Abstract
Cis-regulatory modules are non-protein-coding regions of DNA essential for the control of gene expression. One class of regulatory modules is embryonic enhancers, which drive gene expression during development as a result of transcription factor protein binding at the enhancer sequences. Recent comparative studies have begun to investigate the evolution of the sequence architecture within enhancers. These analyses are illuminating the way that developmental biologists think about enhancers by revealing their molecular mechanism of function.
Introduction
In order to form the tissues of the embryo, the expression patterns of genes must be tightly directed in space and time. This function is largely controlled by cis-regulatory modules (CRMs), specific regions of non-protein coding DNA that regulate genes on the same chromosome. Specifically, early patterns of gene expression regulated by CRMs are required to establish the anterior to posterior axis of the developing embryo. The best-understood CRM is the enhancer, a genetic switch that is bound by specific transcription factors (TFs) and is able to drive distinct patterns of transcription of a target gene in an orientation-independent manner (Dillon and Sabbattini, 2000; Ptashne, 1986) (see Box 1). The binding of TFs to enhancers recruits RNA polymerase II and the associated transcriptional machinery to a gene's promoter to drive the transcription of target genes. Notably, some of the components of the machinery, such as histone acetyltransferases and nucleosome remodelers, have the ability to modify the chromatin environment in order to facilitate transcription (Kim and Dean, 2003; Madisen et al., 1998; Soutoglou and Talianidis, 2002; Utley et al., 1998).
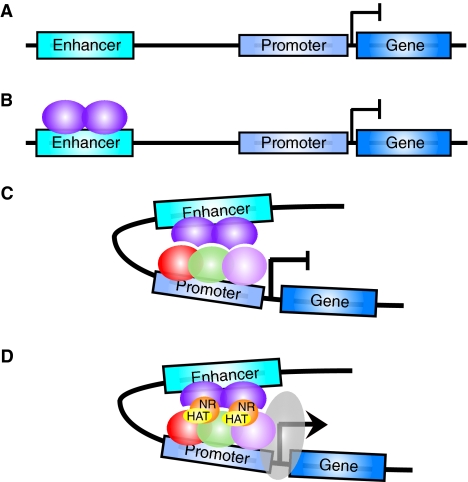
Box 1. Defining a functional enhancer cis-regulatory module
An enhancer is an orientation-independent region of non-protein-coding DNA in the genome that is associated with a promoter and target gene (A). Transcription factor (TF) proteins (purple spheres) bind to the enhancer (B) and interact with the transcriptional machinery (red, green and pink spheres) (C) and eventually recruit RNA polymerase II (gray ellipse) at the promoter of target genes to enhance the transcription of the gene (D). The TFs bound at an enhancer often also recruit chromatin-modifying enzymes such as histone acetyltransferases (HAT, yellow ellipses) and nucleosome remodeling factors (NR, orange spheres) that facilitate changes to the chromatin environment to allow transcription to proceed. Historically, enhancers were often initially identified in genetic screens, as mutation of the enhancer module disrupts TF binding and results in an associated loss of target gene expression. Later, the development of transposon-mediated mutagenesis in Drosophila and plants allowed more sophisticated ‘enhancer trap’ reporter gene constructs to be used. Enhancers identified in these types of genetic screens include those in the bithorax complex of D. melanogaster responsible for regulating the Abdominal B gene (Celniker et al., 1990; Karch et al., 1985) [for a detailed review on enhancer trapping see Bellen (Bellen, 1999)]. The functional sequences of the enhancer module are often resolved in detailed transgenic reporter gene studies [for early resolution of the eve stripe enhancers see papers by Goto and Harding (Goto et al., 1989; Harding et al., 1989)]. More recently, global genome-wide approaches to identifying TF binding sites, such as by chromatin immunoprecipitation combined with a tiled genomic microarray (ChIP on chip) (Visel et al., 2009; Zeitlinger et al., 2007) or bioinformatic analysis using comparative genomics to identify genomic regions with potential cis-regulatory function (Nobrega et al., 2003; Peterson et al., 2009), have been successful at identifying an increasing number of enhancers.
For many years, researchers have sought to identify these modules and their respective target genes. However, recent interest has shifted towards a more complex analysis of individual enhancers in order to elucidate their molecular activity. Key questions include: which TF binding sites are responsible for functional activity; how these binding sites are organized; and what their affinities for different TFs are. The wealth of information that is emerging from the recent sequencing of many different genomes has provided answers to some of these questions, but has also brought controversy to the field. Particularly in insects, as we discuss in this Primer, evolutionary comparisons of enhancers across different species are beginning to show us that slight variations in non-coding regulatory sequences can lead to large phenotypic differences in development (Gompel et al., 2005; Jeong et al., 2006; Prud'homme et al., 2006). Such detailed analyses of enhancer evolution allow for a better understanding of the organizational and structural constraints of a CRM. In turn, the elucidation of such evolutionary constraints should reveal the nature of TF interactions with regulatory DNA sequences, as well as what is required for the transcriptional control of specific genes during development.
In this Primer, we briefly review our current understanding of classical enhancer function and then delve into recent studies that have compared enhancers in Drosophila and related insect species to reveal how evolution acts upon mechanisms of enhancer function. We focus on the even skipped stripe 2 enhancer (S2E) and neurogenic ectoderm enhancers (NEEs) in particular, owing to the lessons that these CRMs can teach us about the general principles of transcriptional control during embryonic development.
The building blocks of gene regulation: the functional characteristics of enhancers
Enhancers were first identified in the SV40 viral genome as regions of non-coding DNA that are crucial for the transcription of adjacent genes (Banerji et al., 1981; Benoist and Chambon, 1981). Functional activity of an enhancer depends upon the binding of specific TF proteins (activators, see Box 2) within the enhancer DNA sequence that help to recruit RNA polymerase II and associated protein factors to the promoter of a target gene (detailed in Box 1) (McEwan et al., 1993; Wildeman et al., 1984). In the years after these initial observations, enhancers were discovered for many genes in a wide range of model organisms (Banerji et al., 1983; Shepherd et al., 1985; Struhl, 1984). In complex eukaryotes, many of the identified enhancers are responsible for directing spatiotemporally restricted patterns of gene expression in the developing embryo (Choi and Engel, 1986; Hiromi et al., 1985). Although a number of enhancers are able to activate transcription at a given eukaryotic gene, the regulation of key genes by a restricted set of tissue-specific enhancers during development must be very carefully controlled because they encode proteins that specify cellular identities in the embryo (Caplan and Ordahl, 1978; Lewis, 1978). These crucial developmental genes cannot, therefore, be expressed ubiquitously. A number of emerging studies indicate that these specific promoter-enhancer interactions are subject to subtle molecular control mechanisms, including the activity of a novel class of regulatory module — the promoter tethering element (Akbari et al., 2008; Akbari et al., 2007; Calhoun et al., 2002; Cande et al., 2009; Fujioka et al., 2009; Kwon et al., 2009).
