Abstract
Understanding the role of microbes in the oceans has focused on taxa that occur in high abundance; yet most of the marine microbial diversity is largely determined by a long tail of low-abundance taxa. This rare biosphere may have a cosmopolitan distribution because of high dispersal and low loss rates, and possibly represents a source of phylotypes that become abundant when environmental conditions change. However, the true ecological role of rare marine microorganisms is still not known. Here, we use pyrosequencing to describe the structure and composition of the rare biosphere and to test whether it represents cosmopolitan taxa or whether, similar to abundant phylotypes, the rare community has a biogeography. Our examination of 740,353 16S rRNA gene sequences from 32 bacterial and archaeal communities from various locations of the Arctic Ocean showed that rare phylotypes did not have a cosmopolitan distribution but, rather, followed patterns similar to those of the most abundant members of the community and of the entire community. The abundance distributions of rare and abundant phylotypes were different, following a log-series and log-normal model, respectively, and the taxonomic composition of the rare biosphere was similar to the composition of the abundant phylotypes. We conclude that the rare biosphere has a biogeography and that its tremendous diversity is most likely subjected to ecological processes such as selection, speciation, and extinction.
Keywords: abundance distribution, bacteria, archaea, pyrosequencing, biogeography
Marine microbes are essential for the functioning of marine ecosystems (1), yet the extent of their diversity remains unclear (2). The early approach of direct cultivation to identify marine microbes gave very low estimates of diversity and abundance. The development and in situ application of molecular tools greatly changed our perception of microbial diversity, revealing that only a very small fraction of microorganisms were detected by culture (3). These cultivation-independent approaches showed that microbes represent the main diversity of life on earth (4). In marine systems, the diversity of microbes seems to increase with increasing sampling effort and resolving power of new molecular tools. Initially the diversity of marine bacteria was predicted to be as low as a few thousand taxa (5); more recent estimates have raised the number to 106–109 taxa (2). The diversity of archaea, in turn, appears generally to be much lower than for bacteria (6), and marine archaea could be 5–10 times less diverse than bacteria (7, 8).
The main focus of marine molecular microbial studies has been on the most abundant members of the communities for practical reasons; the abundant phylotypes are the easiest to detect with the most widely used molecular tools. PCR-based fingerprinting and cloning techniques using universal primers most easily amplify microbes with abundances in the ecosystem of >1% of the total community (2, 9). The most abundant taxa are also thought to be the most active and most important in fluxes of dissolved organic matter (10). However, abundant species represent only a small portion of microbial diversity. Analyses of abundance distribution indicate that diversity is divided into two main components. One component includes the few species that are very abundant, which is the most studied part of the community. The other component, named “the rare biosphere” (11), comprises a very high number of rare species that contains most of the diversity (2). The presence of this long tail of rare microbes was first demonstrated by targeted pyrosequencing of marine bacteria in the deep Atlantic (11), then in deep oceanic vents (8) and in the Arctic Ocean both for bacteria (12, 13) and archaea (7).
As accumulating data confirm the prevalence of the rare biosphere, we still know little about its ecological and functional role in the ocean. The main hypothesis proposes that the rare members of the biosphere have a cosmopolitan distribution because of a low loss rate by viral lysis or predation, combined with potentially unlimited dispersal capacity governed by stochastic immigration (2). The goal of this study was thus to test whether the rare biosphere represents cosmopolitan taxa or whether, similar to abundant phylotypes, the rare community has a biogeography structured by ecological processes such as selection and speciation. We used data from 16S rRNA gene pyrosequencing analysis of 24 bacterial and eight archaeal samples from across the Arctic Ocean to compare the similarity among rare members of the communities. We furthermore applied models for abundance distribution and described the taxonomic composition of both rare and abundant phylotypes.
Results
Similarity Among Communities.
