Abstract
Many biologically active small molecule natural products produced by microorganisms derive their activities from sugar substituents. Changing the structures of these sugars can have a profound impact on the biological properties of the parent compounds. This realization has inspired attempts to derivatize the sugar moieties of these natural products through exploitation of the sugar biosynthetic machinery. This approach requires an understanding of the biosynthetic pathway of each target sugar and detailed mechanistic knowledge of the key enzymes. Scientists have begun to unravel the biosynthetic logic behind the assembly of many glycosylated natural products, and have found that a core set of enzyme activities is mixed and matched to synthesize the diverse sugar structures observed in nature. Remarkably, many of these sugar biosynthetic enzymes and glycosyltransferases also exhibit relaxed substrate specificity. The promiscuity of these enzymes has prompted efforts to modify the sugar structures and/or alter the glycosylation patterns of natural products via metabolic pathway engineering and/or enzymatic glycodiversification. In applied biomedical research, these studies will enable the development of new glycosylation tools and generate novel glycoforms of secondary metabolites with useful biological activity.
Keywords: Biosynthesis, Unusual Sugars, Enzyme Catalysis, Glycodiversification, Enzyme Mechanism
1. Introduction
Glycosylation is one of the most common and important reactions in biological systems and the resulting glycoconjugates have diverse functions, including information storage and transfer, energy storage, maintenance of cell structural integrity, molecular recognition, signaling, virulence, and chemical defense. Several human diseases are associated with aberrant protein glycosylation patterns,[1, 2] and initiation of viral infections often involves recognition of specific cell surface protein glycoforms.[3] Likewise, bacterial virulence is related to cell surface polysaccharides,[4] and many bacteria use glycosylated small molecules as chemical weapons to gain a selective advantage, or as signaling molecules for intra- and inter-species communication.[5] A significant number of these glycosylated small molecules are clinically useful for the treatment of bacterial and fungal infections, cancer, and other human diseases. This class of small molecule glycoconjugates will be the focus of this review. Changes in the structures of the sugar moieties of glycosylated compounds can have profound effects on their activities, selectivities, and pharmacokinetic properties.[6, 7] For all of these reasons, it is desirable to understand the biochemical processes for the formation of glycoconjugates.
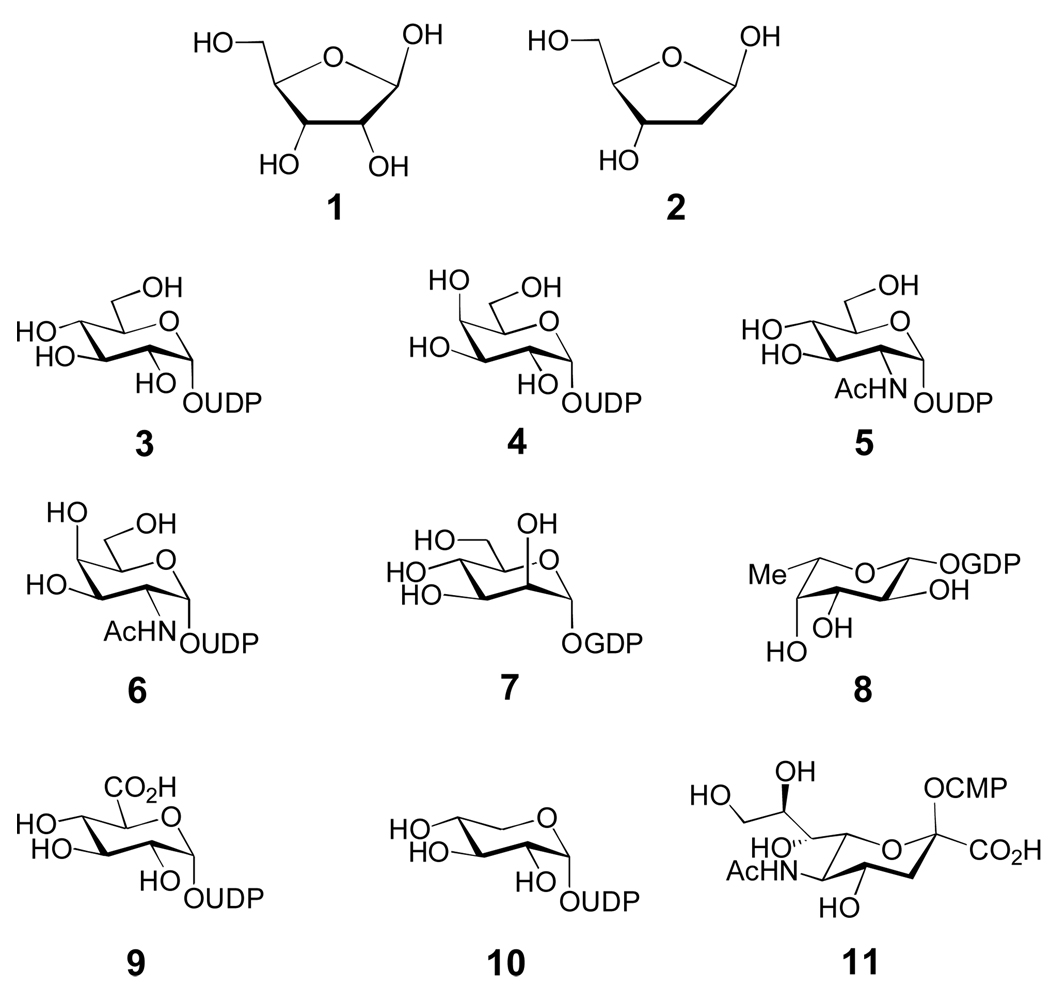
Common glycosylated biomolecules include nucleic acids, polysaccharides, proteins, lipids, and secondary metabolites. The biosynthesis of d-ribose (1), 2-deoxy-d-ribose (2) (Figure 1), and nucleosides will not be covered in this review. Surprisingly, eukaryotic glycoproteins and glycolipids are synthesized from only nine nucleotide sugar donors (3–11, Figure 1).[8] Although several enzymatic tailoring modifications can occur on these sugars after glycosyltransfer, most eukaryotic glycan structural diversity results from variation in the number and type of the sugar moieties, and in the linkages between the sugar components of oligosaccharides. Conversely, prokaryotic polysaccharides and glycosylated natural products contain more than one hundred different sugars, many of which are deoxygenated and highly functionalized. Therefore, prokaryotic glycoconjugates derive most of their structural diversity from the identities of their unusual sugar moieties.
Figure 1. Common sugars of primary metabolism.
d-ribose (1), 2-deoxy-d-ribose (2), UDP-d-glucose (3), UDP-d-galactose (4), UDP-2-N-acetyl-d-glucosamine (5), UDP-2-N-acetyl-d-galactosamine (6), GDP-d-mannose (7), GDP-l-fucose (8), UDP-d-glucuronic acid (9), UDP-d-xylose (10), CMP-N-acetyl-neuraminic acid (sialic acid, 11).
Because these unusual sugar appendages are important for the bioactivities of many bacterial natural products, there has been much interest in developing strategies to alter the sugar structures of these glycoconjugates via biosynthetic engineering approaches.[9] This requires a sound understanding of both the organization of the native biosynthetic machinery and the mechanisms of the encoded enzymes. The advent of modern molecular biological techniques has led to the discovery and sequencing of the biosynthetic gene clusters for many natural 4 products and unusual sugars, and this has made comparative genomic approaches to functional assignment of the encoded enzymes feasible. This, in turn, has enabled the genetic and biochemical characterization of a number of sugar biosynthetic pathways. A key finding from these studies is that many unusual sugar biosynthetic enzymes and glycosyltransferases (GTs, the enzymes that couple activated sugars to an acceptor molecule) have broad substrate specificity, allowing their use both in vivo and in vitro for the attachment of alternative sugars to natural product acceptors (a process termed glycodiversification). In vitro glycodiversification relies on utilizing a GT with broad specificity to couple chemically or enzymatically synthesized non-native sugar donors to acceptor molecules. Gene disruption and heterologous expression of foreign sugar biosynthetic genes has also enabled the manipulation of endogenous sugar biosynthetic pathways in vivo through metabolic pathway engineering and combinatorial biosynthesis. Both in vitro and in vivo strategies have proven effective in generating natural product analogues with modified sugar structures.
In this review, we summarize the current knowledge of the biosynthesis and glycosyltransfer of unusual sugars found in biologically active small molecule natural products of bacterial origin (Section 2). Only those pathways that have been genetically and/or biochemically verified will be discussed in detail. Next, we discuss the catalytic mechanisms of several sugar biosynthetic enzymes, focusing on common themes employed by Nature to generate sugar structural diversity (Section 3). We will also highlight several unusual and not well-understood sugar modifications that merit further investigation. The structure and mechanisms of glycosyltransferases will be presented in Section 4, with a focus on glycosyltransferases involved in bacterial secondary metabolism. Finally, recent attempts to change the sugar components of natural products through enzymatic glycoengineering will be discussed (Section 5). Together, these studies have not only illuminated Nature's stunning ingenuity in using diverse chemical mechanisms and natural combinatorial biosynthetic processes to drive glycodiversity, but have also enabled the development of methods to manipulate sugar biosynthetic machinery in the hope of generating clinically useful agents.
2. Biosynthesis of Unusual Sugars Found in Natural Products
2.1. Sugar Activation
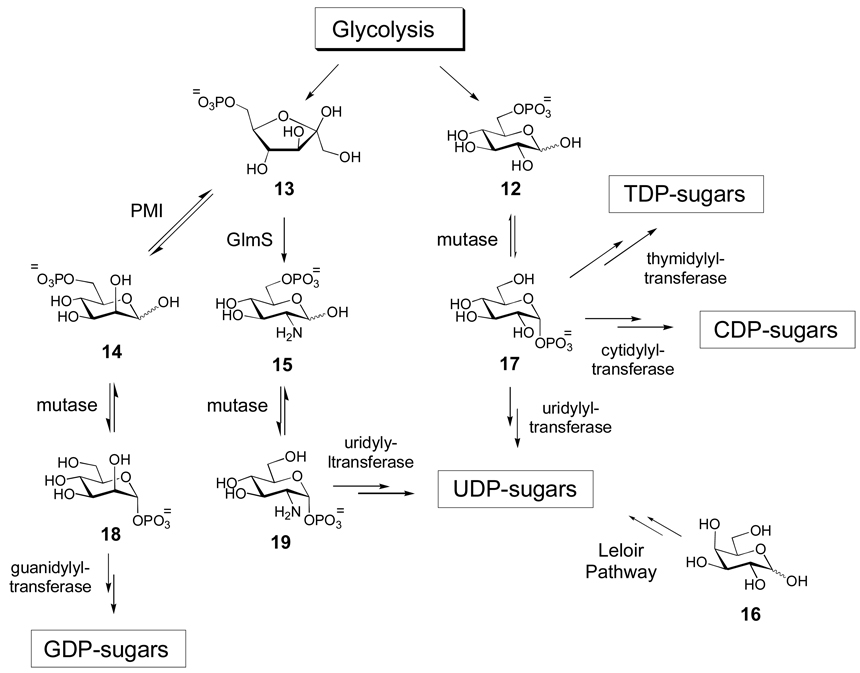
Monosaccharides must first be activated as either nucleotide monophosphate (NMP)- or nucleotide diphosphate (NDP) derivatives so that they can be used by the biosynthetic enzymes and GTs within the cell. Examples of adenylyldiphosphate (ADP)-, thymidylyldiphosphate (TDP)-, guanylyldiphosphate (GDP)-, uridylyldiphosphate (UDP)-, cytidylyldiphosphate (CDP)-, and cytidylylmonophosphate (CMP)-activated monosaccharides are known. The phosphonucleotidyl moiety has dual purposes: it serves as a recognition element for enzymes involved in the biosynthetic pathways, and it functions as a good leaving group during the glycosyltransfer reaction. The glycolytic intermediates, glucose-6-phosphate (12) and fructose-6-phosphate (13), are the sources for most nucleotide sugars (Scheme 1). Fructose-6-phosphate (13) is converted to mannose-6-phosphate (14) by phosphomannoisomerase (PMI) in the biosynthesis of GDP-sugars, and to glucosamine-6-phosphate (15) by glucosamine-6-phosphate synthase (GlmS) in the formation of UDP-sugars. Alternatively, UDP-sugars can be derived from galactose (16) via the Leloir pathway, which ultimately leads to UDP-glucose (3). Glucose-6-phosphate (12) is also a biosynthetic precursor of many UDP-sugars, but is more commonly used in the biosynthesis of TDP- and CDP-sugars. In all cases, the sugar-6-phosphates 12, 14, and 15 are converted to the corresponding sugar-1-phosphates (17, 18, and 19, respectively) by distinct, but related phosphohexose mutases prior to nucleotidylyl transfer.[10] In eukaryotes, salvage pathways that utilize sugars generated via catabolic routes (such as glycoprotein degradation) as biosynthetic precursors also exist for several common sugars such as N-acetylglucosamine, N-acetylgalactosamine, mannose, and fucose.[8] These salvage pathways involve either direct anomeric phosphoryltransfer or 6-phosphorylation followed by a mutase-catalyzed 6→1 migration to yield the sugar-1-phosphate products. The biosynthetic details for the formation of each group of nucleotide sugars will be discussed below.
Scheme 1. Biosynthetic origins of NDP-sugars.
Most NDP-sugars are derived from glycolytic intermediates glucose-6-phosphate (12) and fructose-6-phosphate (13) or from galactose (16). Eventually, all of these sugars are converted into sugar-1-phosphates, which can then be activated by the appropriate nucleotidylyltransferase.
Transfer of nucleotide monophosphates to sugar-1-phosphate substrates is catalyzed by nucleotidylyltransferase enzymes, and this activation reaction usually occurs early in sugar biosynthetic pathways. A notable exception is that nucleotidylyltransfer occurs late in the biosynthesis of CMP-sugars (such as CMP-sialic acid).[11–13] The majority of nucleotidylyltransferases identified so far share modest to high sequence similarity. However, it is not yet possible to reliably predict nucleotide specificity of these enzymes solely based on amino acid sequence, although phylogenetic analysis has had limited success in the identification of subgroups that roughly correlate with nucleotide specificity. The utility of anomeric sugar kinases and nucleotidylyltransferases in the construction of NDP-sugar libraries for in vitro glycoengineering will be discussed below (Section 5.2.1).
2.2. Naturally Occurring TDP-sugars
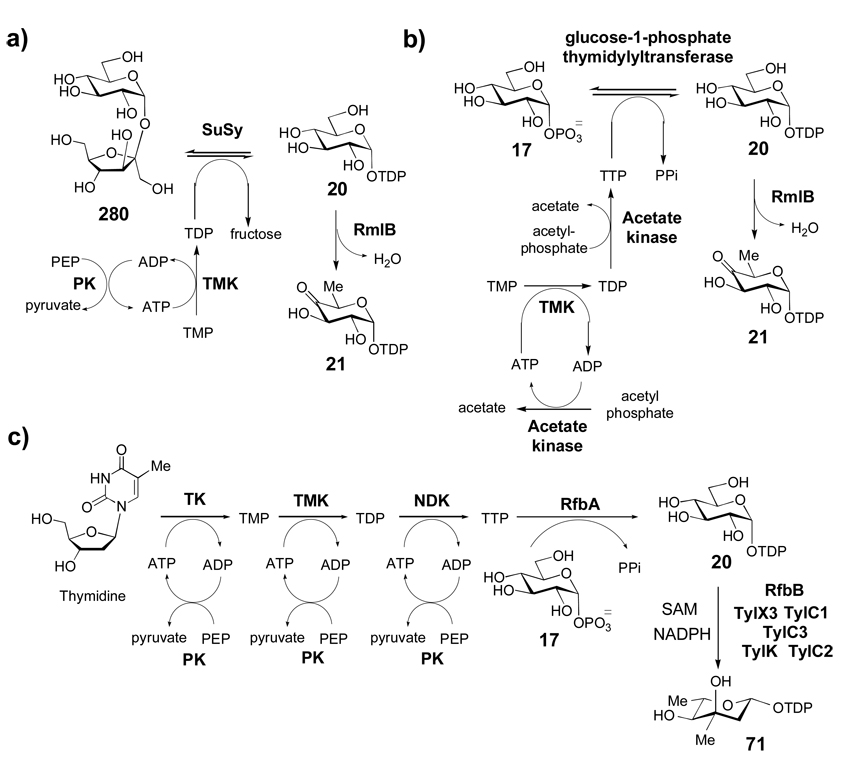
TDP-activated sugars are the most structurally diverse class of nucleotide sugars found in nature. In addition to their uses as building blocks for many bacterial polysaccharides, TDP-sugars are also the preferred sugar donors in the biosynthesis of bacterial glycosylated natural products. Nearly all known TDP-sugars are 6-deoxyhexoses, and many are also deoxygenated at C-2, C-3, or C-4 of the pyranose ring. In fact, TDP-sugars are the only known class of NDP-sugars yet discovered that can be deoxygenated at C-2 or C-4. The combination of deoxygenation at one or more positions and the wide variety of other modifications, many of which are not found in other NDP-sugar classes, leads to the rich diversity of TDP-sugar structures seen in nature.
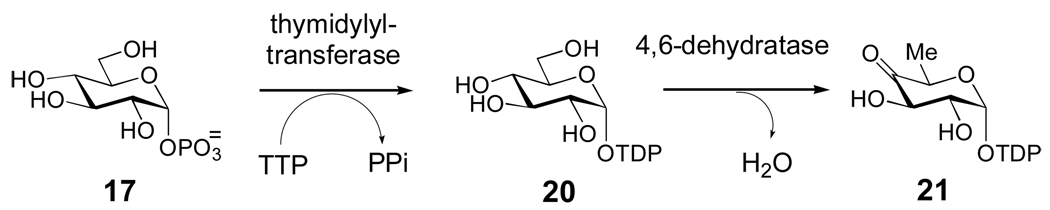
All natural product TDP-sugars whose biosyntheses have been studied are derived from glucose-1-phosphate (17), which is converted to TDP-d-glucose (20) by a thymidylyltransferase and then to TDP-4-keto-6-deoxy-d-glucose (21) by TDP-d-glucose 4,6-dehydratase (Scheme 2). Because 21 is a key intermediate in the biosynthesis of most bacterial deoxysugars, most natural product biosynthetic gene clusters contain genes encoding a dedicated thymidylyltransferase and 4,6-dehydratase, but examples of clusters lacking these genes are not rare. It is presumed that in these latter cases, the enzymes are shared with polysaccharide biosynthesis. To date, the biosynthetic pathways for more than thirty unusual TDP-sugars have been reported. Most of these pathways are proposed based on gene cluster sequence information, and less than half of these pathways are supported by experimental data. However, correlation of phenotypes with specific gene disruptions and/or biochemical characterization of heterologously expressed enzymes have enabled the detailed elucidation of several pathways. These studies have provided an important framework for understanding the molecular logic behind the reaction sequences for the biosynthesis of unusual sugars which has, in turn, allowed better prediction of other pathways based on gene sequence information.
Scheme 2. Entry point into TDP-deoxysugar secondary metabolism in bacteria.
Following thymidylylation of α-d-glucose-1-phosphate (17) by a thymidylyltransferase, a TDP-glucose-4,6-dehydratase enzyme catalyzes the conversion of TDP-d-glucose (20) to TDP-4-keto-6-deoxy-α-d-glucose (21) in the committed step to TDP-deoxysugar biosynthesis.
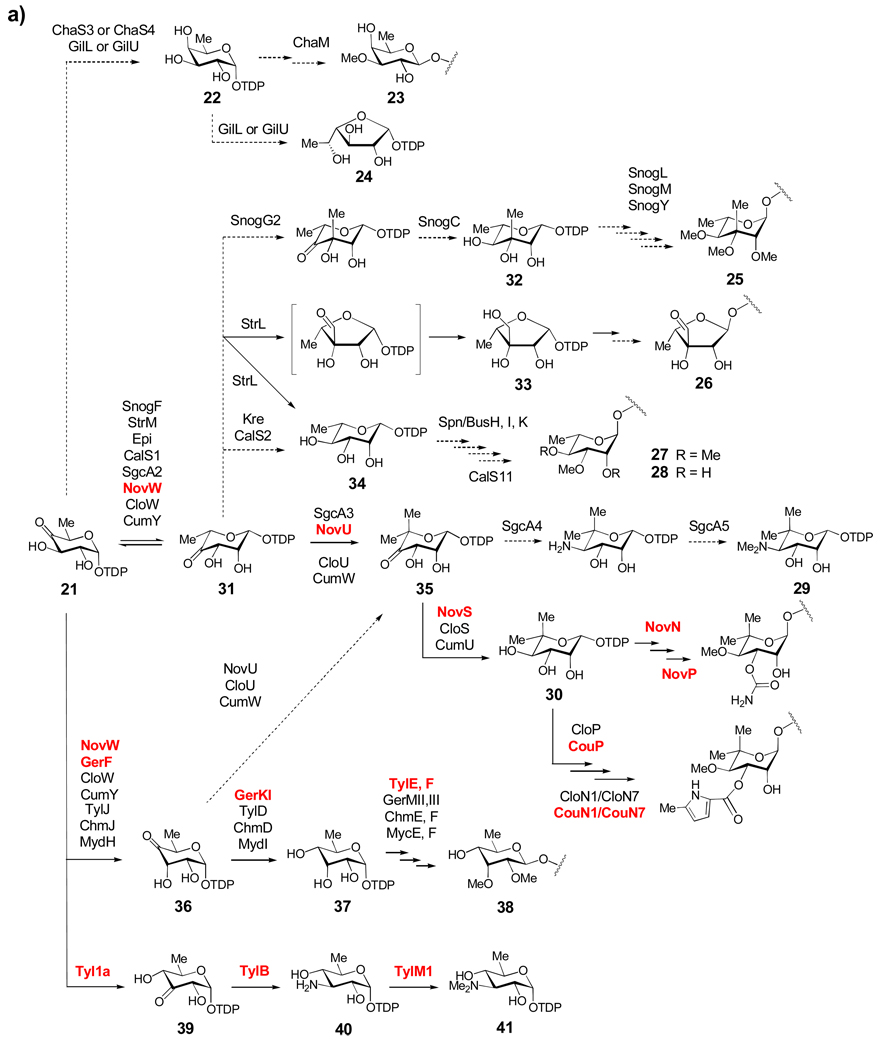
Summarized in Scheme 3 is a nearly comprehensive collection of natural product TDP-sugar biosynthetic pathways, which has been assembled on the basis of at least some biochemical and genetic data. These pathways are divided into three groups based mainly on the degree of deoxygenation. One remarkable aspect of these pathways is that the primary structural differences in the final TDP-sugar products are generated by the action of only five enzyme reaction types, illustrating Nature's economical use of a "combinatorial biosynthesis" strategy to create structural diversity. The mechanistic details of some of these "common" enzymatic activities are discussed in Section 3.1.
Scheme 3.
Scheme 3a: Biosynthesis of Group I TDP-sugars. This group includes 6-deoxysugars (such as 23–28, 30 and 38), as well as the 4-amino-4,6-dideoxysugar (29) and the 3-amino-3,6-dideoxysugar (41). From the common intermediate TDP-4-keto-6-deoxy-α-d-glucose (21), most of the TDP-sugars in this group share an epimerization step (21 → 31) early in their biosynthetic pathways. Solid arrows indicate enzyme-catalyzed reactions that have been verified either in vitro through biochemical experiments with purified enzymes or in vivo through gene disruption/heterologous expression experiments. Dashed arrows indicate reactions that have not been experimentally verified, but have been proposed based on comparison of gene sequences to genes of known function. Names in red indicate enzymes whose functions have been verified biochemically using purified enzymes.
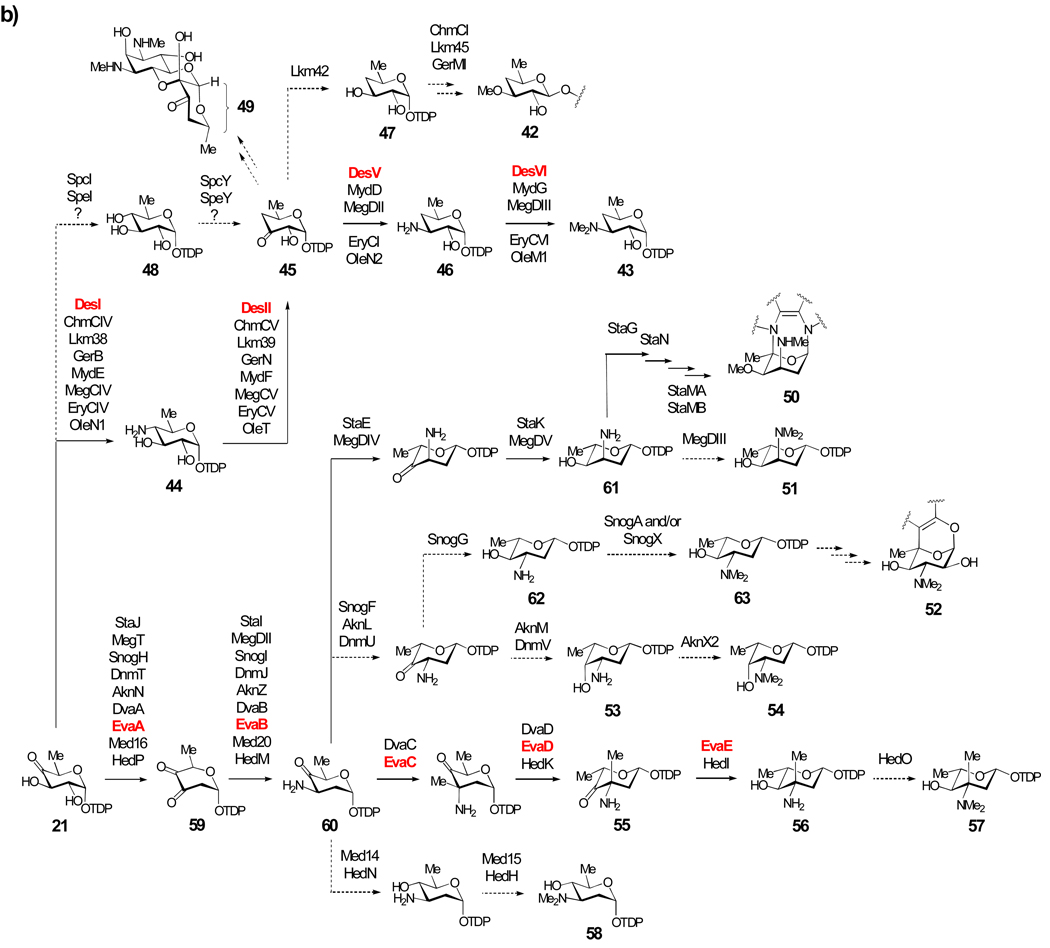
Scheme 3b: Biosynthesis of Group II TDP-Sugars. The extremely rare TDP-4,6-dideoxysugars include TDP-d-desosamine (43), TDP-d-chalcomycin (47), and actinospectose (49). The majority of sugars in Group II are 3-amino-2,3,6-trideoxysugars (50–58 and 60–63) that share a common 2-dehydration/3-aminotransfer reaction sequence (21 → 59 → 60).
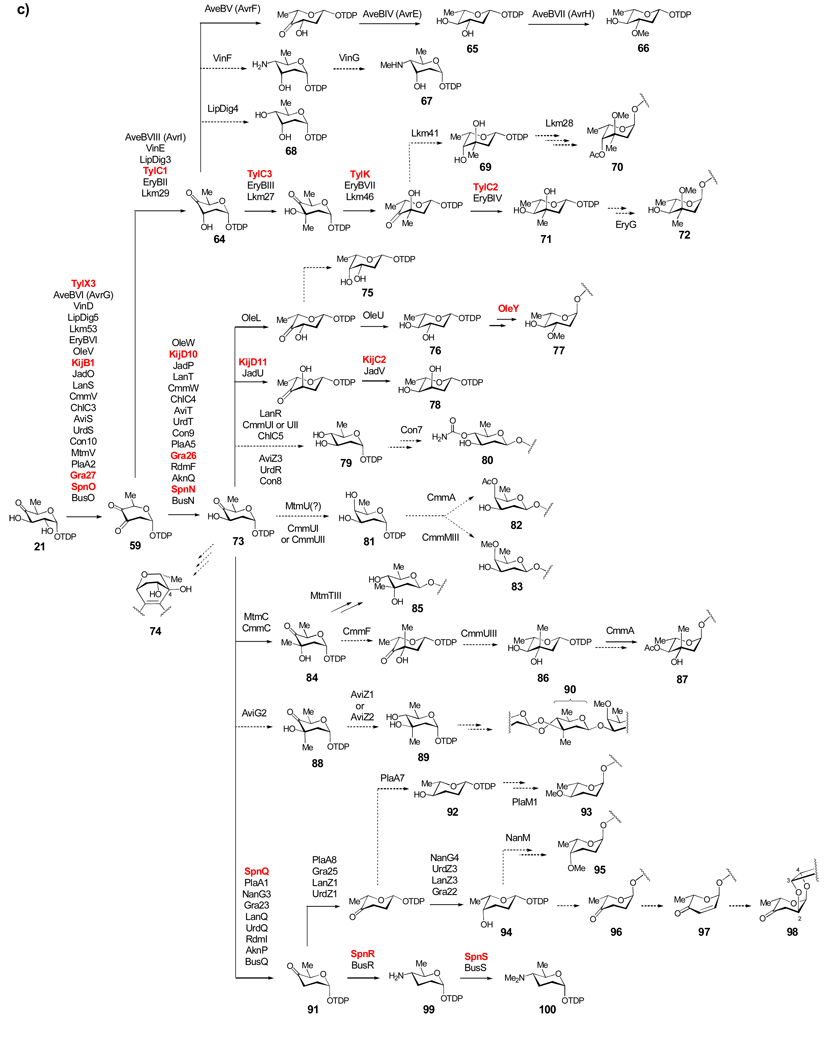
Scheme 3c: Biosynthesis of Group III TDP-sugars. The largest group of TDP-deoxysugars each share 2-dehydration (21 → 59) and 3-ketoreduction steps (59 → 64 or 59 → 73) early in their respective biosynthetic pathways. TDP-sugars derived from 64 are proposed to include 66–72. TDP-sugars proposed to be derived from 73 include numerous 2,6-dideoxysugars (see 75–90) as well as 2,3,6-trideoxysugars (see 91–98), and a TDP-4-amino-2,3,4,6-tetradeoxy sugar (100).
2.2.1. Group I – 6-Deoxy-, 3-Amino-3,6-dideoxy-, and 4-Amino-4,6-dideoxysugars
The d-fucose (see 22) and d-digitalose (23) residues of the antitumor compound chartreusin produced by Streptomyces chartreusis and the d-fucofuranose (see 24) residue of the antibiotic gilvocarcin V produced by Streptomyces griseoflavus are believed to be derived from TDP-d-fucose (22), which is in turn derived from 21 via ketoreduction (Scheme 3a). Compound 22 is also a building block for the capsular polysaccharides in Aneurinibacillus actinomycetemcomitans.[14, 15] The ketoreduction step is likely catalyzed either by ChaS3,[16] a homologue of the ketoreductase Fcd in A. actinomycetemcomitans,[17] or by the short-chain dehydrogenase/reductase (SDR) enzyme ChaS4. d-Fucose is then O-methylated by the methyltransferase ChaM after glycosidic coupling to form d-digitalose (23). In gilvocarcin biosynthesis, GilL and GilU (both of the SDR family) are candidates for catalyzing the conversion of 21 to 22 and the subsequent ring contraction step to form TDP-d-fucofuranose (24), although this is speculative.[18]
Biosyntheses of sugars 25–35 are proposed to share a 3,5-epimerization step converting 21 to TDP-4-keto-6-deoxy-l-mannose (31). Enzymes catalyzing this reaction are homologues of RmlC involved in TDP-l-rhamnose (34) biosynthesis in Salmonella enterica.[17, 19–21] The sugar l-nogalose (25) is present in the anthracycline antibiotic nogalamycin made by Streptomyces nogalater. Formation of 25 was proposed to proceed by a sequential 3,5-epimerization reaction (SnogF), 3-C-methylation (SnogG2), and 4-ketoreduction (SnogC) to afford TDP-6-deoxy-3-C-methyl-l-mannose (32), which is likely the substrate used in the glycosyltransfer reaction.[22] Methylations of the 2-, 3-, and 4-hydroxyl groups by the methyltransferases SnogL, SnogM, and SnogY, to give l-nogalose (25) are presumed to be post-glycosylation events.
