Abstract
Aging is believed to be a primary risk factor for cancer. Interestingly, the sirtuin family of class III histone deacetylases (HDACs) has been implicated in the regulation of longevity and may be a lost link between aging and cancer. SIRT1, a nicotinamide adenine dinucleotide (NAD+)-dependent sirtuin, has been shown to promote cell survival by inhibiting apoptosis or cellular senescence in mammalian cells. Recent studies have provided a link between the cellular metabolic function of SIRT1 and the circadian rhythm (controlled by a clock machinery), which, if deregulated, may lead to an increased risk for some cancers. Interestingly, the loss of the pineal hormone melatonin, a known regulator of circadian rhythm, has been shown to cause deregulation in the circadian rhythm machinery and an increase in susceptibility to cancer. On the basis of scientific evidence, we propose a hypothesis that SIRT1 inhibition will impart an antiproliferative response in age-related cancers via resynchronization of deregulated core clock circuitry at the cellular level. If this hypothesis is found valid, it may ultimately lead to the development of novel approaches toward management of age-related malignancies and possibly other diseases.—Jung-Hynes, B., Ahmad, N. SIRT1 controls circadian clock circuitry and promotes cell survival: a connection with age-related neoplasms.
Keywords: sirtuins, melatonin, cancer, HDACs
According to the American Cancer Society (ACS), age is a primary risk factor for cancer, and ∼77% of all cancers are diagnosed in people aged 55 and older. Prostate cancer (PCa) is an excellent example of an age-related malignancy, as according to estimates from the Centers for Disease Control and Prevention (CDC), a man’s chances of developing PCa grow by age: 1 in 2500 by age 45; 1 in 120 by age 55; 1 in 21 by age 65; and 1 in 9 by age 75. This trend appears to be true for other cancers as well.
In the recent past, increasing emphasis has been placed on defining the mechanistic links between aging and cancer. Interestingly, the sirtuin family of proteins, which decrease with age but rapidly increase in certain cancers, provides a potential connection between aging and cancer (1). It is being appreciated that this family of histone deacetylases (HDACs) may be one of the lost links between aging and cancer (2). SIRT1, the founding and most well-studied member of the sirtuin family, is a multifaceted, nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylase that is involved in a wide variety of cellular processes, including DNA repair, aging, stress response, etc. (2). SIRT1 has been shown to promote cell survival by inhibiting apoptosis or cellular senescence (2). We have recently demonstrated that SIRT1 is overexpressed in PCa, and its chemical inhibition resulted in a FoxO1-mediated inhibition in the growth and viability of human PCa cells (3). Several other laboratories have reported similar results in different cancer model systems (4,5,6,7,8).
On the basis of recent studies, SIRT1 is emerging as a master regulator of metabolic and stress responses with several critical downstream targets, including p53 and the Forkhead (FoxO) transcription factors (reviewed in ref. 9). Recently, a few interesting studies have provided a link between SIRT1 and the circadian rhythm machinery, which is responsible for keeping the biological system on a 24-h cycle with regard to biochemical, physiological, and/or behavioral processes (10,11,12,13). These studies suggested that SIRT1 counteracts the activity of the core clock components (10,11,12,13,14).
According to the circadian disruption hypothesis, factors in the environment (i.e., light disturbances) disrupt the circadian rhythm, which increases the risk for some cancers (ref. 15 and references therein). An example of this is the pineal hormone melatonin that is produced in a circadian pattern, and a decrease in melatonin levels has been linked to disruption in circadian rhythm machinery and an increase in cancer susceptibility or progression (refs. 16, 17 and references therein). The circadian connection of cancer is also evident from studies showing that melatonin possesses antiproliferative effects in cancer cells (refs. 16, 18, 19 and references therein).
On the basis of available information and recent paradigm-shifting scientific literature, we propose a hypothesis that SIRT1 inhibition will impart an antiproliferative response in age-related cancers via resynchronization of deregulated core clock circuitry at the cellular level. This novel hypothesis is experimentally verifiable, and we have presented the possible experimental ways to test the hypothesis.