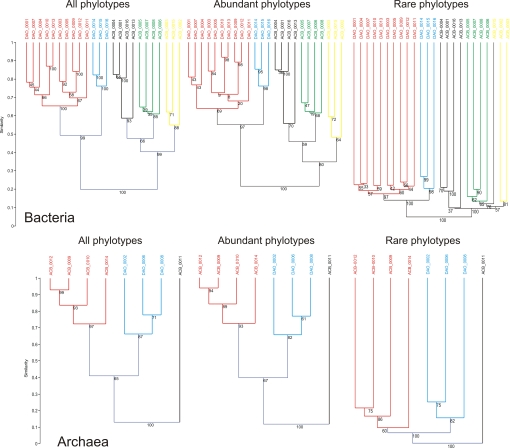
We assessed the community similarity among sites by comparing the relative abundance and distribution of 545,246 bacterial and 195,107 archaeal V6 tag 16S rRNA gene sequences (Table S1). We separated the microbial phylotypes (defined in Materials and Methods) as abundant or rare. Rare phylotypes were arbitrarily defined as having a frequency <0.01% and abundant phylotypes a frequency >1% within a sample. Our data showed that the grouping for the rare bacterial phylotypes was similar to the clustering of the abundant phylotypes, which in turn was similar to the clustering of the entire community (Fig. 1). Archaeal communities gave the same result (Fig. 1). Cluster analysis separated the communities into two main groups. One group contained all communities from surface waters (samples ACB, Fig. 1), and the other group contained all deep water mass communities (samples DAO). Deep communities were further separated into two clusters. One cluster contained communities from the Eurasian Basin (colored red in Fig. 1), and the other cluster contained communities from the Canada Basin (blue) of the Arctic Ocean. Surface samples separated into three groups. The first group comprised samples collected mostly in winter with three from January, but with one sample from July (cluster colored black, Fig. 1). The second contained only summer samples collected in July and August (cluster in green) and the third cluster contained two July and one January sample (cluster in yellow). Archaea separated into three main groups. One contained all surface samples (red, Fig. 1), the other all deep water samples (blue), and the third only one sample (black). This grouping was similar for the abundant and rare samples (the single exception being sample ACB_0015).
Fig. 1.
Dendrograms representing the similarity between the composition of 24 bacterial and eight archaeal communities from deep (DAO) and surface (ACB) water masses of the Arctic Ocean. The clustering pattern including all phylotypes is compared with the clustering obtained for abundant phylotypes only (frequency >1%) and for rare phylotypes only (<0.01%). Colors highlight the clusters conserved through the three analyses. Clustering is based on a distance matrix computed with Bray–Curtis similarity. The dendrogram was inferred with the unweighted pair–group average algorithm. Bootstraps values (in percentages) are given at the nodes.
When phylotypes were defined more stringently (100% identity), the community grouping was still similar between rare and abundant phylotypes for both bacteria and archaea (Fig. S1). The patterns of biogeography were still present for rare phylotype with a frequency <0.001% (Fig. S2). Such a low frequency could be detected only when sequences from all samples were grouped together. This was the lowest threshold that we could apply to define rare phylotype.
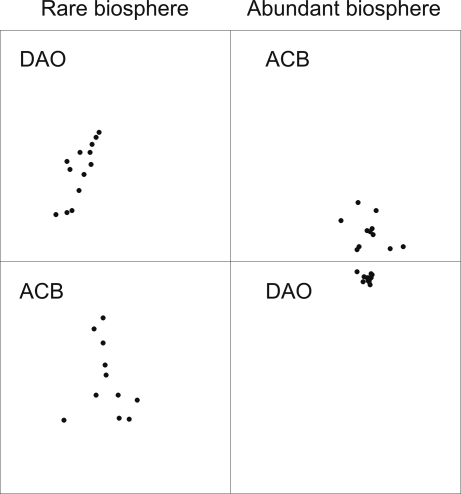
Nonmetric multidimensional scaling analysis showed that the rare phylotypes better separated ACB samples (surface) from DAO samples (deep waters) than the abundant phylotypes did (Fig. 2). This analysis also revealed that the difference in community composition was greater between rare and abundant phylotypes than between surface and deep samples. The result was the same when the more stringent definition (100%) of a phylotype was used (Fig. S3).
Fig. 2.
Nonmetric multidimensional scaling analysis of the phylotype composition of the abundant (>1%) and rare (<0.01%) bacterial biosphere (n = 48). The analysis separated DAO (deep) from ACB (surface) samples, and the abundant from the rare biosphere. Calculated stress for this analysis using Bray–Curtis similarity = 0.202.