The unusual sugar d-streptose (26), found in the aminoglycoside antibiotic streptomycin, is produced by several Streptomyces species, most notably S. griseus. Early biochemical work demonstrated that the immediate donor of the streptose moiety is TDP-d-dihydrostreptose (33),[23] which is formed in two steps from 21: 3,5-epimerization to form 31 followed by NADPH-dependent ring contraction to give 33.[24, 25] The streptomycin gene cluster was later identified in S. griseus, and the epimerization and ring contraction reactions were assigned to be catalyzed by StrM, a RmlC homologue, and StrL, a SDR superfamily member, respectively.[26] Heterologous expression of strL and strM together in a mutant of the methymycin producer S. venezuelae, which accumulates 21, resulted in the production of methymycin derivatives bearing l-rhamnose (see 34).[27] Although no dihydrostreptose was produced, the fact that 21 was converted to TDP-l-rhamnose (34) in this recombinant strain provided strong support that StrM is a 3,5-epimerase and that StrL has 4-ketoreductase activity.[27] The proposed formation of both furanose (33) and pyranose (34) products from 31 by StrL is reminiscent of the reaction catalyzed by UDP-apiose synthase ecoded by AXS1 in Arabidopsis thaliana.[28–30] The ring contraction of 22 to 24 in gilvocarcin biosynthesis may also follow a similar route.
Various O-methylated l-rhamnose moieties exist in nature, such as 27 and 28 found in the macrolide compounds spinosyn and butenylspinosyn, both produced by Saccharopolyspora spinosa,[31] the aromatic polyketide elloramycin produced by Streptomyces olivaceus,[32] and the enediyne calicheamicins of Micromonospora echinospora.[33] The genes spn/busH, spn/busI, and spn/busK encode the O-methyltransferases in the spinosyn/butenylspinosyn pathways, whereas calS11 encodes the 3-O-methyltransferase used in calicheamicin biosynthesis. Interestingly, the genes required for the formation of 34 are absent in the gene clusters of spinosyns/butenylspinosyns. Instead, they are located in other regions of the genome in S. spinosa, and they likely function both in cell wall biosynthesis and in the formation of spinosyns.[34]
TDP-4-N,N-dimethylamino-4-deoxy-5-C-methyl-l-rhamnose (29) and TDP-l-noviose (30) are the predicted sugar donors for the biosynthesis of the enediyne antibiotic C-1027[35] and the aminocoumarin antibiotics novobiocin,[36] clorobiocin,[37] and coumermycin,[38] respectively. These sugars have a 5,5-gem-dimethyl moiety formed by C-methylation at C-5. Their biosynthesis from 21 involves either 3,5- or 3-epimerization catalyzed by RmlC homologues to form 31 or 36, respectively, followed by 5-C-methyltransfer to give TDP-4-keto-6-deoxy-5-C-methyl-l-mannose (35). Results obtained from coupled assays of the purified epimerase NovW and 5-C-methyltransferase NovU from the novobiocin pathway,[39] along with gene disruption studies of cloU from the clorobiocin biosynthesis[40] suggested that the biosynthesis of 30 involves 3,5-epimerization rather than 3-epimerization. However, a recent in vitro study showed that the epimerase NovW is kinetically competent only as a 3-epimerase.[41] The final step of the biosynthesis of 30 is the C-4 reduction of 35 catalyzed by NovS/CloS/CumU.[39] Formation of 29 in C-1027 biosynthesis has been proposed to involve 3,5-epimerization by SgcA2, C-methyltransfer by SgcA3, C-4 aminotransfer by SgcA4, and 4-N,N-dimethyltransfer by SgcA5.
Several post-glycosylation tailoring steps on the l-noviose (see 30) moiety of the aminocoumarin antibiotics have been characterized via gene disruption and in vitro biochemical methods.[42–45] In novobiocin biosynthesis, the carbamoyltransferase NovN modifies the C-3 hydroxyl group of l-noviose, after which the O-methyltransferase NovP acts at the C-4 hydroxyl group to produce the fully elaborated sugar.[44] In clorobiocin and coumermycin biosynthesis, 4-O-methylation catalyzed by CloP/CouP is thought to occur first. The 5-methyl-2-pyrrolylcarbonyl moiety is then transferred from the peptidyl carrier protein (PCP) 11 CloN1/CouN1 to the 3-position of the pendant 4-O-methyl-l-noviose by the acyltransferase CloN7/CouN7.[43, 45]
The sugars d-mycinose (38) and d-mycaminose (see 41) are found in the structures of several macrolide antibiotics, including tylosin, chalcomycin, dihydrochalcomycin, and mycinamicin. Tylosin carries both sugars, whereas chalcomycin, dihydrochalcomycin, and mycinamicin contain 38. The biosynthetic gene clusters for these compounds have been sequenced,[46–50] and recent genetic and biochemical studies performed on the tylosin and dihydrochalcomycin systems have fully established the pathways for the formation of these two sugars.[47, 51–55] The key intermediate, TDP-6-deoxy-d-allose (37), in the pathway of 38 is synthesized from 21 via C-3 epimerization by the RmlC homologues GerF/TylJ/ChmJ/MydH followed by C-4 ketoreduction by GerKI/TylD/ChmD/MydI. In a recent in vitro study, 37 was confirmed to be the sole product formed in incubations of 21 with the purified dihydrochalcomycin biosynthetic enzymes GerF and GerKI.[47] A similar reaction sequence likely occurs in the tylosin, chalcomycin, and mycinamicin pathways. The O-methylation of the two hydroxyl groups occurs after glycosyltransfer, and is catalyzed by GerMII,MIII and Tyl/Chm/MycE,F. During tylosin biosynthesis, TDP-d-mycaminose (41) is constructed in three steps from 21: 3,4-ketoisomerization by Tyl1a to form TDP-3-keto-6-deoxy-d-glucose (39), aminotransfer by TylB to form 40, and N,N-dimethylation by TylM1 to form 41. The functions of Tyl1a,[54] TylB,[53] and TylM1[52] have all been verified biochemically with purified enzymes.
2.2.2. Group II – 4,6-Dideoxy-, 3-Amino-3,4,6-trideoxy-, and 3-Amino-2,3,6-trideoxysugars
The sugars d-chalcose (42) and d-desosamine (see 43) are constituents of many macrolide antibiotics (Scheme 3b). Of those whose gene clusters have been sequenced, lankamycin,[56] chalcomycin,[50] and dihydrochalcomycin[46] contain d-chalcose, while erythromycin,[57] oleandomycin,[58–60] mycinamicin,[49] methymycin/pikromycin,[61] and megalomicin[62, 63] contain d-desosamine. Early gene disruption experiments carried out with the erythromycin producer Saccharopolyspora erythraea led to several possible pathways for TDP-d-desosamine (43) formation.[57, 64–66] Later genetic and biochemical studies of the methymycin/pikromycin system from Streptomyces venezuelae clearly showed that 43 is biosynthesized from 21 in four steps.[67–70] As delineated in Scheme 3b, the reaction is initiated with C-4 aminotransfer catalyzed by DesI to give 44, followed by oxidative deamination by DesII to yield 45, C-3 transamination by DesV to afford 46, and 3-N,N-dimethylation by DesVI to furnish 43.[67–69] The reaction catalyzed by DesII (44 → 45), which is a member of the radical-SAM superfamily, is unique in sugar biosynthesis. Together, DesI and DesII carry out C-4 deoxygenation of 21 to form TDP-3-keto-4,6-dideoxy-d-glucose (45).[70–72]
Homologues of DesI, DesII, DesV, and DesVI are found in the erythromycin, oleandomycin, mycinamicin, and megalomicin pathways, and are presumed to catalyze the equivalent reactions in the biosynthesis of d-desosamine (43) in each pathway. Although the biosynthesis of 43 has now been fully elucidated, that of chalcose (42) remains unexplored. However, genes encoding homologues of DesI and DesII are present in the lankamycin, chalcomycin, and dihydrochalcomycin gene clusters, suggesting that C-4 deoxygenation in chalcose (42) formation occurs in a manner analogous to that of desosamine biosynthesis (21 → 44 → 45). Conversion of 45 to TDP-4,6-dideoxy-d-glucose (47) requires a 3-ketoreductase. An NDP-sugar ketoreductase gene, lkm42, exists in the lankamycin gene cluster, but is absent in the chalcomycin and dihydrochalcomycin clusters. The corresponding gene in the latter cases may be encoded elsewhere in the Streptomyces bikiniensis or Streptomyces sp KCTC 0041BP genomes, respectively. O-Methylation at C-3 to form 42 likely happens after glycosyltransfer, and may be catalyzed by ChmCI/Lkm45/GerMI in chalcomycin, lankomyicn, and dihydrochalcomycin biosynthesis, respectively.
The aminoglycoside antibiotic spectinomycin produced by Streptomyces flavopersicus and Streptomyces spectabilis contains an unusual 3-keto-4,6-dideoxy-glucose moiety, known as actinospectose (49). Partial gene clusters for spectinomycin biosynthesis[73] have been isolated from these two strains. Both clusters contain glucose-1-phosphate thymidylyltransferase and TDP-glucose-4,6-dehydratase genes (spcK and spcJ, respectively, in S. flavopersicus and spcD and spcE, respectively, in S. spectabilis). The activity of SpcE has been verified in vitro,[73] implicating TDP-glucose as the precursor in the actinospectose pathway. Although the mechanism of C-4 deoxygenation is not obvious, both spectinomycin clusters encode a putative radical-SAM enzyme (ScpY in S. flavopersicus and SpeY in S. spectabilis), which may play a role in generating TDP-actinospectose (45). Thus, a pathway involving 4-ketoreduction of 21 to 48 by the SDR enzyme SpcI/SpeI, followed by oxidative dehydroxylation by ScpY/SpeY is conceivable for the biosynthesis of 49. The proposed mechanism (21 → 48 → 45) parallels that of the C-4 deoxygenation step carried out by DesI/DesII during d-desosamine biosynthesis. Interestingly, SpcY and SpeY share no detectable sequence identity with DesII. Their functions clearly warrant further investigation.
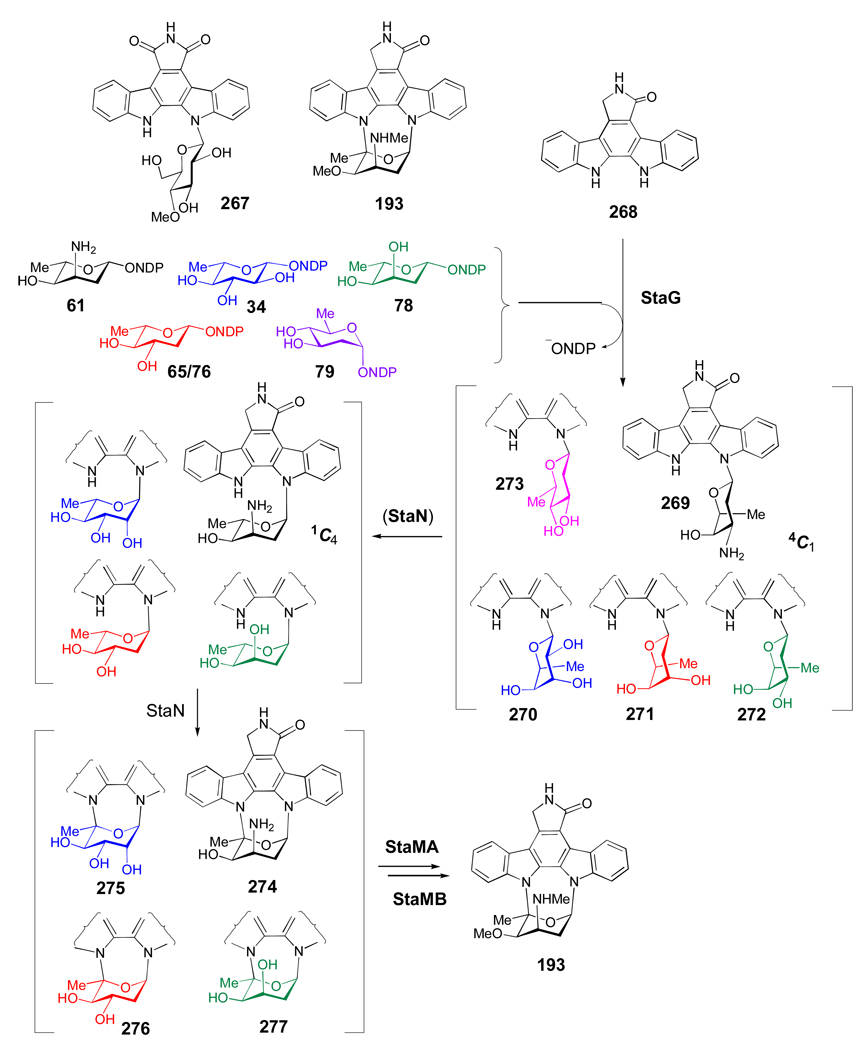
Compounds 50–58 are representatives of 3-amino-2,3,6-trideoxy sugars, whose gene clusters have been sequenced. Each gene cluster encodes a 2,3-dehydratase and a 3-aminotransferase, which catalyze the respective C-2 deoxygenation of 21 to give TDP-3,4-diketo-2,6-dideoxy-d-glucose (59) and the subsequent C-3 aminotransfer to generate TDP-3-amino-4-keto-2,3,6-trideoxy-d-glucose (60). After 60, each individual pathway adopts a distinct combination of epimerization, stereospecific C-4 ketoreduction, and C- and/or N-methyltransfer steps to produce the TDP-sugar product. For example, the key intermediate (61) in the biosynthesis of 3-N-methyl-4-O-methyl-l-ristosamine (50) -the sugar component of the indolocarbazole antibiotic staurosporine -is formed via a StaE-catalyzed C-5 epimerization of 60 followed by StaK-catalyzed C-4 ketoreduction. Transfer of l-ristosamine to the aglycone by StaG is the next step, which is followed by crosslinking between C-5 of ristosamine and the indole nitrogen of the aglycone mediated by StaN, a P450 enzyme. The final 3-N-methylation and 4-O-methylation reactions to give staurosporine result from the action of StaMA and StaMB, respectively.[74] Evidence supporting the proposed pathway for 50 comes from the successful reconstitution of staurosporine biosynthesis in heterologous hosts.[74, 75]
The biosynthesis of l-megosamine (see 51) in the macrolide antibiotic megalomicin is predicted to be analogous to TDP-l-ristosamine (61), involving C-5 epimerization of 60 (MegDIV), C-4 ketoreduction of the resulting l-sugar (MegDV), and 3-N,N-dimethylation of intermediate 61 (MegDIII) to give TDP-l-megosamine (51).[63] Interestingly, megalomicin contains two 3-N,N-dimethylamino sugars, d-desosamine (see 43) and l-megosamine, yet the gene cluster has only one aminotransferase (megDII) and one dimethyltransferase (megDIII) gene. The encoded enzymes likely catalyze the corresponding steps in both sugar biosynthetic pathways.[62]
The sugars l-nogalamine (52),[22] l-daunosamine (see 53),[76] and l-rhodosamine (see 54)[77] are found in the anthracycline antibiotics nogalamycin, daunorubicin, and aclarubicin, respectively. Their common precursor is 60, which undergoes 3,5-epimerization and stereospecific ketoreduction in each pathway. The C-4 hydroxyl group of TDP-l-acosamine (62), produced by the tandem action of SnogF and SnogG, is equatorial, whereas that of TDP-l-daunosamine (53) is axial. These sugars can be 3-N,N-dimethylated to produce TDP-2-deoxy-l-nogalamine (63), the sugar donor in nogalamycin formation, or TDP-l-rhodosamine (54), the sugar donor in aclarubicin and rhodomycin biosynthesis. Once transferred to the aglycone, crosslinking of C-5 of 2-deoxy-l-nogalamine (63) to the aglycone and re-hydroxylation at C-2 are proposed to generate the final compound.[22] The identity of these tailoring enzymes, as well as the logic for having to deoxygenate and then re-hydroxylate at C-2 of the sugar moiety is not clear.
The 3-amino-2,3,6-trideoxysugars, TDP-4-oxo-l-vancosamine (55) and TDP-l-eremosamine (56), are intermediates in the biosynthesis of the vancomycin-type antibiotics balhimycin[78] and chloroeremomycin,[79] respectively. TDP-3-N,N-dimethyl-l-eremosamine (57) along with TDP-d-angolosamine (58) are the two sugar donors in the biosynthesis of hedamycin.[80] Sugar 58 is also involved in the biosynthesis of the benzoisochromanequinone antibiotic medermycin.[81] The complete biosynthetic pathway for 56, starting from 21, has been elucidated through the biochemical analysis of the pathway enzymes.[82] The key intermediate 55, the substrate for glycosyltransfer in the balhimycin pathway, is derived from 60 by C-3 methylation followed by 5-epimerization. Subsequent C-4 ketoreduction of 55 results in 56, the sugar donor in chloroeremomycin biosynthesis. It is unusual for a ketosugar, such as 55, to be a substrate for a glycosyltransferase. However, inspection of the balhimycin gene cluster shows an inactive 4-ketoreductase gene (dvaE), which at one point likely catalyzed the conversion of 55 to 56 in the balhimycin producing strain. This, combined with the extensive conservation observed between balhimycin and chloroeremomycin clusters, suggests a close evolutionary relationship between the two pathways.[83]
l-Vancosamine, the C-4 epimer of l-eremosamine (see 56), is a component of the glycopeptide antibiotic vancomycin. Although analysis of the l-vancosamine biosynthetic genes has not been reported, formation of TDP-l-vancosamine is presumed to be identical to that of 56 except that the stereochemistry of C-4 ketoreduction is reversed. Likewise, TDP-3-N,N-dimethyl-l-eremosamine (57), involved in hedamycin biosynthesis, can be made in an identical manner to that of 56 by the respective Hed biosynthesis enzymes, but with an additional dimethylation step catalyzed by HedO to convert 56 to 57.[80] TDP-d-angolosamine (58), whose genes have been identified in both the hedamycin and medermycin gene clusters, is predicted to be made in two steps from 60: 4-ketoreduction by Med14/HedN, and 3-N,N-dimethyltransfer by Med15/HedH.[80, 81]
2.2.3. Group III – 2,6-Dideoxy-, 4-Amino-2,4,6-trideoxy-, 2,3,6-Trideoxy-, and 4-Amino-2,3,4,6-tetradeoxysugars
TDP-2,6-dideoxysugars and their derivatives, which are formed by 2,3-dehydration of 21 and subsequent 3-ketoreduction, account for the majority of TDP-sugars used in natural product biosynthetic pathways (Scheme 3c). The enzymes catalyzing 2,3-dehydration of 21 to form 59 in each of these pathways are homologous to those catalyzing the same reaction in the biosynthesis of 3-amino-2,3,6-trideoxysugars depicted in Scheme 3b. This group of TDP-sugars can be further divided into two subgroups depending on the configuration of their 3-OH group (see 64 and 73). Interestingly, enzymes catalyzing the axial and equatorial 3-ketoreduction are all NAD(P)H-dependent reductases, but share no detectable sequence similarity, making their coding genes readily distinguishable.
TDP-d-vicenisamine (67), TDP-d-digitoxose (68), 4-O-acetyl-l-arcanose (70), TDP-l-mycarose (71), and l-cladinose (72), all have an axial 3-OH group and each is derived from TDP-4-keto-2,6-dideoxy-d-allose (64), which is formed from 21 by 2,3-dehydration followed by 3-ketoreduction. The enzymes catalyzing these two steps (21 → 59 → 64) in the biosynthesis of 71 (TylX3 and TylC1, respectively) have been characterized in vitro.[84] Compound 66, the sugar donor in the avermectin biosynthetic pathway,[85] is produced from 64 in three steps: 5-epimerization by AveBV (AvrF), 4-ketoreduction by AveBIV (AvrE), and 3-O-methylation by AveBVII (AvrH). Heterologous expression of the complete set of the biosynthetic enzymes supports the proposed pathway of 66.[86] Although the exact order of these steps remains unknown, current data suggest that 3-O-methyltransfer occurs at the TDP-l-olivose (65) stage (65 → 66), rather than as a separate tailoring step.[87]
TDP-d-vicenisamine (67), the sugar donor for the biosynthesis of the macrolactam antibiotic vicenistatin in Streptomyces halstedii, is proposed to be derived from 64 via C-4 transamination by VinF followed by N-monomethylation by VinG.[88] Sugar 67 is the only 4-amino-2,4,6-trideoxysugar whose biosynthetic genes have been identified, and is a rare example of an N-monomethylated aminosugar. The gene cluster encoding the formation of lipomycin, which contains d-digitoxose (see 68), has been located in Streptomyces aureofaciens.[89] This unusual sugar is formed by C-4 ketoreduction of 64 by LipDig4. The sugars 4-O-acetyl-l-arcanose (70), TDP-l-mycarose (71), and the O-methylated l-mycarose derivative l-cladinose (72) are biosynthesized from 64 via similar routes. The biosynthetic pathway for 71, part of the tylosin pathway of Streptomyces fradiae, has been fully characterized in vitro.[90–92] Compound 64 is 3-C-methylated by the SAM-dependent methyltransferase TylC3. Next, TylK epimerizes C-5 and TylC2 reduces C-4 to form 71. In erythromycin biosynthesis, l-cladinose (72) is produced by 3-O-methylation of l-mycarose by EryG after it has been transferred from 71 to the macrolactone. The homologues of 71 biosynthetic enzymes found in the erythromycin pathway must catalyze identical reactions as their counterparts in the tylosin pathway.
The biosynthetic pathway for 4-O-acetyl-l-arcanose (70), which is found in the macrolide antibiotic lankamycin produced by Streptomyces rochei, is expected to be analogous to that of 71.[56] Indeed, genes with high sequence identity (40–75%) to those involved in the biosynthesis of 71 are found in the lankamycin cluster, consistent with a pathway in which all reactions (except the final 4-ketoreduction step) are the same as those found in the biosynthesis of 71. The 4-ketoreduction by Lkm41 would give the C-4 epimer of 71, TDP-l-axenose (69), which is a reasonable substrate for glycosyltransfer. Tailoring reactions involving 3-O-methylation, possibly by Lkm28, and 4-O-acetylation by an unknown enzyme would complete the biosynthesis of 70.
The sugars 2-deoxy-l-fucose (see 75), l-oleandrose (77), l-digitoxose (see 78), d-olivose (see 79), 4-O-carbamoyl-d-olivose (80), d-oliose (see 81), 4-O-acetyl-d-oliose (chromose D, 82), 4-O-methyl-d-oliose (chromose A or olivomose, 83), d-mycarose (85), l-chromose B (or olivomycose, 87), and 2-deoxy-d-evalose (90) are all 2,6-dideoxysugars, most carrying an equatorial 3-OH group. They are biosynthesized from TDP-4-keto-2,6-dideoxy-d-glucose (73), which is derived from 21 via 2,3-dehydration followed by stereospecific 3-ketoreduction. Compound 73 has been suggested to be the substrate for glycosyltransfer in the biosynthesis of mithramycin, an antitumor agent, and granaticin, a benzoisochromanequinone antibiotic. Granaticin contains an unusual aryl-C-l-olivosyl moiety (74), which is likely formed using 73 as the sugar donor followed by oxidative crosslinking between the aglycone and the C-4 carbonyl carbon of the sugar appendage.[93]
Interestingly, mithramycin derivatives bearing a 4-keto-2,6-dideoxy-d-glucose moiety (presumably derived from 73) in place of d-olivose (see 79) were produced by a Streptomyces argillaceus mutant in which a C-methyltransferase gene (mtmC) was inactivated. Curiously, heterologous expression of mtmC in trans in this mutant restored mithramycin production.[94] In a later study, the authors proposed that the MtmC protein may need to be present in order to interact with a 4-ketoreductase (either MtmTI or MtmTII) also encoded in the cluster.[95] They proposed that this 4-ketoreductase may reduce 73 following its transfer to the mithramycin aglycone.
2-Deoxy-l-fucose (see 75) is a sugar component of the anthracycline antibiotics aclarubicin (aclacinomycin) and rhodomycin, and is presumably synthesized as TDP-2-deoxy-l-fucose (75) in two steps from 73: 3,5-epimerization and 4-ketoreduction. Although the gene clusters for both aclarubicin[77] and rhodomycin[96] have been partially sequenced, genes for these activities have not been assigned in either cluster. l-Oleandrose (77) is found in the macrolide antibiotic oleandomycin produced by Streptomyces antibioticus and in avermectin produced by Streptomyces avermitilis. Interestingly, l-oleandrose is constructed via different routes in these two pathways. It was shown via heterologous expression of the oleandomycin biosynthetic genes[97] that 77 is formed from 73 by 3,5-epimerization and 4-ketoreduction catalyzed by OleL and OleU, respectively, resulting in TDP-l-olivose (76), which is the donor for glycosyltransfer. 3-O-Methylation by OleY has been confirmed in vitro to occur after sugar attachment.[98] This is in contrast to the biosynthesis of l-oleandrose in the avermectin pathway, where TDP-Loleandrose (66) is generated from 64 via 5-epimerization, followed by 4-ketoreduction and 3-O-methylation on the nucleotide sugar prior to glycosyltransfer.[86]
TDP-l-digitoxose (78) is the precursor for the l-digitoxose unit found in the antibiotics jadomycin and kijanimicin produced by Streptomyces venezuelae ISP5230 and Actinomadura kijaniata, respectively. Studies of purified A. kijaniata sugar biosynthetic enzymes have fully established the TDP-l-digitoxose (78) pathway. The conversion of 21 to 73 involves KijB1 and KijD10, and that of 73 to 78 is catalyzed by the 5-epimerase KijD11 and the 4-ketoreductase KijC2.[99] The same roles are predicted for the KijD11 and KijC2 counterparts, JadU and JadV, respectively, in the biosynthesis of jadomycin.[100] TDP-d-olivose (79) is a common sugar donor used in the biosynthesis of a variety of natural products, including landomycin,[101] urdamycin,[102] mithramycin,[103] chromomycin,[104] chlorothricin,[105] avilamycin,[106] and concanamycin.[107] The biosynthetic gene clusters for these compounds have been identified. Genes encoding enzymes for the conversion of 21 to 79 have been found in each cluster except that of mithramycin. In the concanamycin cluster, a putative carbamoyltransferase, Con7, catalyzing 4-O-carbamoylation of TDP-d-olivose (79) to make 80 has also been assigned. This reaction could occur prior to or after glycosyltransfer.
TDP-d-oliose (81) is the presumed precursor for the d-oliose moiety in mithramycin, and the chromose D and olivomose moieties in chromomycin. Results of gene disruption studies in the mithramycin producer Streptomyces argillaceus provided indirect evidence that MtmU functions as the 4-ketoreductase converting 73 to 81.[94] However, MtmU shares sequence identity (~ 50%) with sugar 3-ketoreductases rather than 4-ketoreductases. If the proposed activity for MtmU is correct, it would be an interesting example of "regio-promiscuity" of a sugar biosynthetic enzyme. The chromomycin gene cluster encodes two 4-ketoreductase homologues, CmmUI and CmmUII, one of which should catalyze the conversion of 73 to 81. The 4-O-acetylation and 4-O-methylation of the two d-oliose moieties of chromomycin to form chromose D (82) and olivomose (83) may be catalyzed by CmmA and CmmMIII, respectively, and likely occur after glycosyltransfer.[104]
Mithramycin and chromomycin also contain d-mycarose (85) and olivomycose (87), both of which are derived from 3-C-methylation of 73. The methyltransferase MtmC has been assigned this role through gene disruption studies in S. argillaceus.[94] The homologous CmmC encoded in the chromomycin cluster likely functions in the same capacity. The resulting compound, TDP-4-keto-d-mycarose (84) can be used as the substrate in the glycosyltransfer reaction in the mithramycin pathway, since disruption of the gene encoding the C-4 reductase MtmTIII resulted in mithramycin derivatives carrying a 4-keto-d-mycarose moiety (see 84) in place of 85.[95] The olivomycose (87) unit of chromomycin is predicted to be constructed from 84 by 5-epimerization and 4-ketoreduction to give TDP-l-chromose (86), followed by glycosyltransfer and 4-O-acetylation. It is possible that the 4-O-acetylation reaction is also catalyzed by CmmA, as in the proposed pathway for 82.[104]
The 2-deoxy-d-evalose moiety (90) of the heptasaccharide chain of avilamycin A is believed to come from 73 via 3-C-methylation by AviG2 to generate 88. This methylation step is identical to the TylC3/EryBIII/Lkm27/MtmC/CmmC reaction (discussed above). However, in the AviG2-catalyzed reaction, the stereochemistry of the 3-OH group is retained, whereas it is inverted in the TylC3/EryBIII/Lkm27/MtmC/CmmC-catalyzed reactions. Following C-methylation, 4-ketoreduction by either AviZ1 or AviZ2 is expected to produce TDP-2-deoxy-d-evalose (89).[106] After glycosyltransfer, an orthoester linkage is formed between the 2-deoxy-d-evalose moiety and the adjacent d-olivose residue. This step may be catalyzed by one of the three non-heme iron dependent enzymes (AviO1, AviO2, and AviO3) encoded in the avilamycin cluster.
The 2,3,6-trideoxysugars, such as TDP-l-amicetose (92) and TDP-l-rhodinose (94), and the 4-amino-2,3,4,6-tetradeoxysugar, TDP-d-forosamine (100), are another subset of TDP-sugars derived from 73. The key step in their biosynthesis is the C-3 deoxygenation of 73 to form TDP-4-keto-2,3,6-trideoxy-d-glucose (91) as an intermediate.[108] Compound 92 is predicted to be the sugar donor in the biosynthesis of the terpene antibiotic phenalinolactone, which carries a 4-O-methyl-l-amicetose (93) moiety. A pathway consisting of 3-deoxygenation by PlaA1 to form 91, 5-epimerization by PlaA8, and 4-ketoreduction by PlaA7 likely generates 92. O-Methyltransfer by PlaM1, which is assumed to occur after glycosyltransfer, will give 93.[109]
l-Rhodinose (see 94), the C-4 epimer of l-amicetose (see 92), is found in urdamycin,[102] landomycin,[101] aclarubicin (aclacinomycin),[77] rhodomycin,[96] and granaticin,[93] all of whose gene clusters have been sequenced. TDP-l-rhodinose (94) is biosynthesized from 91 via 5-epimerization and 4-ketoreduction. Evidence for the functions of the 5-epimerase (UrdZ1) and the 4-ketoreductase (UrdZ3) in the biosynthesis of 94 was obtained when their corresponding genes were individually disrupted in the urdamycin producer Streptomyces fradiae, which subsequently failed to produce urdaymcin derivatives containing l-rhodinose moieties.[102] Genes encoding enzymes for these steps have been assigned in the landomycin, and granaticin gene clusters, but neither was identified in the rhodomycin or aclacinomycin cluster. The epimerase gene is also not found in the nanchangmycin cluster. These activities may be encoded elsewhere in the genome, or may be carried out by the promiscuous l-rhodosamine biosynthetic enzymes in the case of rhodomycin and aclacinomycin. In the polyether natural product nanchangmycin, l-rhodinose is methylated after attachment by NanM, giving 4-O-methyl-l-rhodinose (95).[110]
The major products of aclacinomycin biosynthesis, aclacinomycin A and B, contain l-cinerulose (96) and l-cinerulose B (98), respectively, while aclacinomycin N contains l-rhodinose (see 94) and is only a minor compound. The available evidence suggests that extracellular oxidases rapidly convert the l-rhodinose moiety to 96, which is further oxidized to l-aculose (97), and finally to 98 outside the cell. An intracellular system for reducing these intermediates back to 96 also exists, but the purpose for these interconversions is not clear.[111] The 4-amino-2,3,4,6-tetradeoxysugar, d-forosamine (see 100), is found in the macrolide antibiotics spinosyn, butenyl spinosyn, and spiramycin. To date, it is the most highly deoxygenated sugar found in nature. The biosynthesis of TDP-d-forosamine (100) in the spinosyn producing strain, Saccharopolyspora spinosa, has been fully elucidated in vitro. In this work, SpnQ was shown to be the 3-dehydrase converting 73 to 91,[112] and SpnR was identified as the 4-transaminase converting the SpnQ product 91 to TDP-4-amino-2,3,4,6-tetradeoxy-d-glucose (99).[113] The N,N-dimethylation of 99 is catalyzed by SpnS. Interestingly, unlike its homologue E1 (see Section 3.1.5 for a mechanistic discussion), SpnQ does not have a dedicated reductase partner encoded in the gene cluster, but instead uses general cellular reductases such as ferredoxin and/or flavodoxin for electron transfer.[112–114]
2.3 UDP-sugars
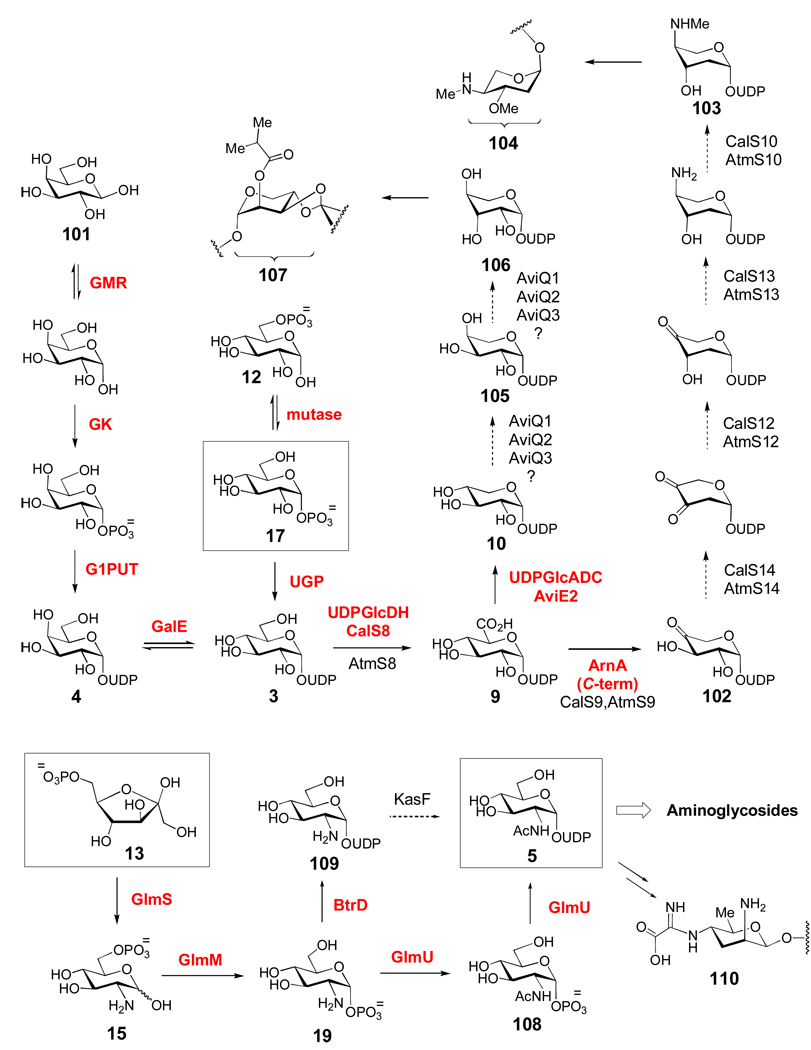
A variety of UDP-sugars exist in nature, including six of the nine common eukaryotic sugar donors and many sugar donors used in the synthesis of bacterial cell surface polysaccharides. Biosynthetically, UDP-activated sugars fall into two groups (Scheme 4): those derived from α-d-glucose-1-phosphate (17) via the glycolytic intermediate α-d-glucose-6-phosphate (12), and those derived from fructose-6-phosphate (13) via UDP-N-acetyl-α-d-glucosamine (5). Compound 17 is converted to UDP-α-d-glucose (3) by α-d-glucose-1-phosphate uridylyltransferase (UGP), an essential enzyme for all organisms. In contrast, the four Leloir pathway enzymes, galactose mutarotase (GMR), galactokinase (GK), galactose-1-phosphate uridylytransferase (G1PUT), and UDP-galactose 4-epimerase (GalE) are responsible for the conversion of β-d-galactose (101) to 3.
