Abstract
Escherichia coli MutS forms a mispair-dependent ternary complex with MutL that is essential for initiating mismatch repair (MMR) but is structurally uncharacterized, in part owing to its dynamic nature. Here, we used hydrogen/deuterium exchange mass spectrometry and other methods to identify a region in the connector domain (domain II) of MutS that binds MutL and is required for mispair-dependent ternary complex formation and MMR. A structurally conserved region in Msh2, the eukaryotic homolog, was required for formation of a mispair-dependent Msh2–Msh6–Mlh1–Pms1 ternary complex. These data indicate that the connector domain of MutS and Msh2 contains the interface for binding MutL and Mlh1–Pms1, respectively, and support a mechanism whereby mispair and ATP binding induces a conformational change that allows the MutS and Msh2 interfaces to interact with their partners.
Keywords: deuterium exchange, mass spectrometry, mismatch repair, Mlh1–Pms1, Msh2–Msh6
Cells have evolved a network of DNA repair pathways that respond to various types of genotypic stress to maintain the stability of their genome. For wild-type cells the mutation rate is extremely low (≈1 × 10−9 to 1 × 10−10 per cell division) (1), which is in part due to DNA mismatch repair (MMR) that removes base–base mismatches and small insertion/deletion mismatches, which arise because of errors in DNA replication, and reduces the error rate of DNA replication by 2 to 3 orders of magnitude (2–5). MMR proteins are also important for recombination and checkpoint responses that lead to the induction of apoptosis in response to some DNA-damaging agents (4, 6, 7). MMR is conserved from bacteria to humans and prevents the development of cancers in humans (8, 9).
The initial stages of MMR are similar in both bacteria and eukaryotes. Mispairs in DNA are recognized by the MutS homodimer in Escherichia coli or by one of two heterodimers of MutS homologs, Msh2–Msh6 or Msh2–Msh3, in eukaryotes (2, 10, 11). This complex then recruits the MutL homodimer in E. coli or, in eukaryotes, one of two MutL heterodimeric complexes, Mlh1–Pms1 or Mlh1–Mlh3, in an ATP-dependent manner (12–16). In E. coli, MutS–MutL–DNA ternary complex stimulates the endonucleolytic activity of MutH, which makes single-strand breaks in the unmethylated DNA strand at transiently hemimethylated GATC sites and thus distinguishes the unmethylated daughter DNA strand from the methylated parental DNA strand during and after DNA replication (17–19). The nick serves to mediate excision and strand resynthesis of the newly synthesized DNA to remove the mispair (20–22). In contrast to E. coli, the downstream events after formation of the ternary complex in eukaryotes, particularly those leading to the initiation of strand-specific MMR, are not well understood.
Despite the numerous reports examining the mechanistic features of the MutS–MutL–DNA complex in the initiation of MMR (2) and the available structures of MutS (23, 24) and the N- and C-terminal domains of MutL (25, 26), little is known about how MutS interacts with MutL. Recently, mutations in the N-terminal domain of Mlh1 were shown to eliminate Msh2-Msh6 binding, although it is unclear whether the mutations affect a region directly involved in complex assembly (27). In addition, the mispair binding domain of Msh6 and the mispair that is being recognized are unlikely to be parts of the interface as revealed by the genetics of an msh6 allele encoding the mispair binding domain of Msh3 (28). There are several reasons for the enigmatic nature of this complex. First, the MutS–MutL–DNA ternary complex and the eukaryotic MSH–MLH–DNA complex are dynamic and exhibit rapid dissociation from DNA (2, 12, 13, 19, 29), which may explain the failure of most large-scale physical interaction studies to identify the Saccharomyces cerevisiae ternary complexes (30–34). Second, both ATP binding and mispair binding are required for the interaction of MutS with MutL and Msh2-Msh6 with Mlh1-Pms1 (2, 12, 13, 19). These cofactors likely transiently induce conformational changes required for ternary complex formation (2). Given the potential complications of understanding this transient complex, we sought an approach that would allow us to identify the protein–protein interface within the context of the full-length proteins in solution.
Results
MutS Has Two Regions of Decreased Solvent Accessibility in the Presence of MutL.
