Abstract
The secondary neurons generated in the thoracic central nervous system of Drosophila arise from a hemisegmental set of 25 neuronal stem cells, the neuroblasts (NBs). Each NB undergoes repeated asymmetric divisions to produce a series of smaller ganglion mother cells (GMCs), which typically divide once to form two daughter neurons. We find that the two daughters of the GMC consistently have distinct fates. Using both loss-of-function and gain-of-function approaches, we examined the role of Notch signaling in establishing neuronal fates within all of the thoracic secondary lineages. In all cases, the ‘A’ (NotchON) sibling assumes one fate and the ‘B’ (NotchOFF) sibling assumes another, and this relationship holds throughout the neurogenic period, resulting in two major neuronal classes: the A and B hemilineages. Apparent monotypic lineages typically result from the death of one sibling throughout the lineage, resulting in a single, surviving hemilineage. Projection neurons are predominantly from the B hemilineages, whereas local interneurons are typically from A hemilineages. Although sibling fate is dependent on Notch signaling, it is not necessarily dependent on numb, a gene classically involved in biasing Notch activation. When Numb was removed at the start of larval neurogenesis, both A and B hemilineages were still generated, but by the start of the third larval instar, the removal of Numb resulted in all neurons assuming the A fate. The need for Numb to direct Notch signaling correlated with a decrease in NB cell cycle time and may be a means for coping with multiple sibling pairs simultaneously undergoing fate decisions.
Keywords: Neurogenesis, Hemilineage, Notch, Numb, Neuroblasts, Drosophila
INTRODUCTION
How such a great diversity of cell types is generated within the nervous system during development remains a major unresolved question in neurobiology. In vertebrates and invertebrates, both inductive (Briscoe, 2009; Edlund and Jessell, 1999) and lineage-based mechanisms are involved in producing this diversity (Desai and McConnell, 2000; Cayoutte et al., 2006), but different regions of the nervous system may be biased towards one end of this spectrum or the other. Within insects, lineage-based mechanisms are responsible for the vast majority of neuronal diversity, with the possible exception of the optic lobes. In the central brain and ventral ganglia, the neuronal stem cells (the neuroblasts, NBs) are identifiable as individuals and each makes a characteristic set of progeny (e.g. Bossing et al., 1996; Schmidt et al., 1997; Schmid et al., 1999). NBs go through asymmetric, self-renewing divisions, each resulting in a neuronal precursor cell, the ganglion mother cell (GMC). Although there are now known to be exceptions (Bello et al., 2008; Bowman et al., 2008; Boone and Doe, 2008), the GMC usually undergoes a terminal division, producing two daughter neurons. The initial progeny made by a NB are often highly diverse and are termed the primary neurons (Hartenstein et al., 2008). Their identities are based on the birth order of the GMCs, and this ordering is determined by a sequence of transcription factors that are passed on to successive GMCs through time and establish neuronal fates within a given lineage (Kambadur et al., 1998; Isshiki et al., 2001; Grosskortenhaus et al., 2005). In the embryo, the daughter neurons produced by the GMC division typically have distinct identities, and this difference is controlled by Notch signaling (Spana and Doe, 1996; Skeath and Doe, 1998).
The bulk of the activity of most NBs is devoted to making secondary neurons. The secondary neurons constitute a more homogeneous population than the initial, primary set. In insects with complete metamorphosis, like Drosophila, most of the secondary neurons are born during a larval phase of neurogenesis. Studies on the generation of secondary neurons in the caterpillar of the tobacco hornworm, Manduca sexta (Witten and Truman, 1991), indicated that the GMC divides to make daughters of divergent phenotypes and that this process is then reiterated scores of times to generate two major classes of interneurons. Similarly, in grasshoppers, Jia and Siegler (Jia and Siegler, 2002) showed that the GMCs from the median neuroblast in the thorax consistently produce an engrailed positive and engrailed negative daughter, which become a local interneuron and a projection cell, respectively. Region-specific cell death of one sibling then sculpts the final lineage composition in a given segment. These examples of diverse classes of cells being generated throughout a lineage is in contrast to neurogenesis in the mushroom bodies, where sibling neurons generated at any given time are morphologically indistinguishable (Lee et al., 1999).
In this paper, we present a comprehensive analysis of the role of Notch signaling in generating neuronal phenotypes within the secondary lineages of the segmental central nervous system (CNS). The universal pattern is for a GMC to produce two neurons of different phenotypes, ‘A’ and ‘B’, with cell death involved in making some lineages monotypic. A clear division of phenotype between these A and B cell types suggest that the circuitry of the thoracic nervous system is generated in developmental units we term ‘hemilineages’. The accompanying paper by Lin et al. (Lin et al., 2010) shows that this pattern also holds the antennal lobes in the brain.
MATERIALS AND METHODS
Fly stocks
Flies were reared on the standard yeast-cornmeal-molasses diet. Mitotic clones were generated using the mosaic analysis using a repressible cell marker (MARCM) technique (Lee and Luo, 1999). We used the pan-neuronal driver, elavC155 GAL4 (Lin and Goodman, 1994) to obtain a range of clones that covered all of the thoracic lineages. Wild-type clones were generated in flies of the genotype: GAL4C155, hsFLP, UAS-mCD8::GFP; FRT2A, tubP-GAL80/FRT2A. Notch null clones were produced using the null allele N55e11 (Heitzler and Simpson, 1991) in the genotype: elavC155,N55e11,FRT 19A/tub-GAL80, hs-flp, FRT 19A; UAS-mCD8::GFP/UAS-mCD8::GFP. Clones that showed constitutive Notch signaling were produced by expressing the intracellular domain of Notch [UAS-NotchCA (Larkin et al., 1996)] using the genotype: elavC155, FRT 19A/tub-GAL80, hs-flp, FRT 19A; UAS-mCD8::GFP/UAS-mCD8::GFP; UAS-NotchCA/+. Numb activity was removed using the numb2 null allele (Frise et al., 1996) in the combination: elavC155, UAS-mCD8::GFP, hs-flp/elavC155, UAS-mCD8::GFP, hs-flp; tub-GAL80, FRT 40A/y+,numb2, ck, FRT40A. Cell death was inhibited in homozygous clones that were mutant for the initiator caspase dronc (Kondo et al., 2006; Williams et al., 2006) using flies of the following genotype: hs-flp, elavC155GAL4, UAS-mCD8::GFP/+; tubP-GAL80, FRT 2A/droncΔA8, FRT2A.