Scheme 4. Biosynthesis of UDP-sugars.
a) Biosynthetically, UDP-α-d-glucose (3) is derived either from glycolytic intermediate 12, or from β-d-galactose (101) via the Leloir pathway (see text for details). The pentose moieties of calicheamicin (104) and avilamycin (107) are likely derived from a UDP-α-d-glucose (3) precursor. A separate group of UDP-sugars are derived from the glycolytic intermediate fructose-6-phosphate (13) via UDP-N-acetyl-d-glucosamine (5, see text for details). Compound 5 is likely the biosynthetic precursor of many aminoglycoside sugars, many of which are 2-aminosugars (see Figure 2).
UDP-α-d-glucuronic acid (9) is formed from 3 by the NAD+-dependent UDP-glucose dehydrogenase (UDPGlcDH). This UDP-sugar is a building block for capsular polysaccharides, which are critical to bacterial virulence.[115] Recently, the activity of a UDP-glucose dehydrogenase, CalS8, was demonstrated to be involved in the synthesis of the deoxyaminopentose moiety (104) of calicheamicin.[116] The formation of the UDP-dideoxyaminopentose (103) used for calicheamicin (Cal) and AT2433 (Atm) biosynthesis was proposed to start with the oxidation of 3 by Cal/AtmS8 to form 9, followed by the oxidative decarboxylation of 9 to form 102 by Cal/AtmS9. This is followed by C-2 deoxygenation (Cal/AtmS14), C-3 ketoreduction (Cal/AtmS12), C-4 transamination (Cal/AtmS13), and 4-N-monomethylation (Cal/AtmS10).[117] Interestingly, the first two steps of this proposed pathway (3 → 9 → 102) have no precedent in TDP-sugar pathways, while the last four steps bear close similarity to reactions which are common in TDP-sugar biosynthesis but are unique for UDP-sugar formation. It was proposed that some (such as CalS8) or all of the enzymes in this pathway are pyrimidine indiscriminant, accepting both UDP- and TDP-sugars as substrates. This is supported by an in vitro analysis of CalS8, which demonstrated that while UDP-glucose (3) is the preferred substrate, TDP-glucose can also be efficiently oxidized.[116]
For the biosynthesis of the l-lyxose-derived moiety (107) of avilamycin in Streptomyces viridochromogenes, compound 9 is converted to UDP-d-xylose (10) by the short-chain dehydrogenase/reductase (SDR) enzyme, AviE2, which is a UDP-glucuronate decarboxylase (or UDP-xylose synthase) homologue.[118] Compound 10 is the common xylose donor used in the biosynthesis of cell wall polysaccharides in plants and fungi, cell surface polysaccharides in bacteria, and for protein glycosylation in animals. Interestingly, with the exception of AviE2, enzymes catalyzing the formation of 10 have not been found in any other secondary metabolite biosynthetic pathways in actinomycetes.[118] Formation of 106 is thought to proceed from 10 by sequential C-4 and C-3 epimerization reactions. These reactions may be catalyzed by two of the three SDR family enzymes (AviQ1, AviQ2, or AviQ3) with unknown functions that are encoded in the cluster.[118] This family of enzymes (discussed in Section 3.2) is known to catalyze the epimerization of hydroxyl groups at unactivated C-2, C-4, and C-6 positions of various NDP-sugar substrates. The involvement of these putative SDR enzymes in the formation of 106 has not yet been established.
A second group of UDP-sugars used in various biosynthetic reactions is derived from fructose-6-phosphate (13) via UDP-N-acetyl-d-glucosamine (5, Scheme 4). The first step in this process is the conversion of 13 to glucosamine-6-phosphate (15) catalyzed by glucosamine-6-phosphate synthase (GlmS). In bacteria, 15 is converted to glucosamine-1-phosphate (19) by phosphoglucosamine mutase (GlmM).[119] This is followed by N-acetyltransfer, catalyzed by the C-terminal domain of GlmU, to generate 108.[120] The final step, resulting in UDP-GlcNAc (5), is catalyzed by the N-terminal domain of GlmU. Recently, a nucleotidyltransferase (BtrD) that catalyzes either uridylylation or thymidylylation of 19 to give 109 was discovered in the biosynthetic pathway of butirosin, an aminoglycoside antibiotic produced by Bacillus circulans.[121] Acetylation of 109 could provide an alternative biosynthetic route to 5 in some bacteria.
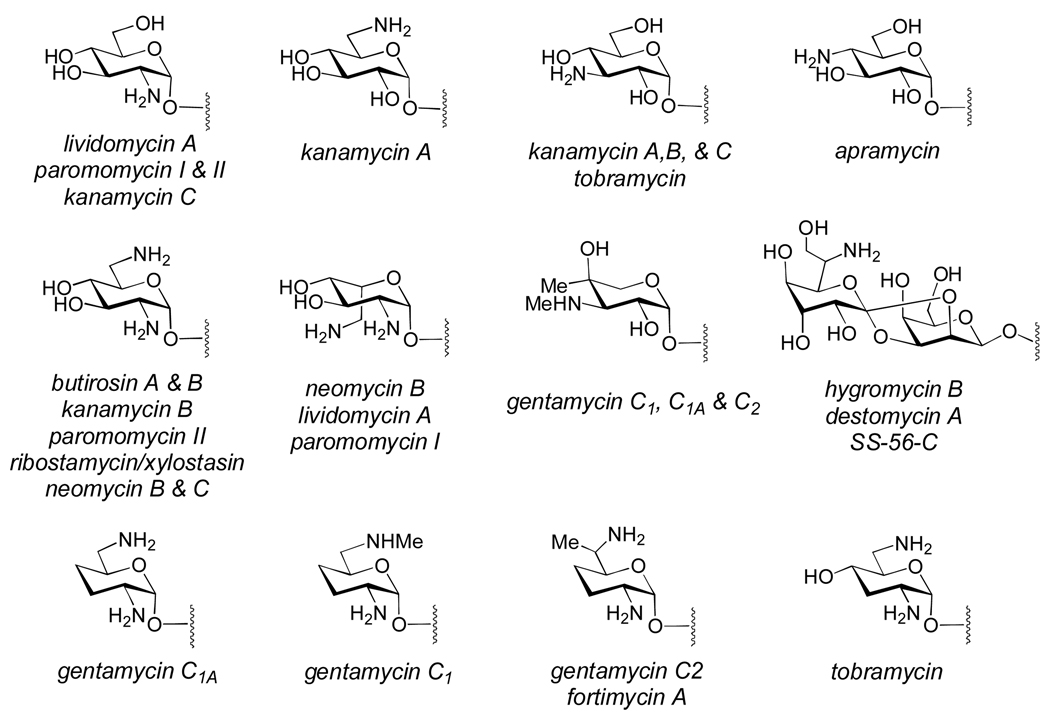
Most aminoglycoside antibiotics containing 2-deoxy-scyllo-inosose or myo-inositol-derived aglycones are decorated with structurally diverse aminosugars (Figure 2).[122] The biosynthetic gene clusters for several members of these classes of aminoglycosides have been identified. These include butirosin, kanamycin, apramycin, lividomycin, paromomycin, neomycin, tobramycin, gentimycin, ribostamycin, fortimicin, and kasugamycin.[122] Since nucleotidylyltransferase genes are absent from most of these gene clusters, the NDP-sugar precursors (such as 3 or 5, Scheme 4) used for the biosynthesis of these sugars are likely derived from the common NDP-sugar pool.[121, 122] As expected, genes encoding NAD(P)-dependent dehydrogenases, oxidoreductases, and PLP-dependent aminotransferase enzymes are abundant in these clusters, and are likely involved in introducing amino groups into the sugar products via various oxidation/transamination reactions. At this point, however, the biosynthesis of most of these sugars is poorly understood, and in most cases it is not clear whether the biosynthetic enzymes perform their reactions on NDP-sugar substrates, or whether they are tailoring reactions after glycosyl coupling. One notable exception is the kasugamine moiety (110) of kasugamycin, whose biosynthetic gene cluster encodes several enzymes with high homology to established UDP- and TDP-sugar modifying enzymes.[123] The aminoglycoside sugars are rich in atypical structural features (Figure 2), such as the unusual patterns of deoxygenation observed in tobramycin, gentamycins, and fortimycins, the unusual C-methyl branches in the gentamycins and fortimicins, and the axial stereochemistry of the C-5 aminomethyl groups of neomycin B, lividomycin A, and paromomycin I. The proposed mechanisms of some of these modifications are discussed in more detail in Section 3.3.
Figure 2. Representative aminoglycoside sugars.
The aminosugar substituents of many aminoglycosides contain unusual modifications, whose biosyntheses are not well understood. See text for details.
2.4. GDP-sugars
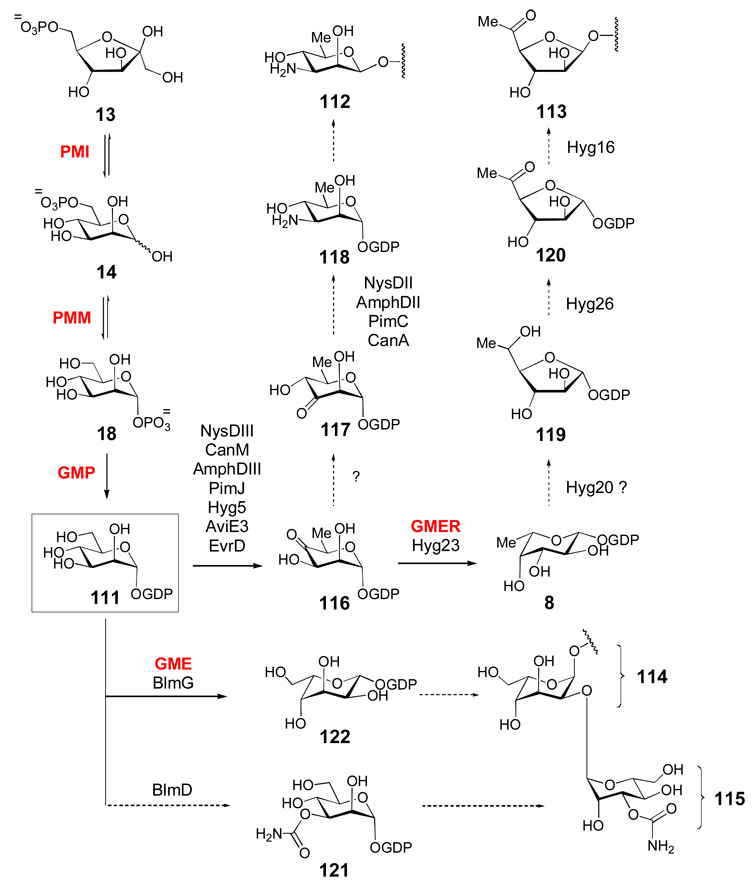
Although GDP-activated sugars (Scheme 5) are generally involved in the biosynthesis of bacterial cell surface polysaccharides and eukaryotic glycans, GDP-mannose (111) is the suggested precursor of the sugar moieties in the polyene macrolide natural products nystatin, amphotericin, pimaricin, and candicidin (which each contain d-mycosamine 112), the aminoglycoside antibiotic hygromycin A (which contains 5-dehydro-α-l-fucofuranose 113), and the antitumor drug bleomycin (which contains both l-gulose 114 and 3-O-carbamoyl-d-mannose 115).[124–126] GDP-α-d-mannose (111) is derived from fructose-6-phosphate (13) by the action of three enzymes: phosphomannose isomerase (PMI) catalyzes the reversible interconversion of 13 and d-mannose-6-phosphate (14); phosphomannomutase (PMM) catalyzes the reversible interconversion of 14 and α-d-mannose-1-phosphate (18), and mannose-1-phosphate guanylyltransferase (also known as GDP-mannose pyrophosphorylase, GMP) catalyzes the GTP-dependent formation of GDP-α-d-mannose (111) from 18. Compound 111 is then converted to GDP-4-keto-6-deoxy-α-d-mannose (116) by GDP-mannose 4,6-dehydratase (GM-4,6-D), a member of the SDR superfamily, which catalyzes essentially the identical reaction as its counterparts in ADP-, CDP-, UDP-, and TDP-sugar biosyntheses.[127] The GM-4,6-D genes (NysDIII/CanM/AmphDIII/PimJ) have been located in the gene clusters of nystatin, candicidin, amphotericin, and pimaricin. Following the conversion of 111 to 116, a 3,4-sugar ketoisomerase, which has not yet been identified, is predicted to convert 116 to 117. Subsequent C-3 transamination catalyzed by an aminotransferase encoded in each gene cluster leads to GDP-d-mycosamine (118),[124] which is the likely sugar donor in the biosynthesis of these compounds.
Scheme 5. Biosynthesis of GDP-sugars.
GDP-sugars are derived from GDP-α-d-mannose (111), which is in turn derived from fructose-6-phosphate (13, see text for details). The biosynthetic gene clusters for nystatin (nys), amphotericin (amph), pimaricin (pim), candicidin (can), hygromycin A (hyg), avilamycin (avi), and evernimicin (evr) each encode putative GDP-mannose-4,6-dehydratase genes that are predicted to convert 111 to 116. The hygromycin A cluster encodes a putative GDP-6-deoxy-4-keto-d-mannose-epimerase/reductase (GMER or GDP-fucose synthase) homologue (Hyg23) -enzymes which are known to convert 116 to 8. The l-gulose (114) and 3-O-carbamoyl-d-mannose (115) residues of bleomycin are proposed to be synthesized from 111 via the GDP-α-d-mannose-3,5-epimerase (GME) homologue BlmG and the carbamoyltransferase BlmD, respectively.
Analysis of the recently sequenced hygromycin A gene cluster has resulted in a proposed biosynthetic route for its 5-dehydro-α-l-fucofuranose moiety (113).[125] The pathway starts with the conversion of 111 to 116 by Hyg5, followed by 3,5-epimerziation and C-4 reduction to GDP-l-fucose (8) by Hyg23, an SDR enzyme. As in the biosynthesis of the d-fucofuranose (24) and d-streptose (26) residues of gilvocarcin V and streptomycin (Scheme 3a), the mechanism for the ring contraction of 8 to 119 is unknown, but the authors proposed that this step could be mediated by Hyg20, which shares sequence identity (31%) with transglucosylases. Though it is not clear how this enzyme would function, a Hyg20 homologue (Ata16) is also present in the gene cluster of the structurally related antibiotic A201A.[125] Following formation of the furanose ring, Hyg26 is predicted to oxidize the C-5 hydroxy group to give 120, which is then coupled to the hygromycin aglycone by Hyg16.
The l-gulose (114) and 3-O-carbamoyl-d-mannose (115) moieties of bleomycin, a hybrid polyketide/non-ribosomal peptide antitumor agent from Streptomyces verticillus, are also derived from GDP-mannose.[126] In addition to the nucleotidylyl transferase (blmC) and glycosyltransferase genes (blmE,F), putative carbamoyl transferase (blmD) and NDP-sugar epimerase (blmG) genes are present in the gene cluster. BlmD likely carbamoylates 111 directly to give 121. BlmG is closely related to the GDP-mannose-3,5-epimerases (GME), which catalyze 3-, 5-, or 3,5-epimerization of GDP-mannose.[128] A 5-epimerization of GDP-mannose (111) would generate GDP-l-gulose (122), which could then be coupled to the aglycone. Finally, putative GDP-mannose-4,6-dehydratase genes (aviE3 and evrD) are also present in the biosynthetic gene clusters for avilamycin A[106] and evernimicin[129] produced by Streptomyces viridochromogenes Tü57 and Micromonospora carbonacea var africana, respectively. It is not known, however, which of the seven sugar residues in each of these heptasaccharide antibiotics are derived from GDP-mannose derivatives, as each cluster also contains a TDP-glucose-4,6-dehydratase gene (aviE1 and evrW).
2.5. CDP-sugars
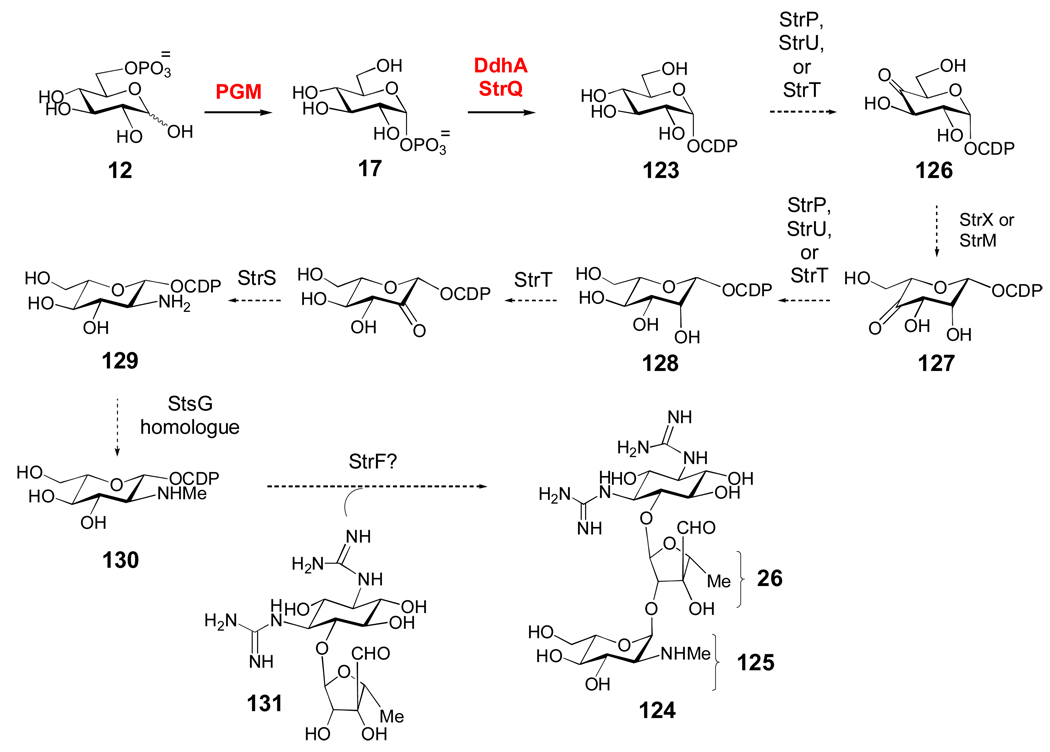
CDP-activated sugars are rare, and are used mostly in the biosynthesis of 3,6-dideoxyhexoses found in the cell wall lipopolysaccharides of certain Gram-negative bacteria, where they are known to be important antigenic determinants. Much like TDP- and UDP-sugars, CDP-sugars are derived from glucose-6-phosphate (12), which is converted to glucose-1-phosphate (17) by phosphoglucomutase (Scheme 6),[130] and then to CDP-α-d-glucose (123) by α-d-glucose-1-phosphate cytidylyltransferase (Ep, DdhA).[131, 132] Interestingly, a DdhA homologue (StrQ) was found in the streptomycin (124) gene cluster of Streptomyces glaucescens, and its cytidylyltransferase activity was verified in vitro.[133] However, TDP-glucose has been implicated as the precursor of the streptose moiety (26) of streptomycin (see also Scheme 3a).[122] Thus, StrQ may participate in the biosynthesis of the N-methyl-l-glucosamine (125) moiety of streptomycin.
Scheme 6. A putative biosynthetic pathway for CDP-l-glucosamine in Streptomyces glaucescens.
The biosynthesis of the N-methyl-l-glucosamine moiety (125) of the aminoglycoside antibiotics streptomycin and bluensomycin is poorly understood. However, the cytidylyltransferase activity (17 → 123) of StrQ encoded by the streptomycin gene cluster of Streptomyces glaucescens was verified in vitro, suggesting that 125 may be derived from a CDP-sugar precursor. The absence of StrQ homologues in other streptomycin and bluensomycin clusters, however, suggests that different biosynthetic routes to 125 may exist (see text for details).
The production of 125 in S. glaucescens has been observed and pathways for its biosynthesis have been proposed, but have not yet been studied in much detail.[122, 134] After the formation of 123 by StrQ (Scheme 6), oxidation of the 4-OH group by StrP, StrT, or StrU could yield 126. These enzymes are all NAD(P)-dependent oxidoreductases/dehydrogenases. The subsequent 3,5-epimerization to 127 is likely carried out by StrX, which has several homologues in TDP- and CDP-sugar biosynthesis including StrM, the TDP-4-keto-6-deoxy-α-d-glucose-3,5-epimerase involved in the biosynthesis of the d-streptose moiety (26) of streptomycin. Following reduction of the 4-keto group of 127 to give 128 by one of the aforementioned NAD(P)-dependent enzymes, the C-2 amine group is likely incorporated by the combined action of the oxidoreductase, StrT, and the PLP-dependent aminotransferase, StrS, to give 129. Interestingly, the strT/S genes are located in tandem in all of the streptomycin and bluensomycin (a closely related antibiotic) gene clusters that have been sequenced.[122] Next, N-monomethylation of 129 to give 130 in S. glaucescens could be carried out by a homologue of StsG, an N-monomethyltransferase which is found in the streptomycin gene cluster of Streptomyces griseus, but which is absent in the S. glaucescens cluster. Coupling of 130 to the d-streptose (26) moiety of 131 may be catalyzed by StrF, which is part of a conserved cassette including the strFGH genes in all streptomycin clusters. Expression of a DNA fragment containing strFG and part of the strH gene led to the restoration of streptomycin production in a Streptomyces bikiniensis mutant strain that otherwise accumulated 131.[135] Interestingly, analysis of the gene clusters for streptomycin and bluensomycin biosynthesis suggest that the pathways for formation of 125 are likely different between the producing strains.[122] Clearly, more work is needed to fully elucidate the biosynthetic pathway for formation of 125.
2.6. Summary of NDP-sugar Biosynthetic Pathways
Through a combination of genetic, biochemical, and bioinformatic efforts, significant advances have been made in our understanding of natural product NDP-sugar biosynthesis. Although many of the steps proposed to occur in the pathways have not been experimentally verified, the following general principles have been gleaned from the work performed on these pathways. First, excluding the few sugars that are not 6-deoxyhexoses such as the N-methyl-l-glucosamine moiety (125) of streptomycin, 4,6-dehydration occurs as the first step after nucleotidylyl transfer in all pathways studied thus far, and is a requisite step for all subsequent reactions. Indeed, many of the following enzymatic modifications (discussed in Section 3) in these pathways either occur directly at the 4-keto site (such as 4-ketoreduction and 4-transamination) or they rely on the activation provided by the 4-keto group to lower the pKa of the C-3 and C-5 protons (3-, 5-, or 3,5-epimerization, 3- and 5-C-methylation, 3,4-ketoisomerization, 3- and 2-dehydration). Second, in all 2,6-dideoxysugar pathways, C-2 deoxygenation occurs after C-6 deoxygenation (21 → 59, Schemes 3b and 3c), and is followed by either 3-ketoreduction (59 → 64 or 59 → 73, Scheme 3c) or 3-aminotransfer (59 → 60, Scheme 3b). The C-3 ketoreductases giving equatorial (59 → 73) or axial (59 → 64) products can be distinguished by amino acid sequence alignments. In the case of 2,3,6-trideoxysugars, C-3 deoxygenation occurs after the C-2 deoxygenation/C-3 ketoreduction step (21 → 59 → 73 → 91, Scheme 3c). For the 4,6-dideoxysugars (e.g., d-desosamine and d-chalcose), C-4 deoxygenation requires prior 4-aminotransfer, and occurs after C-6 deoxygenation (21 → 44 → 45, Scheme 3b). Thus, the order of deoxygenation steps is C-6→C-2 for 2,6-deoxysugars, C-6→C-2→C-3 for 2,3,6-trideoxysugars, and C-6→C-4 for 4,6-dideoxysugars.
Further modifications, such as ketoreduction, C-methylation, epimerization, and transamination (except before C-4 deoxygenation) seem to occur subsequent to all deoxygenation reactions. The C-4 ketoreduction and N-methylation reactions generally occur at late stages of these pathways, while O-methylation usually happens after the TDP-sugar donor has been coupled to its aglycone acceptor. Cumulatively, insight gained from these studies can be used as guidelines for gene cluster-assisted or de novo prediction of natural product sugar biosynthetic pathways. However, this type of sequence-based functional prediction should be performed with caution. In many cases, biochemical characterization of the encoded proteins and mechanistic studies of the key enzymes involved remains necessary to unambiguously establish the overall biosynthetic pathways.
3. The Chemistry of NDP-Sugar Biosynthetic Enzymes
Despite the number of unusual sugar structures present in bacterial secondary metabolites (see Section 2), only five common enzyme reaction types are used by Nature to generate most of this structural variation. Table 1 (supporting information) lists these common reactions along with an illustration of the reaction type and the names of representative enzymes that are known to catalyze these reactions either in vitro or in vivo through gene disruption or heterologous expression experiments. Since several comprehensive reviews on the enzyme chemistry related to deoxysugar biosynthesis are available,[136–139] this section will only highlight the common themes employed by these enzymes to generate sugar diversity.
Because the great majority of natural product sugars are 6-deoxyheoxes, a particularly prevalent theme observed in deoxysugar biosynthesis is the intermediacy of NDP-4-keto-6-deoxyhexose (see 21, Scheme 7) in the pathways. Accordingly, most of the subsequent transformations such as ketoreduction, transamination, epimerization, isomerization, methylation, dehydration, and deoxygenation, have taken advantage of the activation provided by the 4-keto group of this intermediate. The mechanisms of several of the enzymes involved in these transformations will be discussed in Section 3.1. The essential nature of keto-group installation to deoxysuagr biosynthesis is further underscored by the presence of short-chain dehydrogenase/reductase (SDR) enzymes in many sugar biosynthetic pathways. This versatile group of enzymes uses a tightly-bound NAD+ coenzyme to generate a transient NDP-keto-sugar intermediate, which is then further processed within the same active site to achieve a desired chemical transformation. The proposed mechanisms for several selected sugar-modifying SDR enzymes are discussed in Section 3.2. In the final topic of this section, we will investigate several unusual modifications observed in some natural product deoxysugars, whose mechanisms of formation are not well understood. These unusual modifications are also partly responsible for the vast number of different final sugar structures.
Scheme 7. A common theme in the mechanisms of many deoxysugar biosynthetic enzymes.
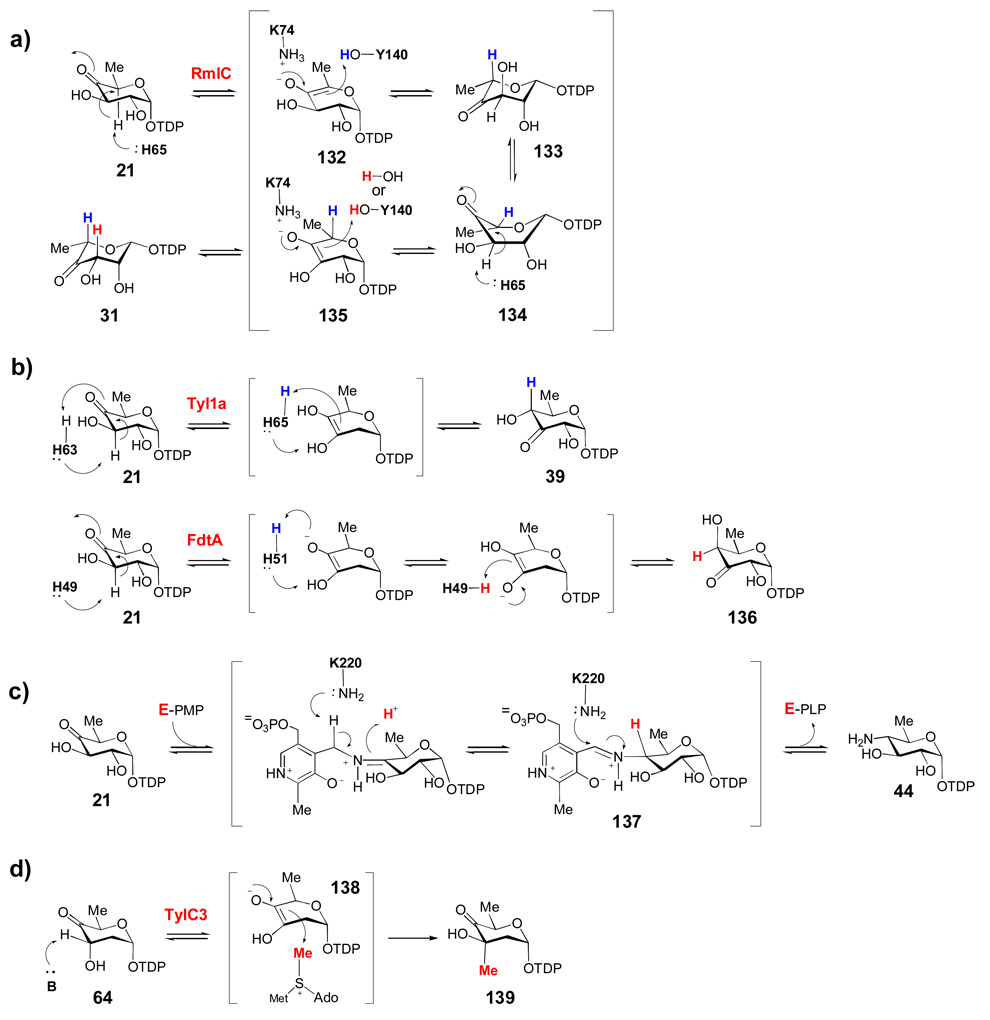
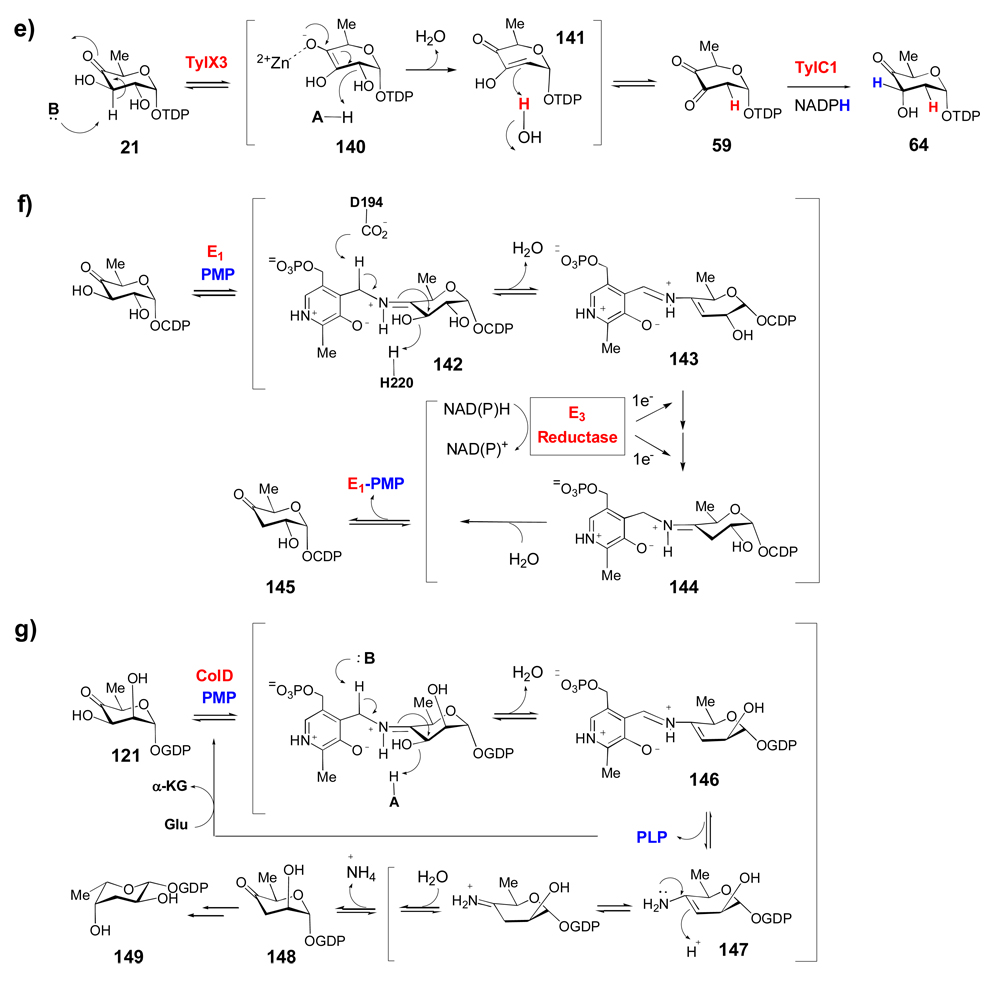
Many deoxysugar biosynthetic enzymes utilize the 4-keto group installed during the first step of deoxysugar biosynthesis (see 20 → 21, Scheme 2) to catalyze their respective reactions. Some examples include the TDP-4-keto-6-deoxy-d-glucose-3,5-epimerase (RmlC) from Pseudomonas aeruginosa (a), the TDP-4-keto-6-deoxy-d-glucose-3,4-ketoisomerases (Tyl1a and FdtA) from Streptomyces fradiae and Aneurinibacillus thermoaerophilus, respectively (b), the TDP-4-keto-6-deoxy-d-glucose-4-aminotransferase (DesI) from Streptomyces venezuelae (c), the TDP-4-keto-2,6-dideoxy-d-glucose-3-C-methyltransferase (TylC3) from Streptomyces fradiae (d), the TDP-4-keto-6-deoxy-d-glucose-2-dehydratase (TylX3) from S. fradiae (e), the CDP-4-keto-6-deoxy-d-glucose-3-dehydrase (E1) from Yersinia pseudotuberculosis (f), and the GDP-4-keto-6-deoxy-d-mannose-3-dehydrase (ColD) from Y. pseudotuberculosis (g). See text for mechanistic details of each reaction.