AGING AND CANCER
Although aging is a highly individualized process, individuals of the same chronologic age can differ greatly in physiological age and other aspects of aging such as functional, social, emotional, and cognitive matters; however, a large number of them unfortunately succumb to cancer (20). It is now well accepted that age is a risk factor for most common cancers and that the incidence and prevalence of cancer will continue to increase with age (21). There are three possible explanations for the increase in cancer incidence with age: the time needed for cancer development and progression, increased susceptibility of aging cells and tissues to environmental toxins, and alterations in bodily conditions that favor tumor growth and metastasis (22). Carcinogenesis is a multistep process involving the activation of cellular oncogenes and the suppression of tumor suppressor genes, and these processes need an extended period of time to reach completion (21). The population of aged cells present directly correlates with the concentration of cells in advanced carcinogenic stages, enhancing the susceptibility of older individuals to various environmental carcinogens (21). These cells and tissues also tend to have molecular changes that favor carcinogenesis (20). Other biological changes of aging that help to promote the growth and the spreading of cancer are immunosenescence, proliferative senescence, and the production of tumor growth factors and proteolytic enzymes (21).
On the molecular and cellular levels, a number of critical changes associated with aging may influence the biology, and therefore the incidence, of cancer. Point mutations, genetic instability, DNA hypermethylation, and DNA adduct formation are critical molecular events that may lead to the activation of oncogenes and suppression of tumor suppressor genes (20, 22, 23). At the cellular level, proliferative and premature senescence associated with the loss of apoptosis, development of immortal cells, and production of tumor growth factors and metalloproteinases are critical events that may influence cancer development and progression (22, 23). Thus, it is now well accepted that an association between aging and carcinogenesis exists; however, the exact molecular mechanisms connecting these two processes are not well understood.
SIRT1: AT THE CROSSROADS OF AGING AND CANCER
Sirtuins (SIRT proteins) are a unique class of type III (NAD+)-dependent histone deacetylases (HDACs), which were originally shown to be involved in gene silencing in yeast (Sir2 family) and now can be grouped into five different classes (I–IV and U) in eukaryotes (24). This family of histone/protein deacetylases mediate post-translational modifications of the N-terminal tails of the histone proteins, which package DNA into chromatin and play key roles in the regulation of gene expression (25). In humans, there are seven reported sirtuins (SIRT1 to SIRT7), with greatly varying functions and locations (24). SIRT1, the mammalian homologue to yeast Sir2, is the most well-characterized member and has been shown to be involved in a number of cellular processes, including gene silencing at telomere and mating loci, DNA repair, recombination, and aging. SIRT1 promotes cell survival by inhibiting apoptosis or cellular senescence induced by stresses, including DNA damage and oxidative stress (2). An increasing number of SIRT1 targets, including p53, FoxO transcription factors, Ku70, and peroxisome proliferator-activated receptor, have been identified (reviewed in ref. 9). In mammalian cells, SIRT1 has been shown to down-regulate stress-induced p53 and FoxO pathways for apoptosis, thus favoring survival (2, 9). The known upstream regulators of SIRT1 that have been identified include active regulator of SIRT1 (AROS), hypermethylated in cancer 1 (HIC-1), and E2F1; however, the exact regulatory mechanism of SIRT1 is not clear (26,27,28).