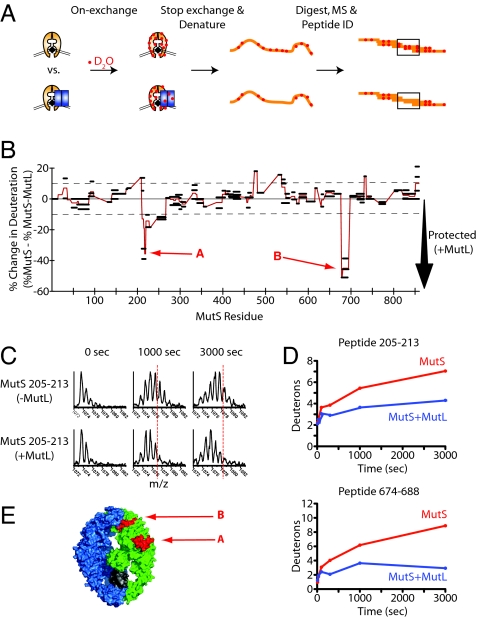
Hydrogen/deuterium exchange mass spectrometry (DXMS) of main-chain amides was used to examine the solvent accessibility of MutS in the presence of ATP and a 71-nt DNA substrate containing a central GT mispair either with or without MutL (Fig. 1A). Regions of MutS directly at the interface were anticipated to have reduced access to solvent and thus reduced levels of deuteration in the presence of MutL. One hundred fifteen high-quality MutS peptides recovered from both sets of reactions were identified by mass spectrometry, resulting in 74% coverage of MutS sequence. Of these, 11 peptides from two distinct regions of MutS, regions A and B (Fig. 1B), had >10% reduction in deuteration when incubated in the presence of MutL (Fig. 1). Region A is located in domain II, the connector domain of MutS, and is exposed on the surface of MutS (23, 24). Two peptides in this region, spanning amino acids 204–213 and 205–213, had a >30% reduction in deuterons incorporated in the presence of MutL after 3,000 sec (Fig. 1B). The centroid of the mass spectra was shifted to a greater extent in the absence of MutL than in its presence; see the 1,000- and 3,000-sec time points (Fig. 1C). Two other overlapping peptides in this region, spanning amino acids 201–210 and 204–208, did not exhibit any change in deuteration in the presence of MutL; thus, it is likely that amino acids 211, 212, and 213, which make up 33% and 37.5% of peptides 204–213 and 205–213, respectively [the hydrogens from the peptide N terminus back-exchange too rapidly to be observed by DXMS (35)], were not deuterated in the presence of MutL. These peptides contain a number of hydrophobic amino acid side chains that are exposed to the solvent (23). An additional five peptides in domain II covering amino acids 214–225 and 241–260 had modest reductions (10–20%) in deuteration in the presence of MutL. Region B (Fig. 1B), located in domain V, was represented by four peptides that had a ≈40–50% reduction in deuterium incorporation and covered amino acids 673–686. This region is near the ATP binding site of the MutS dimer and has few solvent-exposed side chains. The peptides from both regions had similar kinetics of protection (Fig. 1D). Region A from 204–225 and region B from 673–686 are shown mapped onto the MutS—GT structure (23) (Fig. 1E and Fig. S1).
Fig. 1.
DXMS reveals two regions of decreased solvent accessibility on MutS in presence of MutL. (A) Experimental scheme for examining the effect of ternary complex formation with MutL on the solvent accessibility of MutS. (B) The difference in percentage between deuterium incorporation for individual peptides of MutS in the presence of a 71-bp DNA substrate containing a GT mispair and ATP and in the presence or absence of MutL after 3,000 sec. Peptides are shown as black bars spanning over the indicated sequence on the x axis. Negative values represent decreases in deuterium incorporation in the presence of MutL, relative to the absence of MutL. The two regions of significant MutL-dependent reduced deuteration are marked as A and B. (C) The mass spectra of a representative peptide from region A corresponding to residues 205–213 of MutS in the presence of a DNA substrate containing a GT mispair, ATP, and the presence (Bottom) or absence (Top) of MutL. Note the reduction in shifting of the centroid of the peak in the presence of MutL. The vertical dashed lines are arbitrary reference points used to visualize the peak shift. (D) Kinetics of deuteron incorporation for a representative peptide from regions A and B. (E) The two regions of MutS with MutL-dependent reduced deuteration that were denoted A and B, described above, are shown in red on the surface of the non-mispair contacting subunit of MutS [Protein Data Bank (PDB) ID code 1e3m].
MutS-211 Is Defective in Ternary Complex Formation with MutL.
The MutL-dependent reduced solvent accessibility of regions A and B of MutS could be due to direct contacts or induced conformational changes. To differentiate between these possibilities, we constructed a series of mutant MutS proteins with amino acid substitutions of residues that were solvent exposed in the crystal structure (23) in and around regions A and B. Amino acids adjacent to regions A and B were also included because our DXMS experiment cannot measure the rapid side-chain hydrogen exchange (35) and may not identify all of the interface.