For inducing MARCM clones in recently hatched larvae, eggs were collected over a 1- to 2-hour period, maintained at 25°C for 24 hours, and the larvae then heat-shocked at 37°C for 45 minutes to 1 hour. Brief egg collections were also maintained for 72 hours before heat shock to induce clones around the start of the third larval instar.
Immunocytochemistry
Dissected nervous systems were fixed in buffered 3.7% formaldehyde for about an hour at room temperature and then washed three times in PBS-TX (phosphate buffered saline [pH 7.2] with 1% Triton-X100). Fixed samples were blocked in 2% normal donkey serum (Jackson ImmunoResearch Labs, West Grove, PA, USA) in PBS-TX for 30 minutes and then incubated in primary antibodies for 1 to 2 days at 4°C. Primary antibodies were: 1:50 mouse anti-Notch MAb [MAb C17.9C6 (Fehon et al., 1990)]; 1:1000 rat anti-mCD8 (Caltag Laboratories, Burlingame, CA, USA); 1:20 mouse anti-neurotactin MAb [F4A (de la Escalera et al., 1990)]. After three to four rinses to remove the primary antisera, tissues were incubated overnight in combinations of FITC-conjugated and Texas Red-conjugated secondary antibodies at 1:500 dilution (Jackson ImmunoResearch Labs). Nervous systems were then rinsed, mounted on polylysine-coated coverslips, dehydrated through an ethanol series, cleared in xylene and mounted in DPX (Fluka, Bachs, Switzerland).
Image analysis
Confocal image stacks were typically collected at 63× on either a BioRad MRC 600 or a Zeiss 510 confocal microscope. Image stacks were processed using Image J (http://rsb.info.nih.gov/ij/). The z-projections for a given clone (green) included all of the sections from the cell body cluster to the end of the neurite bundle. The z-projection for the reference channel (magenta) typically included only the sections in the neuropil region that showed the neurotactin-positive bundles needed for lineage identification. Images were only globally adjusted for intensity and background.
We collected cell number data in NB clones by placing a mark on z-stack at the center of each cell. Each cell was marked only once and a count of the total marks yielded the total number of cells.
RESULTS
Effects of Notch on neuronal fates of the secondary neurons of the thorax
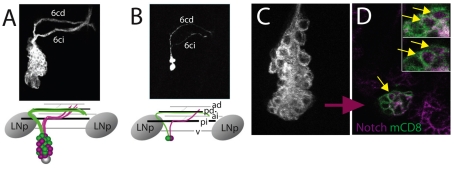
In a given lineage, the primary neurons generated during the embryonic phase of neurogenesis are typically diverse (Bossing et al., 1996; Schmidt et al., 1997; Schimid et al., 1999). By contrast, the secondary neurons produced during the postembryonic neurogenic phase are much more homogeneous (Truman et al., 2004; Pereanu and Hartenstein, 2006; Brown and Truman, 2009; Zhou et al., 2009). As seen in Fig. 1A, a NB typically generates either one or two classes of secondary neurons, as defined by their pattern of neurite projection. In the latter case, the two classes are based the division of the GMC, with the two daughters showing distinct growth decisions (Fig. 1B). This difference is then repeated for all of the GMCs generated during the second neurogenic phase. Skeath and Doe showed that Notch signaling is responsible for the differences in sibling fates during GMC divisions in the embryo (Skeath and Doe, 1998), and we hypothesized that this mechanism probably also extends into the secondary phase of neurogenesis. Immunostaining of neuroblast clones for Notch showed the prominent membrane localization of Notch in the NB, GMCs and an adjacent cluster of young neurons (Fehon et al., 1991), but also, typically, two of the young neurons had nuclear Notch (Fig. 1C,D) suggesting that these were in the process of establishing their sibling fates. Therefore, we examined Notch signaling in the secondary lineages using both loss-of-function and gain-of-function approaches.
Fig. 1.
Relationship of sibling differences to overall lineage phenotypes. (A,B) z-projection of a lineage 6 neuroblast (A) and GMC (B) clone. The primary neurites from the two siblings form the six contralateral intermediate (6ci) and six contralateral dorsal (6cd) bundles. The schematics show how the primary neurites relate to the segmental commissures and leg neuropils (LNp). (C,D) The appearance of Notch immunostaining in the nuclei of two young neurons in a lineage 1 MARCM clone. (C) A z-projection of the cell cluster. (D) An optical section showing two neurons with nuclear Notch immunostaining; insets are magnified views of the neurons in this and the adjacent section. Yellow arrows: nuclear Notch staining. Commissures: ad, anterior dorsal; pd, posterior dorsal; ai, anterior intermediate; pi, posterior intermediate; v, ventral.
MARCM clones were induced postembryonically to include only the secondary neurons born during the larval neurogenic period. Notch loss-of-function clones were homozygous mutant for the null allele N55e11 (Heitzler and Simpson, 1991). During Notch signaling the receptor is cleaved and the intracellular domain translocated to the nucleus (Struhl et al., 1993). We generated Notch gain-of-function clones either by expressing the intracellular domain of Notch, which serves as a constitutive activator [NotchCA (Larkin et al., 1996)], or by making MARCM clones that were null for numb, a negative regulator of Notch (Knoblich et al., 1995; Spana and Doe, 1996). Fig. 2 summarizes the effects of manipulating Notch signaling in the 25 secondary lineages in the ventral CNS. Nervous systems were counterstained for neurotactin and the clones identified by the neurotactin-positive bundle in which their neurites project (Truman at al., 2004). As a control for the Notch loss-of-function analysis, we looked at CNSs that were counterstained for Notch and confirmed the loss of Notch protein in the clones (data not shown).
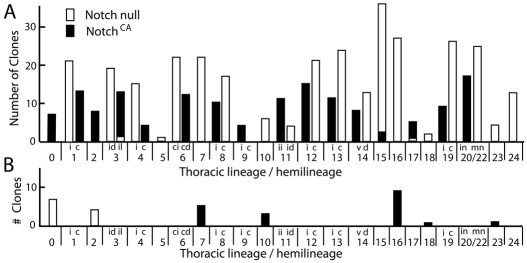
Fig. 2.