While a plethora of studies has suggested a role of SIRT1 in aging, only limited reports are available regarding the role of sirtuin proteins in cancer. Wang et al. (8) have demonstrated an involvement of SIRT1 in human melanoma A375-S2 cell death by evodimine isolated from Evodia rutaecarpa. Ota et al. (7) have shown that the SIRT1 inhibitor sirtinol induces a senescence-like growth arrest in human breast cancer MCF-7 cells and lung cancer H1299 cells. Ford et al. (5) have shown that the silencing of SIRT1 gene caused growth arrest and/or apoptosis of human epithelial cancer cells (colorectal, breast, cervical), without affecting the normal human diploid fibroblasts. In another study, Chu et al. (4) showed that the expression of SIRT1 increased both at the RNA and protein levels in five drug-resistant cell lines compared to their drug-sensitive counterparts. Further, siRNA-mediated down-regulation of SIRT1 was shown to significantly reverse the resistance phenotype and reduce the expression of the multidrug resistance molecule P-glycoprotein (4). Another study reported that treatment of PCa cells with a SIRT1-inhibitor, sirtinol, resulted in an inhibition of cell growth and an increased sensitivity to campothecin and cisplatin (6). In a recent study, we have shown that SIRT1 is significantly overexpressed in human prostate cancer cells (DU145, LNCaP, 22Rν1, and PC3), compared to normal prostate epithelial cells (PrEC) at protein, mRNA and enzymatic activity levels (3). SIRT1 was also found to be overexpressed in human PCa tissues compared to adjacent normal prostate tissue (3). Interestingly, our data also demonstrated that SIRT1 inhibition via nicotinamide and sirtinol (at the activity level) as well as via short hairpin RNA (shRNA)-mediated RNA interference (at the genetic level) resulted in a significant inhibition in the growth and viability of human PCa cells, while having no effect on normal prostate epithelial cells (3). Further, we found that inhibition of SIRT1 caused an increase in FoxO1 acetylation and transcriptional activation in PCa cells (3). Furthermore, in a very recent interesting study, we found that shRNA-mediated SIRT1 inhibition resulted in an increase in senescence in PC3-p53 cells, whereas it resulted in an increase in apoptosis in PC3 cells, suggesting that Sirt1 inhibition may have different downstream targets in cells with active p53 vs. cells where p53 is inactive (29). Collectively, these findings uncovered a proproliferative function of the longevity gene SIRT1.
Interestingly, a few contradictory studies have suggested a tumor suppressor function of SIRT1 (30,31,32). However, the exact reasoning behind these discrepancies is not clear at present. A possible explanation for this may be that different cell types were used, which may, in turn, have different circadian rhythm oscillations with diverse outcomes. Further, multiple SIRT1 inhibitory agents were utilized in these studies, which makes it possible that other pathways may also be affected. Furthermore, it has been shown that SIRT1 plays a critical role in a number of pathways, suggesting that the observed differences may be due to hitherto unknown signaling regulated by SIRT1. Thus, the exact cause of the observed discrepancies is not clear and needs to be investigated in detail.
CIRCADIAN REGULATION AND CORE CLOCK COMPONENTS: CONNECTION WITH SIRT1
The mammalian circadian rhythm is controlled by a number of metabolic and physiological core components, where the circadian clock is composed of and regulated by several genes. The circadian rhythm apparatus is composed of a central pacemaker in the suprachiasmatic nucleus (SCN) of the brain, which acts to synchronize several clock component genes, including Clock, Bmal1, Period (Per), and Cryptochrome (Cry). Clock and Bmal1 are basic helix-loop-helix Per/Arnt/Sim (bHLH-PAS) transcription factors that act as the master regulator (33). Normally, the complex of Clock:Bmal1 binds E-box elements at the promoter region of Per1–3 and Cry1–2 to induce their transcription (33). Interestingly, the transcription of Per1–3 and Cry1–2 acts in a negative feedback loop to inhibit the transcriptional activity of the Clock:Bmal1 complex (33). The circadian clock machinery directs metabolic and physiological rhythms in whole organisms, as well as in some individual cells. It has been reported that 10% of all mammalian mRNA transcripts in various tissues oscillate in their abundance and are involved in a number of processes, such as tissue homeostasis, cellular metabolism, and cell cycle (34).
The cell cycle machinery, which plays a decisive role in the development of neoplasms, has been reported to be under circadian control in many organisms (35). The circadian clock machinery shares common features with the cell cycle, as both rely on sequential phases of transcription-translation and protein modification and degradation, and both are based on interlocked autoregulatory loops (reviewed in ref. 36). In addition, most eukaryotic cells in culture undergo division with a periodicity of roughly 24 h, and several mammalian cell cycle genes, such as c-myc, Cyclin D1, and Wee-1, are regulated in a circadian fashion (36). Further, mutations in various clock genes have been shown to result in the modulation of a number of cell cycle-associated genes (37). For example, Per2-mutant mice, which were shown to be more susceptible to radiation-induced malignant lymphoma, were also found to have alterations in the circadian pattern of cell-cycle regulatory genes such as Cyclin D1, Cyclin A, Mdm-2, and GADD45a (38). Furthermore, Per2-mutant mice have been shown to develop colonic polyps and have an increase in small intestinal mucosa β-catenin and cyclin D protein levels (39). In addition, a study by Zheng et al. (40) found that mice with a Per2 deletion mutation had a shorter circadian period. Also, mutations in Clock have been shown to result in inhibition of several proproliferative genes and up-regulation of genes associated with growth arrest and apoptosis (41). Thus, the scientific literature suggests that clock genes have distinct circadian and noncircadian functions important for proper homeostasis and are linked to proper cell cycle progression.
