations are responsible for relatively subtle biochemical defects, which do not normally pose a threat to the survival and propagation of their carriers. Perhaps, such variations may be the result of a climatic adaptation influenced by the geographic distributions of the different haplogroups (54). However, when a new pathogenic mutation, such as the NARP/MILS, is superimposed, the overall OXPHOS function becomes insufficient, leading to overt symptoms of mitochondrial disease.
In light of our observations, the ATP6 mutation load cannot be considered the only criterion for predicting the clinical and biochemical outcomes. A detailed correlation between mtDNA background and clinical presentation, associated with a more in depth analysis of global OXPHOS function and assembly, will help in providing appropriate genetic counseling and prenatal diagnosis for NARP/MILS.
MATERIALS AND METHODS
Reagents
All reagents used were from Sigma-Aldrich unless indicated otherwise.
Cell culture
Cybrids were generated by fusion of platelets or enucleated fibroblasts from NARP/MILS patients with human osteosarcoma 143B cell line lacking mtDNA (ρ0 cells) as described elsewhere (55). Cells were cultured in Dulbecco Modified Eagle's Medium (DMEM, Invitrogen) supplemented with 5% fetal bovine serum (FBS, Gemini Bio-Products) and 50 µg/ml uridine.
To block mitochondrial protein translation, 15 µg/ml doxycycline was added to the culture medium for 6 days. The cells were harvested at indicated time points during and after the treatment.
Growth rates in glucose or galactose were determined by seeding 0.1 × 106 cells in six-well plates in triplicates in DMEM containing 4.5 mg/ml glucose and 1 mm pyruvate supplemented with 5% FBS and 50 µg/ml uridine or in DMEM without glucose containing pyruvate plus 4.5 mg/ml galactose and supplemented with 5% dialyzed FBS and 50 µg/ml uridine. Cell counts were obtained after 3 days of culture.
MtDNA analyses
Total cell DNA was extracted by standard techniques and PCR/RFLP analysis was performed to detect the T8993G, T8993C and T9176G mutations as described elsewhere (6,56,57).
The entire mtDNA was PCR-amplified and sequenced as previously reported (58). The http://www.mitomap.org/ website was used to reference mtDNA mutations already reported in the literature. Evolutionary amino acid conservation among species was determined by alignment of patient's mtDNA encoded proteins with protein sequences obtained from http://www.ncbi.nlm.nih.gov/. The http://www.phylotree.org/ website was used for the phylogenetic tree editing.
Quantification of mtDNA relative to nuclear DNA (nDNA) was performed by real-time PCR in a LightCycler system (Roche), using COX1 and 18S rRNA primers, respectively, and the LightCycler FastStart DNA Master SYBR Green I (Roche) according to the manufacturer's instructions. Relative amounts of mtDNA and nDNA were calculated using linear amplification standard curves of serially diluted DNA samples.
Respiratory chain analyses
ATP synthesis was measured as in ref. (59) using 2 × 106 cells, 0.04 mg/ml digitonin and pyruvate and malate as substrates. Oxygen consumption was measured in intact cells using 1 mm pyruvate as substrate, with or without 1 µM of the uncoupler carbonyl cynide p-trifluoromethoxyphenylhydrazone (FCCP), and 2 mm KCN as the terminal inhibitor, in an Oxygraph chamber equipped with a Clark-type electrode (Hansatech) at 37°C, as described in ref. (60).
Complex IV and citrate synthase activities were measured in total cell lysates as described (61). Complex I activity was measured in isolated mitochondria. Briefly, 20 × 106 cells were washed in PBS, and homogenized in a buffer containing 225 mm Mannitol, 75 mm sucrose, 1 mm EGTA, 2 mg/ml BSA, 20 mm HEPES (pH 7.2). Homogenates were centrifuged at 600 g for 5 min at 4°C, and the resulting supernatant centrifuged at 12000 g for 10 min at 4°C. Mitochondrial pellets were disrupted by one cycle of freezing and thawing. Complex I activity was measured in 20 mm HEPES (pH 7.8) using 200 µg mitochondrial protein, 50 µM NADH and 40 µM Q1 in a Lambda 35 Spectrophotometer (PerkinElmer), following NADH oxidation at λ = 340. Complex I specific activity was assessed after subtraction of the residual activity insensitive to rotenone (2.5 µM).