Summary of the results of removing Notch or having constitutively active Notch on the phenotypes of NB MARCM clones. (A) The frequency of particular lineages (for monotypic lineages) or hemilineage bundles under the two conditions. (B) The frequency of ‘disembodied’ lineages that included the NBs, GMCs and a few young neurons, but few or no neurites emerging from the cluster. The NB was identified by the location of the cluster and the lack of its characteristic neurotactin-positive, neurite bundle. Notch null, white bars; constitutively active Notch (NotchCA), black bars. Numbers refer to the NBs; 20/22, combined data for lineages 20 to 22; in, local leg interneurons, mn, motoneurons. Other designations are neurite bundles: c, contralateral; ci, contralateral intermediate; cd, contralateral dorsal; d, dorsal; i, ipsilateral; id, ipsilateral dorsal; ii, ipsilateral intermediate; il, ipsilateral lateral; v, ventral.
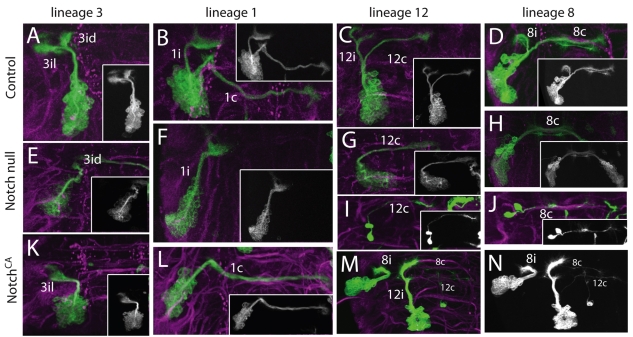
The cell clusters from NBs 1, 3, 6, 8, 11, 12, 13 and 19 have two primary neurite bundles of roughly equivalent size (e.g. Fig. 3A-D). This dichotomy was also seen in their respective GMC clones (Fig. 1B and data not shown), showing that the two neurite trajectories reflect the different fates of the two siblings. When Notch was either removed or constitutively activated in these lineages, we saw only one neurite bundle, characteristic of a single sibling type, but the bundle differed under the two conditions (Fig. 2). For example, as seen in Fig. 3, for lineages 1, 3, 8 and 12 the Notch loss-of-function condition resulted in the presence of the 1i, 3id, 8c and 12c neurite bundles, respectively, whereas the 1c, 3il, 8i and 12i bundles were present in the NotchCA clones. Relative to Notch signaling, the latter sibling is considered the ‘A’ fate (NotchON), whereas the former shows the ‘B’ fate (NotchOFF) (Skeath and Doe, 1998). Notch null clones always produced a single sibling type. However, the NotchCA clones sometimes contained one or two neurons of the ‘B’ type and the remainder of the ‘A’ type (Fig. 3M,N). We assume the presence of a few ‘B’ types in the NotchCA clones is because after clone induction one or two neurons may still commit to a NotchOFF fate before sufficient Notchintra protein is made to suppress the ‘B’ phenotype.
Fig. 3.
MARCM clones showing the effects of manipulating Notch signaling on lineages that produce two major classes of interneurons. Green, anti-CD8; magenta, anti-neurotactin showing the arrangement of lineage bundles. Insets are a reduced grayscale image of each clone. (A-D) Wild-type NB clones. (E-H) NB clones that are homozygous Notch null show a single neurite bundle. (I,J) Examples of Notch null, GMC clones for lineages 12 and 8, showing that both siblings have the same neurite projection. (K-N) NB clones that express NotchCA. M and N contain lineages 8 and 12 clones and the grayscale image is shown at full magnification. Both show one axon (8c, 12c) of the other sibling phenotype. Neurite bundle names are as in Fig. 2.
The shift in cellular composition of these clones was the result of a change in cell fate and not selective cell death. This switch in fate was clearly shown in GMC clones that lacked Notch activity. For the above lineages, wild-type GMC clones always show the two daughters that make different outgrowth decisions (e.g. Fig. 1B). In Notch null GMC clones, by contrast, the two daughters were always the same (e.g. the lineage 8 and 12 GMC clones in Fig. 3I,J). GMC clones that expressed NotchCA typically showed both sibling types, but for reasons stated above, we think that this is because of the delay involved in the GAL4 system so that the daughter cells make their fate decisions before sufficient Notchintra protein has been made. Cell counts support the conclusion that the Notch manipulation results in fate changes throughout a given lineage. In the case of lineage 1, for example, control (125±10 s.e.m.; n=5), Notch null (106±4 s.e.m.; n=5) and NotchCA clones (118±6 s.e.m.; n=5) had roughly the same number of neurons, despite the differences in cell composition under the three conditions.
Neuroblasts 0, 4, 9, 14, 20, 21 and 22 also generate two classes of neurons, but only a few of one type and many of the other. For example, lineages 20, 21 and 22 include one or two motoneurons as well as a large number of local interneurons that supply the leg neuropil (Truman et al., 2004). We had previously thought that NBs 4 and 14 produced only one type of neuron (Truman et al., 2004), but we have since found that they produce one or two interneurons that have trajectories that markedly differ from the remainder of neurons in the lineage. These rare neurons are among the first postembryonic neurons to be born and are often missing from neuroblast clones. The effect of Notch manipulation on these unequal lineages was also straightforward. Under one condition, a large cell cluster containing only the major neuron type was evident, whereas under the other condition, the minor cell type, with numbers typically doubled, was seen along with a slightly separated NB with its cluster of GMCs and young neurons, but the latter apparently dying soon after their birth. For example, NB 14 generates a large number of interneurons that project through the ventral commissure to the contralateral leg neuropil and one or two neurons projecting to the dorsal neuropil (Fig. 4D). In Notch null clones, we saw only the dorsal-projecting neurons accompanied by a detached cell cluster with the NB (Fig. 4H). In such cases, neurotactin staining showed that the lineage 14 ventral bundle was missing from that side (data not shown). In MARCM clones expressing NotchCA, by contrast, lineage 14 clones contained only the ventral projecting neurons. The size of the cell cluster was roughly twice that seen under wild-type conditions.
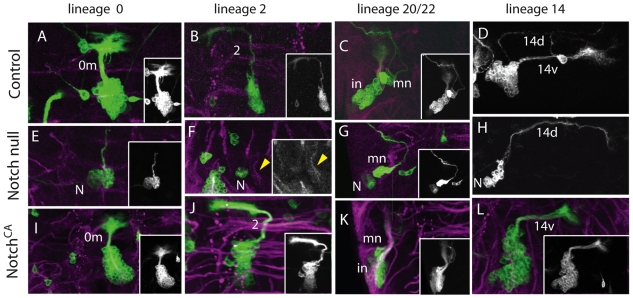
Fig. 4.