We began by making two proteins with multiple amino acid substitutions in and around region A, which had the greatest degree of MutL-dependent protection from solvent exposure (Fig. 1B). Constructs altering extensive stretches of residues in region A, replacement of amino acids 201–217 of MutS with the unstructured amino acids 7–24 of S. cerevisiae Msh6 (36), and the alteration of all nine solvent-exposed amino acids (W202K, E203K, E205K, D207K, R210D, Q211S, Q212S, N214S, and L215E), encoded insoluble proteins, which were not analyzed further. In contrast, constructs altering shorter stretches of solvent-exposed amino acids in region A, mutS-205 (E205S, D207S) and mutS-211 (Q211S, Q212S, N214S, L215S), encoded soluble proteins. These proteins were purified and tested for interaction with MutL on a 236-nt DNA substrate containing a central GT mispair using surface plasmon resonance. Buffer containing only MutS and MutL protein were first injected over the immobilized GT mispair substrate, and only MutS bound to the mispair. After reaching equilibrium, buffer containing MutS, MutL, and ATP was injected. In the case of wild-type MutS, robust MutS–MutL–DNA complex formation was seen with MutL (Fig. 2A). MutL did not bind to DNA in the absence of MutS, consistent with previous studies with MutL and the eukaryotic homolog Mlh1–Pms1 (13, 14). MutS-205 had a modest defect in MutL binding, whereas MutS-211 (Fig. 2 A and B) had a substantial MutL binding defect. Ternary complex with MutS-211 formed at concentrations of MutL greater than 100 nM, but never at the levels observed for wild-type MutS.
Fig. 2.
MutS-211 is defective in ternary complex assembly with MutL. (A) Biosensor analysis of the association of wild-type MutS (solid line), MutS-205 (dashed line), or MutS-211 (dotted line) with a DNA substrate containing a GT mispair. MutL was present in the association (black line) buffer, but the MutL–MutS–DNA ternary complex formed only after addition of ATP (red line). (B) Amino acids affected by the mutS-211 mutation highlighted in red on the MutS–GT mispair structure (PDB ID code 1e3m). (C) MutS-211 binds mispaired DNA and dissociates upon ATP binding. Biosensor analysis of the association of either wild-type MutS or MutS-211 with a DNA substrate containing a GT mispair (black solid line) or a GC base pair (black dashed line) in the absence of nucleotide. Dissociation was observed by switching the flow from the protein-containing buffer to buffer containing ATP (red line). (D) MutS-211 is proficient in sliding clamp formation. Biosensor analysis of the association of the indicated MutS complex in the presence of ATP is shown with a DNA substrate containing a GT mispair either with (solid red line) or without (dashed red line) a LacI-blocked end. The LacI-block was released by adding IPTG (solid green line).
We were unable to generate mutant proteins that had amino acid substitutions affecting region B that had specific MutL binding defects. All of the mutants generated either had additional defects in ATP-induced conformational changes, as demonstrated by the failure of the mutant MutS proteins to dissociate off of a mispair upon ATP binding as described below (T675K, E676K and N679S, H682S, N683S), or were insoluble (T685K, E686K) and not analyzed.
MutS-211 Binds to Mispairs and Slides on DNA upon Binding ATP.
Mutations in residues in S. cerevisiae Msh2 and Msh6 that result in ATP binding defects and sliding defects also generally result in Mlh1–Pms1 binding defects (37). To rule out the possibility that the MutL-binding defect of MutS-211 was not due to a specific MutL interface defect, we determined whether MutS-211 could bind DNA with specificity for a mispair and convert to a DNA-sliding form in an ATP-dependent fashion by surface plasmon resonance.
We found that MutS-211 still bound DNA in the absence of nucleotide, retained mispair specificity similar to wild-type MutS, and rapidly dissociated from the mispair upon binding ATP (Fig. 2C). To verify that rapid ATP-dependent dissociation was due to the conformational changes that induce the sliding form rather than from direct dissociation, we directly tested its ability to slide on DNA using a previously developed method (13) that uses LacI as a DNA end-block that can be readily removed upon addition of isopropyl beta-D-1-thiogalactopyranoside (IPTG) (T1/2 = 1.6 sec). When ATP binds to mispair-bound MutS, MutS is converted to a form that dissociates from the mispair by sliding off of the end of the DNA. In the presence of LacI protein, a higher level of MutS binding is seen on the mispair substrates (Fig. 2D) because the end-block prevents rapid dissociation of the MutS off of the ends of the DNA, and sliding of MutS exposes the mispair, allowing additional MutS to bind (13, 14, 19, 38). Addition of IPTG resulted in rapid dissociation of MutS by sliding off of the DNA ends due to loss of the LacI end-block (Fig. 2D). We found that MutS-211 behaved identically to wild-type MutS protein in its ability to bind DNA, retain mispair specificity, and slide off of the mispair upon ATP binding (Fig. 2 C and D). Thus, the only defect we observed biochemically for MutS-211 was its ability to interact with MutL (Fig. 2A).
mutS-211 Causes Defects in MMR In Vivo.
