Abstract
Endomesoderm is the common progenitor of endoderm and mesoderm early in the development of many animals. In the sea urchin embryo, the Delta/Notch pathway is necessary for the diversification of this tissue, as are two early transcription factors, Gcm and FoxA, which are expressed in mesoderm and endoderm, respectively. Here, we provide a detailed lineage analysis of the cleavages leading to endomesoderm segregation, and examine the expression patterns and the regulatory relationships of three known regulators of this cell fate dichotomy in the context of the lineages. We observed that endomesoderm segregation first occurs at hatched blastula stage. Prior to this stage, Gcm and FoxA are co-expressed in the same cells, whereas at hatching these genes are detected in two distinct cell populations. Gcm remains expressed in the most vegetal endomesoderm descendant cells, while FoxA is downregulated in those cells and activated in the above neighboring cells. Initially, Delta is expressed exclusively in the micromeres, where it is necessary for the most vegetal endomesoderm cell descendants to express Gcm and become mesoderm. Our experiments show a requirement for a continuous Delta input for more than two cleavages (or about 2.5 hours) before Gcm expression continues in those cells independently of further Delta input. Thus, this study provides new insights into the timing mechanisms and the molecular dynamics of endomesoderm segregation during sea urchin embryogenesis and into the mode of action of the Delta/Notch pathway in mediating mesoderm fate.
Keywords: Endomesoderm, Gene regulatory network, Sea urchin, Delta/Notch pathway, Gcm, FoxA
INTRODUCTION
In many triploblastic animals, most of the endoderm and the mesoderm arise from a common progenitor, the endomesoderm (Rodaway and Patient, 2001; Technau and Scholz, 2003). This conserved mechanism has been reported across the animal kingdom, including in ecdysozoans (Maduro, 2006; Sulston et al., 1983), lophotrochozoans (Gillis et al., 2007), echinoderms (Logan et al., 1999), urochordates (Nishida and Satoh, 1985), and vertebrates (Fukuda and Kikuchi, 2005; Kimelman and Griffin, 2000). In C. elegans, for example, the endomesoderm tissue arises at the four-cell stage and segregates into endoderm and mesoderm with the next cell division, mainly through the asymmetric activity of a Wnt/MAPK pathway (Maduro, 2006; Sulston et al., 1983). In sea urchin, the endomesoderm cells emerge later, at the sixth cleavage, in the vegetal half of the embryo, and the mesoderm and endoderm lineages diverge several cleavages later under the primary control of the Delta/Notch signaling pathway (Cameron et al., 1991; Logan et al., 1999; Ruffins and Ettensohn, 1996; Sherwood and McClay, 1999). In vertebrates, cell-labeling experiments suggest that the mesoderm fate segregates from the endoderm during gastrulation through the activity of a TGF-β/Nodal signaling gradient (Warga and Nusslein-Volhard, 1999). Despite a growing molecular understanding of the regulatory mechanisms of endomesoderm segregation, much less is known about the spatiotemporal dynamics of these mechanisms throughout embryogenesis.
During sea urchin development, the endomesoderm tissue gives rise to three derivatives: the skeletogenic mesoderm (SM), the non-skeletogenic mesoderm (NSM) and the endoderm. Precursors of the SM cells, the micromeres, are the first to segregate. They appear at the fourth cleavage at the vegetal pole and their progeny give rise exclusively to the skeleton of the larvae. These cells are a sea urchin-specific derived cell population, and their development will not be addressed in this manuscript. Nevertheless, they play a key role in the subsequent endoderm and NSM diversification, as they provide the initial source of the Delta ligand. After the emergence of the micromeres, cells that are bipotential for the NSM and the endoderm lineages arise at the sixth cleavage. These cells represent the true evolutionary equivalents of the bilaterian endomesodermal cells, as their progeny generate exclusively endoderm and mesoderm. They appear in the vegetal half of the embryo, immediately above the micromeres, and are called the veg2 cells (for cells of the second vegetal tier) (Cameron et al., 1991; Logan et al., 1999). Cell labeling experiments have shown that, by the onset of gastrulation (mesenchyme blastula stage), the progeny of these cells have already segregated into definitive endoderm and mesoderm (Ruffins and Ettensohn, 1996). However, the exact sequence of cellular and molecular events leading to that subdivision is still not well understood.
The circuitry of endomesoderm specification in sea urchin has been studied in detail and has led to the elaboration of a complex gene regulatory network (GRN) model that displays, as a global view from the genome, how endomesoderm development progresses from fertilization until 30 hours post-fertilization (Davidson et al., 2002) (see http://sugp.caltech.edu/endomes/). Within this GRN, three components contribute to the core machinery of endomesoderm segregation: the Delta/Notch pathway, and the transcription factors FoxA and Gcm. The Delta/Notch pathway regulates NSM specification (Sherwood and McClay, 1997; Sherwood and McClay, 1999; Sweet et al., 2002). Delta, the ligand of the pathway, is first expressed in micromeres (Sweet et al., 2002), where experiments suggested that it signals between the eighth and the tenth cleavage to the neighboring veg2 cells and their further vegetal progeny (McClay et al., 2000; Sweet et al., 2002). Later, at the onset of gastrulation, Delta is expressed in additional cells within the vegetal plate, in all likelihood veg2 descendant cells, where it has a second specification function (Sweet et al., 2002). A known direct target of the first Delta/Notch pathway is the transcription factor Gcm (Ransick and Davidson, 2006). The Gcm promoter contains several Suppressor-of-Hairless (SuH) binding sites that are necessary for Gcm activation (Ransick and Davidson, 2006), and the Notch pathway directly activates SuH (Fortini and Artavanis-Tsakonas, 1994). Gcm is required for mesoderm specification, in particular pigment cell development (Ransick and Davidson, 2006). FoxA is another key transcription factor for endomesoderm segregation. Reports on this gene indicate that FoxA is expressed in the definitive endoderm, where it promotes endoderm specification and represses mesoderm development by preventing Gcm expression (Oliveri et al., 2006).