Effects of Notch manipulation on monotypic lineages. Examples of MARCM clones for lineages that are monotypic (A,B) or produce a major and minor class of neuron (C,D). Green, anti-CD8; magenta, anti-neurotactin. Insets are a reduced grayscale image of each clone, except for F, which shows the neurotactin channel. (A-D) Wild-type NB clones. (E-H) NB clones that are homozygous Notch null. (E) A lineage 0 clone with the neuroblast (N) associated with a compact cell cluster containing a neurite bundle that dwindles as it enters the neuropil. (F) A lineage 2 clone consisting of the NB and a few associated cells but no emerging neurites. The neurotactin-positive bundle of its wild-type, contralateral homolog is indicated by the yellow arrowhead; this bundle is missing on the side with the clone. (G,H) NB clones showing just the rare cell-type and a slightly separated NB with a small compact ball of associated cells. (I-L) NB clones that express NotchCA. Neurite bundle names are as in Fig. 2.
Lineages 20, 21 and 22 were difficult to resolve. Each NB produces one to two motoneurons as well as a large number of interneurons that project to the leg neuropil (Truman et al., 2004). For the Notch null clones, we saw many examples of two to four motoneurons that were associated with a detached cell cluster comprised of the NB, GMCs and very young neurons that lacked neurites. We recovered no Notch null clones that contained the interneurons for these three lineages. By contrast, we recovered many NotchCA clones that contained the lineage 21 or 20/22 interneurons. These sometimes had an associated motoneuron, but its presence is probably due to this cell being generated before the production of sufficient Notchintra, as discussed above.
NB 0 makes a midline cluster of local interneurons but wild-type clones occasionally include a projection neuron with a bifurcating axon, the phenotype of the VUM cells produced by this neuroblast during embryogenesis. NotchCA clones showed an expanded set of local interneurons (Fig. 4I). Loss of Notch resulted in a compact cell cluster around the NB, with a neurite bundle that entered the neuropil but then dwindled away (Fig. 4E). As the neurons with the abortive neurites are located close to the NB, we conclude that they are recently born projection neurons that die soon after their neurite enters the neuropil.
The remaining lineages, from NBs 2, 5, 7, 10, 15, 16, 17, 18, 23 and 24, have only a single neuron type. Clones for some of these lineages are recovered only rarely, even under wild-type conditions, so we can make only tentative conclusions about them. However, others, such as NBs 2, 7, 15 and 16, appear at a high frequency. For lineage 2 clones that lacked Notch, we saw only a cell cluster including an NB, GMCs and young neurons without neurites at the appropriate location, but no neurotactin-positive, neurite bundle (Fig. 4F). The lineage 2 cluster and bundle were present in the NotchCA clones (Fig. 4J). The opposite relationship to Notch was seen for lineages 7 and 16, with an enlarged cluster of neurons having the expected projection pattern under the Notch-null condition (Fig. 2B). For lineage 7, the expression of NotchCA resulted in the loss of the 7c neurotactin bundle and a disembodied NB and associated cell cluster in the normal NB 7 position. NotchCA expression in lineage 16 was complicated because this treatment often resulted in an enlarged cell cluster because of the production of additional NBs (see below). Neurites emerged from this cluster and projected along the expected path but then abruptly terminated, similar to the pattern seen in lineage 0 under the Notch null condition. None of the neurons in the small disembodied cell clusters expressed Broad-Z3 (Broad — FlyBase), which is a marker of intermediate neuronal development (Zhou et al., 2009; data not shown). This pattern suggests that one sibling survives to become a neuron, whereas the other sibling probably dies.
Lineage 15 makes exclusively motoneurons that project to the leg imaginal disc (Truman et al., 2004; Baek and Mann, 2009; Brown and Truman, 2009; Brierley et al., 2009). Removal of Notch results in a doubling in the size of this neuron cluster from 28±2 (n=10) cells in control clones to 57±2 (n=7) neurons in Notch null clones. Under the NotchCA condition we occasionally saw a clone with a moderate number of motoneurons, but we were unable to unequivocally ascribe these to lineage 15 (these neurons show very weak expression of neurotactin). Also, under wild-type conditions, we occasionally saw GMC clones with two lineage 15 motoneurons. We have concluded that the majority of lineage 15 motoneurons arise as the ‘B’ sibling, but a few may arise as part of the ‘A’ portion of the lineage. A second, smaller motor lineage that was missed in Truman et al. (Truman et al., 2004) is lineage 24 (Brown and Truman, 2009), which makes the motoneurons for the proximal leg muscles (Baek and Mann, 2009) (D. J. Brierley and D.W.W., unpublished). These motoneurons arise as the ‘B’ fate in lineage 24 (Fig. 2).
The role of cell death in generating monotypic lineages
The majority of the segmental lineages are either monotypic, like lineages 2 and 10, or make a few of one neuron type and an abundance of the other, such as lineages 4 and 14. The Notch data suggest that in these cases, one hemilineage survives while most or all of the cells of the other hemilineage either die or become another cell type, such as glia, which are not revealed by the elav driver. We assessed the role of cell death in sculpting the composition of these lineages by generating MARCM clones that lacked the initiator caspase, dronc (Nedd2-like caspase — FlyBase) (Kondo et al., 2006). As summarized in Fig. 5A-E, blocking cell death resulted in a striking increase in the numbers of the rare neuron types in lineages 0, 4, 14, 20/22 and 21. In addition, in most of the purely monotypic lineages, we saw the appearance of neurons of novel morphology (Fig. 5G-L). In lineage 2, the new cells also projected to the dorsal ipsilateral neuropil, but to a more lateral extent than the normal sibling (Fig. 5H). In the other lineages, the new cells differed markedly from those seen in the wild-type lineage. For the two motor lineages, the new cells were interneurons (Fig. 5G,K). By contrast, with the blockade of cell death, lineage 5 now contained motoneurons as well as its normal type of interneuron (Fig. 5F). We could not resolve the effects of cell death blockade in lineages 7, 16 and 18, but for the remainder (lineages 10, 17 and 23) a new class of interneurons appeared. Taking these results together with the Notch data, we conclude that for the monotypic lineages one sibling consistently lives, resulting in a single hemilineage.
Fig. 5.
Examples of MARCM NB clones that are null for the caspase dronc. (A-E) Lineages in which one cell type is typically represented by only one or two individuals. Blocking cell death results in the addition of more neurites from the cells of the rare phenotype (red arrow). (A) Lineage 0, (B) lineage 4, (C) lineage 14, (D) two examples of lineage 21, (E) lineage 20/22. (F-L) Monotypic lineages having one neurite bundle (yellow arrow) acquire a new neuronal class (red arrow) when cell death is blocked. (F) Lineage 5, (G) lineage 15, (H) lineage 2, (I) lineage 10, (J) lineage 17, (K) lineage 24, (L) lineage 23. Wild-type examples are included in Fig. 9.