Rare and Abundant Phylotypes.
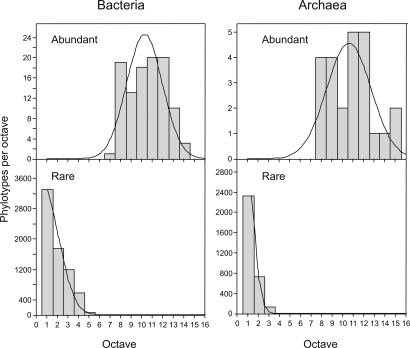
There was a clear difference between the abundance distributions of the rare and abundant phylotypes (Fig. 3). The rare phylotypes followed a log-series distribution, whereas the abundant phylotypes followed a log-normal distribution albeit poorly. This difference in the abundance distribution was observed for both bacteria and archaea (Fig. 3). A similar separation between the abundance distribution of rare and abundant phylotypes was found for ACB (surface) and DAO samples (deep waters) (Fig. S4).
Fig. 3.
Abundance distribution of bacterial and archaeal phylotypes separated as abundant (>1% frequency) and rare (<0.01%). The abundance models predicting the frequency of each abundance class are shown as lines. Abundant phylotypes are predicted by a log-normal model and rare phylotypes by a log-series model. The octaves refer to power-of-2 abundance classes.
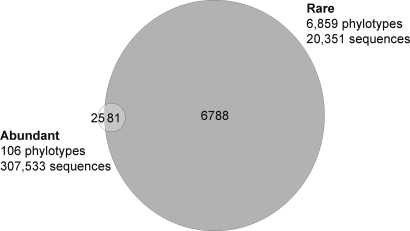
For bacteria, we counted 106 abundant phylotypes that covered 56% of the total number of sequences. On the other end, there were 6,859 rare phylotypes comprising <4% of the sequence abundance (Fig. 4). Overall, 99% of the bacteria phylotypes found to be rare were never detected as abundant, as pointed out by Kirchman et al. (13), who analyzed only the surface bacterial communities. Conversely, 81 phylotypes found to be abundant in some samples were found to be rare in others. Among those 81 phylotypes, 54 were exclusively from ACB samples, whereas 27 were detected only in DAO samples (Fig. S5). For such phylotypes rare in some samples and abundant in others, the phylotypes abundant in surface waters (ACB) were rare in deep waters (DAO), and conversely phylotypes abundant in deep waters were rare in surface samples (Fig. S5).
Fig. 4.
Venn diagram illustrating the number of bacteria phylotypes being rare in all samples (dark gray) vs. phylotypes always abundant and never rare (white area), and phylotypes being rare in some samples but abundant in others (overlap area).
For archaea, there were 22 abundant phylotypes comprising 93% of the sequences, whereas 369 phylotypes were classified as rare and represented 0.3% of the sequences. Up to 6 phylotypes (1.6%) were abundant in some samples and rare in others (Fig. S6).
A large fraction (75%) of rare bacterial DAO phylotypes were only found in deep water samples (3,309 phylotypes of 4,394 total), and likewise 69% of rare ACB phylotypes were only in surface samples (2,465 phylotypes of 3,550 total). Only 16% of the bacteria and 8% of the archaea rare phylotypes were detected in both ACB and DAO samples. Using the more stringent phylotype definition (100% identity), these values were 4% for bacteria and 8% for archaea (Table S2).
Phylogenetic Composition of the Abundant and Rare Phylotypes.
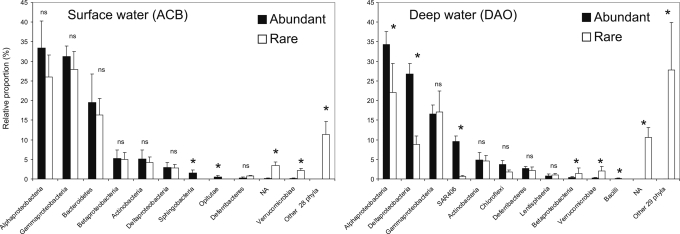
In surface samples (ACB), the most frequent bacterial groups were present in similar proportions in the abundant and the rare phylotypes (Fig. 5). For deep samples (DAO), Alphaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria were the most frequent groups in both abundant and rare phylotypes, but there were some differences between the distributions of the main taxonomic groups (Fig. 5).