We introduced the mutS-205 and mutS-211 alleles onto the E. coli chromosome and tested the mutant strains for increased rates of accumulating rifampicin-resistant mutations, indicative of an MMR defect. The mutS-205 strain was nearly identical to the wild-type strain for MMR (P = 0.0627 for difference) (Table 1). Conversely, the mutS-211 allele caused a much higher mutation rate and was found to be indistinguishable from the MMR-defective mutSΔ11 allele, in which all but the first 11 aa of MutS are deleted (P = 0.8005 for difference) (Table 1). Thus, the mutS-211 allele, which produces a protein that is only biochemically distinguishable from wild-type MutS because of a failure to bind MutL, was completely defective for MMR in vivo.
Table 1.
Effect of mutS mutations on the rate of spontaneous mutations
| Strain | Relevant genotype | Mutation rate (RifR)* |
|---|---|---|
| RDK5011 | Wild-type | 1.8 [0.8–3.0] × 10−8 (1) |
| RDK5014 | mutSΔ11 | 5.5 [4.0–9.4] × 10−7 (30) |
| RDK5012 | mutS205 | 2.5 [1.8–2.9] × 10−8 (1) |
| RDK5013 | mutS211 | 5.9 [5.9–12.2] × 10−7 (32) |
*The numbers in brackets represent low and high values, respectively, for the 95% confidence interval for each rate. The numbers in parentheses indicate rate relative to wild-type rate.
MutS-211,2 Is Defective in Binding MutL.
Two additional mutant proteins, MutS-211,2 (Q211S, Q212S) and MutS-214,5 (N214S, L215S), were purified and studied. MutS-214,5 bound MutL in a manner indistinguishable from wild-type, whereas MutS-211,2 was completely defective in MutL binding (Fig. 3A). Both mutant proteins bound to mispaired DNA and dissociated from the DNA substrate upon ATP binding, similar to wild-type MutS (Fig. 3B). Therefore, Q211 or Q212 or both are essential for binding MutL, and mutating those residues is responsible for the defects found in MutS-211.
Fig. 3.
MutS-211,2, but not MutS-214,5, is defective in ternary complex assembly with MutL. (A) Biosensor analysis of the association of either the wild-type or mutant MutS complexes, as indicated, in the presence of ATP with a DNA substrate containing a GT mispair. Flow was switched at the indicated point to the same buffer containing MutS and ATP but additionally containing either 0 or 50 nM MutL (dashed and solid lines, respectively) at the indicated point and ternary complex formation was monitored. (B) Biosensor analysis of the association of either the wild-type or mutant MutS complexes, as indicated, with a DNA substrate containing a GT mispair (black solid line) or a GC base pair (black dashed line) in the absence of nucleotide. Dissociation was observed by switching the flow from the protein-containing buffer to buffer containing ATP (red line).
Msh2–Msh6 Complexes with Mutations in Amino Acids in Domain II of Msh2 Are Defective for MMR In Vivo and have Mlh1–Pms1 Binding Defects In Vitro.
We next addressed whether the MutL interface identified in MutS was conserved in the S. cerevisiae Msh2–Msh6 mispair binding complex and whether it was present in Msh2 or Msh6 (39). Msh2 domain II showed greater structural conservation with MutS domain II in the region altered in MutS-211 than Msh6 domain II (Fig. 4 and Fig. S2). We nonetheless tested a series of msh2 and msh6 mutants containing mutations affecting residues in the region in domain II that are required for the MutS interaction with MutL. The msh2 mutants were tested for their ability to complement the high mutation rate of an msh2Δ strain when present on a low-copy-number plasmid, and the msh6 mutants were tested for their ability to complement a msh3Δ msh6Δ strain to eliminate the genetic complexity due to the partial redundancy of MSH6 and MSH3 (40).