Box 2. Glossary of specialized terms
Activator. A transcription factor that, when bound within a particular enhancer, is able to recruit the transcriptional machinery to upregulate transcription of a target gene under control of the enhancer.
Cluster. A group of closely spaced transcription factor binding sites thought to be the hub of enhancer activity. Clusters usually include binding sites for both activators and repressors.
Eve stripe 2 enhancer (S2E). An extensively studied embryonic enhancer of the even skipped gene.
Neurogenic ectodermal enhancer (NEE). A specific type of enhancer that directs gene expression in the neurogenic ectoderm. Spatial and temporal expression of NEE-driven genes depends upon the activators Dorsal and Twist, and the repressor Snail.
Orthologous gene. A gene derived from the gene of a common ancestor.
PATSER. A pattern search program that identifies potential transcription factor binding sites in a given DNA sequence using a position-weighted matrix.
Position-weighted matrix (PWM). Generated from a compilation of experimentally verified binding site sequences for a specific transcription factor (TF), it is a mathematical matrix that indicates the probability of finding a specific nucleotide at each position in the binding site.
Repressor. A TF that, when bound within a particular enhancer, blocks transcription of the target gene.
Transcription factor binding site (TFBS). A sequence of DNA known to be bound by a specific TF protein that influences transcription of nearby target genes.
Two-dimensional similarity plots. A graphical method that identifies regions of similarity between two sequences.
In addition to activators, another class of regulatory TFs, repressors (see Box 2), are able to bind directly to enhancers, in this case to prevent target gene transcription (Dearolf et al., 1989; Stanojevic et al., 1989). Although the molecular roles of activators and repressors in promoting or preventing the recruitment of RNA polymerase II to gene promoters are not completely understood, some mechanisms for the functional activity of enhancers have been characterized. The predominant theory is that activators directly interact with components of the basal transcription machinery via specific protein domains, such as glutamine-rich, proline-rich and acidic domains or hydrophobic β sheets (McEwan et al., 1993); for a detailed review, see Kadonaga (Kadonaga, 2004). There is also extensive evidence that sequence-specific binding of activator TFs to individual enhancer CRMs leads to the recruitment, via protein interactions with co-activator enzymes, of an extensive number of components of the RNA polymerase II transcriptional machinery, including the Mediator complex. The Mediator complex is highly conserved in eukaryotes from yeast to humans, although its individual components can vary when assembled at individual enhancers (Kadonaga, 2004). The Mediator complex facilitates interactions between the enhancer and chromatin-modifying enzymes, such as histone acetyltransferases and nucleosome remodeling factors, to establish a chromatin environment that facilitates transcription of the target gene (see Box 1). By contrast, repressors may disrupt enhancer functional activity in one of two ways: (1) competition, where the binding sites for repressors and activators within an enhancer sequence overlap and, as a result, repressor binding excludes activators; and (2) quenching, in which repressors are able to inhibit the regulatory activity of activators bound to nearby sites within the enhancer (Gray et al., 1994; Kirchhamer et al., 1996; Levine and Manley, 1989; Small et al., 1991b).
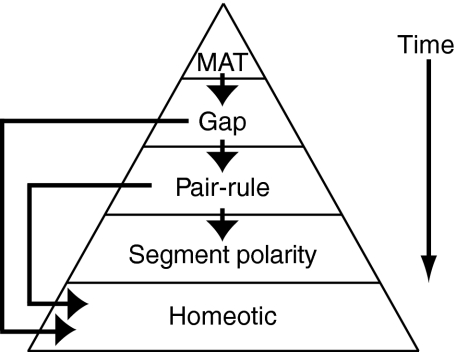
In metazoan species, enhancer CRMs are major players in the developmental cascade that transforms the early embryo from a mass of uniform, undifferentiated cells into a segmented and highly organized structure of differentiated cells. In Drosophila, the dynamic developmental specification of the embryonic body plan and of differentiated cell fates is accomplished by a combination of early spatiotemporal expression gradients of activator and repressor TFs that act upon downstream embryonic enhancers. Based on the timing of their expression during embryonic development, the genes in this cascade represent four basic developmental TF families: maternal, gap, pair-rule and homeotic genes (described in detail in Box 3) (for a review, see Sauer et al., 1996). At the top of the cascade, maternal mRNAs are deposited in the unfertilized egg cell during oogenesis (Berleth et al., 1988; Steward et al., 1988). Spatially restricted translation of localized maternal mRNAs in the fertilized egg establishes TF gradients in the embryo. In turn, maternal TFs bind at target embryonic enhancers for gap genes, directing gap TF expression patterns in the developing embryo (Driever and Nüsslein-Volhard, 1988; Struhl et al., 1989) (Fig. 1A). Gap TFs further regulate downstream target genes, such as those for pair-rule and homeotic TFs (Qian et al., 1991; Stanojevic et al., 1989). At each step in the cascade, gene expression patterns are controlled by specific clusters of activator and repressor binding sites within embryonic enhancers (see Box 2). Fine-tuning of transcription is mediated by the specific molecular properties of individual enhancer CRMs. Whether a given TF acts as an activator or repressor when it binds to an embryonic enhancer can be context dependent (Ip et al., 1991; Small et al., 1996). In addition, DNA sequences within embryonic enhancers may bind TFs with varying affinities (Jiang et al., 1991; Struhl et al., 1989). The functional consequence is that enhancers may require different threshold concentrations of interacting activators and repressors in order to regulate transcription of their target genes. The central role of enhancers in the regulatory cascade responsible for development of the Drosophila embryo makes in-depth analysis of their function crucial to the field of developmental biology.
Box 3. Key players in anterioposterior patterning in early Drosophila development
Early in Drosophila embryonic development, the initial anterior-to-posterior morphogen gradient comprises maternally deposited (MAT) mRNAs in the unfertilized egg, including bicoid and hunchback. Translation produces the MAT transcription factors (TFs), which can activate or repress spatiotemporally restricted patterns of expression of gap genes, such as Kruppel, knirps and giant. In turn, these gap TFs regulate downstream expression of pair-rule genes, such as eve, in stripes in the developing Drosophila embryo. Pair-rule TFs regulate segment polarity genes further downstream in the cascade. In addition, gap and pair-rule TFs both regulate the expression of the homeotic genes, which ultimately define segmental identity.