Fig. 5.
Phylogenetic composition of abundant phylotypes (>1% frequency) compared with rare bacterial phylotypes (<0.01%) in surface (ACB) and deep (DAO) water masses of the Arctic Ocean. NA, not assigned; ns, not significant. *, P < 0.05 (t test results for difference between rare and abundant phylotypes).
For both ACB and DAO samples, there were more nonassigned groups among rare phylotypes, and a much higher number of taxonomic groups were present among rare phylotypes compared with abundant phylotypes (Fig. 5).
Discussion
The recent development of pyrosequencing for studying microbes in their environment has revealed the tremendous diversity of marine bacterial communities and the prevalence of phylotypes in the rare biosphere (11). This long tail of rare microbes represent most of the diversity in the oceans (2); yet little is known about the ecology of this major component of marine microbial ecosystems.
The community composition of both the rare bacterial and archaeal phylotypes differed among sites in the Arctic Ocean, in a way similar to that of the abundant members and the entire community. These results show that the rare biosphere had patterns of biogeography, implying that the rare phylotypes expand or contract their area of distribution after ecological mechanisms such as speciation, extinction, dispersal, or species interactions equivalent to those ruling abundant phylotypes (14). The presence of such processes argues against the hypothesis that the rare marine biosphere has a cosmopolitan distribution governed by stochastic immigration. The cosmopolitan theory proposes that rare phylotypes are recruited through immigration (dispersal) and that they are protected from active loss by both viral lysis and predation because of their low abundance. Consequently, if all microbes are easily and globally dispersed, the rare biosphere would be ubiquitously distributed, and all rare phylotypes would be found everywhere (2). Our results, demonstrating patterns of biogeography for the rare biosphere, clearly show that the dictum “everything is everywhere” does not apply here. It therefore suggests that one or both postulates for the cosmopolitan theory are not valid: Marine microorganisms are not as easily dispersed as previously thought, and so barriers for dispersal exist, and/or the rates of active loses (predation or viral lysis) on the rare biosphere are significant.
An important ecological aspect for understanding biogeography is to determine how patterns of diversity are shaped by the relative influence of allopatric vs. sympatric speciation models. Sympatry will occur under a strong influence of contemporary environmental factors, and microbial diversity would be shaped by the presence of multiple habitats, corresponding to different environmental conditions, within one province (14). The biogeography of bacterial and archaeal diversity has recently been associated with water masses (12, 15, 16), and each water mass could represent a specific habitat within the same province represented by an ocean. Differences in community composition would thus reflect the differences in environmental conditions existing among water masses as proposed for planktonic foraminifers (17, 18). On the other hand, allopatric speciation shaped by the influence of historical events has also been shown for marine plankton (19). A historical isolation of communities and lack of biological dispersal would imply the presence of multiple provinces but one habitat only (14). The density difference between water masses creates strong physical boundaries that can limit the dispersion of microbes because of their small size and planktonic lifestyle. In that context, within the same marine habitat, water masses physically isolated from each other over time would represent different provinces. The present composition of marine microbial community, as for macroorganisms, probably reflects the influences of both historical and contemporary environmental conditions.
The intrinsic multidimensional and dynamic characteristics of the ocean complicate the test of classical ecological models such as the taxa–area or the distance–decay relationship recently applied to microorganisms (20, 21). For instance, in our study, distance was much less important than water mass for explaining differences in community composition. Communities originating from the same water mass but separated by thousand kilometers (e.g., DAO_0003 and DAO_0005 or DAO_0004 and DAO_0007) were much more similar to each other than communities only separated by a few 100 m (e.g., DAO_0003 and DAO_0004) but originating from different water mass. Thus, the concept of spatial scale that has been applied to microorganisms in soils (22), springs (23), or lakes (24) cannot be directly transposed to marine ecosystems where water masses with motion have to be considered.