In this study, we analyze in detail, at a single cell resolution, the expression patterns of Delta, Gcm and FoxA prior to, during and after endomesoderm diversification, to establish the spatial and temporal dynamics of the GRN states that control this cell fate decision during sea urchin embryogenesis. For a short period of time, before hatching, Gcm and FoxA are co-expressed in a subset of the veg2 descendant cells, the Veg2L (lower Veg2) tier. Shortly after, at hatching, expression of these two transcription factors is segregated into two distinct cell populations. Gcm continues to be expressed in the Veg2L cells, while FoxA is downregulated in those cells and activated in the above neighbor cells, the Veg2U (upper Veg2) cells. Delta/Notch signaling is known to be necessary for Gcm expression and mesoderm development. Here, we show that approximately 2.5 hours of continuous reception of the Delta signal is crucial for the maintenance of Gcm expression in the Veg2L cells before Gcm expression continues on its own, independent of Delta activation. Assuming that definitive mesoderm and endoderm paths begin when cells lose the expression of either Gcm or FoxA, respectively, we show that endomesoderm diversification occurs in the veg2 progeny at hatching, and that the requirement for the Delta/Notch signal lasts for a significant time period before the two fates continue their separate ways.
MATERIALS AND METHODS
Animals
Lytechinus variegatus adults were collected from Beaufort, NC (Duke University Marine Laboratory) and maintained in the laboratory in artificial seawater (ASW) between 20°C and 23°C. Gametes were obtained by 0.5 M KCl injections. Embryos were cultured at 21°C in ASW from fertilization up to the various developmental stages (see Fig. 1 for precise timing).
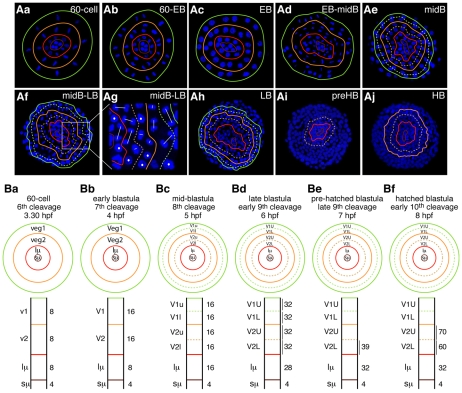
Fig. 1.
Cell lineage and cell fate of the vegetal plate of Lytechinus variegatus embryos from the 60-cell stage to the hatched blastula stage. (Aa-Aj) Single plane projections of confocal sections of vegetal plate embryos. Plain color-coded lines outline the repartition of each of the different cell sub-types that constitute the vegetal plate at the various developmental stages (small micromeres in brown, large micromeres in red, veg2 cells in orange, veg1 cells in green). The dotted colored lines indicate when a cell division occurred perpendicular to the animal-vegetal (AV) axis and created two cell layers of the same cell sub-type, e.g. veg2 layer in EB into Veg2U (Veg2 upper) and Veg2L (Veg2 lower). Nuclei are in blue. EB, early blastula stage; midB, mid-blastula stage; LB, late blastula stage; preHB, pre-hatched blastula stage; HB, hatched blastula stage. (Ba-Bf) Schematics of the distribution of the cell lineages that compose the vegetal plate through time. The circles reproduce schematically the information presented in B at the primary stages for this study. The rectangles provide the same information along the AV axis, with the vegetal pole at the bottom. Numbers on the right are the average number of cells counted for each cell layer, based on observations from this study. In both circles and rectangles, similar codes (color and plain or dotted lines) have been applied to those in A, and each cell layer was named according to its origin and cell division events.
Fluorescent in situ staining
Sequences of all probes used during this study are available in GenBank (LvDelta AAL71861, LvGcm ABY40630, LvFoxA FN392326 and LvT-brain FN392327). For single in situ hybridization (ISH), probes were labeled using digoxigenin-11-UTP (Roche) and revealed with the Cy3-Tyramide Signal Amplification System (Perkin Elmer). For double ISH, digoxigenin-11-UTP (Roche) and fluorescein-12-UTP (Roche) tagged probes were used and detected with Cy3-Tyramide and fluorescein-Tyramide reagents, respectively. For both single and double ISH experiments, probes were used at a final concentration of 1 ng/μl. Expression patterns of each gene studied were investigated by single fluorescent ISH on embryos from two different cultures, and by double fluorescent ISH. For each condition, at least six embryos were examined in detail by confocal microscopy, providing at least 18 samples for each gene at each stage. For the total number of embryos analyzed, the average number of cells expressing the studied genes, and the standard deviation, see Table S1 in the supplementary material.
Embryos, fixation and ISH analyses were performed as previously described (Croce et al., 2003). Fluorescent ISH was carried out using the TSA-plus kit (Perkin Elmer), following manufacturer's recommended protocol. Nuclear stains were obtained by a 5-minute incubation in Hoechst 33342 (2 μg/ml)/TBST.
Imaging and image processing
Two-channel fluorescence and DIC pictures were generated with an upright Zeiss Axioplan2 fluorescence microscope. The three-channel fluorescence images, including the nuclear staining, were obtained using a Zeiss LSM510 confocal microscope. The same embryo was first imaged by fluorescence and then analyzed using the confocal microscope. For confocal microscopy, images were scanned at 1.5-μm intervals, and up to 30 to 35 microns into the embryo. Projections of the vegetal plates were obtained from a stack of a total of 20 to 25 images merged together using the maximum intensity projection tool of the LSM software. To count the number of nuclei, each confocal stack was post-processed using ImageJ and Adobe Photoshop. For each stack, all snaps were extracted, and for each image the three RGB channels were separated. All generated images were then reassembled in chronological order into a unique document as transparent layers in Adobe Photoshop. The outlines of the expression domains were drawn onto that document and the nuclei were counted by hand, layer by layer.
Morpholino antisense oligonucleotides (MASO) and mRNA microinjection
Delta-MASO (5′-GTGCAGCCGATTCGTTATTCCTTT-3′) was obtained from Gene Tools. The morpholinos were designed against the 5′UTR of LvDelta as previously described (Sweet et al., 2002) and used at a final concentration of 0.15 mM. The Delta, ActNotch and dnNotch mRNAs were made and used as previously described (Sweet et al., 2002; Sherwood and McClay, 1999).
Micromanipulation
Micromere-less embryos were obtained by microsurgery at the 16- to 32-cell stage using a fine glass needle and micromanipulators (Narishige). For micromere transplants and half-embryo recombinants, donor control or injected embryos, labeled by microinjection of rhodamine dextran or a brief incubation in CFDA (0.001%; Molecular Probes), were transferred to unlabeled host embryos at the 16- or 32-cell stage in Ca2+-free seawater. Microsurgery was performed with fine glass needles and micromanipulators (Narishige) (for details, see Logan et al., 1999).