Several studies have ascertained the role of circadian rhythm genes in cancer. Per2 inhibition has been shown be involved in β-catenin-induced intestinal epithelial neoplastic transformation in vivo (38, 39, 42). In an interesting study, You et al. (43) compared the tumor size and circadian clock gene expression pattern (among other things) in mouse liver and tumor cells and found that the gene expression of Per1 and Per2 failed to maintain daily rhythms in tumors compared to their normal counterpart. These studies suggest that deregulation of circadian rhythm components may lead to the development and/or progression of cancer.
Extremely important to our hypothesis are a few very recent groundbreaking studies from two independent groups showing that SIRT1 regulates the circadian clock-associated genes. Nakahata et al. (12) demonstrated that SIRT1 was regulated in a circadian fashion and its expression correlated with the acetylation of Bmal1 and histone H3. SIRT1 was also found to directly associate with Clock and was recruited to the Clock:Bmal1 chromatin complex, where it was able to deacetylate histone H3 and Lysine 537 of Bmal1 (12). This study suggested that SIRT1 functions as an enzymatic rheostat of circadian function, transducing signals originated by cellular metabolites to the circadian clock (12). In a separate study, Asher et al. (13) found that SIRT1 is required for transcription of several core clock genes, including Bmal1, Rorγ, Per2, and Cry1. Further, this study demonstrated that SIRT1 binds to the Clock:Bmal1 complex and also promotes the deacetylation and degradation of Per2 (13). These studies suggested that SIRT1 may deacetylate the Clock:Bmal1 complex to counteract the transcriptional activity it possesses. It has been shown previously that the Clock protein contains histone acetyltransferase activity (HAT) and acetylates histone H3 and its dimerization partner Bmal1 at lysine 537, suggesting it may be involved in chromatin remodeling in the circadian regulation of gene expression (44). These studies provide a link between cellular metabolism and the circadian rhythm system.
In addition to SIRT1 being directly linked to the Clock:Bmal1 complex, a couple of recent studies have shown that intracellular NAD+ levels show circadian oscillations driven by the circadian clock (10, 11). Nakahata et al. (11) have reported that the Clock:Bmal1 complex physically associates with the E-boxes on the Nampt promoter in a time-dependent manner, which was consistent with NAMPT (nicotinamide phosphoribosyltransferase) circadian expression. NAMPT is the rate-limiting step enzyme in the NAD+ salvage pathway, as it catalyzes the first step in the biosynthesis of NAD from nicotinamide (45). Nakahata et al. (11) suggested that an interlocking of two autoregulatory systems is occurring, in that a classical transcription circadian loop is coupled to an enzymatic feedback loop. The second study by Ramsey et al. (10) found that NAMPT and the levels of NAD+ display circadian oscillations are regulated by the core clock machinery (10). In addition, the authors found that inhibition of NAMPT allowed for the release of SIRT1 from the Clock:Bmal1 complex, in turn, causing an increase in the oscillations of the Per2 gene machinery. It was suggested that through NAMPT-mediated NAD+ biosynthesis the circadian feedback loop may function to modify the daily cycles of energy storage and utilization, as well as work to coordinate these processes with the rest-activity cycle (10). These studies, as well as the studies that linked metabolism and the circadian rhythm system, suggest an interaction between SIRT1 and the core clock circuitry.
Some studies have also linked the FoxO pathway with circadian regulation. Zheng et al. (46) found that Drosophila lacking the FoxO gene product (FoxO mutants) showed a rapid decline in rest:activity rhythms with age. In another study, Rouyer et al. (47) isolated a new circadian rhythm-regulated gene Crg-1 that had some sequence similarity with the DNA-binding domain of the HNF3/forkhead family of transcription factors. In addition, Crg-1 showed transcript oscillation patterns similar to that of known Drosophila circadian rhythm genes, period (per) and timeless (tim) (47). Some studies have also provided a connection between circadian rhythm machinery with FoxO homologues and/or aging in Caenorhabditis elegans (48, 49). These findings are interesting because FoxOs have been shown to be downstream targets of SIRT1. Taken together, these studies have provided evidence regarding the existence of an association between SIRT1, the FoxO pathway, and the circadian rhythm components.