Our results indicate that the vast majority of the rare phylotypes (99%) were always rare, i.e., never detected as abundant in any of the samples analyzed. Those samples comprised contrasting environmental conditions such as winter and summer, or surface and deep water masses. If rare phylotypes were acting as a source (seed bank), they would be expected to be rare under certain environments and abundant when conditions become adequate. For example, rare deep water phylotypes could become abundant if brought up to the surface. Similarly, a phylotype rare in winter could bloom during summer and become abundant. However, Kirchman et al. (13) recently reported that phylotypes that were rare in winter in surface waters of the Arctic Ocean did not become abundant during summer. Likewise, our results indicate that rare phylotypes remained rare when surface samples were compared with deep water samples. For macroorganisms, data also suggest that species retain their basic status as common or rare up to one million years (25). We therefore hypothesize that regardless of spatial or temporal scales most of the rare phylotypes are always rare within an ecosystem. Under that hypothesis, the few rare phylotypes that are sometimes detected as abundant represent traces of phylotypes that are highly abundant in some habitats (here water masses). A very high abundance implies a probable higher dispersion rate, resulting in abundant phylotypes being present (as rare) in many different habitats. This was illustrated by the higher proportion of phylotypes that were abundant and never rare in deep water (35% of abundant sequences) than in surface waters (19%). Abundant surface phylotypes have higher probability of sinking and be rare at depth than deep water phylotypes have to reach the surface.
The phylogenetic composition of the rare biosphere was similar to the composition of the abundant members of the community. The abundant taxonomic groups are thought to be well adapted to their environment and to contribute the most to biomass production (10, 26). However, some rare species are important for global biogeochemical cycling. Nitrogen fixation in the open ocean, for example, is mediated by rare members of the microbial community (27). However, the similarity between the phylogeny of rare and abundant phylotypes makes us speculate that most rare phylotypes are adapted to and active in their environment in a way similar to that of the abundant phylotypes. In surface waters, for example, many rare phylotypes belong to the class Gammaproteobacteria or the phylum Bacteroidetes, which account for a high proportion of biomass production in surface Arctic waters (28). In the deep waters, members of the rare biosphere are Deltaproteobacteria (SAR324), Chloroflexi (SAR202), and SAR406, all specific to and active in the deep ocean (29, 30). The distribution patterns of the rare biosphere phylogeny suggest that rare members of a community along with the abundant members may represent the pan community able to carry out all of the biogeochemical function of the ecosystem (31).
There were, however, some differences in the taxonomic composition of rare vs. abundant phylotypes, such as the higher number of unassigned phylotypes within the rare biosphere. This is an additional argument indicating that the rare biosphere is composed of microbes that are always rare. If the phylotypes classified as rare in our samples had been abundant in another habitat, they would be expected to have been in databases, given recent global sequencing efforts. This is not the case, as rare phylotypes were not easily identified by comparison with databases.
The abundance distributions of rare and abundant phylotypes were different. Rare phylotypes followed a log-series distribution model and abundant phylotypes a log-normal albeit poorly. Using the same models, core species of macroorganisms may be separated from occasional species (32). Thus, at our sampling scale, microorganisms followed ecological patterns similar to those of macroorganisms. Those different distributions argue for a genuine separation between rare and abundant phylotypes. Other lines of evidences also indicate that we successfully targeted the rare members of the microbial communities. First, the low diversity of archaea communities enabled good coverage of the natural communities by our pyrosequencing effort. We can assume that if the community diversity was well covered, then we were also able to detect the rare phylotypes. Second, our abundance criterion to define rare phylotypes was 10 times more severe than with earlier definitions (33).
There has been some concern that sequencing errors may produce the tremendous microbial diversity detected by pyrosequencing (34). Recent studies have demonstrated that the abundance of genes and taxa can be overestimated in metagenomes (35) and that diversity can be overestimated in 16S rRNA gene studies using pyrosequencing (36, 37). Methods to reduce errors have, however, been proposed. The new program PyroNoise detects errors directly from the light intensity flowgram generated by the pyrosequencer (37), but its computational demands may limit its use (34). Alternatively, quality filters applied to the reads themselves can remove artefactual sequences and greatly improve the quality of the dataset (38, 36). Finally, an additional step to avoid overestimating diversity consists of clustering pyrosequencing reads into phylotypes using a 97% identity threshold (36). Our cautious methodological approach, based on both the use of quality filters to remove erroneous sequences and the clustering of reads into phylotypes, should guarantee that most of the rare biosphere that we observed was real. The quality filters applied to our dataset limit the number of errors by pushing the pyrosequencing accuracy rate to >99.8% (38). In addition, the best-match definition that we used to define phylotypes corresponded to clusters sharing 94% identity (12), which is much lower than the 97% threshold advocated (36). This low identity level should substantially reduce the possibility that any erroneous sequences that may have passed through the quality filter appeared as new phylotypes, as any sequences containing small errors were grouped with error-free phylotypes from which they would have been derived.
