Abstract
New modes of humoral recognition have been identified by studies of antibodies that neutralize human immunodeficiency virus type 1 and influenza A viruses. Understanding how such modes of antibody-antigen recognition can occur in the context of sophisticated mechanisms of humoral evasion has implications for the development of effective vaccines. Here we describe eight modes of antibody recognition first observed with human immunodeficiency virus type 1. Similarities to four of these modes have been identified with antibodies to a conserved ‘stem’ epitope on influenza A viruses. We outline how each of these different modes of antibody recognition is particularly suited to overcoming a specific viral evasion tactic and assess potential routes of re-elicitation in vaccine settings.
Human immunodeficiency virus type 1 (HIV-1) generates a persistent infection, whereas infection with influenza A virus can normally be cleared within a few days. Generally, neutralizing antibodies are readily elicited to either virus, but they target regions of the viral envelope with little functional constraint, and viral evolution outpaces antibody adaptation1,2. The end result is high titers of strain-specific antibodies with little or no broadly neutralizing ability3. However, high-throughput screening of large numbers of HIV-1-infected people indicates that a substantial subpopulation of infected people develop antibodies after 5–10 years that can effectively neutralize diverse strains of primary HIV-1 viruses4–7. In addition, certain monoclonal antibodies derived from HIV-1-infected people are able to neutralize multiple strains and subtypes8. But for influenza, until very recently, the identification of such broadly neutralizing antibodies effective against multiple subtypes has been much more elusive.
The mechanisms by which broadly neutralizing antibodies overcome the formidable immune-evasion mechanisms of the HIV-1 envelope (Env) glycoprotein spike9–12 (extensive glycosylation, hypervariability of amino acid sequences, conformational masking and inaccessibility of conserved sites) have been the subject of intense interest, as elicitation of such antibodies potentially represents a direct route to a vaccine. Such antibodies seem to develop only after several years of infection. Does their rarity indicate that they are ‘freaks of nature’ that might not be re-elicited easily? Is an extended period of antigen exposure required for elicitation? And what actual antigen or antigens led to their initial elicitation? For influenza virus, the annual trivalent vaccine suffices for the most part and usually protects from the viruses circulating at the time. But the isolated, although highly lethal, occurrences of avian influenza or ‘bird flu’ in the human population in the past few years, as well as the present 2009 ‘swine flu’ outbreak, have heightened fears about the next influenza pandemic. Here we describe eight newly identified modes of antibody recognition for HIV-1, which were not anticipated from decades of investigation of antibody-antigen recognition in model systems, such as the mouse. We compare and contrast these with three independent findings of a particular germline family of antibodies that are effective against multiple subtypes of influenza virus. Finally, we discuss how the lessons learned from these new antibody-binding modes with HIV-1 might be applied to vaccine design, not only for vaccines against HIV-1 but also for those against viruses with similar mechanisms of immune evasion.
Glycan recognition by ‘domain-swapped’ antibody 2G12
The HIV-1 gp120 Env glycoprotein is one of the most heavily glycosylated viral proteins identified so far, with approximately 25 N-linked glycans in about 500 residues. The sequence Asn-X-Ser (Thr) (where ‘X’ is any amino acid and ‘Ser (Thr)’ indicates serine or threonine at that position) specifies the addition of a large preformed sugar moiety of about 3 kilodaltons to the nascent polypeptide chain. Processing of this sugar moiety in the endoplasmic reticulum and Golgi first trims the precursor dolichol-linked glycan to a highly restricted core set of sugars (high-mannose glycans) and then adds an enormous diversity of other sugars (for hybrid and complex glycans). Because such N-linked glycans are derived from the host glycan pathways themselves, they are seen by the immune system as ‘self’ (which thus shields underlying protein epitopes from antibody recognition). How then does the human immune system recognize such glycans as ‘non-self’, as the antibody 2G12 has been able to do?