PINEAL HORMONE MELATONIN: LINKING CIRCADIAN RHYTHM WITH CANCER
Melatonin (chemically known as N-acetyl-5-methoxytryptamine) is a small molecule that is essentially secreted by the pineal gland, and its synthesis displays a circadian rhythm that is generated by a circadian clock located in the SCN of the hypothalamus region of the brain. Light signals through a direct retinal pathway to the SCN, which is set to a 24-h day, and then sends circadian signals over a neural pathway to the pineal gland, driving rhythmic melatonin synthesis (reviewed in ref. 16). Thus, the synthesis and release of melatonin are stimulated by darkness and inhibited by light, much the same as the circadian rhythm genes function. A number of studies have suggested that melatonin possesses chemopreventive, oncostatic, and tumor inhibitory effects in a variety of in vitro and in vivo experimental models of neoplasia (reviewed in ref. 16). Recently, epidemiological studies have revealed that the frequency of breast, prostate, endometrial, and colorectal cancers have been reportedly increased in individuals who routinely work at night or whose circadian rhythms are disrupted for other reasons, such as jet lag (reviewed in ref. 15). Interestingly, those individuals who were reported to be exposed to light at night, due to workplace environment or other reasons, were also found to have lower circulating melatonin levels (15). These studies suggested a negative correlation between melatonin levels and cancer risk. In addition, it has been reported that melatonin levels reach a maximum at a young age, remain relatively stable until 35–40 yr of age, and thereafter diminish, gradually reaching levels similar to daytime low concentrations (17). As a result, many aged individuals do not exhibit a day-night difference in melatonin levels. Interestingly, the deterioration of circadian rhythms, which play an important role in homeostasis, is also a characteristic of the elderly population. It has been suggested that the loss of melatonin in the elderly may lead to disorder in the circadian rhythm components, causing a desynchronization of the various genes resulting in a decrease in overall health and possibly an increase in cancer susceptibility and/or progression (17).
Because melatonin is synthesized in a circadian pattern, some interesting studies have attempted to determine whether melatonin has an effect on the circadian rhythm genes. Torres-Farfan et al. (50) assessed the patterns of clock gene proteins in melatonin-proficient mice (C3H) vs. melatonin-deficient mice (C57BL). The authors found Per1-, Cry2-, and Bmal1-protein levels to be consistently lower in the adrenal cortex of C57BL mice compared to C3H mice (50). Another study demonstrated that inhibition of maternal melatonin changed the expression of Bmal1, Per2, and melatonin type 1 (Mela) receptor in the fetal SCN (51). Further, Imbesi et al. (52) recently demonstrated that melatonin differentially regulated the expression of clock genes in striatal neurons. These studies suggested that melatonin controls certain clock genes, and also points toward a possible scenario that melatonin may be controlling circadian rhythm via SIRT1, which may be the mechanism connecting melatonin with age and cancer susceptibility.
Notably, a study by Guiterrez-Cuesta et al. (53) investigated the effect of melatonin on the prosurvival process in an in vivo setting. In this study, the authors treated senescence-accelerated mice (SAMP8) with melatonin (10 mg/kg) for 8 mo and used senescence-associated resistant mice (SAMR1) as controls. The authors found that SIRT1 expression was higher in 10-mo-old SAMR1 mice compared to SAMP8 mice, which is to be expected based on our own studies, as well as other published studies. In addition, this study reported that in SAMR1 mice, the levels of SIRT1 increased from 3 to 12 mo of age, whereas a decrease in SIRT1 was observed in SAMP8 mice over the same time period (53). The authors also assessed the effect of melatonin on the cellular pathways regulated by SIRT1. They found that melatonin increased the protein levels of SIRT1 and subsequently decreased the levels of acetylated p53 and acetylated NF-κB in SAMP8 mice, but not in the SAMR1 mice (53). It was suggested that melatonin suppressed oxidative stress in SAMP8, which allowed for SIRT1 levels to increase to the values of SIRT1 present in SAMR1 mice (53). This is an interesting study, suggesting that SIRT1 could be modulated by melatonin. However, more in-depth mechanistic studies are needed in cancer cells vs. normal cells to reach a firm conclusion. We would argue that both SIRT1 and melatonin have different modes and mechanisms of action in cancer vs. normal cells.