Maternal genes
Bicoid (BCD). The unfertilized egg contains a broad, maternally deposited distribution of bicoid mRNA, which is transported to the anterior pole of the developing embryo prior to translation. After translation, BCD protein diffuses from a high concentration at the anterior end across the embryo and acts as an activator of hunchback.
Hunchback (HB). The hunchback gene is activated by BCD in the anterior half of the embryo. Maternally deposited hunchback mRNAs also help to produce this protein in the embryo. HB protein functions as a regulator of other gap genes and as a threshold level-dependent activator or repressor at different enhancers of the pair-rule genes, including even skipped (eve).
Gap genes
Kruppel (KR). The Kruppel gene is repressed by high levels of HB at the anterior end of the embryo, leading to its localization in the middle of the embryo, where HB levels begin to diminish. KR protein functions as a repressor of downstream target genes, including eve.
Knirps (KNI). The knirps gene is repressed by intermediate levels of HB, and thus KNI protein is present in a peak posterior to that of KR. KNI functions as a repressor of eve.
Giant (GT). The giant gene is repressed by low levels of HB, and is therefore only expressed in one anterior and one posterior region of the embryo, where HB is not present. GT protein functions as a repressor of eve.
Pair-rule genes
Even skipped (EVE). The eve gene is expressed in seven stripes in the developing embryo. eve enhancers are activated by BCD and HB, and repressed by HB, KR, KNI and GT. EVE protein establishes segmentation in the embryo.
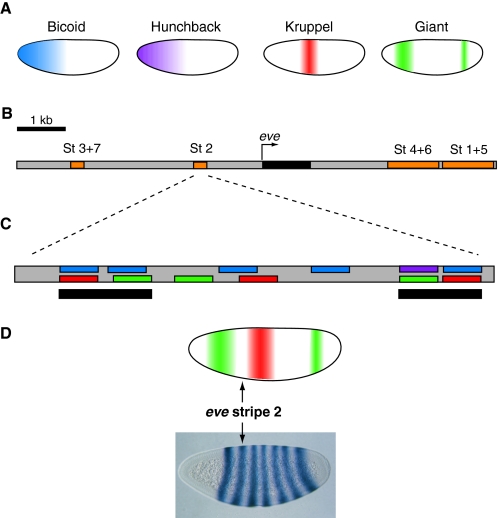
Fig. 1.
Transcription factor gradients activate even skipped expression. (A) The localization patterns of four major transcription factors (TFs) in the early Drosophila embryo are shown. Embryos are oriented anterior to the left and dorsal is up. The Bicoid (blue) and Hunchback (purple) TFs are activators of the even skipped (eve) stripe 2 enhancer, whereas the Kruppel (red) and Giant (green) TFs are repressors. Both activators are broadly expressed in the anterior half of the embryo, but repressor expression is more restricted. (B) Map of the eve genomic locus, including the embryonic enhancers (orange) responsible for eve (black) expression. Each enhancer drives eve expression in one or two developing parasegments of the embryo. Together, the enhancers drive expression in seven stripes (St 1-7) in odd-numbered parasegments. (C) The minimal eve stripe 2 enhancer (St 2) of D. melanogaster contains twelve transcription factor binding sites. The locations of color-coded binding sites for the activators and repressors shown in A are indicated. Two clusters of binding sites (black bars) at each end of the minimal stripe 2 module are thought to be particularly important for functional activity. (D) Sharp borders of eve stripe 2-directed expression are established by high concentrations of Giant (green) at the anterior boundary and Kruppel (red) at the posterior boundary.
A CRM paradigm: even skipped stripe 2 enhancer
Probably the best-characterized embryonic CRM is the even skipped (eve) S2E in Drosophila melanogaster (see Box 2). eve is a pair-rule gene that is expressed under the control of various neighboring CRMs in seven distinct stripes in the developing embryo (Macdonald et al., 1986) (Fig. 1B-D). The pattern of eve expression that S2E directs is entirely derived from the combination of transcription factor binding sites (TFBSs, see Box 2) embedded within its DNA sequence (Small et al., 1991b) (Fig. 1). Pioneering studies in the laboratory of Michael Levine twenty years ago discovered that twelve different binding sites for Kruppel (KR), Giant (GT), Bicoid (BCD) and Hunchback (HB) TFs are located in the minimal S2E, a 480 bp region of the CRM that is sufficient to drive eve stripe 2 expression (Fig. 1C) (Goto et al., 1989; Small et al., 1991b; Stanojevic et al., 1989). Notably, eight of the twelve TFBSs form two tight clusters at each end of the minimal S2E. Each cluster has closely spaced activator binding sites, each of which strongly overlaps with a repressor binding site (Fig. 1C). These observations suggested two important ideas regarding the molecular architecture of an enhancer: (1) clustering of TFBSs is crucial to enhancer function; and (2) the overlap between activator and repressor TFBSs enables the characteristically sharp boundaries between stripes of gene expression that are driven by many early embryonic enhancers (Fig. 1D).
Analysis of the TF gradients present in the developing Drosophila embryo reveals how S2E directs eve expression with such sharp boundaries. Activation of S2E-driven expression of eve occurs through the binding of the HB and BCD activators (see Box 3), which are present at high concentrations in the anterior of the embryo (Fig. 1A). Binding of the KR and GT repressors (see Box 3), which are present in distinct non-overlapping patterns in the anterior half of the embryo, prevents activation of S2E (Small et al., 1991b) and thus delineates the boundaries of the second stripe of eve expression (Fig. 1D). As a result, in the presumptive second abdominal segment (the spatial interval in the embryo where HB and BCD activators are present and KR and GT repressors are absent), HB and BCD are able to occupy activator binding sites in S2E (Small et al., 1991a) (Fig. 1C). With strong activation and no repression, S2E only drives eve expression in a narrow 2- to 3-cell-wide region of the developing embryo (Macdonald et al., 1986) (Fig. 1D). By integrating the study of the spatial distribution of TFs in the embryo, the binding of these TFs at specific enhancers and the resulting patterns of target gene expression, we begin to see how an embryonic enhancer is analogous to a central processing unit. The enhancer must be capable of receiving input signals from TFs and of outputting a response in the form of directing a very specific spatiotemporal pattern of target gene expression.