Fig. 4.
msh2 and msh6 domain II mutants have MMR defects in vivo. (A) Residues mutated in S. cerevisiae Msh2 in the same region as the E. coli mutS-211 mutations (PDB ID code 1e3m, green) are colored in red on the model of H. sapiens Msh2 domain II (PDB ID code 2o8b, violet). (B) MSH2 complementation of a msh2Δ strain. The indicated msh2 alleles were expressed on a low-copy-number plasmid bearing a marker allowing growth on media lacking Ura. Plasmids were transformed into a msh2Δ strain, and isolates were patched onto plates lacking Ura and then replica plated onto plates lacking Ura and Lys or Ura and Thr. (C) Residues mutated in S. cerevisiae Msh6 in the same region as the E. coli mutS-211 mutations (PDB ID code 1e3m, green) are colored red on the model of H. sapiens Msh2 domain II (PDB ID code 2o8b, blue). (D) MSH6 complementation of a msh3Δ msh6Δ strain. The indicated msh6 alleles were expressed on a low-copy-number plasmid bearing a marker allowing growth on media lacking Leu. Plasmids were transformed into a msh3Δ msh6Δ strain, and isolates were patched onto plates lacking Leu and then replica plated onto plates lacking Leu and Lys or Leu and Thr.
Patch tests revealed that only the msh2–235 allele (K235D, V237S) was able to complement the mutator phenotype of the msh2Δ strain, whereas the msh2–237 (V237S, E238S, D240S, L214S), msh2–241 (L241S, K243D), and msh2–249 (L249S, L251S) alleles all resulted in a strong mutator phenotype (Fig. 4B). Only one of the MSH6 alleles, msh6–573, failed to complement the mutator phenotype of the msh3Δ msh6Δ strain and resulted in a partial mutator phenotype (Fig. 4D); however, this mutation affected residues that were removed from the region of domain II predicted to interact with MutL family proteins.
We next biochemically characterized the mutant Msh2–Msh6 complexes that failed to fully complement the MMR deficient strains. We were unable to overexpress Msh2–249–Msh6 or Msh2–Msh6–573, suggesting the MMR defects caused by these mutations might be due to protein folding problems. We were able to overproduce and purify the Msh2–237–Msh6 and Msh2–241–Msh6 complexes, and these complexes were analyzed for their ability to form Msh2–Msh6–Mlh1–Pms1 ternary complexes on a GT mispair, essentially as described for MutS and MutL except that the DNA ends were blocked with LacI protein to prevent ternary complex at the DNA ends (13). In the absence of Mlh1–Pms1, both the Msh2–237–Msh6 and the Msh2–241–Msh6 complexes bound to the mispaired DNA substrate in the presence of ATP at wild-type-like levels (Fig. 5A). However, both mutant complexes showed greatly reduced levels of ternary complex formation as compared with wild-type Msh2–Msh6 (Fig. 5A). Control experiments showed that Msh2–237–Msh6 and Msh2–241–Msh6 bound to mispaired DNA and dissociated from the DNA substrate upon ATP binding, similar to wild-type Msh2–Msh6 (Fig. 5B), and were also proficient for ATP-induced sliding off of the ends of DNA, as revealed by releasing the end-block with IPTG (Fig. 5C). Thus, the msh2–237 and the msh2–241 mutations cause specific defects in binding Mlh1–Pms1.
Fig. 5.
Msh2–Msh6 complexes containing amino acid substitutions in domain II of Msh2 are defective in ternary complex assembly with Mlh1–Pms1. (A) Biosensor analysis of the association of either the wild-type or mutant Msh2–Msh6 complex in the presence of ATP with a DNA substrate containing a GT mispair and end-blocked with 30 nM LacI. 0 or 25 nM Mlh1–Pms1 (dashed and solid lines, respectively) was added at the indicated point, and ternary complex formation was monitored. (B) Biosensor analysis of the association and dissociation of either the wild-type or mutant Msh2–Msh6 complexes, as indicated, with a DNA substrate containing a GT mispair (solid line) or a GC base pair (dashed line). The indicated Msh2–Msh6 complex was bound to the DNA substrate in running buffer in the absence of nucleotide (black line). Dissociation was observed by switching the flow from the protein-containing buffer to buffer containing ATP (red line). (C) Biosensor analysis of the association of the indicated Msh2–Msh6 complex in the presence of ATP is shown with a DNA substrate containing a GT mispair either with (solid red line) or without (dashed red line) a LacI-blocked end. The LacI-block was released by adding IPTG (solid green line).
Domain II of MutS Fused to Maltose Binding Protein (MBP) Binds MutL and Domain II of Msh2 Fused to MBP Binds Mlh1–Pms1.