EXPERIMENTALLY VERIFIABLE HYPOTHESIS
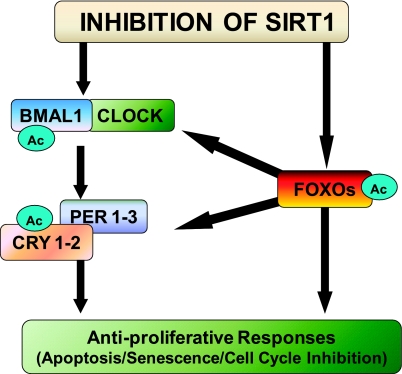
We have provided a compelling argument supported by published scientific literature, which leads us to believe that SIRT1 may be promoting the survival of age-related malignancies through direct or indirect interactions with the core components of the circadian rhythm circuitry. We present a hypothesis that SIRT1 inhibition will impart an antiproliferative response in age-related cancers via resynchronization of deregulated core clock circuitry at the cellular level. Our specific hypothesis, involving a proposed interplay of specific core clock components and FoxO factors, is depicted in Fig. 1.
Figure 1.
Schematic representation of hypothesis. We hypothesize that inhibition of SIRT1 will lead to a resynchronization of the core clock machinery via inhibiting the deacetylation of the Clock:Bmal1 complex. This increase in acetylation will inhibit the deacetylation and subsequent degradation of Per 1–3 and/or Cry 1–2 transcription factors. This will cause an activation of these clock genes which, in turn, will lead to antiproliferative responses by apoptosis, senescence, and/or cell cycle inhibition. In addition, the inhibition of SIRT1 may also be associated with a increase in the acetylation of the FoxO factors, which may modulate the clock genes or directly impart antiproliferative response against cancer cells.
This hypothesis is experimentally verifiable, and some possible experimental scenarios are as follows. One would first need to define the association and deacetylation activity of SIRT1 with the Clock:Bmal1 complex in an age-related malignancy, such as PCa, and the effects of acetylation status of the Clock:Bmal1 complex on its transcriptional activation. It will also be important to identify the transcriptionally regulated targets of the Clock:Baml1 complex (i.e., Per1–3, Cry1–2, etc.). Another important experiment could be to determine whether SIRT1 inhibition or overexpression alters the circadian oscillation patterns (mRNA and/or protein levels) of clock genes such as Per1–3 or Cry1–2. We could also use specific inhibitors or gene-knockdown approaches of SIRT1 to ascertain whether an inhibition of SIRT1 in PCa cells (or other age-related cancers) imparts an antiproliferative response (cell growth/viability/cell cycle inhibition, apoptosis/senescence induction) via changes in the binding and acetylation of circadian rhythm genes (Clock, Bmal1, Per 1–3, Cry 1–2, etc.). Further, we could determine whether the antiproliferative effects of melatonin are mediated via SIRT1 inhibition-mediated modulations in clock machinery. Similar studies could be conducted under in vivo situations in animal models of age-related neoplasms such as PCa. Other experiments that may be useful in exploring our proposed hypothesis include conducting preclinical, clinical, and epidemiological studies to determine whether melatonin or other specific SIRT1 inhibitors prevent, delay, or inhibit development and/or progression of the age-related cancer; determining whether melatonin is able to alter the circadian rhythm genes; and defining the effects of melatonin on the modulation of sirtuins.
We predict that SIRT1 inhibition via melatonin or other chemical/genetic means will impart a decrease in cell growth/viability of various age-related cancer cells, such as PCa. In addition, we believe that inhibition of SIRT1 will lead to modulations in the acetylation of the Clock:Bmal1 complex and the circadian oscillation patterns of various clock genes, thereby inhibiting the progression/susceptibility of age-related cancers. If this hypothesis is found to be valid, it may ultimately lead to the development of novel approaches toward the management of age-related malignancies and possibly other diseases.