Evolution of the eve stripe 2 embryonic enhancer
Recent evolutionary analyses of TFBSs within enhancer sequences and their relative rate of evolutionary turnover are beginning to provide a useful perspective on the molecular mechanisms of enhancer function. Early comparative studies in the Kreitman laboratory concentrated on evolutionary analysis of the sequence of S2E across several Drosophila species, including D. melanogaster, D. yakuba, D. erecta and D. pseudoobscura (see Fig. 2A for phylogenetic relationship). Their studies revealed that all of the Drosophila species investigated possess an orthologous early embryonic enhancer (see Box 2) capable of driving eve expression in a similar spatiotemporal stripe 2 pattern when tested in transgenic D. melanogaster (Ludwig et al., 1998). This functional conservation partially extends to chimeric enhancers created by juxtaposing two halves of the S2E, each half taken from a different Drosophila species, which are capable of directing a similar pattern of gene expression in transgenic D. melanogaster (Ludwig et al., 2000). However, the expression directed by the chimeric enhancers is not identical to the activity of the endogenous eve S2E. In some embryos carrying the chimeric enhancers, reporter gene expression undergoes a posterior shift or expansion (Ludwig et al., 2000), suggesting that nuanced enhancer architecture might be encoding important functional information. Bioinformatic analysis revealed that the TFBSs contained within the S2E are not well-conserved between orthologs (Ludwig and Kreitman, 1995; Ludwig et al., 1998). There is, in fact, significant turnover of TFBSs between the Drosophila S2E orthologs studied: of the seventeen known binding sites analyzed in the 798 bp full-length S2E from D. melanogaster, only three are completely conserved at the sequence level in the other three Drosophila species studied (Ludwig et al., 1998) (Fig. 2B). In addition, none of the sixteen binding sites that were surveyed from the full-length D. melanogaster S2E are conserved in all thirteen sequenced Drosophila species (Ludwig et al., 2000). Detailed sequence alignment revealed that binding sites for BCD, HB, KR and GT identified in D. melanogaster are subject to extensive nucleotide substitutions in other Drosophila species (Ludwig et al., 2000; Ludwig et al., 1998). The key question therefore becomes: how is the functional activity of the enhancer preserved at all, despite the evolutionary turnover of TFBSs within the CRM?
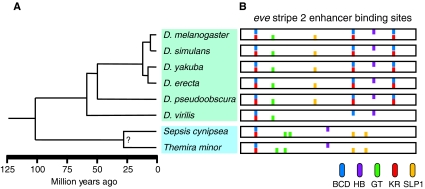
Fig. 2.
Phylogenetic relationship and eve stripe 2 enhancer architecture of Drosophila and Sepsid species. (A) The Drosophila (green box) diverged from the Sepsid family (blue box) approximately 100 million years ago (Mya). The distantly related Drosophila species, D. melanogaster and D. virilis, diverged 60 Mya (Tamura et al., 2004). (B) The organization of a subset of bioinformatically predicted binding sites for the transcription factors Bicoid (BCD, blue), Hunchback (HB, purple), Giant (GT, green), Kruppel (KR, red) and Sloppy paired 1 (SLP1, yellow) within the minimal eve stripe 2 enhancer [summarized from Hare (Hare et al., 2008a)] are shown for eight species: D. melanogaster, D. simulans, D. yakuba, D. erecta, D. pseudoobscura, D. virilis, Sepsis cynipsea and Themira minor.
Some resolution to this intriguing issue is being provided by exciting new studies that have expanded the evolutionary scope of S2E analysis. These studies analyze evolutionarily divergent species outside of the Drosophila genus (Fig. 2). Drosophila species generally have small, compacted genomes with relatively uniformly conserved non-coding DNA. In comparison, species of the true fruit flies (Tephritidae family, Sepsid species) have genomes that are between 4- and 6-times larger than that of D. melanogaster and contain blocks of conserved non-coding sequence flanked by regions that are poorly conserved (Peterson et al., 2009). The increased evolutionary divergence between these species, combined with the differences in overall genome structure, enable a more accurate measure of TFBS turnover and of the functional significance of sequence conservation.
Recent studies have compared the eve regulatory locus in Drosophila with that of Sepsid species, which diverged approximately 100 million years ago (Hare et al., 2008a) (Fig. 2A). Nevertheless, eve is expressed in the same seven transverse stripes in Sepsid embryos as in Drosophila embryos (Hare et al., 2008a). In addition, many of the TFs upstream in the developmental cascade, including HB, GT and KR, are also expressed in conserved embryonic patterns in the Sepsid species Themira minor. Despite the fact that eve regulatory regions are found to be only minimally conserved between these groups, Sepsid eve enhancers identified through clusters of HB, Caudal (CAD), Knirps (KNI), KR and BCD binding sites are able to drive conserved gene expression patterns in transgenic D. melanogaster.
Functional conservation of the Sepsid and Drosophila S2E orthologs despite relatively low sequence conservation indicates that some other shared molecular property is responsible for their activity. The identity of this common molecular mechanism is currently the focus of active research and debate. Bioinformatic analysis in the Eisen laboratory demonstrates that there has been large-scale reorganization of the TFBSs in the eve enhancers from different Drosophila and Sepsid species (Fig. 2B), indicating that the spatial organization of the binding sites within enhancer regions may not be crucial (Hare et al., 2008a). However, detailed analysis of the architecture of S2E orthologs reveals the existence of 20-30 bp blocks of highly conserved sequence enriched in pairs of neighboring or overlapping TFBSs. This suggests that the relative position of binding sites to one another might be more important than their overall spatial arrangement within an enhancer (Hare et al., 2008a) (Fig. 2B).
Several crucial issues were raised following the conclusions drawn from the initial studies of the orthologous insect S2Es (Hare et al., 2008a). A primary concern is that the S2E sequences in Drosophila and Themira are not as diverged as originally indicated, and therefore the lack of sequence homology does not indicate a lack of conserved TF organization. Using two-dimensional similarity plots (see Box 2), Crocker and Erives aligned the S2Es from Drosophila melanogaster and Themira putris and found extensive homology between a series of specific 14-41 bp stretches of sequence. The order of these sequence blocks is conserved across the entire length of the enhancer, suggesting that their position relative to each other is crucial. As a result, these authors argue that these sequences harbor binding sites for KR, BCD and GT and thus that the TFBS architecture is largely unchanged between these insect families (Crocker and Erives, 2008a). In response, Hare et al. note that the plots only align half of the minimal S2E region; by contrast, the other half of S2E exhibits little or no conservation despite its necessity for proper functional activity of the S2E module (Hare et al., 2008b). Furthermore, computational studies using PATSER [a bioinformatics tool that can be used to scan a given DNA sequence with a position-weighted matrix (PWM, see Box 2) representing a consensus binding site for a specific transcription factor] to predict TFBSs within S2E orthologs reveals that an extensive genomic reorganization of binding sites has occurred between Drosophila and Sepsid families. Indeed, 24 of the TFBSs analyzed in this study are not conserved between Drosophila and Sepsids, despite the fact that many of these sites bind conserved proteins known to regulate eve expression through S2E, such as HB, BCD, GT, KR and an additional pair-rule repressor TF, Sloppy paired 1 (SLP1) (Andrioli et al., 2002; Hare et al., 2008b). The studies by Hare et al. suggest that TFBSs are the essential molecular components within enhancer regions that, even when extensively reorganized, can still function to produce the same patterns of expression (Hare et al., 2008a; Hare et al., 2008b).