Numb loss-of-function and neuronal fates
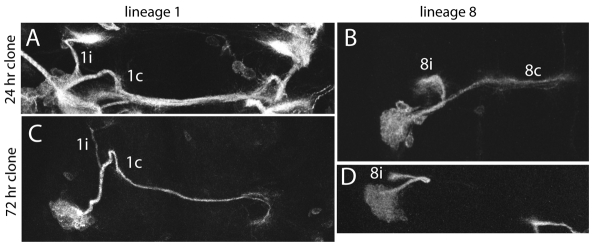
A way to bias the cells to activate Notch signaling is through the loss of numb function (Knoblich et al., 1995; Karacavich and Doe, 2005; Spana and Doe, 1996; Frise et al., 1996; Guo et al., 1996). However, unlike the situation reported for the embryo (Skeath and Doe, 1998), we found that the NBs were refractory to the loss of Numb during early larval growth. For clones induced soon after hatching and then examined in wandering larvae, we typically saw that both types of siblings were present (Fig. 6). Comparison of bundle size between the two siblings was problematic, because in many cases the loss of Numb resulted in a duplication of the NB, and this often resulted in the disproportionate production of ‘A’ siblings (see below).
Fig. 6.
Examples of MARCM NB clones that are homozygous for a numb null mutation (numb2). (A,B) Bundles from both sibling types are evident in numb clones induced at 24 hours AEL and examined at wandering. The lineage 1 clone (A) was among clones from other lineages, but the 1i and 1c bundles are readily distinguishable. (C,D) numb clones induced at 72 hours AEL and examined at wandering showed only the A sibling in lineage 8 (D), or only a few axons of the B type (bundle 1i) and the remainder of the A type (bundle 1c) in lineage 1 (C). Neurite bundle names are as in Fig. 2.
We also generated a smaller set of clones at the start of the third instar, inducing the clones by heat shock at 72 hours after egg laying (AEL) and examining the morphology of the resulting clones at wandering. Lineages such as 1, 8 and 11 now showed clones that responded as expected, with the A phenotype being the sole (Fig. 6D) or predominant (Fig. 6C) phenotype in the clone.
Notch and neuroblast duplication
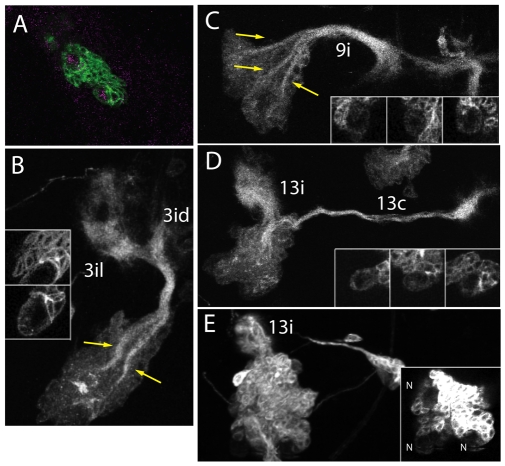
Studies of the Drosophila brain have shown that establishing constitutive Notch activity, either through loss of numb or by expression of Notchintra, results in the generation of supernumerary NBs (Bowman et al., 2008). We found that this phenomenon also occurs in the ventral CNS but on a reduced scale (Fig. 7). The extra NBs in MARCM clones were identified by their large size and their expression of grainy head (Fig. 7A), a marker for NBs (Almeida and Bray, 2005). For NB clones that were induced around hatching and were null for Numb, 35% (n=324) showed supernumerary NBs when examined late in the wandering stage. Unlike the massive increase in NBs seen in some brain lineages, the increase in NBs in the ventral CNS was modest after numb removal, with numbers of extra stem cells ranging from one to eight. When multiple NBs were present, each was typically associated with a set of young neurons with neurites that showed the projection and targeting that were typical of neurons generated at that site. The size and geometry of the cell cluster associated with each NB suggested that the generation of a supernumerary NB typically occurred early in the neurogenic period. For example, Fig. 7B,C show clones for lineages 3 and 9, which have two and three NBs, respectively. Each NB is associated with a separate cluster of progeny and the fasciculated bundles from each cluster join at the base of the clone where they enter the neuropil. Such large and relatively equivalent clusters could only arise if the NB replication occurred early in larval life. The conclusion that NB duplication typically occurs early in postembryonic neurogenesis is also supported by the results of inducing numb clones at 72 hours AEL. We generated only a limited number of such clones, but these included examples for lineages, 8, 9, 11, 16, 19 and 20/22. For early-induced clones in these lineages, 82% (n=67) showed supernumerary NBs, whereas only 9% (n=11) of the late-induced clones had extra NBs. Overall, only 6% (n=34) of the 72-hour clones produced extra NBs.
Fig. 7.
Effects of Notch activation on NB duplication. MARCM NB clones showing that loss of numb (A-D) or constitutive Notch signaling (E) results in clusters with multiple NBs. (A) Optical section through a clone (green) showing the expression of grainy head (magenta) in the supernumerary NBs. (B,C) Examples of lineages 3 and 9, which have supernumerary NBs (insets); each NB is at the end of a discrete cell cluster (arrows) and their fasciculated neurites join at the neuropil into a common bundle that projects to their normal targets. (D,E) Examples of lineage 13 clones that are numb null or express NotchCA, respectively. Both treatments induced supernumerary NBs (insets). With loss of numb (D), the 13c bundle was of normal size, whereas 13i was hypertrophied. With NotchCA expression (E), bundle 13i was hypertrophied but 13c was missing. N, neuroblast.
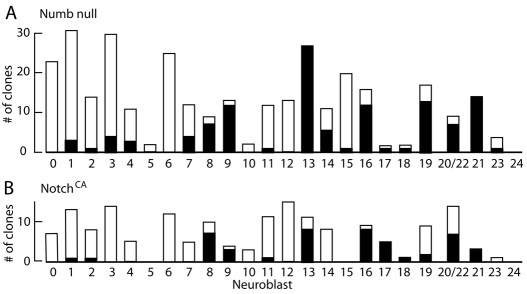
An intriguing feature of the loss of numb function was that the thoracic NBs differed markedly in their response to this loss. For example, as seen Fig. 8A, lineages 0, 6 and 12 rarely, if ever, showed a supernumerary NB, whereas lineages 9, 13 and 16 consistently produced extra NBs. The pattern of extra NBs seen after loss of numb was similar to that seen when the same set of NBs was examined for their response to expression of NotchCA (Fig. 8B). Again, lineages 0, 6 and 12 were unaffected, while 9, 13 and 16 showed multiple NBs. A lineage that responded differently to the two treatments was lineage 19, which often showed supernumerary NBs after loss of numb, but did not do so in seven NB clones that expressed NotchCA.