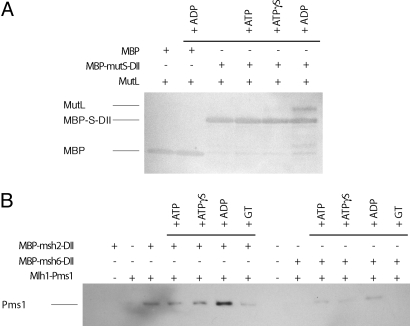
We next determined whether MutS domain II could directly mediate interactions that were sufficiently strong to bind MutL in vitro. We fused domain II of MutS (amino acids 116–266) to the C terminus of MBP. This MBP–MutS-DII fusion retained the ability of wild-type MBP to bind amylose and could be eluted with maltose. MBP or MBP–MutS-DII was incubated with and without MutL and amylose in either the absence of nucleotides or the presence of ADP, ATP, or ATPγS. After washing, the bound proteins were eluted with 10 mM maltose and analyzed by SDS-PAGE. A substoichiometric amount of MutL copurified with MBP–MutS-DII in the presence of ADP and to a lesser extent in the presence of ATP and ATPγS, but did not copurify with MBP under any conditions (Fig. 6A).
Fig. 6.
Domain II of MutS and Msh2 fused to the C terminus of MBP bind MutL and Mlh1–Pms1, respectively. (A) Reactions containing the indicated combinations of MBP, MBP–MutS–DII, MutL, and ADP, ATPγS, or ATP, as indicated, were incubated with amylose resin, washed, eluted with 10 mM maltose, run on an SDS-PAGE gel, and silver-stained. (B) Reactions containing MBP–Msh2-DII, MBP–Msh6-DII, Mlh1–Pms1, and ADP, ATPγS, ATP, or a DNA substrate containing a GT mispair, as indicated, were incubated with amylose resin, washed, eluted with 20 mM maltose, run on an SDS-PAGE gel, and a Western blot was performed using an anti-flag antibody to detect Mlh1–Pms1, which had a flag-tag on the C terminus of Pms1.
We then similarly analyzed domain II of Msh2 and/or Msh6 by fusing them to the C terminus of MBP. Purified MBP–Msh2-DII or MBP–Msh6-DII was incubated with and without Mlh1–Pms1 and amylose. After washing, the bound proteins were eluted with 20 mM maltose, fractionated by SDS-PAGE, and the gel was analyzed by Western blot using an anti-flag antibody that is specific for the flag-tag on the C terminus of Pms1 to detect bound Mlh1–Pms1 complex. The MBP–Msh2-DII protein bound the Mlh1–Pms1 complex, and this binding was enhanced by ADP (Fig. 6B), similar to the interaction between MBP–MutS-DII and MutL (Fig. 6A). The presence of mispair containing DNA, ATP, or ATPγS had no affect or slightly inhibited the interaction. In contrast, the MBP–Msh6-DII protein bound much less Mlh1–Pms1 compared with the MBP–Msh2-DII protein. These results, which indicate that the Msh2–DII is capable of efficiently binding Mlh1–Pms1, taken together with prior observations that mutations altering the critical interface residues of Msh2–DII result in defects in Msh2–Msh6 Mlh1–Pms1 ternary complex formation, indicate that the domain II of Msh2 contains the interface for binding Mlh1–Pms1.
Discussion
Here we have used DXMS to identify two regions of E. coli MutS that show substantial protection from deuteration in the presence of MutL: region A, corresponding to a surface region of domain II (the connector domain), and region B, corresponding to an α-helix in domain V (the ATPase domain). We were able to engineer several proteins with mutations in region A, including MutS-211 and MutS-211,2, that substantially inhibited the ability of MutS to associate with MutL without affecting other biochemical properties of MutS for DNA binding, mispair recognition, and ATP-induced sliding along DNA. This is a demonstration of mutant MutS proteins that are biochemically indistinguishable from wild-type MutS in all respects except MutL binding. Introduction of the mutS-211 allele onto the E. coli chromosome conferred a MMR null phenotype, and an isolated MutS domain II fusion with MBP bound MutL, indicating that domain II of MutS is necessary and sufficient to mediate MutL interactions and that these interactions are required for MMR in vivo. The role of region A seemed to be conserved in eukaryotes, because this region was conserved in Msh2 but not Msh6, and the introduction of mutations altering amino acid residues in this region of the S. cerevisiae MutS homologs Msh2, but not Msh6, generated mutations that were MMR defective in vivo and caused defects in binding Mlh1–Pms1 in vitro but did not cause defects in mispair recognition or ATP-induced sliding along DNA. Moreover, an isolated Msh2 domain II–MBP fusion, but not an Msh6 domain II–MBP fusion, was capable of binding MutL homologs similarly to the binding of MutS domain II–MBP with MutL, suggesting that domain II of Msh2, but not domain II of Msh6, contains conserved MutL homolog binding features of MutS domain II.