RESULTS
Cell lineage and fate of the vegetal plate of Lytechinus variegatus embryos
Our goal was to determine when, prior to mesenchyme blastula, could we detect the first separation of fates of the mesoderm and endoderm lineages and in precisely which cells. This required a highly detailed and accurate cell lineage analysis of the cells in which the endomesoderm genes investigated are expressed. To provide a template for this, we first examined confocal optical sections of the vegetal region of fixed embryos labeled with the nuclear stain Hoechst 33342 (Fig. 1). To orient the embryos, an early skeletogenic mesenchyme (SM) cell marker (T-brain) was used to identify the vegetal pole (marker expression is outlined but not shown in Fig. 1). Vegetal plates were imaged starting at the 60-cell stage, then every 30 minutes until hatching, and from then every hour until ingression of the SM cells.
At the 60-cell stage (sixth cleavage), the vegetal plate comprises four cell lineages, all concentrically distributed around the vegetal pole: the small micromeres (four cells), the large micromeres (eight cells), the endomesoderm cells or veg2 cells (eight cells) and the ectoendodermal cells or veg1 cells (eight cells; Fig. 1Aa,Ba) (Logan et al., 1999). As cell divisions progress toward the mid-blastula stage (240 cells — eighth cleavage), the cleavages are mostly synchronous within the cell lineages and are similarly oriented from cell to cell (Fig. 1Ab-Ae). It is thus relatively easy up to this stage to establish the vegetal plate cell lineage maps by simply following the behavior of the nuclei. From the 60-cell stage to the mid-blastula stage, the small micromeres do not divide, the large micromeres divide once, and the veg2 and veg1 cells undergo two rounds of cell division. In the vegetal half, the seventh cleavage plane occurs longitudinal to the animal-vegetal (AV) axis, and doubles the number of cells per cell tier (Fig. 1Ab,Ac,Bb). Then, the eighth cleavage plane is equatorial to the AV axis and leads to the formation of two cellular layers per cell lineage, one closer to the vegetal pole (lower) and one farther from it (upper) (Fig. 1Ad,Ae,Bc). Thus, at the mid-blastula stage (240 cells — eighth cleavage), the vegetal half of the embryos is composed of six tiers of cells: the small micromeres (four cells), the large micromeres (16 cells), the lower Veg2 cells (Veg2L; 16 cells), the upper Veg2 cells (Veg2U; 16 cells), the lower Veg1 cells (Veg1L; 16 cells), and the upper Veg1 cells (Veg1U; 16 cells; see Fig. 1Ae,Bc).
At the ninth cleavage, mitotic timing becomes asynchronous within the embryo and from cell to cell within the cell layers, and the cleavage plane orientations begin to be randomized within the cell layers. This is especially true for the lineages closest to the vegetal pole, where tiers of cells have smaller diameters than do tiers near the equator. For instance, in the Veg2L layer, cells still in prophase are found next to cells dividing along the AV axis and cells cleaving perpendicularly to it (Fig. 1Af,Ag). Thus, following the eighth cleavage it becomes difficult to assess when the subsequent cleavages are over within the entire embryo, and to retain knowledge of the precise ancestry of the cells. Beginning at the eighth cleavage (5 hours post-fertilization), we chose therefore to identify the succeeding stages in terms of hours post-fertilization (hpf) rather than number of cells or cleavages (for detail, see Fig. 1). In addition, to assign a cell to a particular ring of cells concentrically distributed around the vegetal pole, we used as landmarks the expression patterns of specific known markers, i.e. Delta, FoxA, Gcm and T-Brain. Through these analyses, we found that at hatched blastula stage (8 hpf), the vegetal plate consists of four small micromeres surrounded by 32 large micromeres, an average of 60 Veg2L cells and an average of 70 Veg2U cells, each spread over several sub-tiers, but which for the rest of this study are grouped into either the Veg2L or the Veg2U layers, for simplicity (Fig. 1Aj,Bf).
Although the cell lineage maps presented here through the hatched blastula stage are not complete, the transition to markers provides an accurate reflection of the cell lineages of the vegetal plate, especially for the later stages. Importantly, this lineage analysis provides a suitable framework to follow on a cell-to-cell basis the process of endomesoderm diversification that, based on the molecular data obtained in this study, happens first at the hatched blastula stage.
Endomesoderm diversification begins at hatching
To approximate when the endomesoderm tissue diversifies into definitive endoderm and mesoderm, we investigated the expression pattern of Gcm and FoxA, cell by cell over time. Gcm and FoxA have been shown previously to be specifically expressed by mesoderm and endoderm, respectively, and to be required for their development (Ransick and Davidson, 2006; Oliveri et al., 2006). Thus, if these two genes were co-expressed, the endomesoderm fate would not have yet segregated, whereas once they are detected in different cell populations the separation of the definitive endoderm from the mesoderm would have happened.
In Lytechinus variegatus embryos, Gcm mRNAs are first detected by whole-mount in situ hybridization (ISH) at the mid-blastula stage (5 hpf, eighth cleavage; see below), whereas FoxA mRNA is detected at 7 hpf (pre-hatched blastula stage). At that time, Gcm is expressed in a ring of 39 cells (Veg2L) that surround 36 cells that are not stained (the large and small micromeres; Fig. 2Aa,Ab), and FoxA is observed also in a ring of 39 cells distributed around an average of 36 unlabeled micromeres (Fig. 2Ba,Bb). Thus, at late ninth cleavage (7 hpf), Gcm and FoxA are expressed in the same cells, a result that is confirmed by the co-staining of Gcm and FoxA within the same embryos through double fluorescent ISH (Fig. 2Ca-Cc,G). Careful examination of these co-stained embryos indicates further that, in most cases, the two expression domains do not match exactly. In all embryos examined, a few cells located at the edge of the domains always appear to express either Gcm or FoxA (on average five cells for each gene), and the position of these cells with regard to the AV and oral-aboral axes is random from one embryo to another (Fig. 2Cc). At 7 hpf, just prior to hatching, most of the Veg2L cells are thus still endomesodermal, at least by the definition that places the expression of Gcm and FoxA in the same cells, and a few, in a stochastic manner, have already diverged toward the endoderm or mesoderm fate path.