Fig. 8.
Manipulations that cause NB duplication. Summary of the analysis of NB MARCM clones that were (A) null for numb function or had (B) constitutive Notch signaling. White bars, clones with a single NB; black bars, clones with supernumerary NBs.
DISCUSSION
The role of Notch signaling in establishing sibling difference in secondary neurons and the ‘hemilineage’
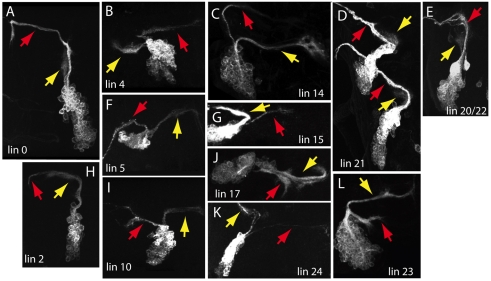
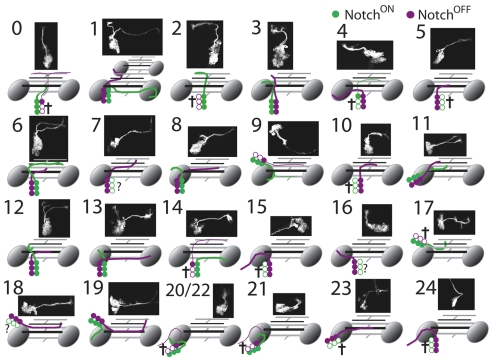
The great diversity of cell types within the nervous system has been appreciated since the studies by Cajal. Understanding the rules that are used for generating such diversity remains one of the major goals of neurodevelopment, and the insect CNS has contributed significantly to understanding this problem. With the possible exception of the optic lobes, the generation of central neurons is strictly a lineage-related process, with no regulation of cell fates seen between lineages (Taghert et al., 1984; Witten and Truman, 1991). In the embryo, the neurons arising from the division of the GMC typically differ from each other (e.g. Skeath and Doe, 1998), but for mushroom body neurons born during larval life, the two siblings are indistinguishable (Lee et al., 1999). The mushroom body pattern, however, is quite different from that inferred from studies in the ventral nervous system of Manduca (Witten and Truman, 1991) and the grasshopper (Jia and Seigler, 2002), which indicate that the two siblings assume different fates. The data in this paper and the study on the antennal lineages by Lin et al. (Lin et al., 2010) indicate that the mushroom body pattern is atypical. As seen in Fig. 9, the pattern across the 25 thoracic lineages is for the GMC to produce two different daughters. In lineages that are monotypic, a situation seemingly similar to that seen in the mushroom body, one of the siblings is consistently removed by programmed cell death soon after its birth.
Fig. 9.
Summary of the role of Notch signaling in the 25 lineages of secondary neurons in the ventral CNS. The images show the wild-type morphology of each lineage. The diagrams shows the path of the neurite bundle from each hemilineage (see Fig. 1). Green, Notch-on fate; magenta, Notch-off fate. Open cell bodies show cell types that do not appear in the wild-type clones. Ones that are known to undergo programmed cell death are designated by a cross. When only one cell is filled, the oldest few neurons in the hemilineage survive while the remainder die.
Previous studies (Almeida and Bray, 2005) showed that Notch was not needed for the maintenance and division of NBs during the larval neurogenic period. Our data are completely consistent with their findings. Notch also seems dispensable for the early differentiation and survival of the young neurons. With Notch loss-of-function, we sometimes saw an NB with a cluster of immature cells but no neurites from maturing neurons exiting from the cluster, but this was only seen in lineages in which the B (NotchOFF) sibling normally dies. Similarly, expressing NotchCA also resulted in ‘disembodied’ NBs and cell clusters, but only in the lineages in which the A (NotchON) sibling is fated for death. These disembodied clusters were especially impressive in lineages, such as lineage 16, that also showed supernumerary NBs due to constitutive Notch activation. Overall, though, these data and those from the loss of the initiator caspase Dronc, show that the abnormal death seen with Notch manipulation is a result of the role of Notch in determining cell fate, rather than a requirement for Notch for survival or early differentiation.
An interesting question is whether there are global rules for fate determination that apply across the lineages. In monotypic or almost monotypic lineages, there is no consistent relationship of the dominant sibling to the state of Notch signaling. In seven of the monotypic thoracic lineages the ‘A’ sibling is the dominant surviving cell type, whereas in nine lineages, the ‘B’ sibling is the dominant cell type. Axonal projection, however, does correlate strongly with whether or not the Notch pathway is activated. Only four bundles (6ci, 7c, 18c and 19c) project into the longitudinal tracts, and these are the B siblings of their respective lineages. Five lineages produce motoneurons (from NBs 15, 20, 21, 22 and 24) and they also represent the ‘B’ fate. In addition, six more lineages [0, 4, 8, 12, 13 and 19] have one sibling that has a local primary target, whereas the other sibling projects to the periphery or across a commissure to the contralateral side of the CNS. Lineage 4 is the only one of this group in which the B sibling stays within its hemineuropil, whereas the A sibling projects across a commissure. The remaining ten lineages are uninformative because they are situations like that in lineage 3, in which both siblings stay local, or lineage 1, in which both siblings are projection cells. These last examples notwithstanding, there is a strong bias for the A (NotchON) sibling to stay local and for the B (NotchOFF) sibling to project to distant targets. Notch signaling works through the Suppressor of hairless [Su(h)] transcription factor (Bailey and Posakony, 1995), so one might suspect that targets downstream of Su(h) might promote features of local interneurons and suppress projection neuron characteristics.
Although the role of Notch in establishing sibling identity is consistent across the lineages and through time, that of numb is not. During embryonic neurogenesis, the loss of numb function results in constitutive activation of Notch and the production of only A siblings (Skeath and Doe, 1998). Early in the postembryonic period, however, we found that the GMCs produce daughters of both fates despite the loss of numb function (Fig. 6A,B), but by the start of the third instar numb becomes essential for directing Notch activity (Fig. 6C,D). Therefore, early in the secondary phase of neurogenesis, Notch signaling is not dependent on numb, and other factors must be at play to allow Notch to establish the differences in sibling identity. We do not know the nature of these factors.