In summary, the patterns of biogeography argued against a cosmopolitan distribution of the rare biosphere, and rather suggested ecological controls of the diversity and limited dispersal potentials. It means that microbes inhabit water masses with physical boundaries, created by density differences, which strongly limit the dispersion of communities. Within those water masses, the rare phylotypes together with the abundant members may constitute an active pan community essential for functioning of the marine ecosystem.
Materials and Methods
Sample Collection.
Samples were collected from surface (samples ACB) and deep water masses (samples DAO) in five geographical regions of the Arctic Ocean in the Chukchi Sea, Beaufort Sea, Franklin Bay, Baffin Bay, and Laptev Sea (Table S1). Samples were collected during cruises conducted through various Arctic research projects (7, 13). Water was sampled with a rosette system fitted with PVC bottles and equipped with conductivity, temperature, and depth (CTD) profiler. Within 30 min of collection into clean PVC bottles, 5–6 L of sea water were filtered successively through a 50-μm mesh (DAO samples), 3-μm (DAO samples), or 0.8-μm (ACB samples) pore size 47-mm polycarbonate (PC) filters, and finally a 0.2-μm pore size Sterivex unit (DAO samples) (Millipore Canada Ltd. ) or 0.2-μm pore size Durapore (Milliopore) filters (ACB samples). The whole cell concentrates were preserved in 1.8 mL of buffer (50 mM Tris·HCl, 0.75 M sucrose, 40 mM EDTA) and frozen at −80 °C.
DNA Extraction and Pyrosequencing.
The DNA extraction methods were described previously for ACB (13) and DAO samples (7). The bacterial hypervariable V6 region of the 16S rRNA gene was amplified using a pool of five forward primers and four reverse primers described earlier (8). The archaeal V6 16S rRNA was amplified with a set of one forward primer and two reverse primers (8). The final amplicon was sequenced with a 454 Life Sciences GS20 sequencer at the Marine Biological Laboratory in Woods Hole, Massachusetts. For each read from the sequencer, the primer bases were trimmed from the beginning and the end, and low-quality sequences were removed (38). Sequences were flagged as low quality when they had <50 nucleotides in length, the start of the sequence did not exactly match a primer sequence, one or more Ns were in the sequence, or the first five nucleotides of a tag did not correspond to the expected 5 nucleotides run key (used to sort the pyrosequencing reads). Pyrotag sequences have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession numbers SRP001222 and SRP001208.
Taxonomic Identification of Pyrosequencing Sequences.
The taxonomic identification of the pyrosequencing reads (“tags”) followed the approach of Sogin et al. (11). The tags were compared by BLASTN to a reference database of hypervariable region tags (RefHVR_V6, http://vamps.mbl.edu/) based on the SILVA database, version 95 (39), and the 100 best matches were aligned to the tag sequences using MUSCLE (40). A reference sequence or sequences were defined as those having the minimum global distance (number of insertions, deletions and mismatches divided by the length of the tag) to the tag sequence, and all reads showing the best match to the same reference V6 tag were grouped together as representing a same phylotype (11). Taxonomy was assigned to each reference sequence (phylotype) with the RDP Classifier (41).
Bacteria from the deep Arctic Ocean for which few reference sequences were available were also taxonomically identified by comparing pyrosequencing reads against nearly full-length sequences obtained by cloning (12).
Defining Abundant and Rare Phylotypes.
Abundant phylotypes were defined as the phylotypes with a representation >1% within a sample, which corresponds to the definition by Pedrós-Alió (2). Rare phylotypes were defined arbitrarily as having an abundance <0.01% within a sample. The 0.01% threshold represents the lowest frequency of occurrence that we could detect with our pyrosequencing effort and corresponds to the frequency of phylotypes found only once in our sample with the fewest tags (sample ACB_0002, n = 9,849). Our definition is much more severe than the <0.1% criteria proposed earlier and corresponds to phylotypes that are difficult to detect by traditional PCR-based techniques (2, 33).