3.1. The General Reaction Types of Sugar Biosynthetic Enzymes
3.1.1. Reduction
Ketoreductases are the most widely distributed group of enzymes in deoxysugar biosynthesis, and a number of their functions have been biochemically established (see Table 1). The ketoreducatses found in NDP-sugar biosynthetic pathways catalyze the NAD(P)H-dependent hydride reduction of 3- and 4-ketosugars to yield the corresponding secondary alcohol. Both 3- and 4-ketoreduction can occur with either stereochemistry. Many 2,6-dideoxysugar biosynthetic gene clusters encode a 3-ketoreductase, whose activity is required to reduce the unstable NDP-3,4-diketosugar produced by the 2-dehydratase-catalyzed reaction (see Section 3.1.5). Another possible explanation for the large number of ketoreductases in these pathways is simply that the biosynthesis of most deoxysugars involves ketosugar intermediates, which are essential for the enzyme-catalyzed reactions described below. Following the necessary chemical transformations, ketoreductases (many of which are believed to act at late stages in NDP-deoxysugar biosynthesis) may serve to stabilize the final NDP-sugar product. Interestingly, multiple amino acid sequence alignments of established and putative NDP-sugar ketoreductases indicate evolutionary divergence between the 3- and 4-ketoreductases, as the two groups do not share any significant sequence similarity. In addition, within the 3-ketoreductase group, enzymes that generate axial and equatorial C-3 hydroxyl groups can be readily distinguished, whereas the stereochemistry of the 4-ketoreductase-catalyzed reaction is more difficult to predict based on amino acid sequence alone. Due to the variety of C-2, C-3, C-4, and C-5 substituents that must be accommodated by individual ketoreductases during their catalyzed reactions, detailed structural and substrate specificity studies of these enzymes should help to assess their usefulness for in vitro NDP-sugar synthesis and glycoengineering applications.
3.1.2. Epimerization/Isomerization
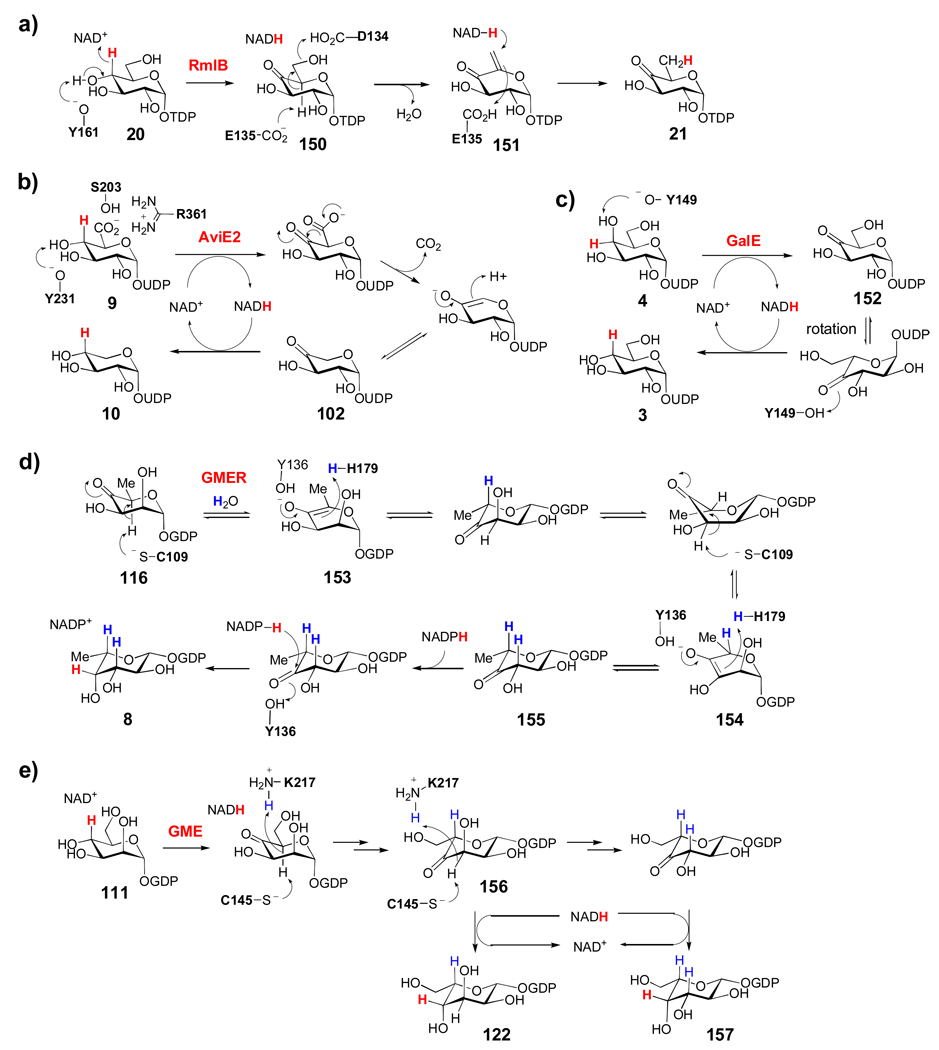
RmlC, which catalyzes the conversion of TDP-4-keto-6-deoxy-α-d-glucose to TDP-4-keto-6-deoxy-l-mannose (21 → 31, Scheme 7a) in the biosynthesis of l-rhamnose in bacteria, is one of the most studied sugar epimerases/isomerases and can serve as the prototype for other NDP-ketosugar-3-, 5-, and 3,5-epimerases. Structural and mechanistic studies of Pseudomonas aeruginosa RmlC led to a mechanism in which both 3- and 5-epimerization of 21 proceed with deprotonation at C-3 and C-5 by His65, which forms a catalytic dyad with a conserved Asp171 residue.[19] The resulting enolate intermediates (132, 135) are stabilized by Lys74, and the subsequent protonation is mediated by Tyr140 (or possibly a water molecule for the C-3 epimerization) to complete each epimerization step. Deuterium exchange studies have shown that epimerization at C-5 is much more facile than at C-3, and thus likely occurs first. After C-5 epimerization, a ring-flipped intermediate (133) in the 1C4 conformation typical of l-sugars, is proposed to form in order to avoid steric clashes between the 5-methyl group, the O1 atom and His65. Intermediate 133 is likely in equilibrium with a twist boat conformation (134), in which the C-3 hydrogen is orthogonal to the plane of the 4-keto group to facilitate C-3 proton abstraction. While most other RmlC homologues involved in natural product biosynthesis (Scheme 3a, the enzymes that catalyze 21 → 31) are not as well characterized as RmlC from Pseudomonas aeruginosa, sequence alignments show that all these enzymes share the conserved His-Lys-Tyr catalytic machinery, so that they likely operate by a similar mechanism.
TDP-4-keto-6-deoxyglucose (21) is also the substrate for Tyl1a (Scheme 7b), the TDP-4-keto-6-deoxy-3,4-ketoisomerase from Streptomyces fradiae, which catalyzes the conversion of 21 to 39 in the d-mycaminose pathway.[54, 55] While few genes encoding Tyl1a homologues are found in natural product biosynthetic gene clusters, they are abundant in the biosynthetic gene clusters for bacterial outer membrane polysaccharides. Among these, FdtA from Aneurinibacillus thermoaerophilus L420-91T, has recently been structurally and mechanistically characterized.[140] A conserved histidine pair (His49 and His51) in FdtA is proposed to catalyze the isomerization (Scheme 7b), with His49 being responsible for C-3 deprotonation and His51 mediating the proton transfer between O3 and O4. Subsequent protonation at C-4 by His49 results in the formation of 136. The corresponding residues His63 and His65 in Tyl1a are expected to play similar roles in the conversion of 20 to 39, as shown in Scheme 7b.
3.1.3. Transamination
Another common enzymatic reaction used in these biosynthetic pathways is the pyridoxal 5'-phosphate (PLP)/pyridoxamine 5'-phosphate (PMP)-dependent transamination reaction. The crystal structures of several sugar aminotransferases have been solved, including those of the 4-aminotransferase, DesI,[72] and the 3-aminotransferase, DesV, involved in d-desosamine biosynthesis in Streptomyces venezuelae.[141, 142] The structure of DesI, in the presence of PLP and the aminosugar product 44 (Scheme 7c), revealed an external aldimine intermediate (137) where Lys220, the residue that normally anchors PLP in the active site via a Schiff base linkage, is in close proximity to both C-4' of PLP (3.4 Å) and the C-4 atom of the sugar substrate (3.0 Å). It likely plays a role in mediating the proton transfers that occur during the transamination. Interestingly, when compared to the structure of PseC from Helicobacter pylori,[143] a 4-aminotransferase that introduces an axial amino group into a 4-ketoketosugar, the hexose moiety observed in DesI is flipped about 180°. This major difference in hexose orientation is likely responsible for the opposite stereochemistry of amino group incorporation catalyzed by these two enzymes.[141]
3.1.4. Methylation
The 3-C-methyl transfer reaction catalyzed by TylC3 in the biosynthesis of the l-mycarose moiety of tylosin in Streptomyces fradiae was the first NDP-sugar C-methyltransferases to be characterized in vitro (Scheme 7d).[90] This enzyme, like many other C-, O-, and N-methyltransferases, requires an S-adenosylmethionine (SAM) cosubstrate for catalysis. Similar to the reactions catalyzed by 3,5-epimerases and 3,4-ketoisomerases, catalysis is initiated by the abstraction of the C-3 proton from 64, which may need to adopt a twisted conformation (similar to the conversion of 133 → 134 in Scheme 7a) to faciliate the deprotonation step by an active site base. The nascent enediolate intermediate (138) then reacts with the electrophilic methyl group of SAM to generate 139 with net inversion of the 3-OH stereochemistry. Interestingly, no metal ion is required for this transformation, suggesting that the TylC3 active site stabilizes the enediolate intermediate (138) mainly by electrostatic interactions. The activities of a few other NDP-sugar C-3 and C-5 methyltransferases have also been verified in vitro (Table 1). They are all believed to employ a similar mechanism to that of TylC3.[39, 40, 82, 94]
3.1.5. Deoxygenation
The 2,6-dideoxysugars depicted in Schemes 3b and 3c represent the largest group of unusual sugars found in natural products. All of these sugars require a 2-deoxygenation step catalyzed by 2-dehydratase enzymes at an early stage of their biosynthesis. Gra Orf27 from the granaticin pathway of Streptomyces violaceoruber Tü22 and the accompanying 3-ketoreductase (Gra Orf26) were the first enzymes involved in NDP-sugar 2-deoxygenation to be studied.[144] Shortly after this initial report, studies on TylX3 and TylC1, the corresponding 2-dehydratase and 3-ketoreductase from the l-mycarose pathway of Streptomyces fradiae, provided additional insights into the mechanism for 2-deoxygenation.[84] It was shown that TylX3 activity required a Zn2+ ion, which is most likely involved in activating a water molecule to serve as the base for C-3 deprotonation or in stabilizing the enolate intermediate (140). Following β-elimination of the 2-OH group, the nascent enol product (141) is ketonized to 59 with a solvent hydrogen stereospecifically incorporated into the equatorial position at C-2. Subsequent reduction of the 3-keto group by the NADPH-dependent TylC1 gives 64 with an axial 3-OH group. In the biosynthesis of granaticin, the 3-ketoreducatse Gra Orf26 transfers the NADPH-derived hydride to the opposite side of the 3-ketohexose (59), resulting in an equatorial 3-OH.
The mechanism of 3-deoxygenation further demonstrates the diverse transformations in NDP-deoxysugar biosynthesis involving 4-keto-6-deoxy-α-d-glucose. This reaction requires two enzymes and the mechanism was originally established for CDP-4-keto-6-deoxy-d-glucose-3-dehydrase (E1) and its reductase (E3) in the ascarylose biosynthetic pathway from Yersinia pseudotuberculosis.[145–148] E1 is homologous to PLP-dependent aminotransferases, but contains PMP instead of PLP as the coenzyme and possesses a histidine in place of the conserved Schiff base-forming lysine found in all aminotransferases. E1 also contains a catalytically essential [2Fe-2S] cluster and requires a [2Fe-2S]-containing flavodoxin-NADP+ reductase partner, E3, for activity. The E1 mechanism begins with Schiff base formation between PMP and the 4-keto group of the substrate to form 142 (Scheme 7f). Next, the C-4' proton of PMP is abstracted, which triggers expulsion of the 3-OH group to form the Δ3,4-glucoseen intermediate 143. This intermediate is then reduced to 144 by two sequential one-electron transfers from the NADH reduced E3-bound FAD via the [2Fe-2S] cluster of E3 and the [2Fe-2S] cluster of E1. Subsequent hydrolysis gives product 145 and regenerates PMP.
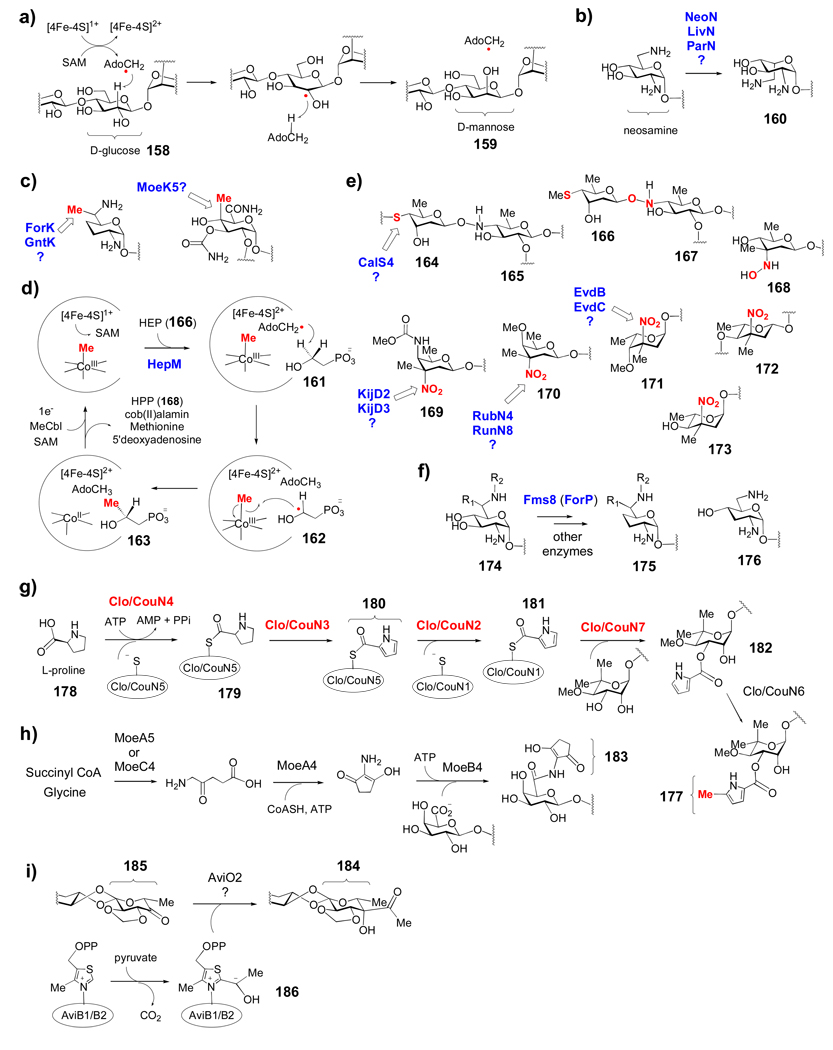
Recently, the 3-dehydrase activity of SpnQ from the TDP-d-forosamine (100) biosynthetic pathway of Saccharopolyspora spinosa was verified in vitro.[112] No E3 homologue is present in the spn gene cluster, and efficient conversion of 59 → 91 (Scheme 3c) was observed only in the presence of various cellular enzymatic reducing systems suggesting that SpnQ, like E1, most likely employs a general reductase from the cellular pool to complete the 3-deoxygenation process. Interestingly, the 3-dehydrase (ColD) from the l-colitose (see 149) biosynthetic pathway of Yersinia pseudotuberculosis, is also a PMP-dependent enzyme, but it lacks the [2Fe-2S] cluster present in E1.[149] The first half of ColD catalysis was shown to mimic the E1 reaction, with Schiff base formation and dehydration to give an intermediate similar to 143 (146, Scheme 7g). The second half of the ColD reaction involves hydrolysis of the Δ3,4-amino-mannoseen intermediate (146) to give PLP and an enamine sugar (147), which then undergoes tautomerization followed by hydrolysis to form the 4-keto-3,6-dideoxymannose product (148), releasing ammonia in the process. In contrast to the E1 reaction, where PMP is regenerated by sequential one-electron reduction from E3, the PMP coenzyme in ColD must be regenerated from PLP after each catalytic cycle by a transamination reaction in the presence of glutamate. The combined deoxygenation-transamination activity makes ColD a unique enzyme.
3.2. The Versatile Roles of SDR Family Enzymes in Unusual Sugar Biosynthesis
The importance of ketosugar intermediates in sugar biosynthesis is further underscored by the extensive use of a subfamily of the short-chain dehydrogenase/reductase (SDR) enzymes in many NDP-sugar biosynthetic pathways.[150] These "nucleotide sugar modifying" SDR enzymes have a conserved protein fold and active site geometry, and commonly use an NAD(P)+ cofactor (or occasionally an NAD(P)H cosubstrate) in a variety of reactions, including ketoreduction, oxidation/dehydration, epimerization at unactivated carbon centers, α-epimerization/ketoreduction, and oxidation/decarboxylation. In the first step of most of these reactions, the SDR enzyme in question oxidizes one of the sugar hydroxyl groups to a keto group, thus generating a reactive intermediate that can be further manipulated in the active site to effect numerous chemical transformations.
The SDR-enzyme-catalyzed 4,6-dehydration is one of the most important reactions in sugar biosynthesis. As illustrated in the previous section, the product of this reaction, NDP-4-keto-6-deoxysugar, is a key intermediate in deoxysugar biosynthesis. The mechanism[124–127] and structure[151–154] of several NDP-d-glucose-4,6-dehydratases have been characterized in great detail. The reaction catalyzed by the 4,6-dehydratases is initiated by deprotonation of the 4-OH group of NDP-glucose (such as TDP-glucose, 20) by a conserved Tyr residue with concomitant transfer of the 4-H as a hydride to NAD+ (Scheme 8a). Crystallographic studies of 4,6-dehydratases from Salmonella enterica,[151] Streptococcus suis,[153] and Streptomyces venezuelae[154] as well as several other sugar-modifying SDR enzymes[128, 155–158] have revealed the presence of conserved Ser/Thr and Lys residues, which likely lower the pKa of the sugar C4-OH and Tyr hydroxyl groups, respectively. Together, this conserved Ser/Thr-Tyr-Lys triad forms one of the signature motifs in SDR enzymes and is believed to be involved in mediating the hydride transfer steps in all of these enzymes. The dehydration of 150 across the C-5/C-6 bond is facilitated by a Glu residue, which is conserved in NDP-sugar-4,6-dehydratases and which deprotonates C-5, resulting in an enone intermediate (151). In many 4,6-dehydratases, such as RmlB of S. enterica, an Asp residue is believed to be responsible for protonating the C6-OH group to facilitate dehydration.[153] This Asp residue, however, is absent in the GDP-α-d-mannose-4,6-dehydratases, implying the involvement of a different catalytic acid group in these enzymes. Compound 151 is then reduced at C-6 with the hydride that was originally derived from 4-H and is protonated at C-5 by the Glu residue to yield the product, 21. Internal hydride return from the transient NAD(P)H to the product is a key mechanistic trait of most SDR enzymes involved in sugar biosynthesis. Thus, in these SDR enzymes, NAD+ is a tightly-bound coenzyme rather that a cosubstrate.
Scheme 8. Mechanisms of selected sugar-modifying short-chain dehydrogenase/reductase (SDR) enzymes.
Proposed mechanisms of TDP-α-d-glucose-4,6-dehydratase (RmlB) from Salmonella enterica (a), UDP-α-d-glucuronate decarboxylase (AviE2) from Streptomyces viridochromogenes (b), UDP-α-d-galactose-4-epimerase (GalE) from E. coli (c), GDP-4-keto-6-deoxy-α-d-mannose-3,5-epimerase-4-reductase (GMER or GDP-fucose synthase) from E. coli (d), and GDP-α-d-mannose-3,5-epimerase (GME) from Arabidopsis thaliana (e). See text for details of each proposed mechanism.
UDP-xylose (10, Scheme 8b) is required for primary metabolism in all organisms, but related pentoses are rare constituents of natural products. In primary metabolism, UDP-xylose results from decarboxylation of UDP-α-d-glucuronic acid (UDP-GlcA, 9) by the NAD+-dependent UDP-glucuronate decarboxylase (also known as UDP-xylose synthase). Early mechanistic studies of this enzyme demonstrated that the reaction is initiated by oxidation of the 4-OH group of 9, followed by decarboxylation and protonation to give 102.[159] Reduction of 102 with the transiently formed NADH yields UDP-xylose (10). Structural studies and amino acid sequence alignments of the ArnA decarboxylase domain of E. coli (a related enzyme that catalyzes 9 → 102) suggest that an Arg/Ser pair that is conserved in this class of enzymes is important in mediating the decarboxylation event.[157] Recently, AviE2, which is involved in the biosynthesis of the l-lyxose-derived moiety (107, Scheme 4) of avilamycin, has been shown to be a UDP-GlcA decarboxylase, making it the first enzyme of this type found in an actinomycete natural product sugar biosynthesis pathway.[118] Genes encoding putative UDP-GlcA decarboxylase homologues (CalS9 and AtmS9, respectively) are also present in the gene clusters for the enediyne antibiotic calicheamycin and the indolocarbazole antibiotic AT2433.[33, 117] Although the activities of CalS9/AtmS9 have not yet been biochemically verified, they are believed to catalyze the formation of 102, rather than 10, which makes their activities more similar to the ArnA decarboxylase domain than to UDP-xylose synthases.
Formation of the l-lyxose-derived moiety of avilamycin requires epimerization at an unactivated carbon atom. This type of reaction is often catalyzed by a group of SDR enzymes homologous to the well-studied UDP-galactose-4-epimerase (GalE) from the Leloir pathway of primary metabolism.[160] GalE homologues that catalyze epimerization of pyranose hydroxyl groups at C-2, C-4, and C-6 have been characterized.[160–165] Structural studies of GalE from E. coli have shown that, following the oxidation of the 4-OH group of UDP-galactose (4 Scheme 8c), the hexose ring of intermediate 152 rotates along the C1-O-P bond in the active site.[156] This allows the transfer of the hydride from NADH to the opposite face of 152 at C-4 to form UDP-glucose (3) and regenerate NAD+. With regard to avilamycin biosynthesis, epimerization at both C-3 and C-4 of UDP-xylose (10) are required to form 106 (Scheme 4). Two putative SDR-enzymes, AviQ1 and AviQ2, that show homology to GalE are encoded in the avilamycin cluster, and may be responsible for the epimerization reactions.[118] If this is found to be the case, it will be the first example of an SDR enzyme-catalyzed epimerization of an NDP-sugar 3-hydroxyl group.
Interestingly, several SDR 3,5-epimerases also have 4-reductase activity (Scheme 8d). GDP-6-deoxy-4-keto-d-mannose-epimerase/reductase (GMER or GDP-fucose synthase), involved in GDP-fucose biosynthesis in all organisms, is a representative of such a dual function enzyme. The E. coli enzyme catalyzes the 3,5-epimerization reaction using a His179/Cys109 pair in the absence of NADPH.[158, 166–168] The conserved Tyr136 residue of the Ser/Thr-Tyr-Lys motif stabilizes the enolate intermediates (153 and 154). In the reductive reaction, Tyr136 protonates the 4-keto group of 155 upon hydride transfer from NADPH. Recently, a GMER homologue (Hyg23) was proposed to catalyze an identical reaction in the hygromycin A biosynthesis pathway of Streptomyces hygroscopicus NRRL 2388 (Scheme 5).[125] Interestingly, the related SDR enzyme, GDP-mannose-3,5-epimerase (GME), uses the NAD+ coenzyme to oxidize the 4-OH group of GDP-d-mannose (111) prior to 3,5-epimerization (Scheme 8e).[128] Overall, catalysis by GME is very similar to that of GMER, except that GME uses a Lys/Cys acid/base pair instead of a His/Cys pair. Following 5-epimerization, the intermediate 156 can be reduced at C-4 to give GDP-β-l-gulose (122), or epimerized at C-3 and then reduced at C-4 to give GDP-β-l-galactose (157). The l-gulose moiety of bleomycin (114, Scheme 5) is believed to be generated in an analogous manner, most likely by BlmG.[126]
3.3. Unusual Modifications in Natural Product Sugar Biosynthesis
While most unusual sugar biosyntheses are accomplished by the "common" enzyme activities listed in Table 1, further structural diversification involving modifications such as epimerization and methylation at unactivated carbon centers, sulfurylation, nitro and hydroxylamino group formation, ring contractions, and others, also happen. However, most of these uncommon tailoring modifications have not been experimental investigated.
Recently, Boll et al. demonstrated that removing a single gene, aviX12, from the avilamycin A gene cluster of Streptomyces viridochromogenes Tü57, led to an inactive avilamycin derivative (gavibamycin N1) where a glucose moiety (see 158) replaced the mannose moiety (see 159) that is normally present (Figure 3a).[169] This surprising observation suggested that the mannose residue is not directly derived from GDP-mannose as previously thought.[106] Instead, it may be formed via C-2 epimerization of a glucose unit (158 → 159) mediated by AviX12, which leads to the final active form of avilamycin A. Examination of the predicted AviX12 amino acid sequence revealed a CxxxCxxC motif, which is characteristic for a radical SAM enzyme.[170] Thus, the reaction catalyzed by AviX12 is proposed to be initiated by hydrogen atom abstraction at C-2 of the glucose unit by the 5′-deoxyadenosyl radical (AdoCH2•) generated by reductive cleavage of SAM with the reduced [4Fe-4S]1+ cluster (Figure 3a), followed by delivery of a hydrogren atom to the opposite side of the sugar ring to give mannose as the product. If verified, this would clearly be an unusual radical initiated epimerization reaction. Interestingly, the aminoglycoside antibiotics neomycin B, lividomycin A, and paromomycin I, all contain a C-5 epimer of neosamine (160, Figure 3b), and each gene cluster contains a putative radical SAM enzyme gene, NeoN, LivN, and ParN, respectively.[122] The encoded enzymes may also catalyze AviX12-like epimerization reactions.
Figure 3. Selected unusual sugar-tailoring modifications.
a) Conversion of d-glucose to d-mannose (158 → 159) in the biosynthesis of avilamycin by AviX12 – a radical SAM-dependent enzyme. b) Putative function of several radical SAM-dependent enzymes encoded in the neomycin, lividomycin, and paromomycin clusters. c) Methylation at unactivated carbon centers in gentamycin, fortimycin, and moenomycin biosynthesis may be carried out by a group of radical SAM/cobalamin-dependent enzymes. d) Proposed mechanism for Fom3, a radical SAM/cobalamin-dependent enzyme from the fosfomycin biosynthetic pathway of Streptomyces wedmorensis. e) Representative thio-, nitro- and hydroxylamine sugars found in several bacterial natural products. f) Putative involvement of Fms8 in 3,4-didehydroxylation of a fortimycin biosynthetic intermediate and the 2,3,6-trideoxysugar residue (176) found in tobramycin. g) Assembly of the 5-methyl-carboxypyrrole substituent (177) of cloromomycin and coumermycin using non-ribosomal peptide synthase biosynthetic logic. h) Biosynthesis of the unsual sugar substituent (183) in moenomycin. i) Unusual modifications required to synthesize the methyleurekanate moiety (184) of avilamycin A.
A new type of methylation mediated by methylcobalamin-dependent radical SAM enzymes is speculated to be involved in the formation of several antibiotics including moenemycin A of Streptomyces ghanaensis,[171] fortimycin A of Micromonospora olivasterospora, and gentamycin of Micromonospora echinospora.[172] Each of these gene clusters contains a gene encoding a putative methylcobalamin-dependent radical SAM enzyme (MoeK5, ForK, and GntK), respectively), which could introduce a methyl group at an unactivated carbon center of the respective sugar substrate (Figure 3c). These enzymes have not been studied, but a similar enzyme (Fom3) in fosfomycin biosynthesis has been identified and its mechanism has been proposed.[173] The enzyme catalyzes the conversion of 2-hydroxyethylphosphonate (HEP, 161) to (S)-2-hydroxypropylphosphonate (HPP, 163). As shown in Figure 3d, the reduced [4Fe-4S]1+ cluster first generates the 5'-deoxyadenosyl radical (AdoCH2•), which abstracts a hydrogen atom from the substrate (161) to generate a radical intermediate 162. The substrate radical then abstracts Me• from the methylcobalamin to form the methylated product (163) and cob(II)alamin. This putative mechanism is metabolically expensive since 2 equivalents of SAM are consumed per cycle. Also the role of methylcobalamin as a methyl radical donor is highly unusual.