It may be important that the lack of a requirement for numb is correlated with rate of division of the NB. In the embryo (Campos-Ortega and Hartenstein, 1997) and in the third larval instar (Truman and Bate, 1988) the cell cycle of the NB is less than an hour. In the latter case, we see two or more neurons in a given cluster showing nuclear-localized Notch (Fig. 1C,D) suggesting that siblings from successive GMCs undergo fate decisions in an overlapping manner. Using Numb protein at this time to bias sibling identity would ensure that a given sibling would not be given ambiguous signals from a cousin. When the NBs first reactivate in the second instar, however, they are dividing much more slowly (Truman and Bate, 1988). Assuming that the dynamics of GMC lifespan are similar to those seen later, we suspect that at these earlier times only one sibling pair in a cluster may be undergoing Notch-dependent decisions at a time. This would permit the types of cell-cell interactions, such as seen in some peripheral sensory precursor cells (Hartenstein and Posakony, 1989) or postulated for grasshopper lineages (Doe et al., 1985), to come into play. The biasing of cells by Numb may be an adaptation for rapid cell cycles when multiple neuron pairs are sensitive at the same time.
Thoracic NBs show similarities to the PAN neuroblasts
Recently, it has been shown that some NBs, the posterior asense-negative (PAN) NBs, differ from the classic scheme in that their GMCs undergo additional divisions before making postmitotic neurons (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). PAN neuroblasts respond dramatically to enhanced Notch signaling; clones that either express NotchCA or are numb negative show a dramatic expansion in the number of NBs in a given cluster, as some of the GMCs transform into NBs (Bowman et al., 2008). In contrast to the PAN NBs, ‘classic’ brain NBs are unaffected by enhanced Notch activity (Bowman et al., 2008).
We also find a dichotomy in how the NBs in the ventral CNS respond to the loss of Numb or expression of NotchCA. Most NBs and GMCs characteristically maintain their normal pattern of division despite constitutive Notch activation (Fig. 7). A few lineages (NBs 8, 9, 13, 16, 17, 19 and the 20s), however, resemble the PAN lineages of the brain in that they respond to constitutive Notch signaling by generating multiple NBs. Their responses are more muted than the brain NBs, however, with only a few extra NBs being generated in a given cluster.
As also seen for an antennal lobe lineage by Lin et al. (Lin et al., 2010), the sensitivity to constitutive Notch signaling is greatest early in the postembryonic life of the NB. This transient sensitivity to Notch activation suggests that some thoracic NBs may have PAN neuroblast characteristics early after their reactivation but later establish a traditional mode of behavior for the remainder of their lineage. We have not, however, found any GMC clones with more than two siblings, which would be expected if some thoracic lineages made a few ‘transiently amplifying’ progeny. We have not systematically induced clones through the early period of larval neurogenesis, however, so we cannot exclude the possibility that a few transiently amplifying GMCs are produced among the earlier GMCs in these lineages.
The production of supernumerary NBs has also been seen in caterpillars of Manduca sexta, after treatment with hydroxyurea (Truman and Booker, 1986; Witten and Truman, 1991), a drug that blocks nucleotide reductase (Timson, 1975). Although the drug treatment was devised to kill cycling NBs, we found that there was a brief window at the start of postembryonic neurogenesis when drug treatment caused some NBs to duplicate, rather than die, and resulted in twice as many neurons of the appropriate phenotype. It may be that some NBs in moths may also have PAN neuroblast characteristics early in larval life and that hydroxyurea causes a transiently amplifying precursor to cross a line that changes it into a fully fledged NB.
It is interesting that the lineages that are the most sensitive (i.e. show extra NBs in over 50% of the clones) to either Numb loss or Notch activation are ones that supply local interneurons to the leg neuropil (lineages 8, 9, 13, 14, 16, 19, 20, 21 and 22). Providing PAN neuroblast characteristics to these NBs may be a strategy to enhance the cell number or cellular diversity in this highly integrative region of the CNS.
Conclusions
In this paper we present a comprehensive analysis of the role of Notch signaling in generating neuronal phenotypes within the secondary lineages of the thoracic ventral CNS. The universal pattern is for a GMC to produce two neurons of different phenotypes, A and B, with cell death involved in making some lineages monotypic. A clear division of labor between these A and B cell types suggest that the components of circuitry of the thoracic nervous system are generated in developmental units we term ‘hemilineages’. We believe that viewing the construction of this part of the nervous system through the lens of the hemilineage will allow us to gain insights into the way by which the genome generates units of connectivity and how these are constructed into circuits underlying behavior.