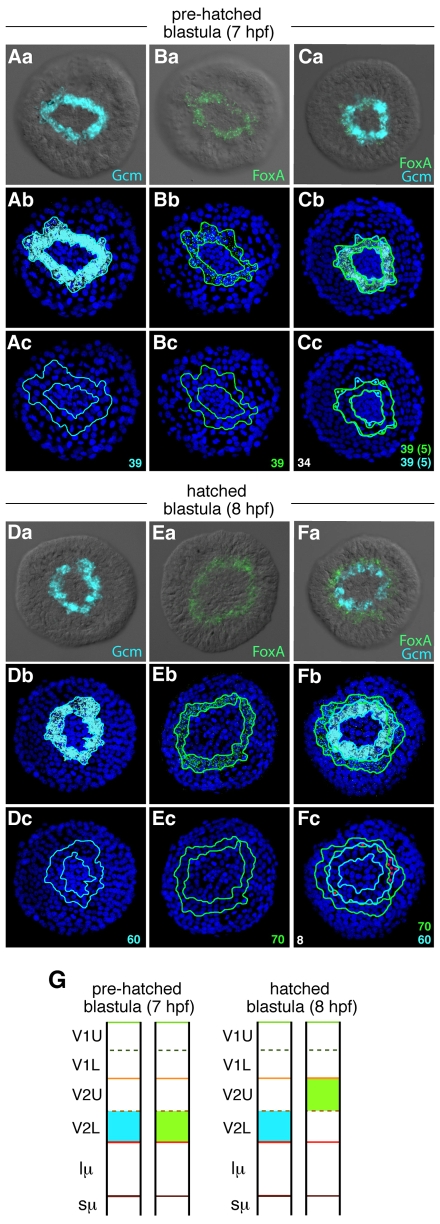
Fig. 2.
FoxA-Gcm relationship on a cell-to-cell basis over time. (Aa-Fc) Single (A,B,D,E) and double (C,F) fluorescent in situ hybridization analyses of Gcm and FoxA expression at pre-hatched blastula stage (7 hpf; A-C) and hatched blastula stage (8 hpf; D-F). For both stages, the first row shows two-channel fluorescence and DIC images of FoxA (light green) and Gcm (blue) expression either individually (Aa,Ba,Da,Ea) or together (Ca,Fa). The second and third rows show confocal projections of the first row embryos with blue-labeled nuclei. On the third rows, only the outlines of the expression domains of FoxA and Gcm have been kept to determine the average number of cells (numbers in the right corner) expressing each gene (Ac,Bc,Dc,Ec) or both genes (Cc,Fc). In Cc, dots reflect cells that express either FoxA or Gcm (average numbers in parentheses). In Fc, red dots highlight cells that express both genes, on average eight cells. (G) Summary of FoxA (light green) and Gcm (blue) expression profiles throughout time and in regard to cell lineage.
An hour later, at hatched blastula stage, Gcm is detected in a ring of 60 cells surrounding 40 cells that are not labeled (the micromere lineages; Fig. 2Da-Dc), and FoxA mRNAs are detected in a ring of 70 cells (Veg2U) that are distributed around 95 FoxA-negative cells (the micromeres + Veg2L; Fig. 2Ea-Ec). A co-staining for Gcm and FoxA carried out at that stage revealed that these two genes are now expressed in two distinct cell populations (Fig. 2F,G), with only a few cells located at the boundary between the two expression domains expressing both genes (Fig. 2Fc). Thus, by hatched blastula stage (8 hpf), the first segregation of the endomesoderm has happened. However, once again, although most of the Veg2L and Veg2U cells have now adopted a definitive germ layer identity, some of them (on average eight cells per embryo at this time) remain in an intermediate molecular endomesoderm state that will undoubtedly be resolved shortly thereafter.
Gcm and Delta mRNA distribution within the vegetal plate over time
To better understand the molecular signature of endomesoderm diversification, we then analyzed at a cellular resolution the spatiotemporal relationship between Delta, the ligand of the pathway, and Gcm, a direct target of Notch signaling, from the 60-cell stage to the hatched blastula stage. Delta mRNAs are first detected at the 32-cell stage in the micromeres (see Fig. S1 in the supplementary material), where they become prominent at the 60-cell (Fig. 3Ba-Bc,H) and early blastula (Fig. 3Ca-Cc,H) stages. Gcm expression is likely to be activated during the seventh cleavage but is first detected at the eighth cleavage (5 hpf), which subdivides the veg2 layer into Veg2L and Veg2U and separates the Veg2U cells from contact with the micromeres. At that stage, Gcm mRNAs are observed in 32 cells arranged in two-cell layers that correspond to the Veg2L and Veg2U layers (Fig. 3Da-Dc,H). These cells surround the 20 micromeres that do not express Gcm but express Delta mRNA (Fig. 3Da-Dc,H). Double fluorescent ISH shows further that these two expression domains are exclusive from one another and, thus, that at this stage Delta is not expressed in endomesodermal cells.
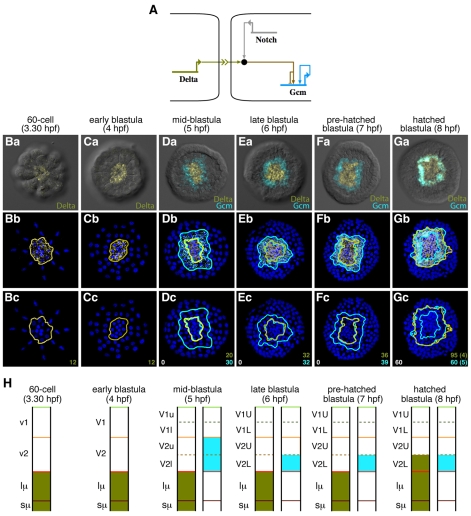
Fig. 3.
Delta-Gcm relationship on a cellular basis prior to and at endomesoderm diversification. (A) Schematic of the known Delta/Notch signaling-Gcm regulatory relationship. Delta through its interaction with Notch activates Gcm in the neighboring cells. Gcm is able at some point to maintain its own expression by a positive feedback loop onto its own promoter. (Ba-Gc) Expression profiles of Delta (olive) and Gcm (blue) by double fluorescent in situ hybridization from the 60-cell to the hatched blastula stage. First row shows two fluorescence channels and DIC pictures of vegetally viewed embryos. Second and third rows show confocal images of first row embryos with the nuclei in blue. On the third row, only the outlines of the gene expression domains have been kept to determine the number of cells expressing each gene (numbers color-coded at the bottom right corner), on how many cell layers those cells are distributed (data reported in H), how many cells express both genes simultaneously (number in white in the bottom left corner), or solely one or the other gene (dots on Gc and numbers in parentheses). (H) Summary of Delta (olive) and Gcm (blue) expression patterns throughout time and with respect to cell lineage.