Phylotype Definition and Data Analysis.
To target different levels of diversity within the rare members of the microbial communities, we investigated both bacteria and archaea communities, archaea having a lower diversity than bacteria (6). Furthermore, to avoid bias resulting from how phylotypes are defined, we used two different phylotype definitions to infer the ecology of the rare biosphere (Fig. S7). The “best match” definition was the less stringent. Reference sequence-based phylotypes were defined as containing all sequences sharing the same RefHVR_V6 database sequence (or group of sequences) as their nearest neighbor. This definition has been used frequently for microbial ecology studies by pyrosequencing (7, 11, 42) and corresponded to a 94% identity clustering threshold (12). If not specified otherwise, this is the phylotype definition used for all analysis throughout our study. For comparison we also used a much more stringent definition in which sequences with 100% identity were grouped as belonging to the same phylotype. We called this the “100% identity” phylotype definition.
To estimate community similarity among samples we applied a hierarchical cluster analysis on the basis of the abundance of phylotypes in the communities using Bray–Curtis similarity and a dendrogram inferred with the unweighted pair–group average algorithm. To determine the robustness of the clustering, data were subjected to bootstrapping with 1,000 resampling and the analysis was rerun after removing the largest and the smallest samples. The difference in phylotype composition was examined by nonmetric multidimensional scaling analysis. The analysis was based on the relative abundance of the phylotypes within each sample and calculated as Bray–Curtis similarity. Analysis of similarity statistics were used to verify the significance of the dendrogram clustering by testing the hypothesis that bacterial communities from the same cluster were more similar in composition with each other than with communities in different clusters. A Bray–Curtis similarity matrix computed from the abundance of phylotypes was used to generate one-way analysis of similarity statistics with 10,000 permutations.
Abundance distributions were plotted following power-of-2 abundance classes (octave classes) and then fit to abundance models. The log-normal and log-series fitting algorithms (43) were applied to each dataset and a significance value was calculated based on a χ2 test. The model with p≠0 was chosen as the one best fitting the abundance distribution.
All statistical and diversity analyses were conducted with the program PAST, version 1.91 (44).
Supplementary Material
Acknowledgments.
We deeply appreciate the assistance and support of the men and women of the Canadian Coast Guard icebreaker CCGS Louis St. Laurent, and the U.S. Coast Guard icebreaker USCGC Healy. We acknowledge Deep Arctic samples collected by K. Scarcella, E. Didierjean, and M-É Garneau. We also thank two anonymous reviewers for suggestions and constructive comments. This work was supported by Marie Curie Grant CRENARC MEIF-CT-2007–040247 (to P.E.G.); Spanish Grant CGL2006–12058-BOS (to E.O.C.); a grant from the Natural Sciences and Engineering Council, Canada, Special Research Opportunity Fund and ArcticNet (C.L.); National Science Foundation Grant OPP0632233 (to D.K.); and a Keck foundation grant to the International Census of Marine Microbes led by M. Sogin and L. Ameral Zettler. Financial and ship time support were also provided by Fisheries and Oceans Canada and the International Polar Year Programs' Canada's Three Oceans project and the Nansen and Amundsen Basins Observational System project. This is a contribution to the International Census of Marine Microbes (ICOMM).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908284106/DCSupplemental.