Although we cannot rule out a direct interaction between MutL and the partially protected region B, several lines of evidence lead us to suspect that this region is not part of the MutL interface. We were not able to identify mutations affecting region B that only affected MutL binding without causing additional defects that would be anticipated to indirectly cause MutL binding defects (37). Region B corresponds to an α-helix that, in the available crystal structures, is not substantially exposed to solvent. This α-helix has a disordered N terminus that contains the highly conserved N2 signature motif that functions in trans in ATP catalysis in the ATPase of the other MutS subunit in the homodimer (23, 24, 39, 41). If the ATPase domains of MutS close upon ATP binding similar to the Rad50 ABC ATPase (42), increased solvent protection could be due to ordering of the helix N terminus, which might be solely due to MutL-dependent stabilization of ATP-bound MutS and/or MutL-induced conformational changes to ATP-bound MutS, although at longer time points (1,000 to 3,000 sec) the disordered loop was fully deuterated even in the presence of MutL.
Localization of an MutL binding interface on domain II of MutS and identification of an equivalent function for domain II of the eukaryotic Msh2 suggests a number of key features for the MutS–MutL–DNA ternary complex. In the context of the MutS dimer, the two domain IIs are on opposite faces of the dimer (23, 24). Although it is possible, given the size and flexibility of MutL, that one MutL dimer might straddle the MutS–DNA complex and recognize both domain IIs simultaneously, evidence presented here for the S. cerevisiae homologs suggests that this is not the case. The crystal structures of the MutS homodimer on mispair-containing DNA revealed that the two subunits of MutS are functionally asymmetric (23, 24); one subunit directly recognizes the mispair, and the other subunit does not. This has been conserved in the Msh2–Msh6 and Msh2–Msh3 eukaryotic heterodimers; Msh6 and Msh3 directly recognize the mispair, and Msh2 does not (28, 39, 43–45). In fact, the domain of Msh2 that is the equivalent of the mispair binding domain is completely dispensable for Msh2–Msh6-mediated repair, although it does play some role in Msh2–Msh3-mediated repair (28, 43). Thus the fact that domain II of Msh2, but not domain II of Msh6, seems to conserve most of the MutL-recognition features is consistent with the stronger conservation of region A in Msh2 relative to Msh6. Moreover, the pattern of eukaryotic conservation is suggestive of additional functional separation of the two subunits of MutS and its homologs, whereby one subunit directly recognizes the mispair and the second subunit recruits the MutL homodimer.
Data presented here demonstrating the interactions between MutL and the isolated MutS domain II suggest that ATP and mispair binding by MutS, which are required for ternary complex formation, facilitate exposure of MutS domain II for MutL recruitment. The most robust domain II–MBP fusion interactions were observed in the presence of ADP, which seemingly contradicts previous observations that mutant MutL proteins with reduced ability to bind and hydrolyze ATP are proficient to bind MutS (14, 19); however, this could be explained if binding of the isolated domain II is stabilized by one of the N-terminal conformations of MutL induced by the ATP binding and hydrolysis required for MMR (25, 46, 47) or induces ADP binding by MutL. Given that the isolated domain II bypasses the MutS ATP binding requirement, ATP and mispair recognition by MutS may primarily drive conformational changes that cause domain II to become accessible for MutL recruitment. Intriguingly, the mispair binding domain and domain II are linked to each other and the remainder of MutS by floppy linkers in the MutS structures (23, 24), which would be consistent with the ability of these domains to reorganize on ATP and mispair binding. All of these structural features are conserved in the Msh2–Msh6 structures (39), and our data show that the Msh2 domain II interactions with Mlh1–Pms1 are remarkably similar genetically and biochemically. Thus, our data also imply that the mechanism controlling the eukaryotic Msh2–Msh6 and Mlh1–Pms1 interaction is conserved with the bacterial homologs.
Materials and Methods
Materials.
All E. coli proteins and MBP fusions had N-terminal His6 tags. See SI Materials and Methods for all plasmid and strain construction, as well as protein expression and purification.
Biochemical Methods.
DXMS analysis, Biacore experiments, and amylose pulldowns were performed as described previously (37, 48–51). These methods are described in SI Materials and Methods.
Genetic Analysis in E. coli and in S. cerevisiae.
Genetic methods for both E. coli and S. cerevisiae were performed as described previously (28, 52) and are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Malcolm Winkler, Manju Hingorani, and Anthony Poteete for plasmids and bacterial strains; and Kathleen Matthews for providing LacI protein. Supported by National Institutes of Health Grants GM50006 and CA92584 (to R.D.K) and CA099835, CA118595, and AI076961 (to V.L.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912250106/DCSupplemental.