At late (6 hpf) and pre-hatched (7 hpf) blastula stages, the Delta-Gcm spatial relationship remains similar in appearance to that observed at the eighth cleavage (5 hpf) with one exception: Gcm is no longer detected in the Veg2U cells after being expressed there only for about 30 minutes. By 6 hpf, Gcm is detected only in the Veg2L layer, the layer that remains in contact with the micromeres and that is composed of 32 and 39 cells at 6 hpf and 7 hpf, respectively (Fig. 3Ea-Ec,Fa-Fc,H). At the hatched blastula stage (8 hpf), the Delta-Gcm relationship then changes. The Delta expression domain spreads out and comprises now 95 cells (on average), of which the 60 most peripheral ones (the Veg2L tier) also express Gcm (Fig. 3Ga-Gc). The 36 cells that express Delta but not Gcm correspond to the micromere progeny and the 60 Delta-Gcm positive cells correlate to the Veg2L descendant cells (Fig. 3H). Thus, at hatched blastula stage, Gcm is exclusively within the Veg2L layer, and Delta is now expressed in the Veg2L cells, in addition to its continuing expression in the micromeres.
Endoderm fate specification is inhibited by the early Delta signal
Although it has been established that Delta from the micromeres directly activates Notch in the veg2 cells to activate Gcm, was that signal also responsible for elimination of FoxA from the Veg2L progeny? FoxA expression was assessed in hatched blastula stage embryos microinjected with Delta-MASO or Delta mRNA. In Delta mRNA-injected embryos, in which the Delta protein is ectopically distributed, FoxA transcripts were not detected (Fig. 4Ab). By contrast, in Delta-MASO-injected embryos, FoxA mRNAs were present in a ring closer to the vegetal pole than in controls (Fig. 4Aa) and that includes both Veg2U and Veg2L cells (Fig. 4Ac). Thus, perturbation of the Delta/Notch signal, thereby of mesoderm specification, also has a reciprocal impact on FoxA expression.
Fig. 4.
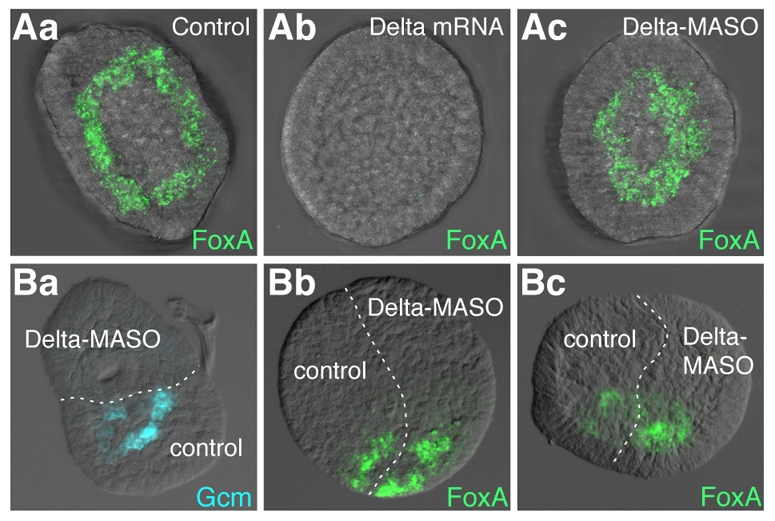
FoxA expression in Notch signaling perturbed embryos. (Aa-Ac) Effects of ectopic activation or repression of Delta/Notch signaling on FoxA expression. (Aa) control embryos, and embryos injected with (Ab) Delta mRNAs or (Ac) Delta-MASO. Embryos were fixed at the hatched blastula stage. (Ba-Bc) Delta-MASO/control chimera embryos generated at the 16-cell stage by the recombination of two AV half embryos; embryos were fixed at the hatched blastula stage. (Ba) Gcm (blue), (Bb,Bc) FoxA (green). All embryos are shown in vegetal view except for Bb and Bc, which are in lateral view.
To further corroborate this conclusion a recombination experiment was performed. Embryos injected with Delta-MASO were bisected along the AV axis at the 16-cell stage and each half was recombined with a similarly bisected control half-embryo. The AV recombinants were fixed at hatched blastula stage and examined by fluorescent ISH for Gcm expression, as control, or FoxA expression. First, as expected, in the Delta-MASO injected half, Gcm mRNAs were not detected (Fig. 4Ba). By contrast, FoxA expression was observed in a more extensive domain in the Delta-MASO half than in the control half (Fig. 4Bb,Bc). These results further corroborate our previous findings and, thus, indicate that the mesoderm GRN activated by Delta somehow also results in the elimination of FoxA.
Duration of the Delta signal is crucial for mesoderm specification
The Delta/Notch pathway is known to act solely between neighboring cells, as it involves a membrane-bound ligand (Delta) on one cell and a receptor (Notch) on its neighbor (Fehon et al., 1990). At the eighth cleavage, however, Gcm mRNAs are detected in both the Veg2L and the Veg2U cells, even though the nascent Veg2U cells are not in contact with the Delta-positive micromeres. It was likely that at that stage the presence of Gcm in the Veg2U layer was due to earlier reception of the Delta/Notch signal by the parental veg2 cells, which had been in contact with the micromeres since their emergence at the sixth cleavage. However, at 6 hpf, 30-60 minutes after no longer being in contact with micromeres, Gcm is no longer detected in the Veg2U descendant cells. We therefore investigated whether this disappearance of Gcm expression from the Veg2U cells was due to loss of contact with micromeres, starting at the eighth cleavage.
As controls, we first showed that an absence of Delta proteins, caused by injection of Delta-MASO, or an absence of Notch signaling, caused by micromere removal at the 16-cell stage, prevented Gcm transcription in the veg2 descendants as expected (Fig. 5Aa-Ac). We found that Delta/Notch signaling is indeed required for Gcm transcription, as had been previously shown in a cis-regulatory analysis of the Gcm promoter (Ransick and Davidson, 2006). To determine then whether the loss of Gcm expression in the Veg2U cells was due to the lack of Delta input, we generated chimeras in which some of the Veg2U cells remained in contact withDelta-positive cells, even through the ninth cleavage and later. A control micromere was transplanted in between the veg1 and veg2 layers of a 60-cell-stage control embryo (Fig. 5Ba) and left there until these chimeras were fixed at hatching (at which time Gcm normally is expressed only in the Veg2L layer). Of the 25 chimera embryos analyzed, 22 (88%) exhibited ectopic Gcm expression in cells located between the endogenous Gcm domain and the transplanted micromere (Fig. 5Bb,Bc). As a control for this experiment, we generated identical chimeras in which the transplanted micromere came from a Delta-MASO-injected embryo (Fig. 5Ca). Out of the three embryos observed, none displayed extra Gcm in the Veg2U cells, even though the transplanted micromeres were identifiable because they expressed Tbr (Fig. 5Cb). Thus, the loss of Gcm expression in the Veg2U tier starting at early ninth cleavage is in all likelihood due to the deprivation of Delta inputs from the neighboring micromeres.
