Abstract
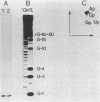
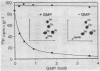
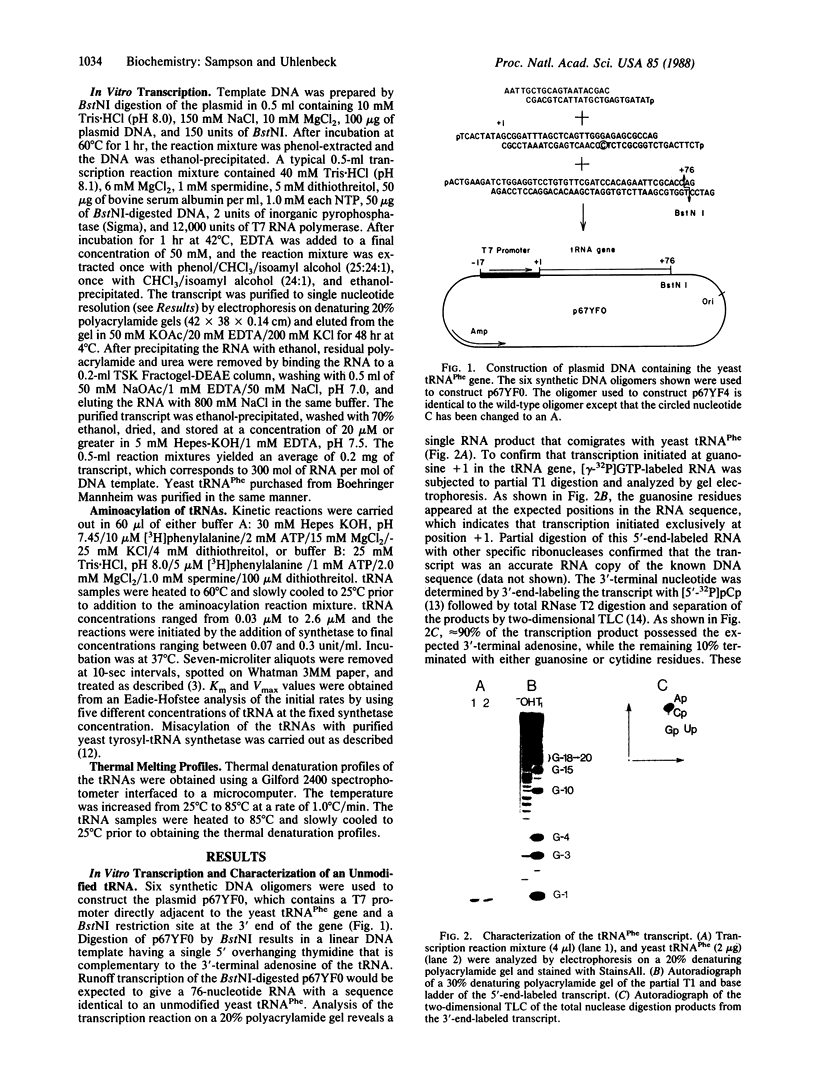
A recombinant plasmid was constructed with six synthetic DNA oligomers such that the DNA sequence corresponding to yeast tRNA(Phe) is flanked by a T7 promoter and a BstNI restriction site. Runoff transcription of the BstNI-digested plasmid with T7 RNA polymerase gives an unmodified tRNA of the expected sequence having correct 5' and 3' termini. This tRNA(Phe) transcript can be specifically aminoacylated by yeast phenylalanyl-tRNA synthetase and has a Km only 4-fold higher than that of the native yeast tRNA(Phe). The Km is independent of Mg2+ concentration, whereas the Vmax is very dependent on Mg2+ concentration. Comparison of the melting profiles of the native and the unmodified tRNA(Phe) at different Mg2+ concentrations suggests that the unmodified tRNA(Phe) has a less stable tertiary structure. Using one additional DNA oligomer, a mutant plasmid was constructed having a guanosine to thymidine change at position 20 in the tRNA gene. A decrease in Vmax/Km by a factor of 14 for aminoacylation of the mutant tRNA(Phe) transcript is observed.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bare L., Uhlenbeck O. C. Aminoacylation of anticodon loop substituted yeast tyrosine transfer RNA. Biochemistry. 1985 Apr 23;24(9):2354–2360. doi: 10.1021/bi00330a034. [DOI] [PubMed] [Google Scholar]
- Been M. D., Cech T. R. Sites of circularization of the Tetrahymena rRNA IVS are determined by sequence and influenced by position and secondary structure. Nucleic Acids Res. 1985 Dec 9;13(23):8389–8408. doi: 10.1093/nar/13.23.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Enzymatic replacement of the anticodon of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1982 Mar 2;21(5):855–861. doi: 10.1021/bi00534a007. [DOI] [PubMed] [Google Scholar]
- Carbon P., Ebel J. P. In vitro construction of yeast tRNAAsp variants: nucleotide substitutions and additions in T-stem and T-loop. Nucleic Acids Res. 1987 Mar 11;15(5):1933–1950. doi: 10.1093/nar/15.5.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Sprinzl M., Watanabe K., Albani M., Kersten H. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 1979 Apr;6(4):1571–1581. doi: 10.1093/nar/6.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher T. W., Bujarski J. J., Hall T. C. Mutant viral RNAs synthesized in vitro show altered aminoacylation and replicase template activities. Nature. 1984 Sep 13;311(5982):171–175. doi: 10.1038/311171a0. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Church G. M., Kim S. H. RNA-ligant interactions. (I) Magnesium binding sites in yeast tRNAPhe. Nucleic Acids Res. 1977 Aug;4(8):2811–2820. doi: 10.1093/nar/4.8.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Litt M., Greenspan C. M. Kethoxal inactivation of three transfer ribonucleic acids chargeable by yeast phenylalanyl transfer ribonucleic acid synthetase. Biochemistry. 1972 Apr 11;11(8):1437–1442. doi: 10.1021/bi00758a017. [DOI] [PubMed] [Google Scholar]
- Loftfield R. B., Eigner E. A., Pastuszyn A. The role of spermine in preventing misacylation by phenylalanyl-tRNA synthetase. J Biol Chem. 1981 Jul 10;256(13):6729–6735. [PubMed] [Google Scholar]
- McCutchan T., Silverman S., Kohli J., Söll D. Nucleotide sequence of phenylalanine transfer RNA from Schizosaccharomyces pombe: implications for transfer RNA recognition by yeast phenylalanyl-tRNA synthetase. Biochemistry. 1978 May 2;17(9):1622–1628. doi: 10.1021/bi00602a007. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka E., Tanaka S., Tanaka T., Miyake T., Markham A. F., Nakagawa E., Wakabayashi T., Taniyama Y., Nishikawa S., Fukumoto R. Total synthesis of a RNA molecule with sequence identical to that of Escherichia coli formylmethionine tRNA. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5493–5497. doi: 10.1073/pnas.78.9.5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B., Michael M., Dudock B. Function of N2 methylguanine in phenylalanine transfer RNA. Nat New Biol. 1973 Dec 5;246(153):135–138. doi: 10.1038/newbio246135a0. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Graeser E. Role of the 5'-terminal phosphate of tRNA for its function during protein biosynthesis elongation cycle. Nucleic Acids Res. 1980 Oct 24;8(20):4737–4744. doi: 10.1093/nar/8.20.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow H., Guthrie C. Structure of intron-containing tRNA precursors. Analysis of solution conformation using chemical and enzymatic probes. J Biol Chem. 1984 Apr 25;259(8):5197–5207. [PubMed] [Google Scholar]
- Vacher J., Springer M., Buckingham R. H. Functional mutants of phenylalanine transfer RNA from Escherichia coli. EMBO J. 1985 Feb;4(2):509–513. doi: 10.1002/j.1460-2075.1985.tb03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. B., Qiu M. S., Liang Z. H., Zheng K. Q., Wu R. L., Wang C. Y., Liu X. Y., Zheng H. D., Bao Y. D., Zhu X. L. Synthesis of the 3'-half molecule of yeast alanine tRNA. Sci Sin B. 1983 May;26(5):482–494. [PubMed] [Google Scholar]
- Zeevi M., Daniel V. Aminoacylation and nucleoside modification of in vitro synthesised transfer RNA. Nature. 1976 Mar 4;260(5546):72–74. doi: 10.1038/260072a0. [DOI] [PubMed] [Google Scholar]