References
- Fraga M F, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- Longo V D, Kennedy B K. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of sirtuin histone deacetylase Sirt1 in prostate cancer: A target for prostate cancer management via its inhibition? J Biol Chem. 2009;284:3823–3832. doi: 10.1074/jbc.M807869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Chou P M, Zheng X, Mirkin B L, Rebbaa A. Control of multidrug resistance gene mdr1 and cancer resistance to chemotherapy by the longevity gene sirt1. Cancer Res. 2005;65:10183–10187. doi: 10.1158/0008-5472.CAN-05-2002. [DOI] [PubMed] [Google Scholar]
- Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- Kojima K, Ohhashi R, Fujita Y, Hamada N, Akao Y, Nozawa Y, Deguchi T, Ito M. A role for SIRT1 in cell growth and chemoresistance in prostate cancer PC3 and DU145 cells. Biochem Biophys Res Commun. 2008;373:423–428. doi: 10.1016/j.bbrc.2008.06.045. [DOI] [PubMed] [Google Scholar]
- Ota H, Tokunaga E, Chang K, Hikasa M, Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y, Kaneki M. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang M W, Tashiro S, Onodera S, Ikejima T. Roles of SIRT1 and phosphoinositide 3-OH kinase/protein kinase C pathways in evodiamine-induced human melanoma A375–S2 cell death. J Pharmacol Sci. 2005;97:494–500. doi: 10.1254/jphs.fpj04055x. [DOI] [PubMed] [Google Scholar]
- Taylor D M, Maxwell M M, Luthi-Carter R, Kazantsev A G. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci. 2008;65:4000–4018. doi: 10.1007/s00018-008-8357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey K M, Yoshino J, Brace C S, Abrassart D, Kobayashi Y, Marcheva B, Hong H K, Chong J L, Buhr E D, Lee C, Takahashi J S, Imai S I, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente L P, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt F W, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Belden W J, Dunlap J C. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter R J, Tan D X, Korkmaz A, Erren T C, Piekarski C, Tamura H, Manchester L C. Light at night, chronodisruption, melatonin suppression, and cancer risk: a review. Crit Rev Oncog. 2007;13:303–328. doi: 10.1615/critrevoncog.v13.i4.30. [DOI] [PubMed] [Google Scholar]
- Jung B, Ahmad N. Melatonin in cancer management: progress and promise. Cancer Res. 2006;66:9789–9793. doi: 10.1158/0008-5472.CAN-06-1776. [DOI] [PubMed] [Google Scholar]
- Karasek M. Does melatonin play a role in aging processes? J Physiol Pharmacol. 2007;58:105–113. [PubMed] [Google Scholar]
- Blask D E, Sauer L A, Dauchy R T. Melatonin as a chronobiotic/anticancer agent: cellular, biochemical, and molecular mechanisms of action and their implications for circadian-based cancer therapy. Curr Top Med Chem. 2002;2:113–132. doi: 10.2174/1568026023394407. [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Spence D W, Pandi-Perumal S R, Trakht I, Cardinali D P. Therapeutic actions of melatonin in cancer: possible mechanisms. Integr Cancer Ther. 2008;7:189–203. doi: 10.1177/1534735408322846. [DOI] [PubMed] [Google Scholar]
- Ershler W B. Cancer: a disease of the elderly. J Support Oncol. 2003;1:5–10. [PubMed] [Google Scholar]
- Balducci L. Epidemiology of cancer and aging. J Oncol Manag. 2005;14:47–50. [PubMed] [Google Scholar]
- Balducci L, Ershler W B. Cancer and ageing: a nexus at several levels. Nat Rev Cancer. 2005;5:655–662. doi: 10.1038/nrc1675. [DOI] [PubMed] [Google Scholar]
- Carreca I, Balducci L, Extermann M. Cancer in the older person. Cancer Treat Rev. 2005;31:380–402. doi: 10.1016/j.ctrv.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Porcu M, Chiarugi A. The emerging therapeutic potential of sirtuin-interacting drugs: from cell death to lifespan extension. Trends Pharmacol Sci. 2005;26:94–103. doi: 10.1016/j.tips.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem Soc Trans. 2004;32:904–909. doi: 10.1042/BST0320904. [DOI] [PubMed] [Google Scholar]