References
- 1.Drake JW. Spontaneous mutation. Annu Rev Genet. 1991;25:125–146. doi: 10.1146/annurev.ge.25.120191.001013. [DOI] [PubMed] [Google Scholar]
- 2.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 3.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 4.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 6.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 7.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 8.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 9.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 10.Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 12.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSalpha. hMutLalpha.heteroduplex complexes. J Biol Chem. 2001;276:33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 13.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 14.Selmane T, Schofield MJ, Nayak S, Du C, Hsieh P. Formation of a DNA mismatch repair complex mediated by ATP. J Mol Biol. 2003;334:949–965. doi: 10.1016/j.jmb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Grilley M, Welsh KM, Su SS, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 16.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsh KM, Lu AL, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 18.Hall MC, Matson SW. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J Biol Chem. 1999;274:1306–1312. doi: 10.1074/jbc.274.3.1306. [DOI] [PubMed] [Google Scholar]
- 19.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 20.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc Natl Acad Sci USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 22.Runyon GT, Bear DG, Lohman TM. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc Natl Acad Sci USA. 1990;87:6383–6387. doi: 10.1073/pnas.87.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamers MH, et al. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 24.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 25.Ban C, Yang W. Crystal structure and ATPase activity of MutL: Implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 26.Guarne A, et al. Structure of the MutL C-terminal domain: A model of intact MutL and its roles in mismatch repair. EMBO J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plotz G, et al. Mutations in the MutSalpha interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res. 2006;34:6574–6586. doi: 10.1093/nar/gkl944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shell SS, Putnam CD, Kolodner RD. Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc Natl Acad Sci USA. 2007;104:10956–10961. doi: 10.1073/pnas.0704148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galio L, Bouquet C, Brooks P. ATP hydrolysis-dependent formation of a dynamic ternary nucleoprotein complex with MutS and MutL. Nucleic Acids Res. 1999;27:2325–2331. doi: 10.1093/nar/27.11.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 31.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 32.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 34.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 35.Garcia RA, Pantazatos D, Villarreal FJ. Hydrogen/deuterium exchange mass spectrometry for investigating protein-ligand interactions. Assay Drug Dev Technol. 2004;2:81–91. doi: 10.1089/154065804322966342. [DOI] [PubMed] [Google Scholar]
- 36.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hess MT, Mendillo ML, Mazur DJ, Kolodner RD. Biochemical basis for dominant mutations in the Saccharomyces cerevisiae MSH6 gene. Proc Natl Acad Sci USA. 2006;103:558–563. doi: 10.1073/pnas.0510078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gradia S, et al. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 39.Warren JJ, et al. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 41.Biswas I, et al. Disruption of the helix-u-turn-helix motif of MutS protein: Loss of subunit dimerization, mismatch binding and ATP hydrolysis. J Mol Biol. 2001;305:805–816. doi: 10.1006/jmbi.2000.4367. [DOI] [PubMed] [Google Scholar]
- 42.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 43.Lee SD, Surtees JA, Alani E. Saccharomyces cerevisiae MSH2-MSH3 and MSH2-MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J Mol Biol. 2007;366:53–66. doi: 10.1016/j.jmb.2006.10.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drotschmann K, Yang W, Brownewell FE, Kool ET, Kunkel TA. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2-Msh6. J Biol Chem. 2001;276:46225–46229. doi: 10.1074/jbc.C100450200. [DOI] [PubMed] [Google Scholar]
- 45.Das Gupta R, Kolodner RD. Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat Genet. 2000;24:53–56. doi: 10.1038/71684. [DOI] [PubMed] [Google Scholar]
- 46.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: A switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 47.Tran PT, Liskay RM. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha. Mol Cell Biol. 2000;20:6390–6398. doi: 10.1128/mcb.20.17.6390-6398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black BE, Brock MA, Bedard S, Woods VL, Jr, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brock M, et al. Conformational analysis of Epac activation using amide hydrogen/deuterium exchange mass spectrometry. J Biol Chem. 2007;282:32256–32263. doi: 10.1074/jbc.M706231200. [DOI] [PubMed] [Google Scholar]
- 50.Melnyk RA, et al. Structural determinants for the binding of anthrax lethal factor to oligomeric protective antigen. J Biol Chem. 2006;281:1630–1635. doi: 10.1074/jbc.M511164200. [DOI] [PubMed] [Google Scholar]
- 51.Pantazatos D, et al. Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci USA. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller J. A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.