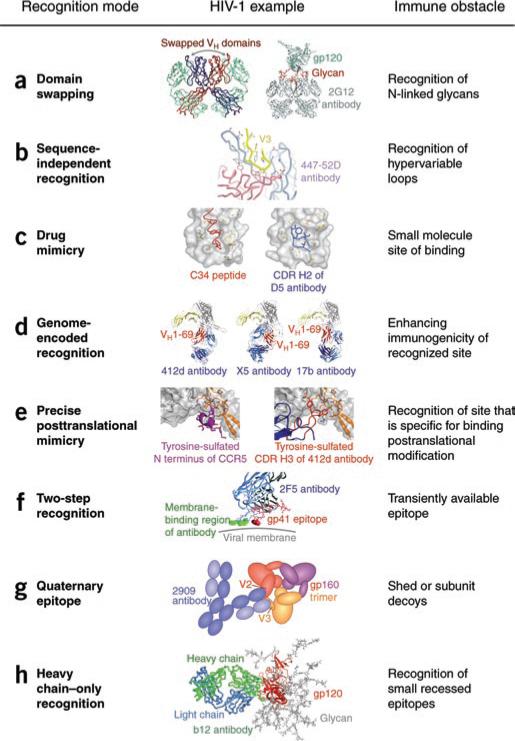
Structural analysis shows that 2G12 adopts a highly unusual ‘domain-swapped’ architecture in which the variable heavy chains are exchanged on adjacent antigen-binding (Fab) arms13 (Fig. 1a). Such intermolecular swapping converts the bivalent arms of the antibody into a single, multivalent surface that expands the antibody combining site from around 20 Å2 × 30 Å2 (suitable for recognition of a single N-linked glycan) to 20 Å2 × 60 Å2 (suitable for recognition of up to three N-linked glycans). The antibody, therefore, not only has acquired the ability to see ‘self’ glycans as foreign because of their clustering on the gp120 surface but also has achieved high-affinity binding (in the nanomolar range) in much the same way that lectins and other carbohydrate-binding proteins do through multivalent recognition. The presence of clustered high-mannose glycans on HIV-1 gp120 seems to be a consequence of the extraordinary high density of N-linked glycans on gp120, which limits glycan processing in the Golgi. These high-mannose glycans are also relatively well conserved, as they are used by HIV-1 to interact with receptors, such as DC-SIGN14, to gain entry into host cells. Analysis of 2G12, therefore, has shown that although N-linked glycans are normally not recognized by antibody as ‘non-self’, a combination of glycan-cluster recognition and high-mannose restriction achieves the required specificity and affinity.
Figure 1.
New modes of antibody recognition demonstrated by HIV-1-reactive antibodies. (a) Domain swapping: exchange of the heavy-chain variable (VH) domains on adjacent antigen-binding arms of the antibody 2G12 (left) rigidifies the interface between the arms, which permits recognition of three glycan moieties of gp120 (right). (b) Sequence-limited recognition: binding of the hypervariable V3 loop of gp120 (yellow) to the antibody 447-52D (light chain, salmon; heavy chain, sky blue), which uses mainly sequence-independent interactions with main-chain atoms. (c) Drug mimicry: binding of the C34 peptide55 (left), a slightly smaller version of the drug Fuzeon, and binding of the CDR H2 of antibody D5 (right) to gp41 (gray). (d) Germline-encoded recognition: binding of the VH1-69 encoded regions (red) of various CD4-induced antibodies to gp120 (gray) and CD4 (yellow). Despite recognition of the similar regions on gp120, the relative position of the VH1-69-encoded region varies. (e) Precise post-translational mimicry: the cleft between gp120 core (gray) and V3 loop (orange) specifically recognizes O-sulfated tyrosine on CCR5 (left) and the antibody 412d (right). (f) Two-step recognition: the epitope for the antibody 2F5 is thought to be only transiently available; corecognition of this epitope in the context of membrane increases binding affinity. (g) Quaternary-specific recognition: the antibody 2909 does not recognize gp120 or gp41 as separate units but only as the assembled functional Env viral spike. (h) Heavy chain–only recognition: binding of the antibody b12 to its gp120 epitope using only its heavy chain (green).
Sequence-independent recognition by V3-reactive antibodies
A second means by which HIV-1 evades humoral recognition involves the use of exposed hypervariable loops that readily elicit an antibody response (one consequence of which is the highly specific antibody response to the inducing strain, at least for the first year or more of infection). The hypervariable V3 region, for example, shows up to 6% sequence divergence in a single infected person in a single year, comparable to the yearly global divergence observed with pandemic strains of flu15. Despite this extensive variation, a few amino acids at the tip of the V3 loop are conserved, as they are required for interaction with the chemokine coreceptor CCR5.