Other novel enzyme activities are required for the biosynthesis of unusual sugars, including the thiosugars found in calicheamicin (164) and esperamicin (166), the hydroxylamine sugars of calicheamycin (165), esperamicin (167) and viriplanin A (168), and the nitrosugars of kijanimicin (169), rubradirin (170), evernimicin (171), cororubicin (172), and decilorubicin (173) (Figure 3e). To date, the enzymes responsible for these modifications are not known, and only the calicheamicin,[33] evernimicin,[129] rubradirin,[174] and kijanimicin[99] biosynthetic gene clusters have been identified. Notably, the calicheamicin gene cluster encodes a putative cysteine desulfurase (CalS4), which may be involved in the biosynthesis of the calicheamicin thiosugar moiety (164).[175] A cytochrome P450 enzyme or a flavin-dependent monooxygenase is expected to be responsible for the formation of the hydroxylamine moiety.[175] The nitrosugars (169–173) are most reasonably derived from oxidation of the corresponding aminosugars. In fact, the clusters for evernimicin,[129] rubradirin,[174] and kijanimicin[99] each contain a three-gene cassette encoding a NDP-3-C-methyltransferase (EvdA/RubN5/KijD1), a NDP-3-aminotransferase (EvdB/RubN4/ KijD2), and a flavin-depedent oxidoreductase (EvdC/RubN8/KijD3) that may be involved in 3-methyl-3-nitrosugar biosynthesis.[99] The O-methylcarbamate moiety of kijanose (169) is also unusual. A series of N-methylation, methyl oxidation to a carboxylate group, and O-methylation mediated by KijD8, KijB3 and KijD9, respectively, has been proposed.[99] As highlighted by the thio, nitro, and hydroxylamine sugars, many interesting modifications in natural product sugar biosynthesis remain to be discovered and explored.
Although initial studies have produced significant advances in our understanding of the mechanisms of enzymes catalyzing C-O bond cleavage in deoxysugar biosynthesis,[136–138] our knowledge is far from complete. For example, quite a few deoxygenations, such as those in the formation of the 2,6-diamino-2,3,4,6-tetradeoxysugar unit (175) of gentimicin and fortimicin, and the 2,6-diamino-2,3,6-trideoxy neosamine moiety (176) in tobramycin (Figure 3f), may proceed via distinct mechanisms. Studies of Micromonospora olivasterospora mutants blocked at various stages of fortimicin A biosynthesis revealed the accumulation of various 3,4-dihydroxy-(174) and 3,4-dideoxysugar (175) intermediates, but no 3- or 4-monohydroxylated compounds.[176] Complementation studies of these mutants using fragments of the fortimicin gene cluster eventually led to identification of the fms8(forP) gene product as the possible catalyst for the didehydroxylation step,[177] because when heterologously expressed in M. olivasterospora, Fms8 restored the didehydroxylation phenotype.[177, 178] How the didehydroxylation occurs and whether other enzymes are needed remain unclear, but phosphorylation of an intermediate in the pathway may be critical, because Fms8 is a homologue of NmrA, a phosphotransferase involved in neomycin B resistance. A similar didehydroxylation mechanism is possible for the formation of gentimicin. The biosynthetic route to the tobramycin sugar (176) is also mysterious, as the characterized mechanisms for 3-deoxygenation require the generation of a 4-keto-6-deoxy sugar intermediate by a 4,6-dehydratase, followed by 3-dehydration catalyzed either by an E1 or a ColD homologue (see Scheme 7f). However, none of the genes for these enzymes are present in the reported tobramycin clusters, suggesting a different mechanism for 3-deoxygenation in the tobramycin pathway.
The attachment of a 5-methylpyrrole-2-carboxyl moiety (177) to the 4-O-methyl-noviose residue in the antibiotics clorobiocin and coumermycin A1 is another remarkable tailoring modification (Figure 3g). This modification greatly enhances the ability of these drugs to inhibit the bacterial type-II topoisomerase DNA gyrase. The biosynthesis of 177 and its attachment has been studied both in vivo and in vitro.[43, 45] It was found that 177 is derived from l-proline (178), which is activated as an acyl-adenylate and linked to the Clo/CouN5 peptidyl carrier protein (PCP) by Clo/CouN4. Subsequent oxidization of 179 by Clo/CouN3 results in a PCP-linked pyrrole-2-carboxyl substituent (180), which is transferred to a separate PCP (Clo/CouN1) by the acyl-ACP-synthase (Clo/CouN2) to give 181. The final steps include the transfer of 181 to the 4-O-methyl-noviose moiety by a thioesterase (Clo/CouN7) to give 182, and C-methylation by a methylcobalamin-dependent radical SAM methyltransferase (Clo/CouN6) to afford 177. Likewise, the biosynthesis of the C5N unit (183) in moenomycin produced by Streptomyces ghanaensis is also intriguing (Figure 3h). An aminolevulinate synthase (MoeA5), an acyl-CoA-ligase (MoeA4), and an amide synthetase (MoeB4), which are located together in the gene cluster, have been proposed to catalyze the conversion of succinyl-CoA and glycine to 183.[171] Indeed, a moeA4− knockout mutant failed to produce moenomycin bearing 183.
The methyleurekanate (184) residue in avilamycin of Streptomyces viridochromogenes also requires several intriguing tailoring steps: the formation of the orthoester linkage at C-1, the incorporation of the methylene unit between O2 and O3, and the attachment of the 4-C-acetyl moiety (Figure 3i). A 4-ketosugar (perhaps 185) is likely the precursor of 184. Condensation of 185 with the thiamine-pyrophosphate (TPP)-bound acetyl carbanion unit (186) would give 184.[179] The acetyl carbanion unit is likely generated from pyruvate by an AviB1/B2 complex whose subunits share homology with the α and β chains, respectively, of pyruvate dehydrogenase. The enzyme(s) responsible for the incorporation of the methylene unit between O2 and O3, and for the formation of the orthoester linkage are unknown, but as noted earlier, two of the three non-heme iron-dependent enzymes encoded in the avilamycin cluster could be responsible for these tailoring reactions.
3.4. Outlook
The work highlighted in the previous sections has revealed Nature's ingenious and judicious utilization of a small set of core enzyme activities to generate significant sugar structural diversity. Most of these enzymes operate on similar ketosugar substrates, but are able to catalyze distinct reactions using unique active site architecture and cofactor requirements. Future structural and mechanistic studies on sugar biosynthetic enzymes may help clarify the potential of these enzymes as synthetic catalysts for glycodiversification. In particular, members of the SDR family of enzymes represent attractive targets for the rational engineering of enzyme function, as they catalyze many different reactions on diverse sugar substrates using a conserved protein fold and similar, yet distinct active site chemistries. Further elaboration of sugar structures could be achieved by employing enzymes catalyzing unusual transformations and/or tailoring reactions. Together, an understanding and appreciation of the unusual sugar biosynthetic pathways and the mechanisms of the enzymes involved, has contributed to the recent explosion in the use of glycoengineering approaches (the subject of Section 5) to generate new glycoforms of natural products.
4. Natural Product Glycosyltransferases
4.1. The Gatekeepers of Glycodiversity
Glycosyltransferases (GTs) form a critical group of enzymes in biological systems that catalyze the attachment of sugar moieties to acceptor molecules. At present, there are more than 15,800 putative glycosyltransferases in the protein databank, but the functions of most of these GTs have not been verified.[180] Several hundred GTs are predicted to be involved in the biosynthesis of natural products found in bacteria and plants. These GT-catalyzed reactions reside at a critical juncture in natural product biosynthesis, where the products of the sugar and aglycone biosynthetic pathways meet. Hence, in recent years, GTs have been the subject of many studies aimed at understanding and finetuning their biochemical properties.[181] Ultimately, these enzymes may be useful as tools to catalyze "unnatural" coupling reactions to generate new glycoforms of natural products. These efforts have been focused in two main areas: the exploitation of the broad substrate specificity found in many wild-type GTs and the alteration of GT specificity through genetic engineering. The success of these endeavors relies on a multi-faceted strategy encompassing genetic, genomic, molecular biological, biochemical, and chemical approaches.
For most characterized GTs, the donor substrate is a nucleotide diphosphate (NDP)-sugar. However, nucleotide monophosphate (NMP)-sugar and polyprenyl diphosphate-sugar donors are also substrates for specific GTs. Interestingly, a phosphoribosyltransferase (PRTase),[182, 183] which uses 5-phosphoribose diphosphate (PRPP) as a donor for the transfer of 5-phosphoribose to an acceptor substrate was found to be involved in the biosynthesis of the aminoglycoside antibiotic butirosin.[184] This represents the first characterized PRTase involved in natural product biosynthesis.
Like the NDP-sugar donor substrates, the acceptor substrates for natural product GTs are also structurally diverse and include many classes of compounds (Figure 4), such as the polyketide-derived aglycones of pikromycin (187), urdamycin A (188), calicheamicin (189), avilamycin (190), and BE-7585A (191),[185] the non-ribosomal peptide (NRP)-derived aglycone of vancomycin (192), the indolocarbazole aglycone of staurosporine (193), the aminocoumarin aglycone of novobiocin (194) and many others. The coupling reaction entails the displacement of the anomeric substituent of the sugar donor by a nucleophilic functional group of the acceptor to form the glycosidic linkage. The nucleophile is most commonly a hydroxyl group. However, N- and aryl-C-glycosidic linkages (as in 193 and 188, respectively), as well as the unusual orthoester (as in 190), hydroxylamine (as in 189), and thio linkages (as in 191) are also seen in some natural products. The mechanisms for the formation of the latter three types of linkages have not been explored.
Figure 4. Representative bacterial natural product glycoforms.
Examples of glycosylated microbial natural products: pikromycin (187), urdamycin A (188), calicheamicin (189), avilamycin (190), BE-7585 (191), vancomycin (192), staurosporine (193), and novobiocin (194). Glycosidic linkages are typically O-linked, but C-glycosides (see 188), N-glycosides (see 193), hydroxylamine linkages (see 189), and orthoester linkages (see 190) also exist in nature. The disaccharide moiety of 191 is coupled to the aromatic aglycone through an unusual thio linkage.
4.2. Structures of Glycosyltransferases
Since the first GT crystal structure was reported in 1994,[186] 35 GTs structures have been solved.[187] With the exception of a bifunctional transpeptidase-glycosyltransferase involved in peptidoglycan biosynthesis (which has a novel structure),[188] these GT structures fall into two classes, the GT-A and GT-B families, whose properties have been previously reviewed.[150, 180, 181, 189–194] The GT-A superfamily is characterized by a single domain with an α/β/α sandwich topology that resembles a Rossmann fold.[191–193] The NDP-sugar-binding region of GT-A enzymes contains a conserved DXD (Asp-X-Asp) motif for binding a divalent metal (usually Mn2+) that anchors the diphosphate moiety of the NDP-sugar,[195, 196] and stabilizes the NDP leaving group during turnover.[195–200] Interestingly, there is a recent example of a GT-A enzyme that is metal ion-independent and lacks the DXD motif.[201] In contrast, GT-B superfamily members have two Rossmann fold-like domains with a deep, interdomain cleft where the donor and acceptor substrates bind.[193, 194] They are metal-ion independent and lack universally conserved amino acid residues, though the C-terminal nucleotide binding domain is more conserved than the N-terminal acceptor binding domain. Despite the low sequence identity (<10%) among GTs, the three dimensional structures of GTs within the same superfamily are quite similar.
Almost all bacterial natural product GTs are predicted to be members of the GT-B superfamily.[194] To date, the crystal structures of only a handful of natural prodcut GTs have been determined.[202–207] The structure of GtfB from the chloroeremomycin pathway was the first reported structure.[202] As is typical for GT-B superfamily members, GtfB has two domains separated by a flexible linker region, forming a deep cleft between the two domains. The N-terminal domain contains the aglycone binding site, and the C-terminal domain contains the sugar binding site. Since these two domains appeared to be well-separated in GtfB, it was proposed that it may be possible to create chimeric GTs containing donor and acceptor binding domains from separate GTs.[202] The structures of the l-epivancosaminyltransferases (GtfA and GtfD) from the chloroeremomycin and vancomycin pathways, respectively, were later determined in the presence of bound acceptor substrate and TDP.[203, 204] GtfA was found to exist in both open and closed conformational states, with few inter-domain contacts in the closed state. The open state was seen when only the acceptor substrate was bound, while the closed state was observed when both acceptor and TDP were bound, suggesting that TDP-sugar binding may trigger the formation of the catalytically active, ternary Michaelis complex. In contrast to GtfA, the structure of GtfD in the closed conformation revealed several critical interdomain contacts. These results indicated that creation of chimeric or engineered GT variants may be more complicated than thought based on the GtfB structure. To date, there are only two examples where engineering of a natural product GT-B enzyme has successfully altered substrate specificity (discussed in Section 5.2.2.),[208–210] whereas rational structure-based engineering efforts in the GT-A family enzymes have been sowewhat more successful.[211]
4.3. Mechanisms of Glycosyltransferases
Understanding the mechanisms of GTs is important for active site engineering strategies to broaden or alter their substrate specificities. Mechanistically, GTs can be classified as inverting or retaining based on the stereochemical course of the glycosyltransfer reaction they catalyze (Figure 5a).[212] Structural, mechanistic, and computational studies on the inverting GTs support an SN2-like mechanism.[197, 213–217] As shown in Figure 5b, the lone pair electrons from the endocyclic oxygen atom facilitate the formation of an oxocarbenium-like intermediate (or transition state, see 195) by donating electron density to the σ* orbital of the anomeric C1-O1 bond prior to the attack of the acceptor nucleophile. The retaining GTs were originally thought to proceed by a double displacement mechanism involving the initial formation of a covalent sugar-enzyme intermediate (196, Figure 5c),[212] analogous to the well-studied retaining glycosidases.[218] However, structural studies of several retaining GTs failed to identify suitable candidates for the putative enzyme nucleophile.[206, 219–224] A general lack of conserved amino acid residues on the β-face of the anomeric carbon in retaining GTs from the GT-A family has also been noted.[225] Hence, an alternative mechanism was proposed, where the nucleophilic acceptor attacks the anomeric carbon from the same side of the sugar ring as the NDP leaving group in an asynchronous, concerted manner with highly dissociative oxocarbenium-like character (197, Figure 5d).[219, 220, 225, 226]
Figure 5. Mechanisms of glycosyltransferases.
a) Possible stereochemical outcomes of GT-catalyzed reactions. b) Direct displacement mechanism proposed for inverting GTs. c) Double displacement mechanism proposed for retaining GTs. d) Alternative mechanism for retaining GTs involving nucleophilic attack from the same face of the sugar molecule as leaving group departure. e) Proposed mechanisms for aryl-C-glycosidic bond formation. f) UrdGT2 catalyzes the formation of both C- and O-glycosidic linkages (198 → 199 and 200 → 201, respectively).
4.4. Summary of Biochemical Work on Natural Product Glycosyltransferases
Despite the importance of GTs in controlling the glycosylation patterns of natural products, surprisingly few natural product GT activities have been verified in vitro, though a number of GT functions have been deduced by gene knockout and heterologous expression experiments. A list of 167 known and putative bacterial small molecule natural product GTs are compiled in Table 2 (see supporting information). The GTs whose functions have been verified are indicated and the corresponding references are provided. The phylogenetic relationships among antibiotic GTs have been previously reviewed,[227] and most of these enzymes fall into glycosyltransferase family 1 (GT-1),[190] which is comprised of GT-B enzymes catalyzing glycosylation with inversion of stereochemistry. The macrolide resistance GT OleD, which uses UDP-glucose as the sugar donor, was the first macrolide-related GT to be characterized in vitro.[228] Studies on OleD suggested an ordered, sequential kinetic mechanism with the acceptor substrate binding prior to the UDP-sugar to form the ternary Michaelis complex. Following glycosyltransfer, UDP is released from the enzyme prior to the glycosylated product. This kinetic mechanism is supported by the crystallographic studies on other inverting GT-B enzymes, in which a conformational change to a closed state occurs upon binding of NDP to the GT-aglycone complex.[202–205] Although detailed kinetic analyses have not yet been performed on most other natural product GTs, a similar kinetic mechanism is expected to be operative for many inverting GT-B enzymes of the GT-1 family.
It has recently been demonstrated that several macrolide GTs require an auxiliary protein for efficient glycosyltransfer.[229] The genes for the glycosyltransferase and its corresponding auxiliary protein are almost always located next to each other in their respective biosynthetic clusters. The translated gene sequences for these auxiliary proteins share moderate homology with cytochrome P450 enzymes, yet lack the conserved Cys residue that coordinates the heme iron. The requirement of a helper protein for a GT involved in natural product biosynthesis was first established for the desosaminyltransferase, DesVII, and its auxiliary protein, DesVIII, from the methymycin/pikromycin pathway of Streptomyces venezuelae.[229] Subsequently, an enhancement of kcat by AknT was observed for the reaction catalyzed by the anthracycline GT, AknS.[230] The authors proposed that AknT could be functioning as a regulatory subunit that transiently interacts with AknS in order to maintain AknS in an active conformation or to stabilize the transition state for glycosyltransfer. More recent studies of the EryCII/EryCIII glycosylation system demonstrated that the erythromycin GT (EryCIII) remains fully active in vitro after removal of its auxiliary protein (EryCII) from the pre-incubation mixture.[231] Experiments with the DesVII/DesVIII system gave similar results.[232] These observations could suggest that the auxiliary proteins have a chaperone-like function to facilitate a one-time conformational change that activates their corresponding GT. Clearly, more work on these systems is required to fully understand the exact role of the GT auxiliary proteins.
Although the vast majority of glycosylated natural products are O-glycosides, aryl-C-glycosides are also present in bacteria and plants.[233] Two possible mechanisms for C-GT catalysis have been proposed (Figure 5e).[80] One mechanism (path A) involves the initial formation of an O-glycoside, followed by an intramolecular rearrangement to an ortho-C-glycoside. The other mechanism (path B) involves the attack of a resonance-stabilized phenolate anion at the anomeric carbon of the NDP-sugar donor to form the C-glycosidic linkage. Although neither mechanism has been experimentally verified, direct formation of the C-glycosidic linkage is particularly appealing for natural products containing C-glycosyl substituents both ortho- and para-to the activating phenolate group.[233] Several natural products such as gilvocarcin and enterobatin contain only para-C-glycosides, which would be difficult to form with an O-glycosylation/rearrangement sequence.[18, 234, 235]
Recent studies of UrdGT2, a C-GT involved in the biosynthesis of urdamycin in Streptomyces fradiae Tü2717, have provided important insights into the mechanism of C-glycosylation (Figure 5f). While the C-GT activity (198 → 199) of UrdGT2 had been previously established, feeding of the alternative aglycone substrate (200) to an S. fradiae mutant that was deficient in wild-type aglycone biosynthesis, but which still expressed UrdGT2, resulted in the production of the O-glycoside, 201.[236] This study was the first to demonstrate that a natural product GT could synthesize both C- and O-glycosidic linkages. In addition, these results support the direct C-glycoslation mechanism, because initial O-glycosylation of 200 followed by O-O rearrangement is not favored. Moreover, the recently solved X-ray crystal structure of UrdGT2 revealed that the anomeric carbon of the NDP-sugar substrate binds in close proximity to C-9 of the aglycone substrate, and is properly positioned for a direct addition to the aromatic ring.[207] Asp137 was also proposed to be the base that deprotonates the aglycone phenolate group via a tightly bound active site water molecule.
A striking discovery from studies of natural product GTs is that many of these enzymes exhibit remarkably broad substrate specificity towards their aglycone and NDP-sugar substrates. For example, VinC, a GT from the vicenistatin biosynthetic pathway of Streptomyces halstedii HC34, has been shown to accept the α- and β-anomers of both d- and l-sugars in vitro.[237] GtfE, the GT from the vancomycin pathway is known to accept over 30 different NDP-sugars, which has facilitated in vitro glycodiversification of the vancomycin aglycone.[238] The DesVII/DesVIII system has also been shown to accept numerous cyclized and linear forms of aglycone substrates[239–241] as well as a number of different NDP-sugars.[232] In addition to these in vitro studies, the substrate flexibility of many natural product GTs has been demonstrated in vivo. Cumulatively, these studies have helped fuel the metabolic pathway engineering, combinatorial biosynthesis, and in vitro enzymatic glycodiversification efforts that have resulted in the construction of new natural product derivatives with altered glycosylation patterns. Some of this recent glycoengineering work is highlighted in the next section.
5. Natural Product Glycoengineering
A large portion of biologically active natural products are glycosylated. Since the sugar moieties are often important for bioactivity,[5, 7] alteration of the glycosylation patterns of the parent structures (a process known as glycodiversification, glycorandomization, or glycooptimization) has the potential to produce modified molecules with new activities. Accordingly, a number of strategies (both in vivo and in vitro) have emerged in recent years, wherein the biosynthetic machinery (i.e. enzymes) is manipulated to produce new natural product glycoforms. These methods have four advantages over traditional chemical synthesis/derivatization approaches. First, the stereo- and regioselectivity of enzyme-catalyzed reactions generally produce single products with defined stereochemistry. Second, the producing organism is a renewable source of the desired compounds. Third, production of targeted compounds by fermentation is readily scaled up. Finally, both the in vivo and in vitro strategies are amenable to the construction of compound libraries in a combinatorial fashion. The following sections will highlight a few selected in vivo and in vitro experiments designed to alter natural product sugar structures. Several excellent reviews on this topic can be consulted for more information about these methods and their applications.[150, 181, 242–253]
5.1. In vivo Glycodiversification
Our improved understanding of unusual sugar biosynthesis has significantly impacted biosynthesis-based glycosdiversification efforts aimed at producing natural products with altered sugar structures. In early studies of tylosin biosynthesis in Streptomyces fradiae, random mutagenesis yielded S. fradiae strains defective in the biosynthesis or the attachment of each of the three tylosin sugars, mycaminose, mycarose and mycinose.[51] The value of these gene disruption experiments and their potential for generating novel glycosylated natural products was soon recognized. Consequently, more sophisticated in vivo glycodiversification strategies involving heterologous expression of genes were developed and applied in metabolic pathway engineering and combinatorial biosynthesis studies.[254–256]
The use of cells as catalysts to carry out chemical reactions on exogenous molecules is referred as biotransformation. Precursor-directed biosynthesis[257] and mutasynthesis[258] are two well established biotransformation processes in which a biosynthetic precursor of a natural product is replaced by a structural analogue through feeding to a wild-type strain (precursor-directed biosynthesis) or to a gene disruption mutant (mutasynthesis). Metabolic pathway engineering[259] and combinatorial biosynthesis[260] are two more recently developed methods. The basic premise of these methods is that genes from different organisms are combined and expressed in a single host strain in an attempt to re-route the biosynthetic intermediates to new final products. These heterologous expression experiments can be carried out either in the wild type strain, or in knock-out mutant strains, where the mutation allows the accumulation of a specific biosynthetic intermediate by disrupting a downstream step in the pathway. This intermediate can then be processed by the heterologously expressed enzyme(s). The success of these methods relies on the substrate promiscuity of sugar biosynthetic enzymes and GTs. The synthetic potential of these techniques can be further elaborated when performed in conjunction with precursor feeding or bioconversion experiments.
An elegant early example of glycoengineering that utilized a combination of gene disruption and heterologous expression was the creation of 4'-epi-daunorubicin (204) and 4'-epi-doxorubicin (or epirubicin, 205),[256] which are therapeutically useful analogues of the antitumor agents daunorubicin (202) and doxorubicin (203), respectively (Figure 6). In this study, the 4-ketoreductase gene dnmV involved in the final step of TDP-l-daunosamine (53, Scheme 3b) biosynthesis was disrupted in the Streptomyces peucetius host[256] and replaced by avrE or eryBIV, which encode the corresponding epimeric 4-ketoreductase from the l-oleandrose (66) and l-mycarose (71) pathways, respectively (Scheme 3c). The latter two sugars have an equatorial 4-OH group resulting from the axial reduction of the 4-keto group by AvrE or EryBIV. Substitution of dnmV with either of these two reductase genes provided a convienent route to epi-daunosamine. This was the first example of a designed in vivo biosynthesis of a non-natural sugar -i.e., a sugar that has not heretofore been found in nature.
Figure 6. Designed synthesis of an unnatural sugar in Streptomyces peucetius.
The axial C4-OH stereochemistry (highlighted in red) of the daunosamine moiety of daunorubicin (202) and doxorubicin (203) was altered to equatorial in 204 and 205 by the replacement of a single S. peucetius l-daunosamine biosynthetic gene (dnmV) with 4-ketoreductase genes from l-oleandrose (avrE) or l-mycarose (eryBIV) biosynthesis.
5.1.1. Erythromycin
The sugar biosynthetic genes in the erythromycin (206) cluster from Saccharopolyspora erythraea were sequenced, and a number of these genes were individually disrupted and classified as "EryB" or "EryC" genes depending on whether erythronolide B (EB, 207) or mycarosyl erythronolide B (MEB, 208) accumulated (Figure 7).[57, 65] In addition to the accumulation of EB or MEB, other minor derivatives were also produced in these gene disruption mutants (Figure 7, path A). For example, small amounts of desosaminyl erythronolide B (209) were found in an eryBVI disruption mutant, indicating that desosaminyltransfer could still occur to some extent in the EryB mutants.[64] Disruption of the 4-ketoreductase gene, eryBIV, led to an erythromycin analogue (210) having 4-keto-mycarose in place of l-mycarose (211),[57] and disruption of the 3-ketoreductase gene, eryBII, resulted in several minor compounds (one of which is 212) carrying a 2,6-dideoxyglucose instead of l-mycarose.[65] Also, disruption of the C-methyltransferase gene eryBIII gave an erythromycin derivative (213) having 3-desmethylmycarose in place of l-mycarose.[66] Several erythromycin analogues obtained in this manner retained bioactivity, albeit with reduced potency in comparison to erythromycin A.
Figure 7. Engineering erythromycin derivatives in Saccharopolyspora erythraea with gene disruption, heterologous expression, and feeding experiments.
Path A – Disruption of individual l-mycarose (eryB) or d-desosamine (eryC) biosynthetic genes afforded 207–210, 212, and 213. Path B – Heterologous expression of the oleandrosyltransferase (OleG2) from Streptomyces antibioticus in an S. erythraea mutant lacking the endogenous mycarosyltransferase (EryBV). Path C – Expression of the mycaminosyl transferase (TylM2) from the tylosin pathway of Streptomyces fradiae in a triple S. erythraea mutant (SGT2) deficient in desosaminyltransfer (eryCIII−), mycarosyltransfer (eryBV−), and polyketide synthesis (eryA−), generated 216 when the strain was fed tylactone (215). Path D – Novel erythromycin derivates (217 and 218) generated when O-methyltransferase genes (spnI and spnK) from Saccharopolyspora spinosa were heterologously expressed in the S. erythraea SGT2 mutant.
Heterologous expression of foreign glycosyltransferases in various S. erythraea mutants was also used to generate new glycosylated forms of macrolides. For example, expression of the gene encoding the desosaminyltransferase (OleG1) from the oleandomyicn pathway in an S. erythraea mutant lacking the endogenous desosaminyltransferase (EryCIII) restored erythromycin A (206) production,[261] establishing the proposed functions for both EryCIII and OleG1. When the gene encoding the oleandrosyltransferase OleG2 from the oleandomyicn pathway was expressed in an S. erythraea mutant lacking the mycarosyltransferase EryBV (Figure 7, path B), new erythronolide derivatives (214) bearing an l-rhamnose moiety linked to O-3 of the aglycone were formed. Interestingly, the 3-O-l-rhamnosyl erythronolide derivatives were also found when OleG2 was expressed in the wild-type strain, indicating that the heterologously expressed OleG2 could compete with the endogenous GT for sugar transfer to the 3-OH position of the aglycone.
In a separate study, the gene encoding the mycaminosyltransferase (TylM2) from the tylosin biosynthetic pathway of Streptomyces fradiae was integrated into the chromosome of a triple S. erythraea mutant (termed SGT2) that lacked the endogenous glycosyltransferases eryCIII and eryBV, as well as the polyketide synthase gene eryA.[262] When the mutant cell cultures were fed tylactone (215), 5-O-desosaminyltylactone (216) was produced (Figure 7, path C), revealing that the heterologously expressed TylM2 recognizes and couples the non-native desosamine sugar produced by S. erythraea onto its natural aglycone (215). Finally, the individual expression of several l-rhamnosyl-O-methyltransferases from the spinosyn biosynthetic pathway of Saccharopolyspora spinosa in the SGT2 triple mutant, demonstrated that two of these methyltransferases (SpnI and SpnK) could sequentially O-methylate the 2′- and 3′-OH groups, respectively, of exogenously fed 3-rhamnosyl erythronolide B (214) to give 217 and 218 (Figure 7, path D).[263]
5.1.2. Methymycin/Pikromycin
TDP-d-desosamine (43) is the sugar donor used for methymycin and pikromycin (219–221, 187) biosynthesis in Streptomyces venezuelae. As shown in Figure 8, disruption of the dimethyltransferase gene desVI resulted in the accumulation of macrolide analogues carrying 3-N-acetylamino-3,4,6-trideoxy-d-glucose (222) in place of d-desosamine.[67] Similarly, disruption of the aminotransferase gene desV led to analogues bearing 4,6-dideoxy-d-glucose (223);[68] disruption of the desII gene led to analogues with 4-N-acetylamino-4,6-dideoxy-d-glucose (224),[72] and disruption of the desI gene resulted in analogues carrying 6-deoxy-d-glucose (d-quinovose) (225).[264] The ketoreduction at C-4 and C-3 to give the corresponding hydroxyl groups in 223 and 225, and the acetylation to give the N-acetylamino group in 222 and 224 are catalyzed by enzymes not encoded by the pik cluster. These enzymes may be part of the cell surface polysaccharide biosynthetic machinery or they could be involved in other natural product pathways in the host. They function when the appropriate "unnatural" intermediates accumulate. Clearly, the opportunistic participation of some enzymes during metabolic pathway engineering further broadens sugar structural diversity.
Figure 8. Metabolic pathway engineering in the methymycin/pikromycin producer Streptomyces venezuelae.
The natural pathway for the biosynthesis of TDP-d-desosamine (43) and the glycosylated methymycin/pikromycin derivatives (187, 219–221) produced by S. venezuelae are shown in the boxed pathway. Disruption of individual des genes (highlighted in blue), combined with heterologus expression of foreign genes (highlighted in red) was used to engineer a variety of novel, glycosylated macrolide derivatives (222–232).
As described above, TDP-4-keto-6-deoxyglucose (21), which is an intermediate in the desosamine pathway, accumulates in the KdesI S. venezuelae mutant. When a predicted 4-aminotransferase (CalH) from the calicheamicin producer Micromonospora echinospora was expressed in the KdesI mutant, derivatives with the 4-N-acetyl-4,6-dideoxysugar (224) were isolated.[265] In a separate study, the same quinovosyl methynolide derivative (225) was obtained when the putative TDP-4-keto-3,5-epimerase (strM) and TDP-streptose synthase (strL) genes from the streptomycin (124) producer Streptomyces griseus were expressed individually in the KdesI mutant.[27] However, when both genes were expressed together in this mutant, new macrolide derivatives containing an l-rhamnose (226) substituent were generated. This study not only revealed an unexpected 4-ketoreductase activity for StrL, but also demonstrated that the desosaminyltransferase DesVII can process both d- and l-sugar donors.
Several new macrolide derivatives were generated when different combinations of d-mycaminose biosynthetic genes (21 →→ 41, Figure 8) from the tylosin producer S. fradiae were heterologously expressed in S. venezuelae mutants.[55] First, the tylM1/B/M2/M3 genes were expressed in an S. venezuelae KdesI/KdesVII mutant, which lacks the desosaminyltransferase (DesVII) and which was predicted to accumulate 21. These four tyl genes were originally believed to comprise a complete set of mycaminose biosynthetic genes. However, when the mutant cultures were fed tylactone (215), a 5-O-quinovosyl-tylactone derivative was obtained.[56] This result not only reflected the relaxed tolerance of the mycaminosyltransferase (TylM2) for its TDP-sugar donor, but it also suggested that the previously proposed mycaminose pathway was incomplete. An orphan orf in the tylosin gene cluster, tyl1a, was subsequently identified and expressed in the S. venezuelae KdesI/KdesVII mutant along with tylM1/B/M2/M3. When tylactone (215) was fed to this strain, a new tylosin derivative (227) containing a 5-O-mycaminosyl substituent was obtained.[56] Interestingly, when tyl1a was expressed individually in the KdesI mutant, new methymycin/pikromycin derivatives (such as 228 and 229) that carried a mycaminosyl moiety were isolated. These experiments conclusively established the TDP-d-mycaminose pathway (21 → 39 → 40 → 41), and revealed the relaxed substrate specificity of DesV, DesVI, and DesVII/DesVIII.
Finally, when tyl1a was replaced with fdtA (a 3,4-ketoisomerase from Aneurinibacillus thermoaerophilus that catalyzes 21 → 136) in a KdesI mutant, new macrolide derivatives bearing either a 4-epi-d-mycaminose (230 and 231) or a 3-N-monomethyl-3-deoxy-d-fucose (232) substituent were obtained.[266] As neither of these sugars are naturally occurring, this work again illustrates the potential for constructing novel sugar structures by using selected natural sugar biosynthetic enzymes. In addition, these results reveal that many desosamine pathway enzymes, including DesV, DesVI, and DesVII/DesVIII, tolerate sugar donors with an axial 4-OH group. As is evident from these and other[232, 239, 240, 267, 268] studies, the DesVII/DesVIII pair clearly exhibits remarkably relaxed substrate specificity towards its sugar and aglycone substrates.