Blue Native-PAGE and SDS–PAGE
Respiratory chain complexes assembly studies were performed by BN-PAGE as described (45). Samples were solubilized with 1.4 µg/µl digitonin for 10 min followed by 0.6% lauryl maltoside (n-dodecyl β-D-maltoside, LM). For immunodetection of protein complexes, monoclonal antibodies (Invitrogen) against the following subunits were used: 39 kDa of complex I, 70 kDa of complex II, core 2 of complex III, subunit I of complex IV and subunit β of complex V. For second dimension gel electrophoresis lanes excised from the first dimension BN-PAGE were first treated in denaturing conditions and then electrophoresed on a 10% tricine SDS–PAGE (45). For immunodetection of proteins in denaturing gels, monoclonal antibodies against the following subunits were used: 70 kDa of complex II, subunit I of complex IV, subunit β of complex V (all from Invitrogen), TIM23 (BD Biosciences), ATP6 (obtained from Dr Eric A. Schon, College of Physicians and Surgeons, Columbia University, New York), subunit II of complex IV (COXII, Invitrogen), VDAC (Invitrogen) and cytochrome c (BD Biosciences). Immunoreactive bands were visualized by horseradish peroxidase labeled secondary antibodies and SuperSignal West Pico Chemiluminescence substrate (Thermo Scientific). Quantification of respiratory chain complexes was performed by densitometric analyses of western blot digital images using the Scion2 image software (NIH).
Statistical analyses
In all of the assays, the values are averages of at least three independent measurements expressed as a percentage of WT cells. Error bars indicate SD. Statistically significant differences between WT and patient cell lines were estimated by unpaired two-tailed Student's t-test.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of interest statement. The authors have no conflicts of interest to declare.
FUNDING
This work was supported by Development Grant 4360 from the Muscular Dystrophy Association (to M.M.D.) and by grant K02 NS047306 from NIH/NINDS (to G.M).
Supplementary Material
REFERENCES
- 1.Stock D., Leslie A.G., Walker J.E. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 2.Kinosita K., Jr, Yasuda R., Noji H., Ishiwata S., Yoshida M. F1-ATPase: a rotary motor made of a single molecule. Cell. 1998;93:21–24. doi: 10.1016/s0092-8674(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 3.Noji H., Yasuda R., Yoshida M., Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 4.Holt I.J., Harding A.E., Petty R.K.H., Morgan-Hughes J.A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990;46:428–433. [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries D.D., van Engelen B.G., Gabreels F.J., Ruitenbeek W., van Oost B.A. A second missense mutation in the mitochondrial ATPase 6 gene in Leigh's syndrome. Ann. Neurol. 1993;34:410–412. doi: 10.1002/ana.410340319. [DOI] [PubMed] [Google Scholar]
- 6.Carrozzo R., Tessa A., Vazquez-Memije M.E., Piemonte F., Patrono C., Malandrini A., Dionisi-Vici C., Vilarinho L., Villanova M., Schagger H., et al. The T9176G mtDNA mutation severely affects ATP production and results in Leigh syndrome. Neurology. 2001;56:687–690. doi: 10.1212/wnl.56.5.687. [DOI] [PubMed] [Google Scholar]
- 7.White S.L., Shanske S., McGill J.J., Mountain H., Geraghty M.T., DiMauro S., Dahl H.H., Thorburn D.R. Mitochondrial DNA mutations at nucleotide 8993 show a lack of tissue- or age-related variation. J. Inherit. Metab. Dis. 1999;22:899–914. doi: 10.1023/a:1005639407166. [DOI] [PubMed] [Google Scholar]