Structural analysis of the antibody 447-52D shows how a single antibody can bind with high affinity to sequence-divergent V3 loops on diverse HIV-1 isolates16 (Fig. 1b). This antibody binds a twelve-residue epitope, but sequence-specific recognition is found only for three residues, Gly-Pro-X-Arg, that are highly conserved at the loop apex. The antibody contacts are mainly to the main chain of the V3 loop, such that a three- to four-stranded β–sheet is formed between the heavy chain of the antibody's third complementarity-determining region (CDR H3) and the gp120 V3 loop. Substitution of the other amino acids in the V3 loop is permitted as long as the β-hairpin structure is maintained.
Antibody F425-B4e8 expands on the general theme described above and can neutralize diverse isolates from HIV-1 clades B, C and D. Analysis of its structure in complex with its V3 peptide epitope shows sequence-specific recognition for only two residues, a highly conserved isoleucine several residues before the tip and another conserved arginine residue at the loop tip17. Interestingly, the V3 loop is crowned by an unusual five-residue α-turn, which is distinct from the normal four-residue β-turn recognized by 447-52D. The ability of these two antibodies to neutralize overlapping populations of viral isolates shows that the tertiary structure of the loop itself is in part induced by the recognizing antibody. Even with some structural variation, as few as two amino acids seem to be sufficient for specific antibody recognition of this hypervariable portion of V3. These antibodies can therefore use, to a large extent, sequence-independent recognition of these otherwise hypervariable loops to attain interaction with a broad range of HIV-1 isolates. This mode of antibody recognition thus allows recognition of traces of functional conservation in the epitope largely through interaction with the polypeptide backbone of the hypervariable V3 loop.
Drug mimicry by antibody D5
The examples described above demonstrate how exposure of even a few conserved elements (amino acids or sugars) can form the basis of successful antibody recognition and virus neutralization. The conserved surfaces on the envelope proteins that are required for receptor recognition and membrane fusion and that facilitate entry into the host cell are also potential targets of neutralization. To shield these surfaces from immune recognition, HIV uses a variety of mechanisms, such as conformational masking, in which the viral envelope spike adopts a structural conformation that minimizes exposure of the conserved surfaces essential for function and maximizes exposure of glycosylated or hypervariable surfaces. Thus, structural rearrangements to generate receptor- and fusion-competent states of Env (gp120-gp41) are triggered only after binding to the cell surface receptor CD4. In the CD4-induced conformation, the conserved surfaces required for function are transiently exposed and can become available as targets for antibody and drugs. The drug Fuzeon, for example, binds to portions of the gp41 surface that in primary isolates of HIV-1 are exposed or assembled only in the CD4-triggered conformation18,19.
In yet another mode of antibody recognition, identified by selection for antibodies that might mimic interaction of a drug with the Env viral antigen (Fig. 1c), antibody D5 was identified by phage-display library screening against a designed antigen to emulate the interaction with Fuzeon20. Mimicry here occurs on several levels: the antibody is shown to ‘mimic’ Fuzeon, and Fuzeon was initially shown to mimic the C-terminal helix of gp41 in its post-fusion state. In the context of antibody recognition, however, the important point is that the drug-binding studies initially identified a region of potential vulnerability on the antigen that enabled selection of a neutralizing antibody. It should be noted, however, that criteria for drug binding differ from those for antibody binding. Thus, for example, the interaction between gp41 on the virus and the host cell seems to be much more sterically constrained; although the drug Fuzeon is able to readily access the site in vivo, the much larger antibody seems to be sterically restricted, which greatly diminishes its potency.