5.1.3. Elloramycin
The first reported example of in vivo glycodiversification that relied on heterologous expression of biosynthetic genes involved the expression of a cosmid (16F4) that contained most of the elloramycin (233, Figure 9a) biosynthetic gene cluster from Streptomyces olivaceus in the urdamycin (188) producer Streptomyces fradiae Tü2717.[254] The resulting strain produced the hybrid elloramycin derivative 8-demethyl-8-β-d-olivosyltetracenomycin C (234, Figure 9b). The sugar donor TDP-d-olivose was supplied by the urdamycin pathway, and the aglycone (8-DMTC, 235) was produced by the heterologously expressed cosmid 16F4. Later experiments established that the substrate flexible GT, responsible for formation of 234, was ElmGT encoded on cosmid 16F4. In this work, cosmid 16F4 was transformed into a mutant of Streptomyces fradiae Tü2717, in which several genes essential for formation of the urdamycin aglycone were deleted (ΔPKS). Cosmid 16F4 was also transformed into the wild-type and a PKS-defective mutant of the mithramycin (236) producer Streptomyces argillaceus.[269] Expression of cosmid 16F4 in S. fradiae ΔPKS led to increased yields of 234 and also to a new hybrid compound (237) containing the urdamycin sugar l-rhodinose. When the cosmid was expressed in wild-type or ΔPKS S. argillaceus, 234 was again formed along with 8-demethyl-8-β-d-mycarosyltetracenomycin C (238) and the disaccharide-containing compound 8-demethyl-8-β-d-olivo-3'-1"-β-d-olivosyltetracenomycin C (239). When S. fradiae Tü2717/ΔPKS and the S. argillaceus strains were fed 235 in the absence of the cosmid 16F4, no glycosylated tetracenomycin derivatives were obtained, firmly establishing that ElmGT (encoded by cosmid 16F4) is the GT responsible for the formation of the tetracenomycin analogues.
Figure 9. Relaxed NDP-sugar substrate specificity of ElmGT, an l-rhamnosyltransferase involved in elloramycin biosynthesis in Streptomyces olivaceus.
a) Several naturally occurring aromatic polyketides: urdamycin A (188) produced by Streptomyces fradiae, elloramycin (233) produced by Streptomyces olivaceus, mithramycin (236) produced by Streptomyces argillaceus and the 8-DMTC aglycone (235) encoded by the Streptomyces olivaceus cosmid 16F4. b) Hybrid aromatic polyketides produced in vivo by the action of ElmGT expressed from cosmid 16F4 in the heterologous hosts Streptomyces fradiae (compounds 234 and 237) and Streptomyces argillaceus (compounds 234, 238, and 239). c) Novel aromatic polyketides produced through combinatorial biosynthesis (see text for details).
ElmGT was subsequently incorporated into the chromosome of Streptomyces albus, a non-producing strain. This strain was transformed with several plasmids encoding the production of different NDP-sugars, and each resulting strain was then fed 8-DMTC (235).[270] In these experiments, ElmGT was shown to attach l-olivose and l-rhamnose (its natural sugar substrate) onto 235 to generate 240 and 241 (Figure 9c). In a different set of combinatorial biosynthesis studies, cosmid 16F4 was transformed into Streptomyces lividans (also a non-producing strain) along with plasmids encoding the production of NDP-l-digitoxose,[271] NDP-4-deacetyl-l-chromose B,[272] and NDP-l-mycarose.[272] Each of these strains produced the corresponding glycosylated 8-DMTC analogue (242–244, respectively). A glucosylated 8-DMTC compound (245) was also obtained, indicating the unusual tolerance of ElmGT for a sugar containing a 6-OH group.[271] Finally, in an impressive experiment, genes from four different deoxysugar biosynthetic pathways were combined on a single vector to generate d- and l-amicetosyl-8-DMTC derivatives (246 and 247, respectively) when co-expressed in S. lividans 16F4.[273] Since biosynthetic gene clusters for d-amicetose are not available, the above experiment illustrates the power of combinatorial biosynthesis to generate a desired sugar structure based solely on the logic observed in other biosynthetic pathways.
5.1.4. Urdamycin
Urdamycin A (188, Figure 10a), produced by Streptomyces fradiae Tü2717, is an angucycline type antibiotic and anticancer agent. The urdamycin aglycone has an O–linked l-rhodinose residue at C-12b and a C–linked d-olivose-l-rhodinose-d-olivose trisaccharide at C-9. To verify the functions of the four GTs encoded in the urdamycin A gene cluster, as well as the order of glycosylation steps, a number of S. fradiae mutants were constructed in which individual GTs or combinations of GTs were disrupted. This led to a number of urdamycin derivatives (248–258) with unnatural glycosylation patterns (Figure 10a).[274, 275] When the urdGT2 gene was disrupted, several urdamycin shunt metabolites (248–250) accumulated, all of which lacked the trisaccharide moiety at C-9, suggesting that UrdGT2 is the C-GT. Interestingly, 250 showed much better anticancer activity than the parent compound, urdamycin A.[274] In similar knockout experiments, UrdGT1a was identified to be the C-12b-l-rhodinosyl transferase, while UrdGT1c and UrdGT1b were found to be the rhodinosyl- and olivosyltransferases, respectively, responsible for the construction of the trisaccharide. When urdGT1c was overexpressed in the urdGT1c knockout strain, a second l-rhodinose moiety was incorporated into the trisaccharide chain by UrdGT1c to give 257 and 258.
Figure 10. Manipulating urdamycin biosynthesis.
a) Acceptor substrate flexibility of the urdamycin GTs revealed in various Streptomyces fradiae GT mutants. Different combinations of urdamycin A (188) GTs were disrupted, resulting in the production of various glycosylated derivatives (199, 248–258) in the corresponding S. fradiae GT mutant strains. The sugar residues are color-coded to indicate which urdamycin GT is responsible for glycosyl coupling: blue – UdrGT1a, green – UrdGT1b, red – UrdGT1c, and purple – UrdGT2. b) Products isolated from Streptomyces fradiae Tü2717 upon disruption of deoxysugar biosynthetic genes. c) When expressed in glycosyltransferase-deficient mutants of the mithramycin producer Streptomyces argillaceus, UrdGT2 catalyzed production of C-glycosides (263 and 264). Heterologous expression of both UrdGT2 and the d-olivosyltransferase from the landomycin pathway of Streptomyces cyanogenus (LanGT1) in this same S. argillaceus mutant led to compound 265. Expression of UrdGT2, LanGT1, and LanGT4 in a glycosyltransferase deficient mutant of S. fradiae Tü2717, led to the production of 266.
In a separate study, several urdamycin A deoxysugar biosynthetic genes were individually disrupted in S. fradiae, and this led to even more new derivatives.[102] The urdZ3, urdQ, and urdZ1 knockout strains each accumulated urdamycinone B (254, see Figure 10a), reflecting the essential roles of these genes in the biosynthesis of l-rhodinose (Figure 10b). Surprisingly, the inactivation of the 4-ketoreducatse (UrdR) needed for TDP-d-olivose (79) synthesis, yielded urdamycin M (259), which contains a d-rhodinose moiety (see 260) as opposed to normally produced l-rhodinose (see 94) attached to C-9 of 248 through a C-glycosidic linkage. Thus, it appears that the rhodinosyl 4-ketoreductase (UrdZ3) can reduce a rhodinosyl intermediate (such as 91) prior to UrdZ1-catalyzed C-5 epimerization. Intermediate 91 may accumulate to unnaturally high levels in the absence of UrdR, leading to increased concentrations of 260, which can then be coupled to 248 by UrdGT2. These results suggested that UrdGT2 is flexible for its NDP-sugar donor substrate, and is able to accept both TDP-d-olivose (79) and TDP-d-rhodinose (260). In a subsequent study with the S. fradiae urdR− mutant, it was also demonstrated that UrdGT1c could transfer an l-rhodinose moiety to 259 to generate urdamycin R (261).[276] This study also revealed that UrdGT2 could attach l-rhodinose (see 94) to C-9 of 248. The resulting compound could then be l-rhodinosylated by UrdGT1c to give urdamycin S (262). Thus, UrdGT2 is clearly capable of synthesizing C-glycosides using both l- and d-rhodinose in vivo.
When heterologously expressed in Streptomyces argillaceus strains lacking the native mithramycin (236, Figure 9a) glycosyltransferases, UrdGT2 was able to couple the mithramycin deoxysugars, d-olivose and d-mycarose, to the premithramycinone aglycone (to give 263 and 264, Figure 10c) through C-glycosidic linkages at positions of the aglycone that are not normally glycosylated.[277] When UrdGT2 was co-expressed with LanGT1 (a d-olivosyltransferase from the landomycin producer Streptomyces cyanogenus S136) in this same S. argillaceus strain,[277] a hybrid compound (265) was formed. This compound was composed of an S. argillaceus-derived aglycone and a disaccharide assembled by the action of both UrdGT2 and LanGT1. In a separate combinatorial biosynthesis study, heterologous expression of LanGT1 and LanGT4 (an l-rhodinosyl transferase) in a S. fradiae triple GT mutant (urdGT1a-/1b-/1c-) was used to generate hybrid urdamycin/landomycin compounds (such as 266) that contained a new trisaccharide moiety.[278] Clearly, like DesVII and ElmGT, UrdGT2 accepts a variety of NDP-sugar and aglycone substrates and, thus, UrdGT2 may prove to be a useful tool for enzymatic glycodiversification of aryl-C-glycosides.
5.1.5. Indolocarbazoles
The indolocarbazole alkaloid N-glycosides, rebeccamycin (267) produced by Saccharothrix aerocolonigenes and staurosporine (193) produced by several Streptomyces species, are antitumor compounds with DNA topoisomerase I and protein kinase inhibition activities, respectively (Figure 11). Heterologous expression of different combinations of reb and sta genes in the non-producing strain Streptomyces albus helped elucidate the biosynthetic pathway for the rebaccamyin and staurosporine aglycones, and also resulted in a number of new derivatives, many of which were N-glucosylated by RebG.[279] Recent bioconversion experiments demonstrated that RebG, when expressed in either E. coli or S. lividans, could N-glucosylate a number of exogenously fed indolocarbazole derivatives, including the staurosporine aglycone (268).[280] Interestingly, RebG catalysis lacks regioselectivity, since it can glycosylate either of the N atoms of the asymmetric indolocarbazoles used in this study.
Figure 11. Novel indolocarbazoles generated by combinatorial biosynthesis in Streptomyces albus.
The indolocarbazoles rebeccamycin (267) and staurosporine (193) both contain unusual N-glycosidic linkages. Staurosporine biosynthesis was reconstituted in Streptomyces albus by expressing genes required for the formation of the staurosporine aglycone (268), genes encoding production of different deoxysugars (34, 61, 65, 78, and 79), along with the N-GT (StaG) and the P450 enzyme (StaN) responsible for the oxidative crosslinking between C5′ of the sugar and the N-12 atom of the aglycone. While StaG coupled both l- and d-sugars to 268, only the l-sugars could be oxidatively crosslinked by StaN to give 193 and 274–277.
In a separate study, staurosporine biosynthesis was reconstituted in S. albus by co-expressing the biosynthetic genes for the staurosporine aglycone (268), along with those for l-ristosamine, and the putative N-GT, StaG (Figure 11).[74] The transformed S. albus mutant produced holyrine A (270), a compound containing an N-linked 3-N-4-O-didemethyl-l-ristosamine moiety in a 4C1 conformation. When staN (a putative cytochrome P450 gene) was expressed in this S. albus mutant, the cell cultures produced staurosporin, establishing StaN as the enzyme responsible for C5′-N bond formation. The substrate flexibility of StaG was then tested by transforming plasmids encoding the production of different deoxysugars (l-rhamnose 34, l-digitoxose 78, l-olivose 65, and d-olivose 79) into the mutant S. albus strain. HPLC analysis showed that each of the strains expressing l-deoxysugar genes produced two new compounds, while the strain expressing the d-olivose genes only produced one new compound. Subsequent MS and NMR analysis revealed that all five deoxysugars tested in this study could be singly linked to the N-13 atom of the staurosporine aglycone (by StaG) to form 269–273, each with an equatorial N-glycosidic bond that places the sugar in the 4C1 conformation. For the l-sugars, the 4C1 conformation is unusual because the bulky substituents at C-3, C-4, and C-5 are in a less favorable axial configuration. The compounds containing l-sugars (269–272) could be further processed by StaN to yield the doubly attached staurosporine analogues 274–277. Interestingly, in the doubly attached compounds, the l-sugars exist exclusively in the 1C4 conformation, suggesting that StaN converts the 4C1 conformation of the l-sugars into a 1C4 conformation prior to the oxidative coupling of C-5′ to the indole N-12.
5.2. In vitro Glycodiversification
Although significant progress has been made towards natural product glycodiversification through in vivo combinatorial biosynthesis and metabolic engineering, there are several inherent disadvantages that limit the applicability of these approaches. First, it is difficult to control the reaction conditions and to prevent undesired side reactions, which lower the efficiency of the desired glycosylation reactions. Also, the newly generated metabolites are potentially toxic to the bacterial strain used as the host for expression of the heterologous genes. Finally, only those aglycone acceptors and sugar donors that can be biosynthesized or fed to the host can be used as potential building blocks, and this ultimately limits the structural diversity of glycoforms that can be generated. To overcome some of these problems, recent efforts have focused on the development of methods for in vitro glycodiversification using purified sugar biosynthetic enzymes and glycosyltransferases. These efforts have benefited from the accumulated body of knowledge on sugar biosynthetic enzymes and the discovery of several substrate flexible anomeric kinases, nucleotidylyltransferases, and glycosyltransferases. These substrate flexible enzymes have been used to generate libraries of NDP-sugars (reviewed in [281]), which can then be tested in vitro as substrates for glycosyltransferases with natural or engineered substrate flexibility. In this section, we will focus only on those glycoengineering efforts which employ purified sugar biosynthetic enzymes and glycosyltransferases to generate glycorandomized natural product libraries. However, the recent development of purely chemical methods for natural product glycodiversification,[248, 249, 282–286] will undoubtedly provide researchers with robust, alternative strategies for their glycoengineering efforts.
5.2.1. Engineering Sugar Anomeric Kinases
The major limitation to the enzymatic synthesis of NDP-sugars is the availability of the specific enzymes required for the construction of the desired NDP-sugars. To facilitate the preparation of NDP-sugars, directed evolution and structure-based protein engineering have been used to create sugar biosynthetic enzymes with broader substrate specificity. For example, a single round of random mutagenesis on the galactokinase (galK) gene from E. coli[287] was sufficient to generate a GalK variant (Y371H) that tolerates substitutions at C-2, C-3, C-5, and C-6 of d-galactose, but which maintains a stringent requirement for the axial 4-OH group. This mutant can also phosphorylate two l-sugars (278 and 279, Figure 12). Based on a structural homology model with galactokinase from Lactococcus lactis, two conserved residues (Asp37 and Tyr223) in the E. coli enzyme were proposed to form hydrogen bonds with the axial 4-OH group.[288] However, mutation of these residues failed to change the C-4 specificity of the E. coli GalK. In constrast, the Y385H (equivalent to E. coli Y371H) mutant of L. lactis GalK could accept d-glucose and a few other d-sugars with equatorial 4-OH groups as substrates.[289] Further analysis of the E. coli/L. lactis GalK homology model suggested that the Met173 residue in the E. coli enzyme (Leu182 in L. lactis) may have prevented the E. coli enzyme from processing d-sugars with an equatorial 4-OH configuration.[290] Indeed, the E. coli M173L mutant was found to accept d-gluco-configured sugars. Furthermore, the M173L/Y371H double mutant retained the substrate flexibility observed for each single mutant and, in addition, also recognized azido sugars, which can be further modified by chemoselective ligation reactions. The sugar-1-phosphates synthesized by the wild type and mutant E. coli GalK are listed in Figure 12.
Figure 12. Sugar-1-phosphates produced by wild type and engineered E. coli galactokinase (GalK) mutants.
The substrate specificity of wild type GalK was broadened by mutation of active site methionine (M173L) and tyrosine (Y371H) residues. The M173L/Y371H double mutant retained the substrate specificity of each single mutant and also accepted a variety of other sugars. The sugar-1-phosphates generated by each enzyme are boxed and the structural deviations from the wild type GalK substrate (d-galactose) are highlighted in red.
5.2.2. Engineering Nucleotidylyltransferases
Preparation of natural and unnatural sugar-1-phosphates represents only the first stage in the synthesis of NDP-sugars. The next challenge is to convert these compounds to the corresponding NDP derivatives. The α-d-glucose thymidylyltransferase from Salmonella enterica LT2 (RmlA or Ep), which couples either TMP or UMP to a set of sugar-1-phosphates, is the most extensively studied NDP-sugar synthase.[291] RmlA prefers pyranosyl phosphates in the 4C1 chair conformation, and is less efficient towards 2-deoxysugars. It can also process amino and acetamido sugars.[292] The position of the amino group has no effect on turnover, while bulky acetamido groups are only tolerated at the C-2 and C-3 positions. The crystal structures of RmlA in complex with UDP-glucose or TTP[293] showed that the active-site residue Trp224 interferes with the thymidylylation of sugars that contain bulky substituents at C-6. The Trp224 residue was subsequently mutated to His to alleviate the steric crowding around C-6 of the substrate. This mutation may also introduce a positive charge that facilitates binding of sugars containing a C-6 carboxylate group. The substrate flexibility of RmlA was further enhanced by the mutation of Leu89 to Thr, which relieves steric crowding around C-2 of the substrate.[294] In all, over 30 different sugar-1-phosphates were found to be substrates of RmlA or its variants in these studies.
In the search for alternative nucleotidylyltransferases with broad substrate specificity, a heatstable nucleotidylyltransferase from the archaeal organism Pyrococcus furiosus DSM 3638 has been shown to have relatively broad substrate specificity, and can even efficiently uridylylate l-fucose-1-phosphate (see 8, Figure 1).[295] The enzyme is bifunctional and catalyzes 2-N-acetyltransfer to glucosamine-1-phosphate prior to the uridylyltransfer reaction.[296] Using N-acetylcysteamine thioesters in place of acetyl-CoA, several new UDP-glucosamine derivatives were synthesized in one pot from glucosamine-1-phosphate by this enzyme. Nucleotidylyl-transferases from twp other thermophilic archaeal organisms have been shown to accept alternative NTP substrates, including both purine and 2'-deoxy-ribonucleotides.[297, 298]
In a more recent study, RmlA (which is a thymidylyltransferase) was found to use each of the eight naturally occurring NTPs (UTP, CTP, ATP, GTP, TTP, dCTP, dATP, and dGTP) to activate 10 different sugar-1-phosphate substrates, albeit with drastically different catalytic efficiencies.[299] Steady state kinetic analysis indicated that the thymidine preference of RmlA is primarily attributable to a low Km for TTP, while both TTP and UTP are processed with much higher kcat values than other NTPs. Mutation of the Gln83 residue of RmlA, which forms hydrogen bonds to the base-pairing face of the uridine/thymidine moiety, to either Asp or Ser resulted in an enzyme favoring purine nucleotides over pyrimidine nucleotides by three orders of magnitude. As more structural information for nucleotidylyltransferases becomes available, it may be possible to alter the substrate specificity for NTPs by protein engineering as a practical means to expand the repertoire of available NDP-sugars for in vitro glycosylation studies.
5.2.3. In vitro Synthesis of NDP-sugars
To facilitate the enzymatic synthesis of highly-modified TDP-sugars, efficient methods for the preparation of TDP-4-keto-6-deoxy-d-glucose (21) -a common intermediate in many deoxysugar biosynthetic pathways -have been developed. For example, purified sucrose synthase (SuSy) from potato, TDP-glucose-4,6-dehydratase (RmlB) from Salmonella typhimurium, and TMP kinase from yeast were used to synthesize 21 from the inexpensive starting materials sucrose (280) and TMP in one-pot (Scheme 9a) with a typical yield of ~70% (relative to TMP).[300] An ATP-regeneration system consisting of pyruvate kinase (PK) and phosphoenol pyruvate (PEP) was included so that only catalytic amounts of ATP were needed. An analogous strategy using SuSy has also been extended to make other NDP-sugars.[281] In a separate biosynthesis-based approach, TMP kinase (TMK), acetate kinase, and glucose-1-phosphate thymidylyltransferase from E. coli, along with RmlB from S. typhimurium, were expressed in E. coli BL21 cells (Scheme 9b).[301] The crude extracts from these cells were incubated with TMP, acetylphosphate, and glucose-1-phosphate to synthesize 21 in 80% yield (from TMP).
Scheme 9. Enzymatic synthesis of NDP-sugars.
a) One-pot synthesis for the common deoxysugar biosynthetic intermediate (TDP-4-keto-6-deoxy-d-glucose, 21) from sucrose (280) and thymidine monophosphate (TMP) using TMP kinase (TMK), sucrose synthase (SuSy), and RmlB. b) Biosynthesis-based approach for the synthesis of 21 (see text for details). c) Two-stage, one-pot synthesis of TDP-l-mycarose (71). In the first stage, thymidine was converted to TTP by thymidylate kinase (TK), TMK, and nucleotide diphosphate kinase (NDK). Following purification of TTP by filtration, 17 was converted to 71 (in 16% yield from 17) by the combined action of seven enzymes in the presence of S-adenosylmethionine (SAM) and NADPH.
To date, only a handful of highly modified natural product TDP-sugars have been synthesized via tandem reactions using purified biosynthetic enzymes. These include TDP-l-mycarose (71) of the tylosin pathway,[92] TDP-l-epivancosamine (56) of the chloroeremomycin pathway,[82] TDP-d-forosamine (100) of the spinosyn pathway,[114] and TDP-l-digitoxose (78) of the kijanimicin pathway.[99] To avoid complications, a two-stage one-pot approach was developed for the synthesis of TDP-l-mycarose (71) from thymidine and glucose-1-phosphate (17, Scheme 9c). The initial reaction mixture contained thymidine, PEP, ATP and four enzymes, thymidine kinase (TK), thymidylate kinase (TMK), nucleoside diphosphate kinase (NDK), and pyruvate kinase (PK). After incubation and subsequent removal of the enzymes by ultrafiltration, the filtrate was then incubated with glucose-1-phosphate, RfbA and RfbB (a thymidylyltransferase and a TDP-glucose-4,6-dehydratase from Salmonella typhi, respectively), and the mycarose biosynthetic enzymes (TylX3, TylC1, TylC3, TylK, and TylC2, Scheme 3c), together with NADPH and SAM. The yield of 71 was 16%. Interestingly, there are no apparent incompatibilities within the reaction conditions for the enzymes used in this multienzyme-synthesis, and there is no obvious cross-inhibition caused by substrates or products generated in the course of this one-pot synthetic scheme. The successful enzymatic preparation of various TDP-sugars sets the stage for exploring the glycosylation of secondary metabolites in vitro.
5.2.4. Protein Engineering of Glycosyltransferases
An elegant example of GT engineering was recently reported. UrdGT1b and UrdGT1c from the urdamycin pathway (discussed in Section 5.1.4.), share 91% amino acid sequence identity but have distinct substrate specificities. The domain of each enzyme that confers the UrdGT1b- or UrdGT1c-specific activity was localized to a region consisting of 31 amino acids near the N-terminus of both enzymes.[208] When this region in UrdGT1c was replaced with the corresponding region in UrdGT1b, the resulting chimeric enzyme exhibited UrdGT1b-like activity. An analogous result was observed when the region in UrdGT1b was replaced with the corresponding region in UrdGT1c. Of the 31 amino acids in this region, 18 are different between the two enzymes. Further studies indicated that only 10 of these 18 variable amino acids were critical for conferring either UrdGT1b- or UrdGT1c-like activities.[209] These residues were subsequently mutated and the resulting constructs were screened for GT activity. In addition to mutants that retained either UrdGT1b or UrdGT1c activity, and those that had both parental activities, mutants that catalyzed a new reaction were also found. In this new reaction, a d-olivose residue was transferred onto the d-olivose-l-rhodinose disaccharide moiety of 12b-derhodinosyl urdamycin G (253, Figure 13a) to produce a compound with a branched sugar chain (urdamycin P, 281). Interestingly, some of the mutants with the new activity also retained the normal UrdGT1b and/or UrdGT1c activity. Clearly, such protein engineering efforts have potential to generate new GTs with broader substrate specificities and the ability to catalyze new reaction(s).
Figure 13. Protein engineering of glycosyltransferases.
a) Novel activity of UrdGT1b/1c chimeras. The biosynthesis of the trisaccharide moiety of urdamycin A involves the tandem action of UrdGT2, UrdGT1c, and UrdGT1b (198 → 199 → 253 → 256). Several chimeric UrdGT1b/1c enzymes catalyzed a new reaction (253 → 281). b) High-throughput screening of GT activity. Random mutagenesis was used to create a library of OleD variants (a macrolide resistance GT that normally catalyzes 282 → 283). The activity of these variants was then screened in a high-throughput fashion using the fluorescent acceptor 284, whose fluorescence is quenched upon glycosyltransfer. Several active mutants were identified in this manner, some of which had broadened substrate specificity (see text for details).
The high sequence identity between the two urdamycin GTs and the rational selection of amino acid residues for mutation reduced the size of the mutant GT library in the previous example. However, more typcial engineering experiments based on directed evolution and/or random mutagenesis would generate far more mutants. Thus, the development of high-throughput assays for screening enzyme activities is a critical component of protein engineering efforts.[210, 302–304] Recently, the directed evolution of CstII, a sialyltransferase of the GT-A family,[303] and OleD, a macrolide resistance glucosyltrasferase of the GT-B family (that catalyzes 282 → 283, Figure 13b),[210] have been reported. A library of over 1000 OleD variants was constructed using error prone PCR and the GT activities of the variants were screened using a fluorescent aglycone substrate (284, Figure 13b) whose fluorescence is quenched upon glycosylation (284→ 285). Three single-site OleD mutants (Pro67Thr, Ser132Phe, Ala242Val) exhibited enhanced activity for 284 relative to that observed for the wild-type OleD. The corresponding triple mutant was then constructed and its substrate specificity was examined using a library of 22 NDP-sugars with 284 as the acceptor. The triple mutant processed 15 of the 22 sugars, whereas the wild-type OleD used only 3 of the 22 sugars. In addition, the triple mutant exhibited enhanced GT activity for 6 other unnatural acceptors. Interestingly, the Pro67 residue of OleD resides in a hypervariable loop region in the acceptor-binding domain near the N-terminus of the protein. Mutation at the equivalent position in the UrdGTs also altered substrate specificity. Finally, in a very recent study, a high-throughput GT assay was developed wherein the proton released from the acceptor nucleophile upon glycosyltransfer is detected by a pH indicator in weakly buffered solutions.[304] This assay is attractive because it can theoretically be applied to any GT (or GT variant) while using native GT substrates instead of chromophoric substrate analogues. Although the engineering of natural product GTs is still in its infancy, the studies with the urdamycin GTs and OleD clearly demonstrate the significant potential of this approach to generate enzymes with enhanced catalytic efficiency, broadened substrate promiscuity, and/or novel activities.
5.2.5. In vitro Natural Product Glycoengineering
With the rapid expansion of the NDP-sugar pools and the availability of promiscuous GTs, it is now possible to derivatize structurally diverse natural product aglycones with a variety of sugar moieties in order to make different glycoforms. This approach has emerged as an exciting method that provides ready access to new glycosylated natural products. This strategy, termed in vitro glycodiversification or glycorandomization,[248, 249, 286, 305] has recently been used to produce a large number of methymycin/pikromycin (187, 219–221, Figure 8) derivatives[232] and vancomycin (192, Figure 14a) derivatives.[238] The vancomycin aglycone (286) is sequentially glycosylated at the 4-hydroxyphenylglycine residue (286 → 287 → 192) by the glucosyltransferase, GtfE, and the vancosaminyltransferase, GtfD (Figure 14a).[306] GtfD and GtfE had previously been shown to have a relaxed substrate specificity.[255, 306, 307] To exploit these properties, a library of TDP-sugars (prepared using chemical synthesis and the GalK and RmlA mutants described in Sections 5.2.1. and 5.2.2.) was incubated with the vancomycin aglycone and GtfE, and the reaction contents were analyzed by LC-MS (Figure 14b).[238] Twenty-one of the 23 TDP-sugars analyzed in this study were found to be substrates for GtfE, including the TDP-azidosugar, 289, which could be further modified in the presence of alkynes via the Huisgen cycloaddition reaction to generate 39 additional vancomycin derivatives.[238, 308] One of the new compounds displayed improved antibiotic activity against Staphylococcus aureus and Enterococcus faecium. Following these initial studies on vancomycin glycorandomiztion, several other natural product GTs with relaxed substrate specificity were used for similar in vitro glycodiversification studies.[87, 232, 237, 309–316]
Figure 14. Chemoenzymatic glycorandomization of the vancomycin aglycone.
a) The disaccharide moiety of vancomycin (192) is constructed by the tandem addition of d-glucose and l-vancosamine residues to 286 by the glycosyltransferases GtfE and GtfD, respectively. b) A library of reducing sugars was converted to a library of sugar-1-phosphates by either chemical synthesis or by incubation with engineered GalK mutants and ATP. This library was, in turn, converted into a library of NDP-sugars using engineered RmlA nucleotidylyltransferase. These NDP-sugars were then screened as substrates for GtfE in vitro, leading to a glycorandomized library of 21 vancomycin analogues. One of these analogues contained an azido sugar (see 289) that could be further modified by a variety of acetylene compounds using the CuI-catalyzed Huisgen [3+2] cycloaddition to yield 39 additional vancomycin derivatives.
The versatility of in vitro glycodiversification was recently expanded during a calicheamicin (189, Figure 4) glycorandomization study.[310] It was first demonstrated that the calicheamicin GT, CalG1, accepts ten different TDP-sugars as substrates. One of the sugars, TDP-3-deoxy-α-d-glucose (294, Figure 15a), was then incubated with CalG1 and the 3-O-methylrhamnosylated aglycone (290). Since the glycosylation site for CalG1 was already occupied in 290, no reaction was expected. However, a new product (292) carrying a 3-deoxy-α-d-glucose moiety was identified. Interestingly, analysis of the control reactions revealed that CalG1 had catalyzed a reverse GT reaction in the presence of TDP to generate TDP-3-O-methyl-β-l-rhamnose (293) and a de-glycosylated aglycone (291). Glycosylation of 291 by CalG1 could then proceed using the alternative TDP-sugar (294) present in the reaction mixture to give 292. The calicheamicin aminopentosyl transferase (CalG4) and the vancomycin GTs (GtfD and GtfE) were also shown to catalyze reversible reactions in this study, suggesting that reaction reversibility may be a general property of GTs in vitro.
Figure 15. Exploiting GT reversibility in vitro.
a) The calicheamicin GT, CalG1, was shown to synthesize both 293 and 291 from 290 in the presence of TDP through a reverse glycosyltransfer reaction. Alternative TDP-sugars (such as 294) present in the reaction mixture could also be coupled to the aglycone (291) generated in situ to yield a new glycoside, 292. b) CalG1 was used in sugar exchange reactions to rapidly glycorandomize 8 different glycosylated calicheamicin analogues with 10 different sugars to generate a library of 72 compounds. c) One-enzyme aglycone exchange reactions involve a single GT, which transfers a sugar moiety from one aglycone to a structurally similar aglycone. d) Using two enzymes that both recognize the same TDP-sugar, a sugar moiety can be moved from one aglycone to a structurally unrelated aglycone. e) In the presence of excess TDP and a glycosylated natural product, reverse GT catalysis can be used to synthesize TDP-sugars.
The reversible reactions catalyzed by these GTs were then exploited in a number of glycorandomization applications (Figure 15b–e). Using a set of eight calicheamicin derivatives and the ten established CalG1 TDP-sugar substrates, CalG1 catalyzed several "sugar exchange" reactions, yielding a glycorandomized calicheamicin library of over 70 compounds (Figure 15b). CalG1, CalG4, and GtfD were also individually used in one-enzyme "aglycone exchange" reactions, where the 3-O-methyl-β-l-rhamnosyl, aminopentosyl, or vancosaminyl moieties were transferred by CalG1, CalG4, or GtfD, respectively, from one calicheamycin or vancomycin aglycone to another (Figure 15c). Later, a one-pot two-enzyme aglycone exchange reaction was developed, wherein GtfE was used to excise an unnatural azido sugar moiety from a vancomycin aglycone in order to generate a TDP-azidosugar intermediate which was then coupled by CalG1 to a calicheamicin aglycone (Figure 15d). GT reversibility can also be exploited to synthesize NDP-sugars (Figure 15e) and to verify the biological functions of GTs.[87, 252, 317]
6. Summary and Outlook
In this review, we have highlighted our current understanding of unusual sugar biosynthesis. While the final structures of these sugars vary considerably, only a handful enzyme activities are used for their biosynthesis. This process of "natural combinatorial biosynthesis" can account for much of the sugar structural diversity observed in nature. Investigations into the sequence of biosynthetic events and the mechanisms of the pathway enzymes have revealed that most of these enzymes rely on their unique arrangements of catalytic residues, coenzymes, and cofactor requirements in order to carry out specific chemical transformations on chemically similar NDP-ketosugar intermediates. The fact that the family of sugar-modifying SDR enzymes has evolved not only to generate NDP-ketosugar intermediates, but also to manipulate this intermediate in one active site reflects the elegance of Nature's strategy for creating structural diversity. The variation in sugar structure imparted by "natural combinatorial biosynthesis" and SDR enzymes can be further augmented by unusual enzyme activities, many of which remain to be characterized.