- 8.White S.L., Collins V.R., Wolfe R., Cleary M.A., Shanske S., DiMauro S., Dahl H.H., Thorburn D.R. Genetic counseling and prenatal diagnosis for the mitochondrial DNA mutations at nucleotide 8993. Am. J. Hum. Genet. 1999;65:474–482. doi: 10.1086/302488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makela-Bengs P., Suomalainen A., Majander A., Rapola J., Kalimo H., Nuutila A., Pihko H. Correlation between the clinical symptoms and the proportion of mitochondrial DNA carrying the 8993 point mutation in the NARP syndrome. Pediatr. Res. 1995;37:634–639. doi: 10.1203/00006450-199505000-00014. [DOI] [PubMed] [Google Scholar]
- 10.Uziel G., Moroni I., Lamantea E., Fratta G.M., Ciceri E., Carrara F., Zeviani M. Mitochondrial disease associated with the T8993G mutation of the mitochondrial ATPase 6 gene: a clinical, biochemical, and molecular study in six families. J. Neurol. Neurosurg. Psychiatry. 1997;63:16–22. doi: 10.1136/jnnp.63.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enns G.M., Bai R.K., Beck A.E., Wong L.J. Molecular-clinical correlations in a family with variable tissue mitochondrial DNA T8993G mutant load. Mol. Genet. Metab. 2006;88:364–371. doi: 10.1016/j.ymgme.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Tsao C.Y., Mendell J.R., Bartholomew D. High mitochondrial DNA T8993G mutation (<90%) without typical features of Leigh's and NARP syndromes. J. Child Neurol. 2001;16:533–535. doi: 10.1177/088307380101600716. [DOI] [PubMed] [Google Scholar]
- 13.Santorelli F.M., Mak S.C., Vazquez-Memije E., Shanske S., Kranz-Eble P., Jain K.D., Bluestone D.L., De Vivo D.C., DiMauro S. Clinical heterogeneity associated with the mitochondrial DNA T8993C point mutation. Pediatr. Res. 1996;39:914–917. doi: 10.1203/00006450-199605000-00028. [DOI] [PubMed] [Google Scholar]
- 14.Schon E.A., Santra S., Pallotti F., Girvin M.E. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin. Cell. Dev. Biol. 2001;12:441–448. doi: 10.1006/scdb.2001.0281. [DOI] [PubMed] [Google Scholar]
- 15.Baracca A., Barogi S., Carelli V., Lenaz G., Solaini G. Catalytic activities of mitochondrial ATP synthase in patients with mitochondrial DNA T8993G mutation in the ATPase 6 gene encoding subunit a. J. Biol. Chem. 2000;275:4177–4182. doi: 10.1074/jbc.275.6.4177. [DOI] [PubMed] [Google Scholar]
- 16.Mattiazzi M., Vijayvergiya C., Gajewski C.D., DeVivo D.C., Lenaz G., Wiedmann M., Manfredi G. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum. Mol. Genet. 2004;13:869–879. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 17.Sgarbi G., Baracca A., Lenaz G., Valentino L.M., Carelli V., Solaini G. Inefficient coupling between proton transport and ATP synthesis may be the pathogenic mechanism for NARP and Leigh syndrome resulting from the T8993G mutation in mtDNA. Biochem. J. 2006;395:493–500. doi: 10.1042/BJ20051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houstek J., Klement P., Hermanska J., Houstkova H., Hansikova H., Van den Bogert C., Zeman J. Altered properties of mitochondrial ATP-synthase in patients with a T– > G mutation in the ATPase 6 (subunit a) gene at position 8993 of mtDNA. Biochim. Biophys. Acta. 1995;1271:349–357. doi: 10.1016/0925-4439(95)00063-a. [DOI] [PubMed] [Google Scholar]
- 19.Nijtmans L.G., Henderson N.S., Attardi G., Holt I.J. Impaired ATP synthase assembly associated with a mutation in the human ATP synthase subunit 6 gene. J. Biol. Chem. 2001;276:6755–6762. doi: 10.1074/jbc.M008114200. [DOI] [PubMed] [Google Scholar]