References
- 1.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Micro. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 2.Pedrós-Alió C. Marine microbial diversity: Can it be determined? Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Ann Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 4.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 5.Hagstrom A, et al. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl Environ Microbiol. 2002;68:3628–3633. doi: 10.1128/AEM.68.7.3628-3633.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aller JY, Kemp PF. Are archaea inherently less diverse than bacteria in the same environments? FEMS Microbiol Ecol. 2008;65:74–87. doi: 10.1111/j.1574-6941.2008.00498.x. [DOI] [PubMed] [Google Scholar]
- 7.Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- 8.Huber JA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 9.Casamayor EO, Schafer H, Baneras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: Comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cottrell MT, Kirchman DL. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol Oceanogr. 2003;48:168–178. [Google Scholar]
- 11.Sogin ML, et al. Microbial diversity in the deep sea and the under explored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galand PE, Potvin M, Casamayor EO, Lovejoy C. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 2009 doi: 10.1038/ismej.2009.134. [DOI] [PubMed] [Google Scholar]
- 13.Kirchman DL, Cottrell MT, Lovejoy C. Bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2010.02154.x. in press. [DOI] [PubMed] [Google Scholar]
- 14.Martiny JBH, et al. Microbial biogeography: Putting microorganisms on the map. Nat Rev Micro. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 15.Agogué H, Brink M, Dinasquet J, Herndl GJ. Major gradients in putatively nitrifying and non-nitrifying archaea in the deep North Atlantic. Nature. 2008;456:788–791. doi: 10.1038/nature07535. [DOI] [PubMed] [Google Scholar]
- 16.Galand PE, et al. Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ Microbiol. 2009;11:971–980. doi: 10.1111/j.1462-2920.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- 17.Darling KF, et al. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature. 2000;405:43–47. doi: 10.1038/35011002. [DOI] [PubMed] [Google Scholar]
- 18.de Vargas C, Norris R, Zaninetti L, Gibb SW, Pawlowski J. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc Natl Acad Sci USA. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darling KF, Kucera M, Pudsey CJ, Wade CM. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc Natl Acad Sci USA. 2004;101:7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green J, Bohannan BJM. Spatial scaling of microbial biodiversity. Trends Ecol Evol. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM. A taxa-area relationship for bacteria. Nature. 2004;432:750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- 22.Franklin RB, Mills AL. Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol Ecol. 2003;44:335–346. doi: 10.1016/S0168-6496(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 23.Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 24.Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO. Does ecosystem size determine aquatic bacterial richness? Ecology. 2005;86:1715–1722. [Google Scholar]
- 25.McGill BJ, Hadly EA, Maurer BA. Community inertia of Quaternary small mammal assemblages in North America. Proc Natl Acad Sci USA. 2005;102:16701–16706. doi: 10.1073/pnas.0504225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Jiao N, Cottrell MT, Kirchman DL. Contribution of major bacterial groups to bacterial biomass production along a salinity gradient in the South China Sea. Aqua Microbial Ecol. 2006;43:233–241. [Google Scholar]
- 27.Montoya JP, et al. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature. 2004;430:1027–1032. doi: 10.1038/nature02824. [DOI] [PubMed] [Google Scholar]
- 28.Malmstrom RR, Straza TRA, Cottrell MT, Kirchman DL. Diversity, abundance, and biomass production of bacterial groups in the western Arctic Ocean. Aqua Microbial Ecol. 2007;47:45–55. [Google Scholar]
- 29.Varela MM, van Aken HM, Herndl GJ. Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub)tropical Atlantic. Environ Microbiol. 2008;10:1903–1911. doi: 10.1111/j.1462-2920.2008.01627.x. [DOI] [PubMed] [Google Scholar]
- 30.Giovannoni SJ, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437:343–348. doi: 10.1038/nature04158. [DOI] [PubMed] [Google Scholar]
- 31.Höfle MG, Kirchman DL, Christen R, Brettar I. Molecular diversity of bacterioplankton: Link to a predictive biogeochemistry of pelagic ecosystems. Aqua Microbial Ecol. 2008;53:39–58. [Google Scholar]
- 32.Magurran AE, Henderson PA. Explaining the excess of rare species in natural species abundance distributions. Nature. 2003;422:714–716. doi: 10.1038/nature01547. [DOI] [PubMed] [Google Scholar]
- 33.Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- 34.Reeder J, Knight R. The “rare biosphere”: A reality check. Nat Methods. 2009;6:636–637. doi: 10.1038/nmeth0909-636. [DOI] [PubMed] [Google Scholar]
- 35.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1417. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 36.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: Pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2009 Aug 27; doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 37.Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- 38.Huse S, Huber J, Morrison H, Sogin M, Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krebs CJ. Ecological Methodology. New York: Harper & Row; 1989. [Google Scholar]
- 44.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological Statistics Software Package for Education and Data Analysis. 2001. [Accessed May 20, 2009]. Available at http://folk.uio.no/ohammer/past/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.