Genomic VH1-69 recognition by CD4-induced antibodies
The ability to restrict accessibility as a means of immune evasion is especially evident for CD4-induced antibodies that bind to the coreceptor-binding site on gp120. In terms of sequence conservation, the coreceptor-binding site is the most highly conserved surface on gp120 but is only properly configured after CD4 induction, when the virus encounters CD4 at the cell surface21. Furthermore, at the virus-cell interface, molecular modeling suggests that steric hindrance prevents antibody access12. Despite such restrictions, high titers of CD4-induced antibodies are elicited in most HIV-1-infected people22. A partial rationale for this apparent anomaly has been derived by sequence and structural analyses of CD4-induced antibodies, which have shown that most of these antibodies arise from the heavy-chain genomic precursor VH1-69 (ref. 23). Of all the human heavy-chain genomic precursors, only VH1-69 contains a hydrophobic, second complementarity-determining region (CDR H2), and structural analysis of CD4-induced antibodies in complex with gp120 has shown that CD4-induced antibody recognition seems to have two main requirements: a hydrophobic CDR H2 and an acidic CDR H3 (ref. 23). This decrease in the requirement for the generation of diversity in recognition by variable-diversity-joining recombination, through direct encoding of a beneficial germline recognition element in a genomic precursor (Fig. 1d), allows both VH-use bias and greater ease of elicitation of CD4-induced antibodies.
Precise post-translational mimicry by antibody 412d
The primary coreceptor used by HIV-1 in entry, CCR5, is modified on at least two N-terminal residues by post-translational O-sulfation of tyrosine24. Such tyrosine sulfation is essential for gp120-CCR5 interactions and, interestingly, for interaction of gp120 with several CD4-induced antibodies25. Structural analysis of gp120 in complex with the CD4-induced antibody 412d has identified tyrosine-sulfated residues in the CDR H3 region of 412d, which bind to a highly conserved cleft in gp120 (ref. 26). Docking studies, meanwhile, have shown that a tyrosine-sulfated residue on CCR5 binds to the same cleft26. These studies define yet another mode of recognition: precise post-translational mimicry (Fig. 1e). As the sequence signal that specifies O-sulfation is complex (involving tyrosine residues, negatively charged aspartic and glutamic acid residues, and an absence of phenylalanine), such post-translational mimicry requires the coordinated participation of one of the antibody CDRs, which, not unexpectedly, is the most diverse CDR H3. This discovery again provides a novel and unexpected mechanism for an antibody to mimic the natural receptor of the virus and, thus, to acquire the requisite specificity and affinity for recognition of a functionally conserved site on the virus.
Membrane binding by antibodies 2F5 and 4E10
The membrane-proximal external region of HIV-1 Env is the site of recognition for antibodies 2F5 and 4E10, two of the most broadly neutralizing antibodies to HIV identified so far27,28. Analysis of these antibodies has shown that not only is the membrane-proximal external region recognized on gp41 but also involvement of a strong membrane-binding component is indicated: when the membrane-proximal external region epitope is presented in the context of membrane, antibody binding is enhanced by more than an order of magnitude29. Subsequent deconvolution has identified dual binding sites for the antibody to the sequence of the membrane-proximal external region and directly to the membrane30. Such a two-component interaction (Fig. 1f) allows the energy of binding to be shared between sequence-specific features on the epitope and invariant features of the host cell membrane. In addition, it is possible that the 2F5– and 4E10–membrane-proximal external region epitopes are only transiently available for antibody interaction and membrane binding by antibody would thus enable efficient two-dimensional scanning for this transient epitope.
Quaternary epitope recognition by antibody 2909
The converse of an epitope that requires two-component antibody recognition is a two-component epitope recognized by a single antibody. Antibody 2909 is such an antibody31. It recognizes a quaternary epitope, which is thought to consist of part of the V2 loop on one gp120 and part of the V3 loop on a neighboring gp120 protomer in the Env spike (Fig. 1g). As the appropriate binding surface is assembled only on the functional spike, this antibody has the interesting property of being unreactive with soluble forms of the Env spike, including gp120, gp41 and soluble versions of the spike ectodomain. Although such quaternary specificity allows antibody 2909 to escape decoy mechanisms of viral evasion, the antibody itself is extremely strain specific, perhaps because its epitope encompasses two highly variable loops (V2 and V3). It is, nonetheless, extremely potent in its neutralization, with a picomolar 50% inhibitory concentration against the SF162 isolate31, roughly 1,000 times more potent than the broadly neutralizing antibodies b12 or 4E10. Whether quaternary-specific antibodies such as 2909 can be identified with greater neutralization breadth is being explored; tantalizing data concerning new broadly neutralizing antibodies that bind oligomeric spikes better than monomeric units (gp120 or gp41) have been reported32.