- 20.Morava E., Rodenburg R.J., Hol F., de Vries M., Janssen A., van den Heuvel L., Nijtmans L., Smeitink J. Clinical and biochemical characteristics in patients with a high mutant load of the mitochondrial T8993G/C mutations. Am. J. Med. Genet. A. 2006;140:863–868. doi: 10.1002/ajmg.a.31194. [DOI] [PubMed] [Google Scholar]
- 21.Carr1ozzo R., Wittig I., Santorelli F.M., Bertini E., Hofmann S., Brandt U., Schagger H. Subcomplexes of human ATP synthase mark mitochondrial biosynthesis disorders. Ann. Neurol. 2006;59:265–275. doi: 10.1002/ana.20729. [DOI] [PubMed] [Google Scholar]
- 22.Cortes-Hernandez P., Vazquez-Memije M.E., Garcia J.J. ATP6 homoplasmic mutations inhibit and destabilize the human F1F0 ATP-synthase without preventing enzyme assembly and oligomerization. J. Biol. Chem. 2007;282:1051–1058. doi: 10.1074/jbc.M606828200. [DOI] [PubMed] [Google Scholar]
- 23.Garcia J.J., Ogilvie I., Robinson B.H., Capaldi R.A. Structure, functioning, and assembly of the ATP synthase in cells from patients with the T8993G mitochondrial DNA mutation. Comparison with the enzyme in Rho0 cells completely lacking mtDNA. J. Biol. Chem. 2000;275:11075–11081. doi: 10.1074/jbc.275.15.11075. [DOI] [PubMed] [Google Scholar]
- 24.Vazquez-Memije M.E., Shanske S., Santorelli F.M., Kranz-Eble P., DeVivo D.C., DiMauro S. Comparative biochemical studies of ATPases in cells from patients with the T8993G or T8993C mitochondrial DNA mutations. J. Inherit. Metab. Dis. 1998;21:829–836. doi: 10.1023/a:1005418718299. [DOI] [PubMed] [Google Scholar]
- 25.Pallotti F., Baracca A., Hernandez-Rosa E., Walker W.F., Solaini G., Lenaz G., Melzi D'Eril G.V., Dimauro S., Schon E.A., Davidson M.M. Biochemical analysis of respiratory function in cybrid cell lines harbouring mitochondrial DNA mutations. Biochem. J. 2004;384:287–293. doi: 10.1042/BJ20040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baracca A., Sgarbi G., Mattiazzi M., Casalena G., Pagnotta E., Valentino M.L., Moggio M., Lenaz G., Carelli V., Solaini G. Biochemical phenotypes associated with the mitochondrial ATP6 gene mutations at nt8993. Biochim. Biophys. Acta. 2007;1767:913–919. doi: 10.1016/j.bbabio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Manfredi G., Yang L., Gajewski C.D., Mattiazzi M. Measurements of ATP in mammalian cells. Methods. 2002;26:317–326. doi: 10.1016/S1046-2023(02)00037-3. [DOI] [PubMed] [Google Scholar]
- 28.Rana M., de Coo I., Diaz F., Smeets H., Moraes C.T. An out-of-frame cytochrome b gene deletion from a patient with parkinsonism is associated with impaired complex III assembly and an increase in free radical production. Ann. Neurol. 2000;48:774–781. [PubMed] [Google Scholar]
- 29.Bruno C., Martinuzzi A., Tang Y., Andreu A.L., Pallotti F., Bonilla E., Shanske S., Fu J., Sue C.M., Angelini C., et al. A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am. J. Hum. Genet. 1999;65:611–620. doi: 10.1086/302546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vives-Bauza C., Anand M., Shirazi A.K., Magrane J., Gao J., Vollmer-Snarr H.R., Manfredi G., Finnemann S.C. The age lipid A2E and mitochondrial dysfunction synergistically impair phagocytosis by retinal pigment epithelial cells. J. Biol. Chem. 2008;283:24770–24780. doi: 10.1074/jbc.M800706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Pesini E., Lott M.T., Procaccio V., Poole J.C., Brandon M.C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D.C. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrnstadt C., Elson J.L., Fahy E., Preston G., Turnbull D.M., Anderson C., Ghosh S.S., Olefsky J.M., Beal M.F., Davis R.E., et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am. J. Hum. Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torroni A., Huoponen K., Francalacci P., Petrozzi M., Morelli L., Scozzari R., Obinu D., Savontaus M.L., Wallace D.C. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feder J., Blech I., Ovadia O., Amar S., Wainstein J., Raz I., Dadon S., Arking D.E., Glaser B., Mishmar D. Differences in mtDNA haplogroup distribution among 3 Jewish populations alter susceptibility to T2DM complications. BMC Genomics. 2008;9:198. doi: 10.1186/1471-2164-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savontaus M.L. mtDNA mutations in Leber's hereditary optic neuropathy. Biochim. Biophys. Acta. 1995;1271:261–263. doi: 10.1016/0925-4439(95)00037-5. [DOI] [PubMed] [Google Scholar]
- 37.Huoponen K., Lamminen T., Juvonen V., Aula P., Nikoskelainen E., Savontaus M.L. The spectrum of mitochondrial DNA mutations in families with Leber hereditary optic neuroretinopathy. Hum. Genet. 1993;92:379–384. doi: 10.1007/BF01247339. [DOI] [PubMed] [Google Scholar]
- 38.Okura T., Koda M., Ando F., Niino N., Tanaka M., Shimokata H. Association of the mitochondrial DNA 15497G/A polymorphism with obesity in a middle-aged and elderly Japanese population. Hum. Genet. 2003;113:432–436. doi: 10.1007/s00439-003-0983-8. [DOI] [PubMed] [Google Scholar]
- 39.Andreu A.L., Checcarelli N., Iwata S., Shanske S., DiMauro S. A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy. Pediatr. Res. 2000;48:311–314. doi: 10.1203/00006450-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 40.Petros J.A., Baumann A.K., Ruiz-Pesini E., Amin M.B., Sun C.Q., Hall J., Lim S., Issa M.M., Flanders W.D., Hosseini S.H., et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl Acad. Sci. USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johns D.R., Berman J. Alternative, simultaneous complex I mitochondrial DNA mutations in Leber's hereditary optic neuropathy. Biochem. Biophys. Res. Commun. 1991;174:1324–1330. doi: 10.1016/0006-291x(91)91567-v. [DOI] [PubMed] [Google Scholar]
- 42.Ugalde C., Vogel R., Huijbens R., Van Den Heuvel B., Smeitink J., Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 2004;13:2461–2472. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- 43.Robinson B.H. Use of fibroblast and lymphoblast cultures for detection of respiratory chain defects. Methods Enzymol. 1996;264:454–464. doi: 10.1016/s0076-6879(96)64041-5. [DOI] [PubMed] [Google Scholar]
- 44.Parfait B., de Lonlay P., von Kleist-Retzow J.C., Cormier-Daire V., Chretien D., Rotig A., Rabier D., Saudubray J.M., Rustin P., Munnich A. The neurogenic weakness, ataxia and retinitis pigmentosa (NARP) syndrome mtDNA mutation (T8993G) triggers muscle ATPase deficiency and hypocitrullinaemia. Eur. J. Pediatr. 1999;158:55–58. doi: 10.1007/s004310051009. [DOI] [PubMed] [Google Scholar]
- 45.D'Aurelio M., Gajewski C.D., Lenaz G., Manfredi G. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum. Mol. Genet. 2006;15:2157–2169. doi: 10.1093/hmg/ddl141. [DOI] [PubMed] [Google Scholar]
- 46.Yis U., Seneca S., Dirik E., Kurul S.H., Ozer E., Cakmakci H., De Meirleir L. Unusual findings in Leigh syndrome caused by T8993C mutation. Eur. J. Paediatr. Neurol. 2009;13:550–552. doi: 10.1016/j.ejpn.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Dionisi-Vici C., Seneca S., Zeviani M., Fariello G., Rimoldi M., Bertini E., De Meirleir L. Fulminant Leigh syndrome and sudden unexpected death in a family with the T9176C mutation of the mitochondrial ATPase 6 gene. J. Inherit. Metab. Dis. 1998;21:2–8. doi: 10.1023/a:1005397227996. [DOI] [PubMed] [Google Scholar]