One observation that has emerged during studies of natural product glycosylation is that many sugar biosynthetic enzymes and glycosyltransferases exhibit some degree of substrate flexibility towards their NDP-sugar and/or aglycone substrates. The metabolic pathway engineering and combinatorial biosynthesis studies carried out over the past decade have illustrated the potential utility of these enzymes for the generation of new glycoforms. Armed with our understanding of biosynthetic logic and a solid foundation of mechanistic and structural work, we are entering a new era of natural product glycodiversification, where these substrate-flexible enzymes can be further manipulated through protein engineering and used to generate libraries of substrates for in vitro glycosylation reactions, or to rapidy glycorandomize a given natural product scaffold. Such efforts have the potential to produce new compounds that could mitigate the ubiquitous and daunting threat to human health imposed by drug-resistant pathogens.
Supplementary Material
Biographies
Christopher J. Thibodeaux was born and raised in Louisiana (USA), where he earned Bachelor's degrees in both Biochemistry and Plant Biology from the Louisiana State University. He has since joined the research group of Prof. Hung-wen Liu at the University of Texas, Austin, where he is working towards his Ph.D. in Cellular and Molecular Biology. His primary research interests include studying the kinetics and mechanisms of unusual enzyme-catalyzed reactions. In his spare time, he enjoys the outdoors and spending time with his family.
Charles E. (Chad) Melançon grew up in New Orleans, LA (USA), where he earned Bachelor's degrees in both Chemistry and Biology from The University of New Orleans in 2001. Chad did his doctoral work in Biochemistry in Professor Liu's lab at the University of Texas at Austin, where he focused on investigating and engineering macrolide antibiotic sugar biosynthesis and glycosylation pathways of Actinomycetes. In June of 2007, Chad began an NIH postdoctoral fellowship in Dr. Peter Schultz' lab at the Scripps Research Institute in La Jolla, CA, where he works on application of diversity-based strategies to creating engineered organisms with expanded genetic codes.
Hung-wen (Ben) Liu was born in Taipei, Taiwan (ROC). Following his undergraduate, graduate, and postdoctoral studies, he joined the faculty of Chemistry at the University of Minnesota in 1984. In 2000, he moved to the University of Texas, Austin, where he now holds the George H. Hitchings Regents Chair in Drug Design and is a Professor of Medicinal Chemistry, Chemistry, and Biochemistry. His research lies at the crossroads of organic and biological chemistry, with particular emphasis on enzymatic reaction mechanisms, natural product biosynthesis, protein function regulation, inhibitor design and synthesis, and metabolic pathway engineering. He enjoys reading, music, and travelling.
Footnotes
The authors gratefully acknowledge financial support provided by the National Institutes of Health (GM35906 and GM54346).
References
- 1.Martin-Rendon E, Blake DJ. Trends Pharmacol. Sci. 2003;24:178. doi: 10.1016/S0165-6147(03)00050-6. [DOI] [PubMed] [Google Scholar]
- 2.Durand G, Seta N. Clin. Chem. 2000;46:795. [PubMed] [Google Scholar]
- 3.Este JA. Curr. Med. Chem. 2003;10:1617. doi: 10.2174/0929867033457098. [DOI] [PubMed] [Google Scholar]
- 4.Schnaitman CA, Klena JD. Microbiol. Rev. 1993;57:655. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorson JS, Hosted TJ, Jr, Jiang J, Biggins JB, Ahlert J. Curr. Org. Chem. 2001;5:139. [Google Scholar]
- 6.Kren V, Martinkova L. Curr. Med. Chem. 2001;8:1303. doi: 10.2174/0929867013372193. [DOI] [PubMed] [Google Scholar]
- 7.Weymouth-Wilson AC. Nat. Prod. Rep. 1997;14:99. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 8.Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1999. [PubMed] [Google Scholar]
- 9.Blanchard S, Thorson JS. Curr. Opin. Chem. Biol. 2006;10:263. doi: 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Shackleford GS, Regni CA, Beamer LJ. Protein Sci. 2004;13:2130. doi: 10.1110/ps.04801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jelakovic S, Schulz GE. J. Mol. Biol. 2001;312:143. doi: 10.1006/jmbi.2001.4948. [DOI] [PubMed] [Google Scholar]
- 12.Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson J-R, Logan SM. J. Biol. Chem. 2006;281:723. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- 13.Tanner ME. Bioorg. Chem. 2002;33:216. doi: 10.1016/j.bioorg.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Maki M, Jarvinen N, Rabina J, Maaheimo H, Mattila P, Renkonen R. Glycobiology. 2003;13:295. doi: 10.1093/glycob/cwg035. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki N, Nakano Y, Yoshida Y, Nezu T, Terada Y, Yamashita Y, Koga T. Eur. J. Biochem. 2002;269:5963. doi: 10.1046/j.1432-1033.2002.03331.x. [DOI] [PubMed] [Google Scholar]
- 16.Xu Z, Jakobi K, Welzel K, Hertweck C. Chem. Biol. 2005;12:579. doi: 10.1016/j.chembiol.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida Y, Nakano Y, Nezu T, Yamashita Y, Koga T. J. Biol. Chem. 1999;274:16933. doi: 10.1074/jbc.274.24.16933. [DOI] [PubMed] [Google Scholar]
- 18.Fischer C, Lipata F, Rohr J. J. Am. Chem. Soc. 2003;125:7818. doi: 10.1021/ja034781q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong C, Major LL, Srikannathasan V, Errey JC, Giraud MF, Lam JS, Graninger M, Messner P, McNeil MR, Field RA, Whitfield C, Naismith JH. J. Mol. Biol. 2007;365:146. doi: 10.1016/j.jmb.2006.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christendat D, Saridakis V, Dharamsi A, Bochkarev A, Pai EF, Arrowsmith CH, Edwards AM. J. Biol. Chem. 2000;275:24608. doi: 10.1074/jbc.C000238200. [DOI] [PubMed] [Google Scholar]
- 21.Graninger M, Nidetzky B, Heinrichs DE, Whitfield C, Messner P. J. Biol. Chem. 1999;274:25069. doi: 10.1074/jbc.274.35.25069. [DOI] [PubMed] [Google Scholar]
- 22.Torkkell S, Kunnari T, Palmu K, Mantsala P, Hakala J, Ylihonko K. Mol. Genet. Genomics. 2001;266:276. doi: 10.1007/s004380100554. [DOI] [PubMed] [Google Scholar]
- 23.Kniep B, Grisebach H. Eur. J. Biochem. 1980;105:139. doi: 10.1111/j.1432-1033.1980.tb04483.x. [DOI] [PubMed] [Google Scholar]
- 24.Wahl HP, Matern U, Grisebach H. Biochem. Biophys. Res. Commun. 1975;64:1041. doi: 10.1016/0006-291x(75)90152-7. [DOI] [PubMed] [Google Scholar]
- 25.Wahl HP, Grisebach H. Biochim. Biophys. Acta. 1979;568:243. doi: 10.1016/0005-2744(79)90291-2. [DOI] [PubMed] [Google Scholar]
- 26.Pissowotzki K, Mansouri K, Piepersberg W. Mol. Gen. Genet. 1991;231:113. doi: 10.1007/BF00293829. [DOI] [PubMed] [Google Scholar]
- 27.Yamase H, Zhao L, Liu H-w. J. Am. Chem. Soc. 2000;122:12397. [Google Scholar]
- 28.Baron D, Wellman E, Grisebach H. Biochim. Biophys. Acta. 1972;258:310. doi: 10.1016/0005-2744(72)90988-6. [DOI] [PubMed] [Google Scholar]
- 29.Mendecino J, Abou-Issa H. Biochim. Biophys. Acta. 1974;364:159. doi: 10.1016/0005-2744(74)90143-0. [DOI] [PubMed] [Google Scholar]
- 30.Molhoj M, Verma R, Reiter W-D. Plant J. 2003;35:693. doi: 10.1046/j.1365-313x.2003.01841.x. [DOI] [PubMed] [Google Scholar]
- 31.Waldron C, Matsushima P, Rosteck PR, Jr, Broughton MC, Turner J, Madduri K, Crawford KP, Merlo DJ, Baltz RH. Chem. Biol. 2001;8:487. doi: 10.1016/s1074-5521(01)00029-1. [DOI] [PubMed] [Google Scholar]
- 32.Patallo EP, Blanco G, Fischer C, Brana AF, Rohr J, Mendez C, Salas JA. J. Biol. Chem. 2001;276:18765. doi: 10.1074/jbc.M101225200. [DOI] [PubMed] [Google Scholar]
- 33.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. Science. 2002;297:1173. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 34.Madduri K, Waldron C, Merlo DJ. J. Bacteriol. 2001;183:5632. doi: 10.1128/JB.183.19.5632-5638.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W, Christenson SD, Standage S, Shen B. Science. 2002;297:1170. doi: 10.1126/science.1072110. [DOI] [PubMed] [Google Scholar]
- 36.Steffensky M, Muhlenweg A, Wang ZX, Li SM, Heide L. Antimicrob. Agents Chemother. 2000;44:1214. doi: 10.1128/aac.44.5.1214-1222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pojer F, Li SM, Heide L. Microbiology (Reading, U.K.) 2002;148:3901. doi: 10.1099/00221287-148-12-3901. [DOI] [PubMed] [Google Scholar]
- 38.Wang ZX, Li SM, Heide L. Antimicrob. Agents Chemother. 2000;44:3040. doi: 10.1128/aac.44.11.3040-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thuy TT, Lee HC, Kim CG, Heide L, Sohng JK. Arch. Biochem. Biophys. 2005;436:161. doi: 10.1016/j.abb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Freitag A, Li SM, Heide L. Microbiology (Reading, U.K.) 2006;152:2433. doi: 10.1099/mic.0.28931-0. [DOI] [PubMed] [Google Scholar]
- 41.Tello M, Jakimowicz P, Errey JC, Freel Meyers CL, Walsh CT, Buttner MJ, Lawson DM, Field RA. Chem. Commun. (Cambridge, U.K.) 2006:1079. doi: 10.1039/b515763c. [DOI] [PubMed] [Google Scholar]
- 42.Freel Meyers CL, Oberthur M, Heide L, Kahne D, Walsh CT. Biochemistry. 2004;43:15022. doi: 10.1021/bi048457z. [DOI] [PubMed] [Google Scholar]
- 43.Freitag A, Wemakor E, Li SM, Heide L. Chembiochem. 2005;6:2316. doi: 10.1002/cbic.200500252. [DOI] [PubMed] [Google Scholar]
- 44.Freel Meyers CL, Oberthur M, Xu H, Heide L, Kahne D, Walsh CT. Angew. Chem. Int. Ed. 2004;43:67. doi: 10.1002/anie.200352626. Angew. Chem. 2003, 116, 69. [DOI] [PubMed] [Google Scholar]
- 45.Garneau-Tsodikova S, Stapon A, Kahne D, Walsh CT. Biochemistry. 2006;45:8568. doi: 10.1021/bi060784e. [DOI] [PubMed] [Google Scholar]
- 46.Jaishy BP, Lim SK, Yoo ID, Yoo JC, Sohng JK, Nam DH. J. Microbiol. Biotechnol. 2006;16:764. [Google Scholar]
- 47.Thuy TTT, Liou K, Oh T-J, Kim DH, Nam DH, Yoo JC, Sohng JK. Glycobiology. 2007;17:119. doi: 10.1093/glycob/cwl060. [DOI] [PubMed] [Google Scholar]
- 48.Cundliffe E, Bate N, Butler A, Fish S, Gandecha A, Merson-Davies L. Antonie Van Leeuwenhoek. 2001;79:229. doi: 10.1023/a:1012065300116. [DOI] [PubMed] [Google Scholar]
- 49.Anzai Y, Saito N, Tanaka M, Kinoshita K, Koyama Y, Kato F. FEMS Microbiol. Lett. 2003;218:135. doi: 10.1111/j.1574-6968.2003.tb11509.x. [DOI] [PubMed] [Google Scholar]
- 50.Ward SL, Hu Z, Schirmer A, Reid R, Revill WP, Reeves CD, Petrakovsky OV, Dong SD, Katz L. Antimicrob. Agents. Chemother. 2004;48:4703. doi: 10.1128/AAC.48.12.4703-4712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baltz RH, Seno ET. Annu. Rev. Microbiol. 1988;42:547. doi: 10.1146/annurev.mi.42.100188.002555. [DOI] [PubMed] [Google Scholar]
- 52.Chen H, Guo Z, Liu H-w. J. Am. Chem. Soc. 1998;120:9951. [Google Scholar]
- 53.Chen H, Yeung S-M, Que NLS, Muller T, Schmidt RR, Liu H-w. J. Am. Chem. Soc. 1999;121:7166. [Google Scholar]
- 54.Melancon CE, 3rd, Hong L, White JA, Liu Y-n, Liu H-w. Biochemistry. 2007;46:577. doi: 10.1021/bi061907y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melancon CE, 3rd, Yu W-L, Liu H-w. J. Am. Chem. Soc. 2005;127:12240. doi: 10.1021/ja053835o. [DOI] [PubMed] [Google Scholar]
- 56.Mochizuki S, Hiratsu K, Suwa M, Ishii T, Sugino F, Yamada K, Kinashi H. Mol. Microbiol. 2003;48:1501. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- 57.Summers RG, Donadio S, Staver MJ, Wendt-Pienkowski E, Hutchinson CR, Katz L. Microbiology (Reading, U.K.) 1997;143:3251. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 58.Quiros LM, Aguirrezabalaga I, Olano C, Mendez C, Salas JA. Mol. Microbiol. 1998;28:1177. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- 59.Olano C, Rodriguez AM, Michel JM, Mendez C, Raynal MC, Salas JA. Mol. Gen. Genet. 1998;259:299. doi: 10.1007/s004380050816. [DOI] [PubMed] [Google Scholar]
- 60.Aguirrezabalaga I, Olano C, Allende N, Rodriguez L, Brana AF, Mendez C, Salas JA. Antimicrob. Agents Chemother. 2000;44:1266. doi: 10.1128/aac.44.5.1266-1275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xue Y, Zhao L, Liu H-w, Sherman DH. Proc. Natl. Acad. Sci. USA. 1998;95:12111. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peiru S, Menzella HG, Rodriguez E, Carney J, Gramajo H. Appl. Environ. Microbiol. 2005;71:2539. doi: 10.1128/AEM.71.5.2539-2547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Volchegursky Y, Hu Z, Katz L, McDaniel R. Mol. Microbiol. 2000;37:752. doi: 10.1046/j.1365-2958.2000.02059.x. [DOI] [PubMed] [Google Scholar]
- 64.Gaisser S, Bohm GA, Cortes J, Leadlay PF. Mol. Gen. Genet. 1997;256:239. doi: 10.1007/s004380050566. [DOI] [PubMed] [Google Scholar]
- 65.Salah-Bey K, Doumith M, Michel JM, Haydock S, Cortes J, Leadlay PF, Raynal MC. Mol. Gen. Genet. 1998;257:542. doi: 10.1007/s004380050680. [DOI] [PubMed] [Google Scholar]
- 66.Gaisser S, Bohm GA, Doumith M, Raynal MC, Dhillon N, Cortes J, Leadlay PF. Mol. Gen. Genet. 1998;258:78. doi: 10.1007/s004380050709. [DOI] [PubMed] [Google Scholar]
- 67.Zhao L, Sherman DH, Liu H-w. J. Am. Chem. Soc. 1998;120:10256. [Google Scholar]
- 68.Zhao L, Que NLS, Xue Y, Sherman DH, Liu H-w. J. Am. Chem. Soc. 1998;120:12159. [Google Scholar]
- 69.Chang C, Zhao L, Yamase H, Liu H-w. Angew. Chem. Int. Ed. 2000;39:2160. doi: 10.1002/1521-3773(20000616)39:12<2160::aid-anie2160>3.0.co;2-e. Angew. Chem. 2000, 112, 2244. [DOI] [PubMed] [Google Scholar]
- 70.Szu P-H, He X, Zhao L, Liu H-w. Angew. Chem. Int. Ed. 2005;44:6742. doi: 10.1002/anie.200501998. Angew. Chem. 2005, 117, 6900. [DOI] [PubMed] [Google Scholar]
- 71.Borisova SA, Zhao L, Sherman DH, Liu HW. Org Lett. 1999;1:133. doi: 10.1021/ol9906007. [DOI] [PubMed] [Google Scholar]
- 72.Zhao L, Borisova S, Yeung SM, Liu H-w. J. Am. Chem. Soc. 2001;123:7909. doi: 10.1021/ja010587x. [DOI] [PubMed] [Google Scholar]
- 73.Hyun CG, Kim SS, Sohng JK, Hahn JJ, Kim JW, Suh JW. FEMS Microbiol. Lett. 2000;183:183. doi: 10.1111/j.1574-6968.2000.tb08955.x. [DOI] [PubMed] [Google Scholar]
- 74.Salas AP, Zhu L, Sanchez C, Brana AF, Rohr J, Mendez C, Salas JA. Mol. Microbiol. 2005;58:17. doi: 10.1111/j.1365-2958.2005.04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Onaka H, Taniguchi S, Igarashi Y, Fumari T. J. Antibiot. 2002;55:1063. doi: 10.7164/antibiotics.55.1063. [DOI] [PubMed] [Google Scholar]
- 76.Olano C, Lomovskaya N, Fonstein L, Roll JT, Hutchinson CR. Chem. Biol. 1999;6:845. doi: 10.1016/s1074-5521(00)80004-6. [DOI] [PubMed] [Google Scholar]
- 77.Raty K, Kunnari T, Hakala J, Mantsala P, Ylihonko K. Mol. Gen. Genet. 2000;264:164. doi: 10.1007/s004380000306. [DOI] [PubMed] [Google Scholar]
- 78.Pelzer S, Sussmuth R, Heckmann D, Recktenwald J, Huber P, Jung G, Wohlleben W. Antimicrob. Agents Chemother. 1999;43:1565. doi: 10.1128/aac.43.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Wageningen AMA, Kirkpatrick PN, Williams DH, Harris BR, Kershaw JK, Lennard NJ, Jones M, Jones SJM, Solenberg PJ. Chem. Biol. 1998;5:155. doi: 10.1016/s1074-5521(98)90060-6. [DOI] [PubMed] [Google Scholar]
- 80.Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS. Chem. Biol. 2004;11:959. doi: 10.1016/j.chembiol.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 81.Ichinose K, Ozawa M, Itou K, Kunieda K, Ebizuka Y. Microbiology (Reading, U.K.) 2003;149:1633. doi: 10.1099/mic.0.26310-0. [DOI] [PubMed] [Google Scholar]
- 82.Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Proc. Natl. Acad. Sci. USA. 2000;97:11942. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. Mol. Genet. Gen. 2005;274:40. doi: 10.1007/s00438-005-1156-3. [DOI] [PubMed] [Google Scholar]
- 84.Chen H, Agnihotri G, Guo Z, Que NLS, Chen X, Liu H-w. J. Am. Chem. Soc. 1999;121:8124. [Google Scholar]
- 85.Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S. Proc. Natl. Acad. Sci. USA. 1999;96:9509. doi: 10.1073/pnas.96.17.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wohlert S-E, Lomovskaya N, Kulowski K, Fonstein L, Occi JL, Gewain KM, MacNeil DJ, Hutchinson CR. Chem. Biol. 2001;8:681. doi: 10.1016/s1074-5521(01)00043-6. [DOI] [PubMed] [Google Scholar]
- 87.Zhang C, Albermann C, Fu X, Thorson JS. J. Am. Chem. Soc. 2006;128:16420. doi: 10.1021/ja065950k. [DOI] [PubMed] [Google Scholar]
- 88.Ogasawara Y, Katayama K, Minami A, Otsuka M, Eguchi T, Kakinuma K. Chem. Biol. 2004;11:79. doi: 10.1016/j.chembiol.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 89.Bihlmaier C, Welle E, Hofmann C, Welzel K, Vente A, Breitling E, Muller M, Glaser S, Bechthold A. Antimicrob. Agents Chemother. 2006;50:2113. doi: 10.1128/AAC.00007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen H, Zhao Z, Hallis TM, Guo Z, Liu H-w. Angew. Chem. Int. Ed. 2001;40:607. Angew. Chem. 2001, 113, 627. [PubMed] [Google Scholar]
- 91.Takahashi H, Liu Y-n, Chen H, Liu H-w. J. Am. Chem. Soc. 2005;127:9340. doi: 10.1021/ja051409x. [DOI] [PubMed] [Google Scholar]
- 92.Takahashi H, Liu Y-n, Liu H-w. J. Am. Chem. Soc. 2006;128:1432. doi: 10.1021/ja0562144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ichinose K, Bedford DJ, Tornus D, Bechthold A, Bibb MJ, Revill WP, Floss HG, Hopwood DA. Chem. Biol. 1998;5:647. doi: 10.1016/s1074-5521(98)90292-7. [DOI] [PubMed] [Google Scholar]
- 94.Gonzalez A, Remsing LL, Lombo F, Fernandez MJ, Prado L, Brana AF, Kunzel E, Rohr J, Mendez C, Salas JA. Mol. Gen. Genet. 2001;264:827. doi: 10.1007/s004380000372. [DOI] [PubMed] [Google Scholar]
- 95.Remsing LL, Garcia-Bernardo J, Gonzalez A, Kunzel E, Rix U, Brana AF, Bearden DW, Mendez C, Salas JA, Rohr J. J. Am. Chem. Soc. 2002;124:1606. doi: 10.1021/ja0105156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Niemi J, Mantsala P. J. Bacteriol. 1995;177:2942. doi: 10.1128/jb.177.10.2942-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodriguez L, Aguirrezabalaga I, Allende N, Brana AF, Mendez C, Salas JA. Chem. Biol. 2002;9:721. doi: 10.1016/s1074-5521(02)00154-0. [DOI] [PubMed] [Google Scholar]
- 98.Rodriguez L, Rodriguez D, Olano C, Brana AF, Mendez C, Salas JA. J. Bacteriol. 2001;183:5358. doi: 10.1128/JB.183.18.5358-5363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang H, White-Phillip JA, Melancon CE, 3rd, Kwon H-j, Yu W-l, Liu H-w. J. Am. Chem. Soc. 2007;129:14670. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, White RL, Vining LC. Microbiology (Reading, U.K.) 2002;148:1091. doi: 10.1099/00221287-148-4-1091. [DOI] [PubMed] [Google Scholar]
- 101.Westrich L, Domann S, Faust B, Bedford D, Hopwood DA, Bechthold A. FEMS Microbiol. Lett. 1999;170:381. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 102.Hoffmeister D, Ichinose K, Domann S, Faust B, Trefzer A, Drager G, Kirschning A, Fischer C, Kunzel E, Bearden D, Rohr J, Bechthold A. Chem. Biol. 2000;7:821. doi: 10.1016/s1074-5521(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 103.Lombo F, Menendez N, Salas JA, Mendez C. Appl. Microbiol. Biotechnol. 2006;73:1. doi: 10.1007/s00253-006-0511-6. [DOI] [PubMed] [Google Scholar]
- 104.Menendez N, Nur-e-Alam M, Brana AF, Rohr J, Salas JA, Mendez C. Chem. Biol. 2004;11:21. doi: 10.1016/j.chembiol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 105.Jia XY, Tian ZH, Shao L, Qu XD, Zhao QF, Tang J, Tang GL, Liu W. Chem. Biol. 2006;13:575. doi: 10.1016/j.chembiol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 106.Weitnauer G, Muhlenweg A, Trefzer A, Hoffmeister D, Sussmuth RD, Jung G, Welzel K, Vente A, Girreser U, Bechthold A. Chem. Biol. 2001;8:569. doi: 10.1016/s1074-5521(01)00040-0. [DOI] [PubMed] [Google Scholar]
- 107.Haydock SF, Appleyard AN, Mironenko T, Lester J, Scott N, Leadlay PF. Microbiology (Reading, U.K.) 2005;151:3161. doi: 10.1099/mic.0.28194-0. [DOI] [PubMed] [Google Scholar]
- 108.Thorson JS, Lo SF, Liu H-w. J. Am. Chem. Soc. 1993;115:5827. [Google Scholar]
- 109.Durr C, Schnell HJ, Luzhetskyy A, Murillo R, Weber M, Welzel K, Vente A, Bechthold A. Chem. Biol. 2006;13:365. doi: 10.1016/j.chembiol.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 110.Sun Y, Zhou X, Dong H, Tu G, Wang M, Wang B, Deng Z. Chem. Biol. 2003;10:431. doi: 10.1016/s1074-5521(03)00092-9. [DOI] [PubMed] [Google Scholar]
- 111.Raty K, Hautala A, Torkkell S, Kantola J, Mantsala P, Hakala J, Ylihonko K. Microbiology (Reading, U.K.) 2002;148:3375. doi: 10.1099/00221287-148-11-3375. [DOI] [PubMed] [Google Scholar]
- 112.Hong L, Zhao Z, Liu H-w. J. Am. Chem. Soc. 2006;128:14262. doi: 10.1021/ja0649670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao Z, Hong L, Liu H-w. J. Am. Chem. Soc. 2005;127:7692. doi: 10.1021/ja042702k. [DOI] [PubMed] [Google Scholar]
- 114.Hong L, Zhao Z, Melancon CE, 3rd, Zhang H, Liu H-w. J. Am. Chem. Soc. 2008;130 doi: 10.1021/ja0771383. in presss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Campbell RE, Mosimann SC, van de Rijn I, Tanner ME, Strynadka NCJ. Biochemistry. 2000;39:7012. [PubMed] [Google Scholar]
- 116.Bililign T, Shepard EM, Ahlert J, Thorson JS. Chembiochem. 2002;3:1143. doi: 10.1002/1439-7633(20021104)3:11<1143::AID-CBIC1143>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 117.Gao Q, Zhang C, Blanchard S, Thorson JS. Chem. Biol. 2006;13:733. doi: 10.1016/j.chembiol.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 118.Hofmann C, Boll R, Heitmann B, Hauser G, Durr C, Frerich A, Weitnauer G, Glaser SJ, Bechthold A. Chem. Biol. 2005;12:1137. doi: 10.1016/j.chembiol.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 119.Jolly L, Ferrari P, Blanot D, Van Heijenoort J, Fassy F, Mengin-Lecreulx D. Eur. J. Biochem. 1999;262:202. doi: 10.1046/j.1432-1327.1999.00373.x. [DOI] [PubMed] [Google Scholar]
- 120.Pompeo F, Bourne Y, van Heijenoort J, Fassy F, Mengin-Lecreulx D. J. Biol. Chem. 2001;276:3833. doi: 10.1074/jbc.M004788200. [DOI] [PubMed] [Google Scholar]
- 121.Kudo F, Kawabe K, Kuriki H, Eguchi T, Kakinuma K. J. Am. Chem. Soc. 2005;127:1711. doi: 10.1021/ja044921b. [DOI] [PubMed] [Google Scholar]
- 122.Flatt PM, Mahmud T. Nat. Prod. Rep. 2007;24:358. doi: 10.1039/b603816f. [DOI] [PubMed] [Google Scholar]
- 123.Ikeno S, Aoki D, Hamada M, Hori M, Tsuchiya KS. J. Antibiot. 2006;59:18. doi: 10.1038/ja.2006.4. [DOI] [PubMed] [Google Scholar]
- 124.Aparicio JF, Caffrey P, Gil JA, Zotchev SB. Appl. Microbiol. Biotechnol. 2003;61:179. doi: 10.1007/s00253-002-1183-5. [DOI] [PubMed] [Google Scholar]
- 125.Palaniappan N, Ayers S, Gupta S, Habib ES, Reynolds KA. Chem. Biol. 2006;13:753. doi: 10.1016/j.chembiol.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 126.Du L, Sanchez C, Chen M, Edwards DJ, Shen B. Chem. Biol. 2000;7:623. doi: 10.1016/s1074-5521(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 127.Webb NA, Mulichak AM, Lam JS, Rocchetta HL, Garavito RM. Protein Sci. 2004;13:529. doi: 10.1110/ps.03393904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Major LL, Wolucka BA, Naismith JH. J. Am. Chem. Soc. 2005;127:18309. doi: 10.1021/ja056490i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hosted TJ, Wang TX, Alexander DC, Horan AC. J. Indust. Microbiol. Biotechnol. 2001;27:386. doi: 10.1038/sj.jim.7000189. [DOI] [PubMed] [Google Scholar]
- 130.Shackelford GS, Regni CA, Beamer LJ. Protein Sci. 2004;13:2130. doi: 10.1110/ps.04801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lindqvist L, Kaiser R, Reeves PR, Lindberg AA. J. Biol. Chem. 1994;269:122. [PubMed] [Google Scholar]
- 132.Thorson JS, Kelly TM, Liu H-w. J. Bacteriol. 1994;176:1840. doi: 10.1128/jb.176.7.1840-1849.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beyer S, Mayer G, Piepersberg W. Eur. J. Biochem. 1998;258:1059. doi: 10.1046/j.1432-1327.1998.2581059.x. [DOI] [PubMed] [Google Scholar]
- 134.Piepersberg W. In: Biotechnology of Antibiotics. Strohl WR, editor. New York: M. Dekker; 1997. p. 81. [Google Scholar]
- 135.Kumada Y, Horinouchi S, Uozumi T, Beppu T. Gene. 1986;42:221. doi: 10.1016/0378-1119(86)90300-8. [DOI] [PubMed] [Google Scholar]
- 136.He X, Agnihotri G, Liu H-w. Chem. Rev. 2000;100:4615. doi: 10.1021/cr9902998. [DOI] [PubMed] [Google Scholar]
- 137.He XM, Liu H-w. Annu. Rev. Biochem. 2002;71:701. doi: 10.1146/annurev.biochem.71.110601.135339. [DOI] [PubMed] [Google Scholar]
- 138.He X, Liu H-w. Curr. Opin. Chem. Biol. 2002;6:590. doi: 10.1016/s1367-5931(02)00367-8. [DOI] [PubMed] [Google Scholar]
- 139.Johnson DA, Liu H-w. In: Comprehensive natural product chemistry. Barton DHR, Meth-Cohn O, Nakanishi K, editors. Amsterdam: Elsevier; 1999. p. 311. [Google Scholar]
- 140.Davis ML, Thoden JB, Holden HM. J. Biol. Chem. 2007;282:19227. doi: 10.1074/jbc.M702529200. [DOI] [PubMed] [Google Scholar]
- 141.Burgie ES, Holden HM. Biochemistry. 2007;46:8999. doi: 10.1021/bi700751d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Burgie ES, Thoden JB, Holden HM. Protein Sci. 2007;16:887. doi: 10.1110/ps.062711007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM, Bhatia S, Wakarchuk WW, Young NM. J. Biol. Chem. 2006;281:8907. doi: 10.1074/jbc.M512987200. [DOI] [PubMed] [Google Scholar]
- 144.Draeger G, Park S-H, Floss HG. J. Am. Chem. Soc. 1999;121:2611. [Google Scholar]
- 145.Johnson DA, Gassner GT, Bandarian V, Ruzicka FJ, Ballou DP, Reed GH, Liu H-w. Biochemistry. 1996;35:15846. doi: 10.1021/bi961370w. [DOI] [PubMed] [Google Scholar]
- 146.Chen XMH, Ploux O, Liu H-w. Biochemistry. 1996;35:16412. doi: 10.1021/bi961921i. [DOI] [PubMed] [Google Scholar]
- 147.Chang C-wT, Johnson DA, Bandarian V, Zhou H, LoBrutto R, Reed GH, Liu H-w. J. Am. Chem. Soc. 2000;122:4239. [Google Scholar]
- 148.Agnihotri G, Liu Y-n, Paschal BM, Liu H-w. Biochemistry. 2004;43:14265. doi: 10.1021/bi048841w. [DOI] [PubMed] [Google Scholar]
- 149.Beyer N, Alam J, Hallis TM, Guo Z, Liu H-w. J. Am. Chem. Soc. 2003;125:5584. doi: 10.1021/ja030088r. [DOI] [PubMed] [Google Scholar]
- 150.Thibodeaux CJ, Melancon CE, 3rd, Liu H-w. Nature. 2007;446:1008. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 151.Allard ST, Giraud MF, Whitfield C, Graninger M, Messner P, Naismith JH. J. Mol. Biol. 2001;307:283. doi: 10.1006/jmbi.2000.4470. [DOI] [PubMed] [Google Scholar]
- 152.Allard ST, Beis K, Giraud MF, Hegeman AD, Gross JW, Wilmouth RC, Whitfield C, Graninger M, Messner P, Allen AG, Maskell DJ, Naismith JH. Structure. 2002;10:81. doi: 10.1016/s0969-2126(01)00694-3. [DOI] [PubMed] [Google Scholar]
- 153.Beis K, Allard ST, Hegeman AD, Murshudov G, Philp D, Naismith JH. J. Am. Chem. Soc. 2003;125:11872. doi: 10.1021/ja035796r. [DOI] [PubMed] [Google Scholar]
- 154.Allard ST, Cleland WW, Holden HM. J. Biol. Chem. 2004;279:2211. doi: 10.1074/jbc.M310134200. [DOI] [PubMed] [Google Scholar]
- 155.Liu Y, Thoden JB, Kim J, Berger E, Gulick AM, Ruzicka FJ, Holden HM, Frey PA. Biochemistry. 1997;36:10675. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 156.Thoden JB, Holden HM. Biochemistry. 1998;37:11469. doi: 10.1021/bi9808969. [DOI] [PubMed] [Google Scholar]
- 157.Gatzeva-Topalova PZ, May AP, Sousa MC. Biochemistry. 2004;43:13370. doi: 10.1021/bi048551f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Somers WS, Stahl ML, Sullivan FX. Structure. 1998;6:1601. doi: 10.1016/s0969-2126(98)00157-9. [DOI] [PubMed] [Google Scholar]
- 159.Schutzbach JS, Feingold DS. J. Biol. Chem. 1970;245:2476. [PubMed] [Google Scholar]
- 160.Frey PA. FASEB J. 1996;10:461. [PubMed] [Google Scholar]
- 161.Hallis TM, Zhao Z, Liu H-w. J. Am. Chem. Soc. 2000;43:10493. [Google Scholar]
- 162.Hallis TM, Liu H-w. J. Am. Chem. Soc. 1999;121:6765. [Google Scholar]
- 163.Morrison JP, Read JA, Coleman WG, Jr, Tanner ME. Biochemistry. 2005;44:5907. doi: 10.1021/bi050106c. [DOI] [PubMed] [Google Scholar]
- 164.Read JA, Ahmed RA, Morrison JP, Coleman WG, Jr, Tanner ME. J. Am. Chem. Soc. 2004;126:8878. doi: 10.1021/ja0485659. [DOI] [PubMed] [Google Scholar]
- 165.Liu Y, Thoden JB, Kim J, Berger E, Gulick AM, Ruzicka FJ, Holden HM, Frey PA. Biochemistry. 1998;36:10675. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 166.Rizzi M, Tonetti M, Vigevani P, Sturla L, Bisso A, De Flora A, Bordo D, Bolognesi M. Structure. 1998;15:1453. doi: 10.1016/s0969-2126(98)00144-0. [DOI] [PubMed] [Google Scholar]
- 167.Menon S, Stahl ML, Kumar R, Xu G-Y, Sullivan F. J. Biol. Chem. 1999;274:26743. doi: 10.1074/jbc.274.38.26743. [DOI] [PubMed] [Google Scholar]
- 168.Rosano C, Bisso A, Izzo G, Tonetti M, Sturla L, De Flora A, Bolognesi M. J. Mol. Biol. 2000;303:77. doi: 10.1006/jmbi.2000.4106. [DOI] [PubMed] [Google Scholar]
- 169.Boll R, Hofmann C, Heitmann B, Hauser G, Glaser S, Koslowski T, Friedrich T, Bechthold A. J. Biol. Chem. 2006;281:14756. doi: 10.1074/jbc.M601508200. [DOI] [PubMed] [Google Scholar]
- 170.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nuc. Acids Res. 2001;29:1097. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ostash B, Saghatelian A, Walker S. Chem. Biol. 2007;14:257. doi: 10.1016/j.chembiol.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kharel MK, Basnet DB, Lee HC, Liou K, Moon YH, Kim J-J, Woo JS, Sohng JK. Mol. Cells. 2004;18:71. [PubMed] [Google Scholar]
- 173.Woodyer RD, Li G, Zhao H, van der Donk WA. Chem. Commun. (Cambridge, U.K.) 2007:359. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- 174.Sohng JK, Oh T-J, Lee J-J, Kim S-G. Mol. Cells. 1997;7:674. [PubMed] [Google Scholar]
- 175.Thorson JS, Sievers EL, Ahlert J, Shepard E, Whitwam RE, Onwueme KC, Ruppen M. Curr. Pharm. Des. 2000;6:1841. doi: 10.2174/1381612003398564. [DOI] [PubMed] [Google Scholar]
- 176.Odakura Y, Kase S, Itoh S, Satoh S, Takasawa S, Takahashi K, Shirahata K. J. Antibiot. 1984;37:1670. doi: 10.7164/antibiotics.37.1670. [DOI] [PubMed] [Google Scholar]
- 177.Dairi T, Ohta T, Hashimoto E, Hasegawa M. Mol. Gen. Genet. 1992;236:39. doi: 10.1007/BF00279641. [DOI] [PubMed] [Google Scholar]
- 178.Dairi T, Ohta T, Hashimoto E, Hasegawa M. Mol. Gen. Genet. 1992;232:262. doi: 10.1007/BF00280005. [DOI] [PubMed] [Google Scholar]
- 179.Treese I, Hauser G, Muhlenweg A, Hofmann C, Schmidt M, Weitnauer G, Glaser S, Bechthold A. Appl. Environ. Microbiol. 2005;71:400. doi: 10.1128/AEM.71.1.400-406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Davies GJ, Gloster TM, Henrissat B. Curr. Opin. Struct. Biol. 2005;15:637. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 181.Luzhetskyy A, Mendez C, Salas JA, Bechthold A. Curr. Topics Med. Chem. 2008;8:680. doi: 10.2174/156802608784221514. [DOI] [PubMed] [Google Scholar]
- 182.Craig SP, 3rd, Eakin AE. J. Biol. Chem. 2000;275:20231. doi: 10.1074/jbc.R000002200. [DOI] [PubMed] [Google Scholar]
- 183.Sinha SC, Smith JL. Curr. Opin. Struct. Biol. 2001;11:733. doi: 10.1016/s0959-440x(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 184.Kudo F, Fujii T, Kinoshita S, Eguchi T. Bioorg. Med. Chem. 2007;15:4360. doi: 10.1016/j.bmc.2007.04.040. [DOI] [PubMed] [Google Scholar]
- 185.Okabe T, Suda H, Sato F, Okanishi M. Banyu Pharmaceutical Co., Ltd., Japan. 1990:8. [Google Scholar]
- 186.Vrielink A, Ruger W, Driessen HPC, Freemont PS. EMBO J. 1994;13:3413. doi: 10.1002/j.1460-2075.1994.tb06646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. http://afmb.cnrsmrs.fr/~cazy/CAZY/index.html.