Heavy chain–only recognition by antibody b12
Because CD4 is the primary receptor for HIV-1, unlike the secondary sites for coreceptor or Fuzeon binding, the initial site of contact on gp120 for CD4 must always be exposed and available to form the initial encounter complex. However, the overall size of the initial site of contact is small and confined mainly to the outer domain of gp120 (ref. 33). Although this surface is partially recessed on the Env viral spike, antibody b12 nonetheless manages to target this primary contact site and effectively neutralize HIV-1.
Structural analysis has shown that to effect recognition, b12 uses yet another trick, that of heavy chain–only recognition33 (Fig. 1h). Most epitopes are bound at the juncture between heavy and light chains of the antibody, which has the advantage of potentially using all six CDRs of the antibody, as well as the natural binding cleft between heavy- and light-chain surfaces. However, if the target epitope is small and fairly recessed, this type of double-headed Fab recognition may encounter steric barriers. Such ideas have been put forward before (for example, the famous ‘canyon hypothesis’34), but the beauty of the immune system is its ability to find solutions to even the most daunting problem. One potential way to reach into a ‘canyon’ involves the use of extended CDR H3 regions, as has been directly observed in the structure of a rhinovirus-antibody complex35. Indeed, human antibodies that recognize viral pathogens have CDR H3 regions that are longer than average36, and b12, for example, has a rigid CDR H3 that protrudes 15 Å above the rest of the antibody paratope37. However, the criteria for accessing the b12 epitope seem to be strict enough to require a second means of accessing a canyon, that of heavy chain–only recognition.
Crystallographic analysis of the interaction between antibodies and viral envelopes has shown that although heavy-chain recognition often predominates, some level of light-chain recognition is always observed. Heavy chain–only recognition indicates antibody variability is confined to only three CDRs, two of which are germline encoded and together have only about 50 variants in humans. Heavy chain–only recognition thus greatly limits combinatorial diversity but perhaps generates a smaller footprint on the antigen to prevent immune escape. Such variation, however, can be generated by other means—for example, through somatic mutations. Indeed, analysis of the b12 sequence shows more somatic mutations than are obtained with CD4-induced antibodies (45, but for b12, versus 19 ± 5, for the various CD4-induced antibodies)23, and all of the main residues involved in the b12-gp120 interaction seem to be somatically mutated from the germline sequence33. Although CDR recognition using a maximum of two to three loops is possible, as has been shown with shark and camelid antibodies, respectively, which naturally have unpaired immunoglobulin heavy chains, such antibodies have additional means of generating diversity38,39. It will be useful to determine if such exclusive heavy-chain recognition also requires concomitant selection of extended CDR H3 regions in such antibodies. Another case involving influenza virus is discussed below.
Influenza virus ‘stem-reactive’ broadly neutralizing antibodies
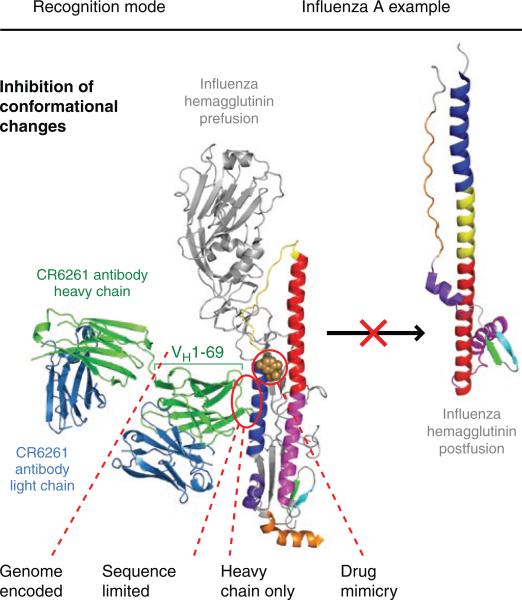
Despite decades of analysis of the influenza virus, antibodies that broadly neutralize different strains and subtypes of influenza virus have been scarce to nonexistent40. Almost all known influenza antibodies bind to the globular head region of the hemagglutinin Env spikes and seem to neutralize by preventing receptor binding. Like most antibodies to HIV-1, the response is confined to the hypervariable loops or regions of the hemagglutinin heads that can easily mutate to escape recognition. In an unexpected series of discoveries, three independent studies have identified ‘stem’ antibodies that cross-react with multiple influenza subtypes41–43. It is not entirely clear why such