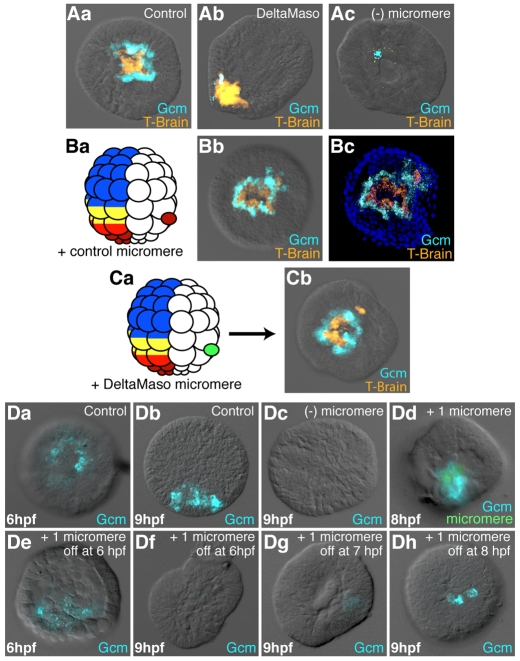
Fig. 5.
Regulation of Gcm expression by the Delta/Notch signal. (Aa-Ac) Effects of the loss of the Delta signal on Gcm expression. Co-staining by double-fluorescent in situ hybridization for T-brain (a micromere progeny marker, orange) and Gcm (blue) on control embryos (Aa), Delta-MASO-injected embryos (Ab) and micromereless embryos (Ac). (Ba,Bb,Ca,Cb) Role of Delta in the loss of Gcm expression in the Veg2U cells. (Ba,Ca) Schematics of the experimental design. A control micromere (Ba) or a DeltaMaso-injected micromere (Ca) was transplanted in between the veg1 and veg2 cell layers of a control 60-cell stage embryo. Gcm, blue; Tbr, orange. At pre-hatched blastula stage, 22 out of the 25 embryos (88%) transplanted with a control micromere showed ectopic expression of Gcm (Bb), whereas none of the Delta-MASO-injected micromeres induced ectopic Gcm expression (Cb). Bb and Cb are two-channel fluorescence and DIC images; Bc is a confocal projection of Bb with nuclear staining in blue. (Da-Dh) Duration of the Delta signal is crucial for the maintenance of Gcm expression. (Da,Db) Control embryos at 6 and 9 hours post-fertilization (hfp). (Dc) Micromereless embryos at 9 hpf. (Dd,De) Embryos in which all micromeres have been replaced by a single green dyed micromere at the 16-cell stage. In Dd, the transplanted micromere has been maintained up to fixation, in De it has been removed just before fixation. In Df, Dg and Dh, the transplanted micromere was eliminated at 6, 7 or 8 hpf, respectively, and the embryos were fixed at 9 hpf. (A-D) All embryos are in vegetal view, except for Ab, Db, Dd and De, which are in lateral view.
As the Veg2U cells first express, then lose Gcm when they are separated from a Delta source following the eighth cleavage, we next asked for how long the Delta signal was necessary before Gcm expression becomes independent of the Delta input. Obviously, for the Veg2U cells reception of Delta for up to about an hour or so was not long enough to maintain Gcm expression. A delayed micromere removal experiment was designed to examine this question. At the 16-cell stage, the four endogenous micromeres of a control embryo were removed and in their place a single micromere stained with CFDA, a green vital fluorescent dye, was inserted. That single inserted micromere was sufficient to induce reasonably normal Gcm expression at the eighth cleavage (6 hpf; Fig. 5De) or later (8 hpf; Fig. 5Dd). Furthermore, if that transplanted micromere was removed at the eighth cleavage and the embryo immediately fixed and stained for Gcm, Gcm expression was similar to control levels (Fig. 5De). By contrast, if after removal at the eighth cleavage (6 hpf) the embryos were further incubated until 9 hpf and then analyzed, Gcm mRNAs were no longer detected, even in the Veg2L cells (Fig. 5Df). Thus, by 6 hpf the Veg2L cells, despite normally expressing Gcm, had not yet received Delta for long enough to stabilize their expression of Gcm. If instead, the transplanted micromere was removed at 7 hpf, a slight expression of Gcm was retained (Fig. 5Dg), and if taken away at 8 hpf, the retention of Gcm expression was at levels approaching those of controls (Fig. 5Dh). Thus, not only is Delta required for Gcm induction, it is required as a continuous input for an extended period of time, about 2.5 hours (more than two cell cycles), before Gcm expression can continue without further additional Delta/Notch inputs.
DISCUSSION
The results presented here provide new insights into the mechanisms that underlie endomesoderm diversification during sea urchin embryogenesis. By investigating the spatial distribution of the transcripts of three known regulators of this process (Delta, Gcm and FoxA), throughout time, and conjointly with cell lineage, we illustrate for the first time the spatiotemporal dynamics of molecular events that define part of the GRN that conducts endomesoderm segregation. This key cell fate decision process occurs for at least one group of blastomeres at hatched blastula stage, when Gcm and FoxA mRNAs are first detected in two distinct cell populations, the Veg2L and the Veg2U cells, respectively. Another major component of this study is the discovery that the delivery duration of the Delta signal plays a crucial role in maintaining mesoderm fate. It is only after an extended period of delivery of the Delta signal that the mesoderm GRN reaches a state of autonomy, although even then transfating experiments have shown that the Veg2L progeny remain capable of switching fates for an extended period of time (Ettensohn and McClay, 1988). The normal fate of the Veg2L cells appears to be the entire non-skeletal mesoderm. Ruffins and Ettensohn showed in a cell lineage study at mesenchyme blastula that there were 66 non-skeletogenic mesoderm cells plus 8 small micromeres. At hatched blastula after 9th cleavage this study counted 60 non-skeletogenic mesoderm cells in the Veg2L and four small micromeres (Ruffins and Ettensohn, 1996). Assuming that a few cells divide between the hatched blastula and the mesenchyme blastula stages, and that the small micromeres divide once, these data suggest that the Veg2L layer contains all non-skeletogenic mesoderm cells.