Acknowledgements
We are grateful to S. Bray, S. Artavanis-Tsakonas, R. Fehon, J. Hirsch, Y. Hiromi, S. Kondo, M. Piovant and H. Ruohola-Baker for fly strains and for antibodies. Work was supported by NIH Grant NS13079 and by the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
References
- Almeida M. S., Bray S. J. (2005). Regulation of post-embryonic neurogenesis by Drosophila Grainyhead. Mech. Dev. 1212, 1282-1293 [DOI] [PubMed] [Google Scholar]
- Baek M., Mann R. S. (2009). Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. J. Neurosci. 29, 6904-6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A. M., Posakony J. W. (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609-2622 [DOI] [PubMed] [Google Scholar]
- Bello B. C., Izergina N., Caussinus E., Reichert H. (2008). Amplification of neuronal stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J. Q., Doe C. Q. (2008). Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185-1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T., Udolph G., Doe C. Q., Technau G. M. (1996). The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 179, 41-64 [DOI] [PubMed] [Google Scholar]
- Bowman S. K., Rolland V., Betschinger J., Kinsey K. A., Emery G., Knoblich J. A. (2008). The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535-546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley D. J., Blanc E., Reddy O. V., Vijayraghavan K., Williams D. W. (2009). Dendritic targeting in the leg neuropil of Drosophila: the role of midline signalling molecules in generating a myotopic map. PLoS Biol. 7, e1000199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J. (2009). Making a grade: Sonic Hedgehog signalling and the control of neural cell fate. EMBO J. 28, 457-465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H., Truman J. W. (2009). Fine-tuning of secondary arbor development: the effects of the ecdysone receptor, EcR, on the adult neuronal lineages of the Drosophila thoracic CNS. Development 136, 3247-3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega J. A., Hartenstein V. (1997). The Embryonic Development of Drosophila melanogaster 2nd edn, pp. 405 Berlin: Springer-Verlag; [Google Scholar]
- Cayouette M., Poggi L., Harris W. A. (2006). Lineage in the vertebrate retina. Trends Neurosci. 29, 563-570 [DOI] [PubMed] [Google Scholar]
- de la Escalera S., Bockamp E. O., Moya F., Piovant M., Jimenez F. (1990). Characterization and gene cloning of neurotactin, a Drosophila transmembrane protein related to cholinesterases. EMBO J. 9, 3593-3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A. R., McConnell S. K. (2000). Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development 127, 2863-2872 [DOI] [PubMed] [Google Scholar]
- Doe C. Q. (2008). Neural stem cells: balancing self-renewal with differentiation. Development 135, 1575-1587 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Kuwada J. Y., Goodman C. S. (1985). From epithelium to neuroblasts to neurons: the role of cell interactions and cell lineage during insect neurogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 312, 67-81 [DOI] [PubMed] [Google Scholar]
- Edlund T., Jessell T. M. (1999). Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell 96, 211-224 [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., Muskavitch M. A., Artavanis-Tsakonas S. (1990). Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61, 523-534 [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Johansen K., Rebay I., Artavanis-Tsakonas S. (1991). Complex cellular and subcellular regulation of notch expression during embryonic and imaginal development of Drosophila: implications for notch function. J. Cell Biol. 113, 657-669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frise E., Knoblich J. A., Younger-Shepherd S., Jan L. Y., Jan Y. N. (1996). The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interactions in sensory organ lineage. Proc. Natl. Acad. Sci. USA 93, 11925-11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R., Pearson B. J., Marusich A., Doe C. Q. (2005). Regulation of temporal identity transitions in Drosophila neuroblasts. Dev. Cell 8, 193-202 [DOI] [PubMed] [Google Scholar]
- Guo M., Jan L. Y., Jan Y. N. (1996). Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27-41 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Posakony J. W. (1989). Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development 107, 389-405 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Spindler S., Pereanu W., Fung S. (2008). The development of the Drosophila larval brain. Adv. Exp. Med. Biol. 628, 1-31 [DOI] [PubMed] [Google Scholar]
- Heitzler P., Simpson P. (1991). The choice of cell fate in the epidermis of Drosophila. Cell 64, 1083-1092 [DOI] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S., Doe C. Q. (2001). Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511-521 [DOI] [PubMed] [Google Scholar]
- Jia X. X., Siegler M. V. S. (2002). Midline lineages in grasshopper produce neuronal siblings with asymmetric expression of Engrailed. Development 129, 5181-5193 [DOI] [PubMed] [Google Scholar]
- Kambadur R., Koizumi K., Stivers C., Nagle J., Poole S. J., Odenwald W. F. (1998). Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 12, 246-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcavich R., Doe C. Q. (2005). Drosophila neuroblast 7-3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J. Comp. Neurol. 481, 240-251 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A., Jan L. Y., Jan Y. N. (1995). Asymmetric segregation of Numb and Prospero during cell division. Nature 377, 624-627 [DOI] [PubMed] [Google Scholar]
- Kondo S., Senoo-Matsuda N., Hiromi Y., Miura M. (2006). DRONC coordinates cell death and compensatory proliferation. Mol. Cell. Biol. 26, 7258-7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. K., Holder K., Yost C., Giniger E., Ruohola-Baker H. (1996). Expression of constitutively active Notch arrests follicle cells at a precursor stage during Drosophila oogenesis and disrupts the anterior-posterior axis of the oocyte. Development 122, 3639-3650 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451-461 [DOI] [PubMed] [Google Scholar]
- Lee T., Lee A., Luo L. (1999). Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126, 4065-4076 [DOI] [PubMed] [Google Scholar]
- Lin D. M., Goodman C. S. (1994). Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron 13, 507-523 [DOI] [PubMed] [Google Scholar]
- Lin S., Lai S.-L., Yu H.-H., Chihara T., Luo L., Lee T. (2010). Lineage-specific effects of Notch/Numb signaling in postembryonic development of Drosophila brain. Development 137, 43-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W., Hartenstein V. (2006). Neural lineages of the Drosophila brain: a three dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci. 26, 5534-5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A., Chiba A., Doe C. Q. (1999). Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126, 4653-4689 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Rickert C., Bossing T., Vef O., Urban J., Technau G. M. (1997). The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev. Biol. 189, 186-204 [DOI] [PubMed] [Google Scholar]
- Skeath J. B., Doe C. Q. (1998). Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125, 1857-1865 [DOI] [PubMed] [Google Scholar]
- Spana E. P., Doe C. Q. (1996). Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21-26 [DOI] [PubMed] [Google Scholar]
- Struhl G., Fitzgerald K., Greenwald I. (1993). Intrinsic activity of the lin-12 and Notch intracellular domains in vivo. Cell 74, 331-345 [DOI] [PubMed] [Google Scholar]
- Taghert P. H., Doe C. Q., Goodman C. S. (1984). Cell determination and regulation during development of neuroblasts and neurones in grasshopper embryo. Nature 307, 163-165 [DOI] [PubMed] [Google Scholar]
- Timson J. (1975). Hydroxyurea. Mutation Res. 32, 115-132 [DOI] [PubMed] [Google Scholar]
- Truman J. W., Booker R. (1986). Adult-specific neurons in the nervous system of the moth, Manduca sexta: selective chemical ablation using hydroxyurea. J. Neurobiol. 17, 613-625 [DOI] [PubMed] [Google Scholar]
- Truman J. W., Bate M. (1988). Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 125, 145-157 [DOI] [PubMed] [Google Scholar]
- Truman J. W., Schuppe H., Shepherd D., Williams D. W. (2004). Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development 131, 5167-5184 [DOI] [PubMed] [Google Scholar]
- Williams D. W., Kondo S., Krzyzanowska A., Hiromi Y., Truman J. W. (2006). Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9, 1234-1236 [DOI] [PubMed] [Google Scholar]
- Witten J. L., Truman J. W. (1991). The regulation of transmitter expression in postembryonic lineages in the moth Manduca sexta. II. Role of cell lineage and birth order. J. Neurosci. 11, 1990-1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Williams D. W., Altman J., Riddiford L. M., Truman J. W. (2009). Temporal patterns of Broad isoform expression during the generation of neuronal lineages in Drosophila. Neural Dev. 4, 39 [DOI] [PMC free article] [PubMed] [Google Scholar]