Overall, there exist caveats to the use of sequence alignments, including similarity plots, which can overemphasize weak homology across short stretches of sequence that are sometimes near the threshold for statistical significance. Although sequence analysis and alignment approaches can be useful in the identification of potential regions of conservation, the biological relevance of such alignments is not easily deciphered. To date, no functional synthetic S2E has been built, which could support the idea that this CRM is a loose cluster of TFBSs with as yet undefined organizational requirements. Further analysis of S2E will continue to shed light on the molecular mechanisms of enhancer regulation. In the meantime, studies of additional Drosophila CRMs may be beginning to elucidate this issue.
Molecular motifs in neurogenic ectodermal enhancer architecture
The functional significance of TFBSs has also been investigated in NEEs (see Box 2), which are crucial for dorsoventral patterning of the neurogenic ectoderm in the developing Drosophila embryo (Erives and Levine, 2004). The gene expression patterns directed by this class of CRMs are mediated largely by three TFs: the activators Dorsal (DL) and Twist (TWI) and the repressor Snail (SNA) (reviewed by Rusch and Levine, 1996). The genomes of D. melanogaster, D. pseudoobscura and D. virilis were analyzed for the presence of a consensus NEE motif comprising several sequences, including closely spaced binding sites for DL and TWI and a SNA binding site that overlapped with the TWI binding site (Crocker et al., 2008b). Using their consensus motif, the authors identified five NEEs in D. melanogaster, five in D. virilis and four in D. pseudoobscura. In agreement with the findings of Hare et al. for the eve gene (Hare et al., 2008a), orthologous NEE-driven genes from different Drosophila species show nearly identical embryonic expression patterns. However, when the bioinformatically identified NEEs were tested in transgenic D. melanogaster, they produced different patterns of expression. Although the predicted NEEs directed expression in a band on the ventral side of the embryo extending along the entire anterioposterior axis (as is normal), the number of nuclei in the band was not consistent. As the computational criteria by which the NEEs were identified depended on homology to a consensus sequence, the orthologous NEEs were very similar in their organization of DL, TWI and SNA TFBSs. These results raise a fundamental question: how are the endogenous NEE-driven patterns of gene expression the same in all species, whereas the identified NEE orthologs direct different patterns of reporter gene transcription in transgenic D. melanogaster?
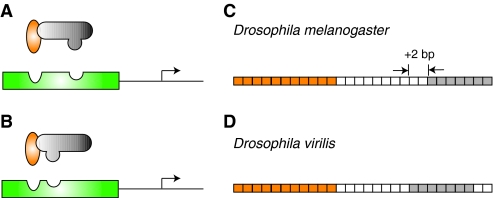
A crucial difference between NEE orthologs that appears to regulate the distinct functional activity of these enhancers from different Drosophila species is the spacing between DL and TWI activator binding sites. For example, in the D. melanogaster ventral nervous system defective (vnd) NEE, the DL and TWI TFBSs are separated by 10 bp (Fig. 3A,C). In the orthologous D. virilis vnd NEE, the distance between the sites is shortened to 8 bp (Crocker et al., 2008b) (Fig. 3B,D). Could this relatively minor modulation of TFBS architecture have a pronounced influence on functional activity? In several experiments, Crocker and colleagues found that adding or deleting just a few base pairs between the two TFBSs in an NEE from one species to mimic the spacing in a second species (or another NEE from the same species) is in fact enough to recreate the pattern of gene expression directed by the second NEE (Crocker et al., 2008a; Crocker et al., 2008b). What could be directing these subtle modifications in CRM architecture? Previous studies suggest that slight differences in the trans environment of different species can drive evolutionary changes at the sequence level in CRMs (Gasch et al., 2004; Ronshaugen et al., 2002). A close investigation of DL and TWI protein sequences has revealed that several changes have occurred between the species, including one mutation in D. melanogaster DL that is known to affect DL-TWI gene activation (Jia et al., 2002). Therefore, the way in which these two proteins interact with their DNA binding sites, and thus with each other indirectly, has probably slightly changed between D. virilis and D. melanogaster (Fig. 3). These modifications at the protein level in turn lead to the co-evolution of the regulatory sequences bound by these TFs (Fig. 3). The pattern of expression of NEE-driven genes is crucial for the development of the fly body plan, so we can assume that any change in expression of NEE-target genes would probably be strongly selected against in evolution. Thus, as the trans environment changes, natural selection also demands compensatory changes in the cis-regulatory sequences. In the case of the NEEs, cis-evolution appears to act by modulating the spacing of binding sites to ensure that functional activity of the CRM is maintained in a given species (Fig. 3). Accordingly, when NEEs are tested in a different species the trans environment will be different and thus the NEEs will not respond to the upstream TFs with the same transcriptional outputs.
Fig. 3.
Changing regulatory environments can drive compensatory changes in enhancer organization. In (A) D. melanogaster and (B) D. virilis, the Dorsal (orange) and Twist (gray) activators cooperatively bind to neurogenic ectoderm enhancer (NEE) sequences (green) to upregulate the transcription of target genes (arrow). This interaction is dependent upon compatibility between the transcription factors and their respective binding sites. In the ~60 million years of evolution separating D. virilis and D. melanogaster (Tamura et al., 2004), protein sequences have diverged only slightly, but enough to affect the DNA-binding domains of the proteins. As a result, in the vnd NEEs of (C) D. melanogaster and (D) D. virilis, the spacing between a conserved Dorsal binding site (orange boxes) and a conserved Twist binding site (gray boxes) is different. The intervening sequence between the binding sites is only 37.5% conserved, but there is also a deletion of two nucleotides (white boxes) in the D. virilis NEE. Even small changes in transcription factor protein domains between different species can drive the architectural reorganization of DNA binding sites. In this case, a decrease in the spacing between sites preserves enhancer functional activity.