- 188.Lovering AL, De Castro LH, Lim D, Strynadka NC. Science. 2007;315:1402. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 189.Campbell JE, Davies GJ, Bulone V, Henrissat B. Biochem. J. 1997;326:929. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Coutinho PM, Deleury E, Davies GJ, Henrissat B. J. Mol. Biol. 2003;328:307. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 191.Unligil UM, Rini JM. Curr. Opin. Struct. Biol. 2000;10:510. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 192.Bourne Y, Henrissat B. Curr. Opin. Struct. Biol. 2001;11:593. doi: 10.1016/s0959-440x(00)00253-0. [DOI] [PubMed] [Google Scholar]
- 193.Breton C, Mucha J, Jeanneau C. Biochimie. 2001;83:713. doi: 10.1016/s0300-9084(01)01298-6. [DOI] [PubMed] [Google Scholar]
- 194.Hu Y, Walker S. Chem. Biol. 2002;9:1287. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 195.Wiggins CAR, Munro S. Proc. Nat. Acad. Sci. USA. 1998;95:7945. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hagen FK, Hazes B, Raffo R, deSa D, Tabak LA. J. Biol. Chem. 1999;274:6797. doi: 10.1074/jbc.274.10.6797. [DOI] [PubMed] [Google Scholar]
- 197.Murray BW, Takayama S, Schultz J, Wong CH. Biochemistry. 1996;35:11183. doi: 10.1021/bi961065a. [DOI] [PubMed] [Google Scholar]
- 198.Busch C, Hofmann F, Selzer J, Munro S, Jeckel D, Aktories K. J. Biol. Chem. 1998;273:19566. doi: 10.1074/jbc.273.31.19566. [DOI] [PubMed] [Google Scholar]
- 199.Shibayama K, Ohsuka S, Tanaka T, Arakawa Y, Ohta M. J. Bacteriol. 1998;180:5313. doi: 10.1128/jb.180.20.5313-5318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Uitdehaag JCM, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. Nat. Struct. Biol. 1999;6:432. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 201.Pak JE, Arnoux P, Zhou S, Sivarajah P, Satkunarajah M, Xing X, Rini JM. J. Biol. Chem. 2006;281:26693. doi: 10.1074/jbc.M603534200. [DOI] [PubMed] [Google Scholar]
- 202.Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure. 2001;9:547. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 203.Mulichak AM, Losey HC, Lu W, Wawrzak Z, Walsh CT, Garavito RM. Proc. Nat. Acad. Sci. USA. 2003;100:9238. doi: 10.1073/pnas.1233577100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mulichak AM, Lu W, Losey HC, Walsh CT, Garavito RM. Biochemistry. 2004;43:5170. doi: 10.1021/bi036130c. [DOI] [PubMed] [Google Scholar]
- 205.Bolam DN, Roberts S, Proctor MR, Turkenburg JP, Dodson EJ, Martinez-Fleites C, Yang M, Davis BG, Davies GJ, Gilbert HJ. Proc. Nat. Acad. Sci. USA. 2007;104:5336. doi: 10.1073/pnas.0607897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Martinez-Fleites C, Proctor M, Roberts S, Bolam DN, Gilbert HJ, Davies GJ. Chem. Biol. 2006;13:1143. doi: 10.1016/j.chembiol.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 207.Mittler M, Bechthold A, Schulz GE. J. Mol. Biol. 2007;372:67. doi: 10.1016/j.jmb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 208.Hoffmeister D, Ichinose K, Bechthold A. Chem. Biol. 2001;8:557. doi: 10.1016/s1074-5521(01)00039-4. [DOI] [PubMed] [Google Scholar]
- 209.Hoffmeister D, Wilkinson B, Foster G, Sidebottom PJ, Ichinose K, Bechthold A. Chem. Biol. 2002;9:287. doi: 10.1016/s1074-5521(02)00114-x. [DOI] [PubMed] [Google Scholar]
- 210.Williams GJ, Zhang C, Thorson JS. Nat. Chem. Biol. 2007;3:657. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]
- 211.Hancock SM, Vaughan MD, Withers SG. Curr. Opin. Chem. Biol. 2006;10:509. doi: 10.1016/j.cbpa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 212.Sinnott ML. Chem. Rev. 1990;90:1171. [Google Scholar]
- 213.Pedersen LC, Darden TA, Negishi M. J. Biol. Chem. 2002;277:21869. doi: 10.1074/jbc.M112343200. [DOI] [PubMed] [Google Scholar]
- 214.Charnock SJ, Davies GJ. Biochemistry. 1999;38:6380. doi: 10.1021/bi990270y. [DOI] [PubMed] [Google Scholar]
- 215.Tarbouriech N, Charnock SJ, Davies GJ. J. Mol. Biol. 2001;314:655. doi: 10.1006/jmbi.2001.5159. [DOI] [PubMed] [Google Scholar]
- 216.Qiao L, Murray BW, Shimazaki M, Schultz J, Wong CH. J. Am. Chem. Soc. 1996;118:7653. [Google Scholar]
- 217.Tvaroska I, Andre I, Carver JP. J. Am. Chem. Soc. 2000;122:8762. [Google Scholar]
- 218.Zechel DL, Withers SG. Acc. Chem. Res. 2000;33:11. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
- 219.Persson K, Ly HD, Dieckelmann M, Wakarchuk WW, Withers SG, Strynadka NC. Nat. Struct. Biol. 2001;8:166. doi: 10.1038/84168. [DOI] [PubMed] [Google Scholar]
- 220.Pedersen LC, Dong J, Taniguchi F, Kitagawa H, Krahn JM, Pedersen LG, Sugahara K, Negishi M. J. Biol. Chem. 2003;278:14420. doi: 10.1074/jbc.M210532200. [DOI] [PubMed] [Google Scholar]
- 221.Boix E, Swaminathan GJ, Zhang Y, Natesh R, Brew K, Acharya KR. J. Biol. Chem. 2001;276:48608. doi: 10.1074/jbc.M108828200. [DOI] [PubMed] [Google Scholar]
- 222.Boix E, Zhang Y, Swaminathan GJ, Brew K, Acharya KR. J. Biol. Chem. 2002;277:28310. doi: 10.1074/jbc.M202631200. [DOI] [PubMed] [Google Scholar]
- 223.Ly HD, Lougheed B, Wakarchuk WW, Withers SG. Biochemistry. 2002;41:5075. doi: 10.1021/bi012031s. [DOI] [PubMed] [Google Scholar]
- 224.Zhang Y, Swaminathan GJ, Deshpande A, Boix E, Natesh R, Xie Z, Acharya KR, Brew K. Biochemistry. 2003;42:13512. doi: 10.1021/bi035430r. [DOI] [PubMed] [Google Scholar]
- 225.Flint J, Taylor E, Yang M, Bolam DN, Tailford LE, Martinez-Fleites C, Dodson EJ, Davis BG, Gilbert HJ, Davies GJ. Nat. Struct. Mol. Biol. 2005;12:608. doi: 10.1038/nsmb950. [DOI] [PubMed] [Google Scholar]
- 226.Gibson RP, Turkenburg JP, Charnock SJ, Lloyd R, Davies GJ. Chem. Biol. 2002;9:1337. doi: 10.1016/s1074-5521(02)00292-2. [DOI] [PubMed] [Google Scholar]
- 227.Liang D, Qiao J. J. Mol. Evol. 2007;64:342. doi: 10.1007/s00239-006-0110-2. [DOI] [PubMed] [Google Scholar]
- 228.Quiros LM, Carbajo RJ, Brana AF, Salas JA. J. Biol. Chem. 2000;275:11713. doi: 10.1074/jbc.275.16.11713. [DOI] [PubMed] [Google Scholar]
- 229.Borisova SA, Zhao L, Melancon CE, 3rd, Kao C-L, Liu H-w. J. Am. Chem. Soc. 2004;126:6534. doi: 10.1021/ja049967j. [DOI] [PubMed] [Google Scholar]
- 230.Leimkuhler C, Fridman M, Lupoli T, Walker S, Walsh CT, Kahne D. J. Am. Chem. Soc. 2007;129:10546. doi: 10.1021/ja072909o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yuan Y, Chung HS, Leimkuhler C, Walsh CT, Kahne D, Walker S. J. Am. Chem. Soc. 2005;127:14128. doi: 10.1021/ja053704n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Borisova SA, Zhang C, Takahashi H, Zhang H, Wong A, Thorson JS, Liu H-w. Angew. Chem. Int. Ed. 2006;45:2748. doi: 10.1002/anie.200503195. Angew. Chem. 2006, 118, 2814. [DOI] [PubMed] [Google Scholar]
- 233.Hultin PG. Curr. Topics Med. Chem. 2005;5:1299. doi: 10.2174/156802605774643015. [DOI] [PubMed] [Google Scholar]
- 234.Fischbach MA, Lin H, Liu DR, Walsh CT. Proc. Nat. Acad. Sci. USA. 2005;102:571. doi: 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Liu T, Kharel MK, Fischer C, McCormick A, Rohr J. Chembiochem. 2006;7:1070. doi: 10.1002/cbic.200600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Durr C, Hoffmeister D, Wohlert S-E, Ichinose K, Weber M, von Mulert U, Thorson JS, Bechthold A. Angew. Chem. Int. Ed. 2004;43:2962. doi: 10.1002/anie.200453758. Angew. Chem. 2004, 116, 3022. [DOI] [PubMed] [Google Scholar]
- 237.Minami A, Eguchi T. J. Am. Chem. Soc. 2007;129:5102. doi: 10.1021/ja0685250. [DOI] [PubMed] [Google Scholar]
- 238.Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS. Nat. Biotechnol. 2003;21:1467. doi: 10.1038/nbt909. [DOI] [PubMed] [Google Scholar]
- 239.Kao C-L, Borisova SA, Kim HJ, Liu H-w. J. Am. Chem. Soc. 2006;128:5606. doi: 10.1021/ja058433v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tang L, McDaniel R. Chem. Biol. 2001;8:547. doi: 10.1016/s1074-5521(01)00032-1. [DOI] [PubMed] [Google Scholar]
- 241.Borisova SA, Kim HJ, Pu X, Liu H-w. ChemBiochem. 2008;9 doi: 10.1002/cbic.200800155. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hutchinson CR. Curr. Opin. Microbiol. 1998;1:319. doi: 10.1016/s1369-5274(98)80036-2. [DOI] [PubMed] [Google Scholar]
- 243.Hallis TM, Liu H-w. Acc. Chem. Res. 1999;32:579. [Google Scholar]
- 244.Floss HG. J. Indust. Microbiol. Biotechnol. 2001;27:183. doi: 10.1038/sj.jim.7000069. [DOI] [PubMed] [Google Scholar]
- 245.Mendez C, Salas JA. Trends Biotechnol. 2001;19:449. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]
- 246.Butler AR, Bate N, Kiehl DE, Kirst HA, Cundliffe E. Nat. Biotechnol. 2002;20:713. doi: 10.1038/nbt0702-713. [DOI] [PubMed] [Google Scholar]
- 247.Walsh CT. Chembiochem. 2002;3:124. [Google Scholar]
- 248.Langenhan JM, Griffith BR, Thorson JS. J. Nat. Prod. 2005;68:1696. doi: 10.1021/np0502084. [DOI] [PubMed] [Google Scholar]
- 249.Thibodeaux CJ, Liu H-w, Thorson JS. In: Comprehensive glycoscience. Kamerling JP, Boons G-J, Lee Y, Suzuki A, Taniguchi N, Voragen AGJ, editors. Vol. 3. Amsterdam: Elsevier; 2007. p. 373. [Google Scholar]
- 250.Thibodeaux CJ, Liu H-w. Pure Appl. Chem. 2007;79:785. [Google Scholar]
- 251.Salas JA, Mendez C. Trends Microbiol. 2007;15:219. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 252.Bode HB, Muller R. Angew. Chem. Int. Ed. 2007;46:2147. doi: 10.1002/anie.200604671. Angew. Chem. 2007, 119, 2195. [DOI] [PubMed] [Google Scholar]
- 253.Mendez C, Luzhetskyy A, Bechthold A, Salas JA. Curr. Topics Med. Chem. 2008;8:710. doi: 10.2174/156802608784221532. [DOI] [PubMed] [Google Scholar]
- 254.Decker H, Haag S, Udvarnoki G, Rohr J. Angew. Chem. Int. Ed. Engl. 1995;34:1107. Angew. Chem. 1995, 107, 1214. [Google Scholar]
- 255.Solenberg PJ, Matsushima P, Stack DR, Wilkie SC, Thompson RC, Baltz RH. Chem. Biol. 1997;4:195. doi: 10.1016/s1074-5521(97)90288-x. [DOI] [PubMed] [Google Scholar]
- 256.Madduri K, Kennedy J, Rivola G, Inventi-Solari A, Filippini S, Zanuso G, Colombo AL, Gewain KM, Occi JL, MacNeil DJ, Hutchinson CR. Nat. Biotechnol. 1998;16:69. doi: 10.1038/nbt0198-69. [DOI] [PubMed] [Google Scholar]
- 257.Thiericke R, Rohr J. Nat. Prod. Rep. 1993;10:265. doi: 10.1039/np9931000265. [DOI] [PubMed] [Google Scholar]
- 258.Weist S, Sussmuth RD. Appl. Microbiol. Biotechnol. 2005;68:141. doi: 10.1007/s00253-005-1891-8. [DOI] [PubMed] [Google Scholar]
- 259.Webster LC, Anastas PT, Williamson TC. ACS Symopsium Series. 1996;626:198. [Google Scholar]
- 260.Rohr J. Angew. Chem. Int. Ed. Engl. 1995;34:881. Angew. Chem. 1995, 107, 963. [Google Scholar]
- 261.Doumith M, Legrand R, Lang C, Salas JA, Raynal MC. Mol. Microbiol. 1999;34:1039. doi: 10.1046/j.1365-2958.1999.01666.x. [DOI] [PubMed] [Google Scholar]
- 262.Gaisser S, Reather J, Wirtz G, Kellenberger L, Staunton J, Leadlay PF. Mol. Microbiol. 2000;36:391. doi: 10.1046/j.1365-2958.2000.01856.x. [DOI] [PubMed] [Google Scholar]
- 263.Gaisser S, Lill R, Wirtz G, Grolle F, Staunton J, Leadlay PF. Mol. Microbiol. 2001;41:1223. doi: 10.1046/j.1365-2958.2001.02594.x. [DOI] [PubMed] [Google Scholar]
- 264.Borisova SA, Zhao L, Sherman DH, Liu H-w. Org. Lett. 1999;1:133. doi: 10.1021/ol9906007. [DOI] [PubMed] [Google Scholar]
- 265.Zhao L, Ahlert J, Xue Y, Thorson JS, Sherman DH, Liu H-w. J. Am. Chem. Soc. 1999;121:9881. [Google Scholar]
- 266.Melancon CE, 3rd, Liu H-w. J. Am. Chem. Soc. 2007;129:4896. doi: 10.1021/ja068254t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Jung WS, Han AR, Hong JSJ, Park SR, Choi CY, Park JW, Yoon YJ. Appl. Microbiol. Biotechnol. 2007;76:1373. doi: 10.1007/s00253-007-1101-y. [DOI] [PubMed] [Google Scholar]
- 268.Pageni BB, Oh T-J, Liou K, Yoon YJ, Sohng JK. J. Microbiol. Biotechnol. 2008;18:88. [PubMed] [Google Scholar]
- 269.Wohlert S-E, Blanco G, Lombo F, Fernandez E, Brana AF, Reich S, Udvarnoki G, Mendez C, Decker H, Frevert J, Salas JA, Rohr J. J. Am. Chem. Soc. 1998;120:10596. [Google Scholar]
- 270.Blanco G, Patallo EP, Brana AF, Trefzer A, Bechthold A, Rohr J, Mendez C, Salas AP. Chem. Biol. 2001;8:253. doi: 10.1016/s1074-5521(01)00010-2. [DOI] [PubMed] [Google Scholar]
- 271.Fischer C, Rodriguez L, Patallo EP, Lipata F, Brana AF, Mendez C, Salas JA, Rohr J. J. Nat. Prod. 2002;65:1685. doi: 10.1021/np020112z. [DOI] [PubMed] [Google Scholar]
- 272.Lombo F, Gibson M, Greenwell L, Brana AF, Rohr J, Salas JA, Mendez C. Chem. Biol. 2004;1709:1709. doi: 10.1016/j.chembiol.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 273.Perez M, Lombo F, Zhu L, Gibson M, Brana AF, Rohr J, Salas JA, Mendez C. Chem. Commun. (Camb) 2005;12:1604. doi: 10.1039/b417815g. [DOI] [PubMed] [Google Scholar]
- 274.Kunzel E, Faust B, Oelkers C, Weissbach U, Bearden DW, Weitnauer G, Westrich L, Bechthold A, Rohr J. J. Am. Chem. Soc. 1999;121:11058. [Google Scholar]
- 275.Trefzer A, Hoffmeister D, Kunzel E, Stockert S, Weitnauer G, Westrich L, Rix U, Fuchser J, Bindseil KU, Rohr J, Bechthold A. Chem. Biol. 2000;7:133. doi: 10.1016/s1074-5521(00)00079-x. [DOI] [PubMed] [Google Scholar]
- 276.Hoffmeister D, Drager G, Ichinose K, Rohr J, Bechthold A. J. Am. Chem. Soc. 2003;125:4678. doi: 10.1021/ja029645k. [DOI] [PubMed] [Google Scholar]
- 277.Trefzer A, Blanco G, Remsing L, Kunzel E, Rix U, Lipata F, Brana AF, Mendez C, Rohr J, Bechthold A, Salas JA. J. Am. Chem. Soc. 2003;124:6056. doi: 10.1021/ja017385l. [DOI] [PubMed] [Google Scholar]
- 278.Trefzer A, Fischer C, Stockert S, Westrich L, Kunzel E, Girreser U, Rohr J, Bechthold A. Chem. Biol. 2001;8:1239. doi: 10.1016/s1074-5521(01)00091-6. [DOI] [PubMed] [Google Scholar]
- 279.Sanchez C, Zhu L, Brana AF, Salas AP, Rohr J, Mendez C, Salas JA. Proc. Nat. Acad. Sci. USA. 2005;102:461. doi: 10.1073/pnas.0407809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang GP, Van Vranken DL, Thorson JS. Chembiochem. 2006;7:795. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]
- 281.Rupprath C, Schumacher T, Elling L. Curr. Med. Chem. 2005;12:1637. doi: 10.2174/0929867054367167. [DOI] [PubMed] [Google Scholar]
- 282.Langenhan JM, Peters NR, Guzei IA, Hoffmann FM, Thorson JS. Proc. Nat. Acad. Sci. USA. 2005;102:12305. doi: 10.1073/pnas.0503270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ahmed A, Peters NR, Fitzgerald MK, Watson JA, Jr, Hoffmann FM, Thorson JS. J. Am. Chem. Soc. 2006;128:14224. doi: 10.1021/ja064686s. [DOI] [PubMed] [Google Scholar]
- 284.Lin H, Walsh CT. J. Am. Chem. Soc. 2004;126:13998. doi: 10.1021/ja045147v. [DOI] [PubMed] [Google Scholar]
- 285.Wang J, Li J, Chen H-N, Chang H, Tanifum CT, Liu H-H, Czyryca PG, Chang C-WT. J. Med. Chem. 2005;48:6271. doi: 10.1021/jm050368c. [DOI] [PubMed] [Google Scholar]
- 286.Griffith BR, Langenhan JM, Thorson JS. Curr. Opin. Biotechnol. 2005;16:622. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 287.Hoffmeister D, Yang J, Liu L, Thorson JS. Proc. Nat. Acad. Sci. USA. 2003;100:13184. doi: 10.1073/pnas.2235011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hoffmeister D, Thorson JS. Chembiochem. 2004;5:989. doi: 10.1002/cbic.200400003. [DOI] [PubMed] [Google Scholar]
- 289.Yang J, Liu L, Thorson JS. Chembiochem. 2004;5:992. doi: 10.1002/cbic.200400041. [DOI] [PubMed] [Google Scholar]
- 290.Yang J, Fu X, Liao J, Liu L, Thorson JS. Chem. Biol. 2005;12:657. doi: 10.1016/j.chembiol.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 291.Jiang J, Biggins JB, Thorson JS. J. Am. Chem. Soc. 2000;122:6803. [Google Scholar]
- 292.Jiang J, Biggins JB, Thorson JS. Angew. Chem. Int. Ed. 2001;40:1502. Angew. Chem. 2001, 113, 1550. [PubMed] [Google Scholar]
- 293.Barton WA, Lesniak J, Biggins JB, Jeffrey PD, Jiang J, Rajashankar KR, Thorson JS, Nikolov DB. Nat. Struct. Biol. 2001;8:545. doi: 10.1038/88618. [DOI] [PubMed] [Google Scholar]
- 294.Barton WA, Biggins JB, Jiang J, Thorson JS, Nikolov DB. Proc. Nat. Acad. Sci. USA. 2002;99:13397. doi: 10.1073/pnas.192468299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Mizanur RM, Zea CJ, Pohl NL. J. Am. Chem. Soc. 2004;126:15993. doi: 10.1021/ja046070d. [DOI] [PubMed] [Google Scholar]
- 296.Mizanur RM, Jaipuri FA, Pohl NL. J. Am. Chem. Soc. 2005;127:836. doi: 10.1021/ja044117p. [DOI] [PubMed] [Google Scholar]
- 297.Zhang Z, Tsujimura M, Akutsu J-i, Sasaki M, Tajima H, Kawarabayasi Y. J. Biol. Chem. 2005;280:9698. doi: 10.1074/jbc.M411211200. [DOI] [PubMed] [Google Scholar]
- 298.Bae J, Kim K-H, Kim D, Choi Y, Kim J-S, Koh S, Hong S-I, Lee D-S. Chembiochem. 2005;6:1963. doi: 10.1002/cbic.200500183. [DOI] [PubMed] [Google Scholar]
- 299.Moretti R, Thorson JS. J. Biol. Chem. 2007;282:16942. doi: 10.1074/jbc.M701951200. [DOI] [PubMed] [Google Scholar]
- 300.Elling L, Rupprath C, Gunther N, Romer U, Verseck S, Weingarten P, Drager G, Kirschning A, Piepersberg W. Chembiochem. 2005;6:1423. doi: 10.1002/cbic.200500037. [DOI] [PubMed] [Google Scholar]
- 301.Oh J, Lee S-G, Kim B-G, Sohng JK, Liou K, Lee HC. Biotechnol. Bioeng. 2003;84:452. doi: 10.1002/bit.10789. [DOI] [PubMed] [Google Scholar]
- 302.Love KR, Swoboda JG, Noren CJ, Walker S. Chembiochem. 2006;7:753. doi: 10.1002/cbic.200600018. [DOI] [PubMed] [Google Scholar]
- 303.Aharoni A, Thieme K, Chiu CPC, Buchini S, Lairson LL, Chen H, Strynadka NCJ, Wakarchuk WW, Withers SG. Nat. Methods. 2006;3:609. doi: 10.1038/nmeth899. [DOI] [PubMed] [Google Scholar]
- 304.Persson M, Palcic MM. Anal. Biochem. 2008;378:1. doi: 10.1016/j.ab.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 305.Yang J, Hoffmeister D, Liu L, Fu X, Thorson JS. Bioorg. Med. Chem. 2004;12:1577. doi: 10.1016/j.bmc.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 306.Losey HC, Peczuh MW, Chen Z, Eggert US, Dong SD, Pelczer I, Kahne D, Walsh CT. Biochemistry. 2001;40:4745. doi: 10.1021/bi010050w. [DOI] [PubMed] [Google Scholar]
- 307.Losey HC, Jiang J, Biggins JB, Oberthur M, Ye X-Y, Dong SD, Kahne D, Thorson JS, Walsh CT. Chem. Biol. 2002;9:1305. doi: 10.1016/s1074-5521(02)00270-3. [DOI] [PubMed] [Google Scholar]
- 308.Fu X, Albermann C, Zhang C, Thorson JS. Org. Lett. 2005;7:1513. doi: 10.1021/ol0501626. [DOI] [PubMed] [Google Scholar]
- 309.Zhang C, Fu Q, Albermann C, Li L, Thorson JS. Chembiochem. 2007;8:385. doi: 10.1002/cbic.200600509. [DOI] [PubMed] [Google Scholar]
- 310.Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee I-K, Li L, Thorson JS. Science. 2006;313:1291. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]
- 311.Minami A, Uchida R, Eguchi T, Kakinuma K. J. Am. Chem. Soc. 2005;127:6148. doi: 10.1021/ja042848j. [DOI] [PubMed] [Google Scholar]
- 312.Kopp M, Rupprath C, Irschik H, Bechthold A, Elling L, Muller M. Chembiochem. 2007;8:813. doi: 10.1002/cbic.200700024. [DOI] [PubMed] [Google Scholar]
- 313.Freel Meyers CL, Oberthur M, Anderson JW, Kahne D, Walsh CT. Biochemistry. 2003;42:4179. doi: 10.1021/bi0340088. [DOI] [PubMed] [Google Scholar]
- 314.Albermann C, Soriano A, Jiang J, Volmer H, Biggins JB, Barton WA, Lesniak J, Nikolov DB, Thorson JS. Org. Lett. 2003;5:933. doi: 10.1021/ol0341086. [DOI] [PubMed] [Google Scholar]
- 315.Yang M, Proctor MR, Bolam DN, Errey JC, Field RA, Gilbert HJ, Davis BG. J. Am. Chem. Soc. 2005;127:9336. doi: 10.1021/ja051482n. [DOI] [PubMed] [Google Scholar]
- 316.Oberthur M, Leimkuhler C, Kruger RG, Lu W, Walsh CT, Kahne D. J. Am. Chem. Soc. 2005;127:10747. doi: 10.1021/ja052945s. [DOI] [PubMed] [Google Scholar]
- 317.Melancon CE, 3rd, Thibodeaux CJ, Liu H-w. ACS Chem. Biol. 2006;1:499. doi: 10.1021/cb600365q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.