The molecular dynamics of endomesoderm segregation
Endomesoderm classically was defined based on cell lineage analyses showing that at some point a diversification occurs so that the progeny of this tissue segregates into mesoderm and endoderm. At a molecular level, endomesoderm cells can be defined by the co-expression of genes known to be specifically expressed by, and required later for, definitive endoderm and mesoderm (Rodaway et al., 1999). In such cases, the diversification mechanism usually results in elimination of the expression of one set of genes in one lineage and of a different set in the other lineage. In sea urchin, endomesoderm diversification contains an example of this molecular divergence pattern. Gcm is involved in the specification and differentiation of the pigment cells, a non-skeletogenic mesenchyme cell derivative (Ransick et al., 2002), and FoxA is expressed in, and is required for, endoderm development (Oliveri et al., 2006). When these two genes are co-expressed it can be argued thus that the cells expressing the two are still endomesoderm. Of course it also could be argued that other genes in the GRN of that cell already have biased the cell in one direction or another, but even if that were the case the co-expression of Gcm and FoxA indicates that the cell is proximal to the time of diversification. A similar situation has been reported in zebrafish where the transcription factors Gata5 and Her5 are known as endodermal molecules, and no tail (ntl) and gsc are mesodermal genes. In two independent studies on endomesoderm segregation, the authors report the co-expression of gata5 and ntl in blastula embryos (Rodaway et al., 1999), and the detection in the same cells of her5 and gsc prior to the onset of gastrulation (Bally-Cuif et al., 2000). In both cases, the co-expression takes place prior to the subsequent segregation of the endoderm and mesoderm genes into different cell populations, as we report here for the sea urchin. Furthermore, in zebrafish, Her5 is a negative regulator of Gsc expression (Bally-Cuif et al., 2000), and FoxA is reported to be a repressor of Gcm transcription in sea urchin (Oliveri et al., 2006). Thus, as well as the existence of an analogous intermediate GRN state, an analogous regulatory feature might help to control the endomesoderm diversification process in both a vertebrate and an invertebrate.
FoxA clearance from the Veg2L cells
At hatched blastula stage, FoxA mRNAs disappear from the Veg2L cells, where they have been present for up to an hour. At that stage many events occur in those cells; Delta signaling from the micromeres continues, thereby activating Gcm and the mesoderm GRN, nuclear β-catenin is gradually and stochastically lost (Logan et al., 1999), Nlk (Nemo-like kinase), an inhibitor of the canonical Wnt pathway, becomes expressed (Rottinger et al., 2006), and, finally, Delta appears in the Veg2L cells (Fig. 3). Exactly how FoxA is eliminated from the Veg2L cells remains a detail to be clarified. Fig. 4 shows that ectopic expression of Delta prevents FoxA expression. However, it is unlikely that the endogenous expression of Delta in the Veg2L cells at hatched blastula is the signal that eliminates FoxA expression. FoxA mRNA in the Veg2L disappears, with our time scale, at the same time as Delta expression is activated in those Veg2L cells. Fig. 3 shows that Delta is first expressed in micromeres at the fifth cleavage but the first sign of Gcm expression in response to that early Delta signal is two cleavages later, or more than an hour later. Because FoxA disappears at the time the later Delta is activated in Veg2L, there simply is not enough time for the Veg2L Delta production and signal transduction to cause FoxA repression. Rather, it is far more likely that the same apparatus that controls Delta expression in the Veg2L also controls FoxA repression. Analysis of the FoxA cis-regulatory apparatus is likely to resolve this puzzle.
The Delta/Notch pathway is required to initiate and maintain mesoderm fate
In sea urchin, the Delta/Notch pathway plays a central role in mediating mesoderm development. Functional analyses carried out on both the ligand and the receptor have shown that loss of Notch pathway activity leads to a significant decrease or a complete elimination of mesoderm derivatives, while ectopic activation of Notch signaling increases the number of mesoderm cells (Sherwood and McClay, 1999; Sweet et al., 2002). In this study, Delta mRNAs are detected as early as the fifth cleavage (32-cell stage; see Fig. S1 in the supplementary material), and at both the sixth and seventh cleavage stages (60-cell and 120-cell stage, respectively) transcripts are observed in the small and the large micromere progeny. Here, ISH analyses were performed using fluorescent technologies that are more sensitive than the conventional chromogenic NBT/BCIP method used in an earlier study that did not detect Delta until later (Sweet et al., 2002). This initial expression of Delta at the fifth cleavage is consistent with quantitative PCR and ISH data obtained elsewhere in support of the endomesoderm gene regulatory network model at http://sugp.caltech.edu/endomes/#EndomesNetwork and described by Smith and Davidson (Smith and Davidson, 2008).
To transmit its signal the Delta/Notch pathway relies on the interaction between a receptor and a ligand that are both single-pass transmembrane proteins. A direct cell-to-cell contact is thus required for the Notch pathway to be activated (Fehon et al., 1990). In the sea urchin, Gcm is identified as a direct target of Delta/Notch signaling (Ransick and Davidson, 2006). At the mid-blastula stage, though, the first stage at which Gcm mRNAs are detected, Gcm is found in two cell layers, the Veg2U and the Veg2L layers, one of which (the former) is no longer in direct contact with the Delta-positive cells. To explain this observation, Gcm transcription ought to be activated by the Notch pathway prior to the eighth cleavage (the cleavage that forms Veg2U and Veg2L). Because transcription of Delta begins at the fifth cleavage, the Delta protein is likely to begin signaling at least by early seventh cleavage when the veg2 cells still form only one layer. Assuming there is some delay in Gcm mRNA appearance after activation of the Notch receptor, activation of Suppressor of Hairless (SuH), and finally the activation of Gcm by SuH, it is likely that Gcm mRNA begins to accumulate before the end of cleavage, but is first detected during the eighth cleavage. The Veg2U cells, by expressing Gcm, support the conclusion that Delta-Notch signaling began during the seventh cleavage or even earlier. Moreover, in Strongylocentrotus purpuratus, Gcm expression is first detected at the seventh cleavage in the veg2 layer (Ransick et al., 2002). Thus, we conservatively assume that the Delta signal is first delivered by the middle of the seventh cleavage cycle. This conservative estimate allows for over an hour for Delta mRNA to be transcribed, translated and transported to the membrane of micromeres, and for Notch signal transduction to activate Gcm expression.