Is transcription factor binding site organization crucial for enhancer function?
The current debate over the functional significance of TFBS organization in CRMs highlights a key mystery in developmental biology: how exactly does an enhancer function at the molecular level? Without a doubt, the TF binding sites within an enhancer are crucially important. However, studies of the eve stripe 2 regulatory module suggest that binding site turnover occurs frequently in evolution, resulting in changes to the overall number and architecture of TFBSs within orthologous enhancers. Surprisingly, these pronounced differences appear to have no significant effect on the regulation of target gene transcription, suggesting that enhancers are under selective pressure to be functionally robust to evolutionary changes at the sequence level. Our own studies on early embryonic enhancers from the homeotic genes in Drosophila (Ho et al., 2009) have corroborated these discoveries. In general, enhancers from different species maintain their conserved functional activity despite significant modulation of their sequence architecture during evolution.
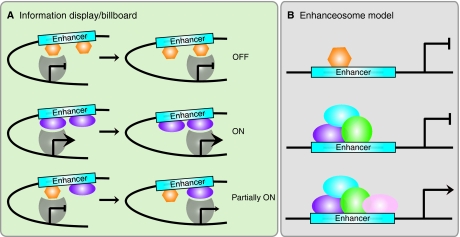
Can we use these observations of enhancer activity to develop a clearer idea of the molecular function of CRMs? Recent studies do in fact lend support to the information display/billboard model of enhancer function (Fig. 4A). The information display/billboard model suggests that an enhancer functions not as a single large processor, but as a series of autonomous signals, each of which can have an effect on target gene transcription (Arnosti et al., 1996; Kulkarni and Arnosti, 2003). In this model, as long as there is never a net decrease in activator and repressor TFBSs, the enhancer can continue to function normally. The only requirement is for the presence of the correct combination of TFBSs somewhere in the enhancer sequence (Fig. 4A). Could this loose requirement be all that is needed to establish a precise response to the complex binding of multiple TFs at a CRM? Hare and colleagues concede that certain binding site clusters may not be so flexible (Hare et al., 2008a). Functionally linked sites, such as the tight clusters at either end of the minimal S2E, may not be subject to an equivalent rate of evolutionary turnover as more independent sites within the CRM. This is certainly the case with the NEEs, where even slight changes in the spacing between binding sites can produce large differences in target gene expression in the same trans environment (Crocker et al., 2008b). The most parsimonious explanation for these conflicting discoveries is that individual CRMs may in fact be very idiosyncratic. Enhancers may function differently depending upon the TFs by which they are bound and the genes they regulate. This might reflect the unique properties of specific TFs. Different TFs will use distinct molecular interactions to bind to DNA and the mechanisms by which they promote transcription will also vary. Innovative work from the Carroll laboratory on CRMs that regulate yellow gene expression in wing pigmentation in different Drosophila species has demonstrated that enhancers with novel regulatory activity can in fact be generated by modifications to TF binding (Gompel et al., 2005) (reviewed by Prud'homme et al., 2007). In a recent study investigating the evolution of Dorsal target enhancers in Drosophila, the mosquito Anopheles gambiae and the flour beetle Tribolium castaneum, putative orthologous enhancers were found to lack sequence conservation. However, what does appear to be under evolutionary constraint is the position of these enhancers relative to their target gene. As a result, as these enhancers are modified over evolutionary time, they become capable of directing divergent patterns of gene expression (Cande et al., 2009).
Fig. 4.
Information display/billboard and enhanceosome models of cis-regulatory module function. (A) In the information display/billboard model, the transcriptional machinery (gray shape) samples the regulatory landscape found at an enhancer module (blue box) (Kulkarni and Arnosti, 2003). If the basal transcriptional machinery encounters only repressors (orange hexagons), the target gene will not be upregulated. The binding of only activators (purple ellipses) at the regulatory module will result in strong target gene expression. A combination of signals from specific DNA sites being bound by repressors and activators will lead to an intermediate (or more probably spatiotemporally restricted) level of target gene activity. An information display enhancer is tolerant of evolutionary binding site turnover because the transcriptional machinery samples only discrete regions of the enhancer and the output signal will not necessarily differ if transcription factor binding sites are located in different regions within the enhancer sequence. (B) In an enhanceosome, the enhancer can only function when a large regulatory protein complex has assembled (purple, blue, green and pink ellipses). If a repressor (orange hexagon) occludes binding sites, or if one of the components of the complex is absent, the enhancer will not drive target gene expression. An enhanceosome will not tolerate binding site turnover, as the protein complex is extremely stereospecific.
In support of the idea of idiosyncratic CRMs, there are examples of enhancers that do not appear to fit well with the information display/billboard model. In vertebrates, the number of well-characterized enhancers is low and few examples support the information display model perfectly. For example, mammalian sequences with multiple binding sites for the GATA1 TF will continue to function as an enhancer if only a single binding site is intact. However, a very particular site must remain intact: eliminating two of the three GATA1 sites in an enhancer may not have any effect on its function, but the single loss of the third crucial site can completely abolish enhancer activity in mice (Cheng et al., 2008). This example stresses the functional importance of a subset of possible binding sites, although the issue of overall TFBS architecture at mammalian enhancers remains largely unexplored.
Perhaps the best-studied counter-example to the information display/billboard model in vertebrates is the interferon β (IFNB1) enhanceosome. The IFNB1 gene is upregulated in mammals in response to viral infection and the enhancer that directs this expression requires a very specific set of TFs to carry out this regulatory activity (Thanos and Maniatis, 1995). Every TF component is required for the enhanceosome to successfully form and to mediate the upregulation of the IFNB1 gene (Panne et al., 2007) (Fig. 4B). As a result of this highly structured enhancer-TF complex, there has been virtually no evolutionary binding site turnover at this CRM throughout 100 million years of mammalian evolution. The architectural constraints on the Drosophila NEEs may be somewhere between the two extremes presented by the IFNB1 enhanceosome and the S2E orthologs. Although the NEEs do not appear to have significant binding site turnover, these CRMs are able to tolerate slight shifts in TFBS position within the module. Indeed, it seems that evolution may have favored these shifts in response to a changing trans environment between different species. In the case of the NEEs, the binding site architecture of a CRM is able to co-evolve with a changing regulatory environment, demonstrating that molecular mechanisms of enhancer function can be plastic. These recent studies demonstrate that evolution can impact enhancers via a number of distinct mechanisms including, but not limited to, reorganization of the total number, binding affinity and spatial arrangement of TFBSs. These discoveries have increased our understanding of the precise molecular mechanisms of enhancer-mediated transcriptional control for a number of key genes during development.