antibodies have been so difficult to identify, but this may relate to the less-than-optimal accessibility of the hemagglutinin stem on native influenza virions. A report several years ago provided rather compelling evidence that the broadly neutralizing C179 antibody40 might bind to the extended stem region of the hemagglutinin, but no structural work has yet confirmed that finding. Crystal structures of the antibodies CR6261 and F10 (Fab and single-chain variable fragment, respectively), bound to the hemagglutinin from the H5NI subtype of influenza virus42,43 and to the hemagglutinin from the pandemic 1918 H1N1 virus43, have shown that they both recognize a highly conserved functional site in the stem region and block membrane fusion by preventing the pH-induced conformational change (Fig. 2). Interestingly, these antibodies belong to the same VH1-69 family of germline genes as the HIV-1 CD4-induced antibodies and, furthermore, use only the heavy chain for interaction, as does antibody b12 to bind to the CD4-binding site on gp120. However, what is distinctly different is that most of the contacts are provided by residues encoded by the original, unmutated germline gene segments with almost no interaction with CDR H3, except for the tyrosine at position 98, which is highly conserved in the D regions and their various reading frames. Sequence conservation with antibodies extracted from a library made from survivors of a Turkish H5N1 avian influenza infection41 suggests that these antibodies also recognize the same hemagglutinin epitope. Thus, these antibodies are very unusual in that they represent essentially an immediate and effective germline response, with almost no somatic mutations, that is very different from most of the highly evolved broadly neutralizing antibodies to HIV-1, such as b12, which accumulate up to 50 or more mutations15, presumably over a long period of time. These antibodies also stabilize a functional site on the hemagglutinin and prevent the switch to the conformation required for membrane fusion.
Figure 2.
Features of HIV-1 antibody recognition used by influenza hemagglutinin-reactive antibodies. Several of the features of the HIV-1 antibodies are recapitulated in the antibodies CR6261 and F10, including heavy chain–only binding, germline-encoded recognition through a VH1-69 response, binding to recessed hydrophobic pockets that could be potential drug targets56 and sequence-independent recognition, although mainly on the antibody side for the formation of main-chain hydrogen bonds. The actual recognition mode, stabilization of the prefusion conformation that prevents the conformational rearrangement of HA2 required for membrane fusion, has been observed in other contexts: with HIV-1, the antibody D5 binds to a fusion intermediate, which prevents subsequent conformational changes for fusion20; and with Ebola virus, the antibody KZ52 binds to the Ebola viral stem and locks the spike into its prefusion conformation57.
These new influenza antibodies have additional similarities to and differences from their HIV-1 counterparts. In some aspects, they emulate 4E10 and 2F5, which recognize the conserved stem region that has been linked to membrane fusion. The heavy chain–only binding is also similar to that adopted by b12, but the extensive use of the germline framework region 3 in CR6261 expands the surface on the heavy chain that can be used for antigen recognition. Although the epitope is highly conserved and mostly helical (70% of the total buried surface), as for the 4E10 epitope, carbohydrate masking prevents interaction with group 2 influenza viruses, characterized by the H3 and H7 subtypes. Finally, the accessibility problem is probably more of an issue for the membrane-proximal site on influenza virus than that on HIV-1 because of the close packing and greater density of the hemagglutinin and neuraminidase spikes (around 450 on average) on the influenza envelope compared with the paucity of the Env proteins (five to ten spikes) on HIV-1 (refs. 44,45).