- 48.Debray F.G., Lambert M., Lortie A., Vanasse M., Mitchell G.A. Long-term outcome of Leigh syndrome caused by the NARP-T8993C mtDNA mutation. Am. J. Med. Genet. A. 2007;143A:2046–2051. doi: 10.1002/ajmg.a.31880. [DOI] [PubMed] [Google Scholar]
- 49.Brown M.D., Torroni A., Reckord C.L., Wallace D.C. Phylogenetic analysis of Leber's hereditary optic neuropathy mitochondrial DNA's indicates multiple independent occurrences of the common mutations. Hum. Mutat. 1995;6:311–325. doi: 10.1002/humu.1380060405. [DOI] [PubMed] [Google Scholar]
- 50.Torroni A., Petrozzi M., D'Urbano L., Sellitto D., Zeviani M., Carrara F., Carducci C., Leuzzi V., Carelli V., Barboni P., et al. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am. J. Hum. Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 51.Brown M.D., Starikovskaya E., Derbeneva O., Hosseini S., Allen J.C., Mikhailovskaya I.E., Sukernik R.I., Wallace D.C. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup. J. Hum. Genet. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- 52.Hudson G., Carelli V., Spruijt L., Gerards M., Mowbray C., Achilli A., Pyle A., Elson J., Howell N., La Morgia C., et al. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am. J. Hum. Genet. 2007;81:228–233. doi: 10.1086/519394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pello R., Martin M.A., Carelli V., Nijtmans L.G., Achilli A., Pala M., Torroni A., Gomez-Duran A., Ruiz-Pesini E., Martinuzzi A., et al. Mitochondrial DNA background modulates the assembly kinetics of OXPHOS complexes in a cellular model of mitochondrial disease. Hum. Mol. Genet. 2008;17:4001–4011. doi: 10.1093/hmg/ddn303. [DOI] [PubMed] [Google Scholar]
- 54.Wallace D.C. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 55.King M.P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 56.Manfredi G., Gupta N., Vazquez-Memije M.E., Sadlock J.E., Spinazzola A., De Vivo D.C., Schon E.A. Oligomycin induces a decrease in the cellular content of a pathogenic mutation in the human mitochondrial ATPase 6 gene. J. Biol. Chem. 1999;274:9386–9391. doi: 10.1074/jbc.274.14.9386. [DOI] [PubMed] [Google Scholar]
- 57.Sciacco M., Prelle A., D'Adda E., Lamperti C., Bordoni A., Rango M., Crimi M., Comi G.P., Bresolin N., Moggio M. Familial mtDNA T8993C transition causing both the NARP and the MILS phenotype in the same generation. A morphological, genetic and spectroscopic study. J. Neurol. 2003;250:1498–1500. doi: 10.1007/s00415-003-0246-6. [DOI] [PubMed] [Google Scholar]
- 58.Vives-Bauza C., Andreu A.L., Manfredi G., Beal M.F., Janetzky B., Gruenewald T.H., Lin M.T. Sequence analysis of the entire mitochondrial genome in Parkinson's disease. Biochem. Biophys. Res. Commun. 2002;290:1593–1601. doi: 10.1006/bbrc.2002.6388. [DOI] [PubMed] [Google Scholar]
- 59.Vives-Bauza C., Yang L., Manfredi G. Assay of mitochondrial ATP synthesis in animal cells and tissues. Methods Cell. Biol. 2007;80:155–171. doi: 10.1016/S0091-679X(06)80007-5. [DOI] [PubMed] [Google Scholar]
- 60.D'Aurelio M., Pallotti F., Barrientos A., Gajewski C.D., Kwong J.Q., Bruno C., Beal M.F., Manfredi G. In vivo regulation of oxidative phosphorylation in cells harboring a stop-codon mutation in mitochondrial DNA-encoded cytochrome c oxidase subunit I. J. Biol. Chem. 2001;276:46925–46932. doi: 10.1074/jbc.M106429200. [DOI] [PubMed] [Google Scholar]
- 61.Trounce I.A., Kim Y.L., Jun A.S., Wallace D.C. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.