Conclusions
Future studies now need to ask precisely what it is within the enhancer sequence, or the genomic context, or the TFs themselves, that contributes to the differences in functional activity between enhancers. These questions will be answered largely through a combination of detailed bioinformatic analyses of genomic sequences and functional tests across related eukaryotic species. As our knowledge of TF binding activity during embryogenesis increases, computational tools, including position-weight matrices, can be used to find binding sites that are conserved between species and to compare their distribution in genomic DNA sequences. In vivo functional studies should follow such investigations, guided by rapid cell-based assays to screen potential sequences. Although these types of experiments have been performed on a few enhancers, the next step is to apply the same techniques to the ever-growing array of other known enhancers. In addition, expanding our current catalog of known enhancer CRM orthologs to other insects and mammals/vertebrates would greatly contribute to our understanding of the evolution of CRM function. Only by exploring the evolution of a wide range of different enhancers can we hope to understand exactly how these CRMs function and direct the precise patterns of gene expression that give rise to the elegant complexity of embryonic development.
Acknowledgements
Research in our laboratory is supported by funding to R.A.D. from the National Science Foundation and National Institutes of Health and a Howard Hughes Medical Institute Undergraduate Science Education Program grant to the Biology department at Harvey Mudd College. M.J.B. was supported by an Engman award. D.A.T. was supported as an Arnold and Mabel Beckman Foundation Scholar. M.C.W.H. received support from the Merck-American Association for the Advancement of Science (AAAS) Undergraduate Science Research Program. Deposited in PMC for release after 12 months.
References
- Akbari O. S., Schiller B. J., Goetz S. G., Ho M. C. W., Bae E., Drewell R. A. (2007). The Abdominal-B promoter tethering element mediates promoter-enhancer specificity at the Drosophila bithorax complex. Fly 1, 337-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Bae E., Johnsen H., Villaluz A., Wong D., Drewell R. A. (2008). A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development 135, 123-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrioli L. P., Vasisht V., Theodosopoulou E., Oberstein A., Small S. (2002). Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development 129, 4931-4940 [DOI] [PubMed] [Google Scholar]
- Arnosti D. N., Barolo S., Levine M., Small S. (1996). The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122, 205-214 [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. (1981). Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299-308 [DOI] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. (1983). A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33, 729-740 [DOI] [PubMed] [Google Scholar]
- Bellen H. J. (1999). Ten years of enhancer detection: lessons from the fly. Plant Cell 11, 2271-2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. (1981). In vivo sequence requirements of the SV40 early promoter region. Nature 290, 304-310 [DOI] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M., Nüsslein-Volhard C. (1988). The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7, 1749-1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V. C., Stathopoulos A., Levine M. (2002). Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA 99, 9243-9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande J. D., Chopra V. S., Levine M. (2009). Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development 136, 3153-3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. I., Ordahl C. P. (1978). Irreversible gene repression model for control of development. Science 201, 120-130 [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Sharma S., Keelan D. J., Lewis E. B. (1990). The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9, 4277-4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., King D. C., Dore L. C., Zhang X., Zhou Y., Zhang Y., Dorman C., Abebe D., Kumar S. A., Chiaromonte F., et al. (2008). Transcriptional enhancement by GATA1-occupied DNA segments is strongly associated with evolutionary constraint on the binding site motif. Genome Res. 18, 1896-1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi O. R., Engel J. D. (1986). A 3′ enhancer is required for temporal and tissue-specific transcriptional activation of the chicken adult beta-globin gene. Nature 323, 731-734 [DOI] [PubMed] [Google Scholar]
- Crocker J., Erives A. (2008a). A closer look at the eve stripe 2 enhancers of Drosophila and Themira. PLoS Genet. 4, e1000276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J., Tamori Y., Erives A. (2008b). Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol. 6, e263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearolf C. R., Topol J., Parker C. S. (1989). Transcriptional control of Drosophila fushi tarazu zebra stripe expression. Genes Dev. 3, 384-398 [DOI] [PubMed] [Google Scholar]
- Dillon N., Sabbattini P. (2000). Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. BioEssays 22, 657-665 [DOI] [PubMed] [Google Scholar]
- Driever W., Nüsslein-Volhard C. (1988). A gradient of bicoid protein in Drosophila embryos. Cell 54, 83-93 [DOI] [PubMed] [Google Scholar]
- Erives A., Levine M. (2004). Coordinate enhancers share common organizational features in the Drosophila genome. Proc. Natl. Acad. Sci. USA 101, 3851-3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Wu X., Jaynes J. B. (2009). A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development 136, 3077-3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Moses A. M., Chiang D. Y., Fraser H. B., Berardini M., Eisen M. B. (2004). Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2, e398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N., Prud'homme B., Wittkopp P. J., Kassner V. A., Carroll S. B. (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481-487 [DOI] [PubMed] [Google Scholar]
- Goto T., Macdonald P., Maniatis T. (1989). Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57, 413-422 [DOI] [PubMed] [Google Scholar]
- Gray S., Szymanski P., Levine M. (1994). Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 8, 1829-1838 [DOI] [PubMed] [Google Scholar]
- Harding K., Hoey T., Warrior R., Levine M. (1989). Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 8, 1205-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare E. E., Peterson B. K., Iyer V. N., Meier R., Eisen M. B. (2008a). Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 4, e1000106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare E. E., Peterson B. K., Eisen M. B. (2008b). A careful look at binding site reorganization in the even-skipped enhancers of Drosophila and sepsids. PLoS Genet. 4, e1000268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y., Kuroiwa A., Gehring W. J. (1985). Control elements of the Drosophila segmentation gene fushi tarazu. Cell 43, 603-613 [DOI] [PubMed] [Google Scholar]