At the ninth cleavage (late blastula stage), Gcm is no longer present in the Veg2U cell descendants. This disappearance is a consequence of the loss of contact between these cells and the Delta-producing cells. In support of this notion, transplantation of a Delta-positive source in direct contact with Veg2U cells was sufficient to maintain Gcm expression in these cells up to hatched blastula stage. In addition, the delayed micromere removal experiment reveals that the Delta signal is necessary for an extended period of time before mesoderm fate continues in the absence of further Delta input. Continuous Delta activation for around 2.5 hours after we calculate the Delta signal is initiated is required before Gcm expression can continue on its own, and the mesoderm fate can therefore be stabilized. It remains to be seen, however, how the Delta signal shapes the detailed GRN of the mesoderm cells. Even though the direct target for Delta-Notch, via SuH, is Gcm, it is likely that other components of the mesoderm GRN also are necessary for stabilized activation of this GRN state. It is possible, as suggested by Ransick and Davidson (Ransick and Davidson, 2006), that Gcm regulates itself, but other regulatory mechanisms might well operate to stabilize the system as well. Work in progress that expands our understanding of the NSM GRN will demonstrate how the stabilization actually happens.
Supplementary Material
Acknowledgements
The authors are grateful to D. McIntyre and G. Lhomond for reviewing this manuscript and for fruitful interactions. We thank Nina Hu for tirelessly counting cells. This work was supported by National Institutes of Health grants NIH HD 14483 and GM 81883 to D.R.M. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.044149/-/DC1
References
- Bally-Cuif L., Goutel C., Wassef M., Wurst W., Rosa F. (2000). Coregulation of anterior and posterior mesendodermal development by a hairy-related transcriptional repressor. Genes Dev. 14, 1664-1677 [PMC free article] [PubMed] [Google Scholar]
- Cameron R. A., Fraser S. E., Britten R. J., Davidson E. H. (1991). Macromere cell fates during sea urchin development. Development 113, 1085-1091 [DOI] [PubMed] [Google Scholar]
- Croce J., Lhomond G., Gache C. (2003). Coquillette, a sea urchin T-box gene of the Tbx2 subfamily, is expressed asymmetrically along the oral-aboral axis of the embryo and is involved in skeletogenesis. Mech. Dev. 120, 561-572 [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Rast J. P., Oliveri P., Ransick A., Calestani C., Yuh C. H., Minokawa T., Amore G., Hinman V., Arenas-Mena C., et al. (2002). A genomic regulatory network for development. Science 295, 1669-1678 [DOI] [PubMed] [Google Scholar]
- Ettensohn C. A., McClay D. R. (1988). Cell lineage conversion in the sea urchin embryo. Dev. Biol. 125, 396-409 [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., Muskavitch M. A., Artavanis-Tsakonas S. (1990). Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61, 523-534 [DOI] [PubMed] [Google Scholar]
- Fortini M. E., Artavanis-Tsakonas S. (1994). The suppressor of hairless protein participates in notch receptor signaling. Cell 79, 273-282 [DOI] [PubMed] [Google Scholar]
- Fukuda K., Kikuchi Y. (2005). Endoderm development in vertebrates: fate mapping, induction and regional specification. Dev. Growth Differ. 47, 343-355 [DOI] [PubMed] [Google Scholar]
- Gillis W. J., Bowerman B., Schneider S. Q. (2007). Ectoderm- and endomesoderm-specific GATA transcription factors in the marine annelid Platynereis dumerilli. Evol. Dev. 9, 39-50 [DOI] [PubMed] [Google Scholar]
- Kimelman D., Griffin K. J. (2000). Vertebrate mesendoderm induction and patterning. Curr. Opin. Genet. Dev. 10, 350-356 [DOI] [PubMed] [Google Scholar]
- Logan C. Y., Miller J. R., Ferkowicz M. J., McClay D. R. (1999). Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development 126, 345-357 [DOI] [PubMed] [Google Scholar]
- Maduro M. F. (2006). Endomesoderm specification in Caenorhabditis elegans and other nematodes. BioEssays 28, 1010-1022 [DOI] [PubMed] [Google Scholar]
- McClay D. R., Peterson R. E., Range R. C., Winter-Vann A. M., Ferkowicz M. J. (2000). A micromere induction signal is activated by β-cateninand acts through notch to initiate specification of secondary mesenchyme cells in the sea urchin embryo. Development 127, 5113-5122 [DOI] [PubMed] [Google Scholar]
- Nishida H., Satoh N. (1985). Cell lineage analysis in ascidian embryos by intracellular injection of a tracer enzyme. II. The 16- and 32-cell stages. Dev. Biol. 110, 440-454 [DOI] [PubMed] [Google Scholar]
- Oliveri P., Walton K. D., Davidson E. H., McClay D. R. (2006). Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development 133, 4173-4181 [DOI] [PubMed] [Google Scholar]
- Ransick A., Davidson E. H. (2006). cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev. Biol. 297, 587-602 [DOI] [PubMed] [Google Scholar]
- Ransick A., Rast J. P., Minokawa T., Calestani C., Davidson E. H. (2002). New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev. Biol. 246, 132-147 [DOI] [PubMed] [Google Scholar]
- Rodaway A., Patient R. (2001). Mesendoderm. an ancient germ layer? Cell 105, 169-172 [DOI] [PubMed] [Google Scholar]
- Rodaway A., Takeda H., Koshida S., Broadbent J., Price B., Smith J. C., Patient R., Holder N. (1999). Induction of the mesendoderm in the zebrafish germ ring by yolk cell-derived TGF-beta family signals and discrimination of mesoderm and endoderm by FGF. Development 126, 3067-3078 [DOI] [PubMed] [Google Scholar]
- Rottinger E., Croce J., Lhomond G., Besnardeau L., Gache C., Lepage T. (2006). Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development 133, 4341-4353 [DOI] [PubMed] [Google Scholar]
- Ruffins S. W., Ettensohn C. A. (1996). A fate map of the vegetal plate of the sea urchin (Lytechinus variegatus) mesenchyme blastula. Development 122, 253-263 [DOI] [PubMed] [Google Scholar]
- Sherwood D. R., McClay D. R. (1997). Identification and localization of a sea urchin Notch homologue: insights into vegetal plate regionalization and Notch receptor regulation. Development 124, 3363-3374 [DOI] [PubMed] [Google Scholar]
- Sherwood D. R., McClay D. R. (1999). LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development 126, 1703-1713 [DOI] [PubMed] [Google Scholar]
- Smith J., Davidson E. H. (2008). Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc. Natl. Acad. Sci. USA 105, 20089-20094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119 [DOI] [PubMed] [Google Scholar]
- Sweet H. C., Gehring M., Ettensohn C. A. (2002). LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development 129, 1945-1955 [DOI] [PubMed] [Google Scholar]
- Technau U., Scholz C. B. (2003). Origin and evolution of endoderm and mesoderm. Int. J. Dev. Biol. 47, 531-539 [PubMed] [Google Scholar]
- Warga R. M., Nusslein-Volhard C. (1999). Origin and development of the zebrafish endoderm. Development 126, 827-838 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.