- Chen W Y, Wang D H, Yen R C, Luo J, Gu W, Baylin S B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Kim E J, Kho J H, Kang M R, Um S J. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress W D, Chen J. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- Jung-Hynes B, Ahmad N. Role of p53 in the anti-proliferative effects of Sirt1 inhibition in prostate cancer cells. Cell Cycle. 2009;8 doi: 10.4161/cc.8.10.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon J M, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano P S, Huber L J. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Ngo D, Forman L W, Qin D C, Jacob J, Faller D V. Sirtuin 1 is required for antagonist-induced transcriptional repression of androgen-responsive genes by the androgen receptor. Mol Endocrinol. 2007;21:1807–1821. doi: 10.1210/me.2006-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn W C, Guarente L P, Sinclair D A. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth [Online] PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray J P, Cagampang F R, Loudon A S, Reppert S M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch M P, Miller B H, Su A I, Schook A B, Straume M, Schultz P G, Kay S A, Takahashi J S, Hogenesch J B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pittendrigh C S. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Kondratov R V, Antoch M P. The clock proteins, aging, and tumorigenesis. Cold Spring Harbor Symp Quant Biol. 2007;72:477–482. doi: 10.1101/sqb.2007.72.050. [DOI] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- Wood P A, Yang X, Taber A, Oh E Y, Ansell C, Ayers S E, Al-Assaad Z, Carnevale K, Berger F G, Pena M M, Hrushesky W J. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008;6:1786–1793. doi: 10.1158/1541-7786.MCR-08-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Larkin D W, Albrecht U, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Miller B H, McDearmon E L, Panda S, Hayes K R, Zhang J, Andrews J L, Antoch M P, Walker J R, Esser K A, Hogenesch J B, Takahashi J S. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wood P A, Ansell C M, Ohmori M, Oh E Y, Xiong Y, Berger F G, Pena M M, Hrushesky W J. Beta-catenin induces β-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009;145:289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- You S, Wood P A, Xiong Y, Kobayashi M, Du-Quiton J, Hrushesky W J. Daily coordination of cancer growth and circadian clock gene expression. Breast Cancer Res Treat. 2005;91:47–60. doi: 10.1007/s10549-004-6603-z. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Revollo J R, Grimm A A, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Zheng X, Yang Z, Yue Z, Alvarez J D, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouyer F, Rachidi M, Pikielny C, Rosbash M. A new gene encoding a putative transcription factor regulated by the Drosophila circadian clock. EMBO J. 1997;16:3944–3954. doi: 10.1093/emboj/16.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Johnson T E. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren J R, Braeckman B P. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol Aging. 1999;20:487–502. doi: 10.1016/s0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- Torres-Farfan C, Seron-Ferre M, Dinet V, Korf H W. Immunocytochemical demonstration of day/night changes of clock gene protein levels in the murine adrenal gland: differences between melatonin-proficient (C3H) and melatonin-deficient (C57BL) mice. J Pineal Res. 2006;40:64–70. doi: 10.1111/j.1600-079X.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- Torres-Farfan C, Rocco V, Monso C, Valenzuela F J, Campino C, Germain A, Torrealba F, Valenzuela G J, Seron-Ferre M. Maternal melatonin effects on clock gene expression in a nonhuman primate fetus. Endocrinology. 2006;147:4618–4626. doi: 10.1210/en.2006-0628. [DOI] [PubMed] [Google Scholar]
- Imbesi M, Arslan A D, Yildiz S, Sharma R, Gavin D, Tun N, Manev H, Uz T. The melatonin receptor MT1 is required for the differential regulatory actions of melatonin on neuronal ‘clock’ gene expression in striatal neurons in vitro. J Pineal Res. 2009;46:87–94. doi: 10.1111/j.1600-079X.2008.00634.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Cuesta J, Tajes M, Jimenez A, Coto-Montes A, Camins A, Pallas M. Evaluation of potential pro-survival pathways regulated by melatonin in a murine senescence model. J Pineal Res. 2008;45:497–505. doi: 10.1111/j.1600-079X.2008.00626.x. [DOI] [PubMed] [Google Scholar]