- Ho M. C. W., Johnsen H., Goetz S. E., Schiller B. J., Bae E., Tran D. A., Shur A. S., Allen J. M., Rau C., Bender W., et al. (2009). Functional evolution of cis-regulatory modules at a homeotic gene in Drosophila. PLoS Genet. 5, e1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip Y. T., Kraut R., Levine M., Rushlow C. A. (1991). The dorsal morphogen is a sequence-specific DNA-binding protein that interacts with a long-range repression element in Drosophila. Cell 64, 439-446 [DOI] [PubMed] [Google Scholar]
- Jeong S., Rokas A., Carroll S. B. (2006). Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 125, 1387-1399 [DOI] [PubMed] [Google Scholar]
- Jia S., Flores-Saaib R. D., Courey A. J. (2002). The Dorsal Rel homology domain plays an active role in transcriptional regulation. Mol. Cell. Biol. 22, 5089-5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Kosman D., Ip Y. T., Levine M. (1991). The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 5, 1881-1891 [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T. (2004). Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116, 247-257 [DOI] [PubMed] [Google Scholar]
- Karch F., Weiffenbach B., Peifer M., Bender W., Duncan I., Celniker S., Crosby M., Lewis E. B. (1985). The abdominal region of the bithorax complex. Cell 43, 81-96 [DOI] [PubMed] [Google Scholar]
- Kim A., Dean A. (2003). A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 23, 8099-8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhamer C. V., Yuh C. H., Davidson E. H. (1996). Modular cis-regulatory organization of developmentally expressed genes: two genes transcribed territorially in the sea urchin embryo, and additional examples. Proc. Natl. Acad. Sci. USA 93, 9322-9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M. M., Arnosti D. N. (2003). Information display by transcriptional enhancers. Development 130, 6569-6575 [DOI] [PubMed] [Google Scholar]
- Kwon D., Mucci D., Langlais K., Americo J. L., DeVido S. K., Cheng Y., Kassis J. A. (2009). Enhancer-promoter communication at the Drosophila engrailed locus. Development, 136, 3067-3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Manley J. L. (1989). Transcriptional repression of eukaryotic promoters. Cell 59, 405-408 [DOI] [PubMed] [Google Scholar]
- Lewis E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565-570 [DOI] [PubMed] [Google Scholar]
- Ludwig M. Z., Kreitman M. (1995). Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Mol. Biol. Evol. 12, 1002-1011 [DOI] [PubMed] [Google Scholar]
- Ludwig M. Z., Patel N. H., Kreitman M. (1998). Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125, 949-958 [DOI] [PubMed] [Google Scholar]
- Ludwig M. Z., Bergman C., Patel N. H., Kreitman M. (2000). Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564-567 [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Ingham P., Struhl G. (1986). Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell 47, 721-734 [DOI] [PubMed] [Google Scholar]
- Madisen L., Krumm A., Hebbes T. R., Groudine M. (1998). Immunoglobulin heavy chain locus control region increases histone acetylation along linked c-myc genes. Mol. Cell. Biol. 18, 6281-6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan I. J., Wright A. P., Dahlman-Wright K., Carlstedt-Duke J., Gustafsson J. A. (1993). Direct interaction of the tau 1 transactivation domain of the human glucocorticoid receptor with the basal transcriptional machinery. Mol. Cell. Biol. 13, 399-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega M. A., Ovcharenko I., Afzal V., Rubin E. M. (2003). Scanning human gene deserts for long-range enhancers. Science 302, 413 [DOI] [PubMed] [Google Scholar]
- Panne D., Maniatis T., Harrison S. C. (2007). An atomic model of the interferon-beta enhanceosome. Cell 129, 1111-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson B. K., Hare E. E., Iyer V. N., Storage S., Conner L., Papaj D. R., Kurashima R., Jang E., Eisen M. B. (2009). Big genomes facilitate the comparative identification of regulatory elements. PLoS ONE 4, e4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme B., Gompel N., Rokas A., Kassner V. A., Williams T. M., Yeh S. D., True J. R., Carroll S. B. (2006). Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440, 1050-1053 [DOI] [PubMed] [Google Scholar]
- Prud'homme B., Gompel N., Carroll S. B. (2007). Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. USA 104, 8605-8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. (1986). Gene regulation by proteins acting nearby and at a distance. Nature 322, 697-701 [DOI] [PubMed] [Google Scholar]
- Qian S., Capovilla M., Pirrotta V. (1991). The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 10, 1415-1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M., McGinnis N., McGinnis W. (2002). Hox protein mutation and macroevolution of the insect body plan. Nature 415, 914-917 [DOI] [PubMed] [Google Scholar]
- Rusch J., Levine M. (1996). Threshold responses to the dorsal regulatory gradient and the subdivision of primary tissue territories in the Drosophila embryo. Curr. Opin. Genet. Dev. 6, 416-423 [DOI] [PubMed] [Google Scholar]
- Sauer F., Rivera-Pomar R., Hoch M., Jäckle H. (1996). Gene regulation in the Drosophila embryo. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 351, 579-587 [DOI] [PubMed] [Google Scholar]
- Shepherd B., Garabedian M. J., Hung M. C., Wensink P. C. (1985). Developmental control of Drosophila yolk protein 1 gene by cis-acting DNA elements. Cold Spring Harb. Symp. Quant. Biol. 50, 521-526 [DOI] [PubMed] [Google Scholar]
- Small S., Blair A., Levine M. (1991a). Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11, 4047-4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S., Kraut R., Hoey T., Warrior R., Levine M. (1991b). Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 5, 827-839 [DOI] [PubMed] [Google Scholar]
- Small S., Blair A., Levine M. (1996). Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol. 175, 314-324 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Talianidis I. (2002). Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295, 1901-1904 [DOI] [PubMed] [Google Scholar]
- Stanojevic D., Hoey T., Levine M. (1989). Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature 341, 331-335 [DOI] [PubMed] [Google Scholar]
- Steward R., Zusman S. B., Huang L. H., Schedl P. (1988). The dorsal protein is distributed in a gradient in early Drosophila embryos. Cell 55, 487-495 [DOI] [PubMed] [Google Scholar]
- Struhl G., Struhl K., Macdonald P. M. (1989). The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57, 1259-1273 [DOI] [PubMed] [Google Scholar]
- Struhl K. (1984). Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc. Natl. Acad. Sci. USA 81, 7865-7869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S. (2004). Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21, 36-44 [DOI] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. (1995). Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83, 1091-1100 [DOI] [PubMed] [Google Scholar]
- Utley R. T., Ikeda K., Grant P. A., Côté J., Steger D. J., Eberharter A., John S., Workman J. L. (1998). Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394, 498-502 [DOI] [PubMed] [Google Scholar]
- Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman A. G., Sassone-Corsi P., Grundström T., Zenke M., Chambon P. (1984). Stimulation of in vitro transcription from the SV40 early promoter by the enhancer involves a specific trans-acting factor. EMBO J. 3, 3129-3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., Young R. A., Levine M. (2007). Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 21, 385-390 [DOI] [PMC free article] [PubMed] [Google Scholar]