Elicitation strategies
The discovery of additional modes of antibody recognition naturally leads to the question of utility: how does such knowledge inform attempts to re-elicit such responses in vivo? The ability of structure-based efforts to assist in vaccine design is beginning to have an effect46, and in these and related efforts, antibody ‘clues’ can provide useful insight. In the case of the unusual, domain-swapped recognition by 2G12, it is clear that a relatively small change in the pairing of residues in the VH-VL interface, combined with substitutions in the hinge region (such as insertion of a single proline residue) and the new VH-VH interface, can foster domain swapping. More important than the actual logistics of how a particular mode of recognition might be recreated may be the underlying criteria for recognition: for 2G12, this involves recognition of self sugars that form dense clusters on the viral surface that are not present on the host cell. Thus, a re-elicitation strategy based on clustered arrays of high-mannose sugars seems logical, although it remains to be seen if this will result in the elicitation of domain-swapped antibodies.
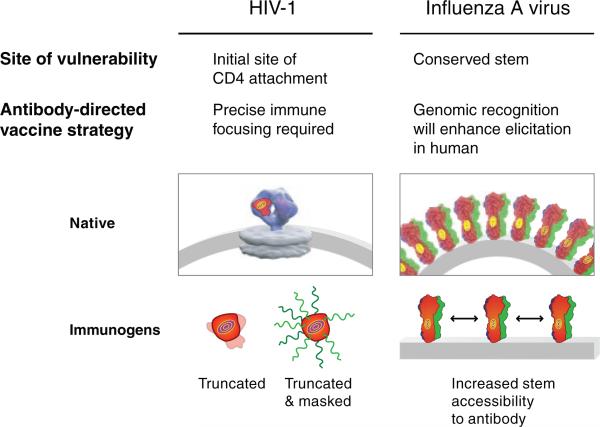
In the same manner, for recognition of intermediate steps of the viral entry pathway, as discussed above, the particulars of drug mimicry or of tyrosine sulfation may not be as important as the knowledge that a particular conformation is formed only transiently. Fixing or stabilizing the immunogen into that particular conformation could assist with the re-elicitation of similar types of antibodies. Similarly, with the two-component recognition of 2F5 and 4E10, the particulars of membrane recognition may not be as important as the use of immunogens that place the appropriate gp41 epitope into a more native, membrane context. For the elicitation of antibodies able to target a small recessed surface, heavy chain–only recognition may not be as important as devising methods of immune focusing to achieve very precise target recognition (Fig. 3). Immunization strategies may also need to take into account much greater than average (or previously anticipated) affinity maturation, as noted for b12 recognition, perhaps by increasing the immunization time frames to allow longer durations of affinity maturation.
Figure 3.
Antibody-directed strategies of vaccine design. Unusual features of antibody recognition may provide clues to the immunological characteristics that allow elicitation of antibodies to a particular site. For example, precise immune focusing may be needed to elicit antibodies to the epitopes recognized by the HIV-1 and influenza antibodies b12, F10 and CR6261, which use heavy chain–only recognition. Immunofocusing techniques, such as glycan masking, in which all surfaces other than the site of vulnerability (‘bulls-eye’) are covered by N-linked glycosylation58, and truncation, in which surfaces other than the site are deleted59,60, may have utility in this context (electron tomogram image of the native HIV-1 spike adapted from ref. 61). With the influenza antibodies, additional clues, such as VH1-69 use with low somatic mutation, suggest that a portion of the target site is recessed and hydrophobic in nature and that elicitation of appropriate antibodies in humans will be enhanced by the direct genomic encoding of recognition elements. Elicitation in vivo may require spacing of spikes far enough apart to increase access of antibody to the conserved stem target, although truncation and masking strategies of immunofocusing should also have utility.
The targeting of conserved functional sites, such as the receptor-binding site on the membrane-distal regions of the Env spikes seems in principle to be more amenable to elicitation of the appropriate antibodies, and many strategies are now being investigated. For influenza viruses, the sialic acid receptor–binding site is very small and shallow and difficult to target without encountering the surrounding hypervariable loops. However, even for influenza, some antigenic regions may be more conserved across subtypes and give rise to cross-protective antibodies47.
Conclusions
Analysis of interactions of the immune system with HIV-1 and influenza has clearly succeeded in identifying additional modes of antibody recognition not anticipated from the plethora of previous studies mainly using mouse antibodies. Such modes in HIV-1 may arise because the persistent nature of a typical HIV-1 infection, often accompanied by a concomitant high antigen load, drives immune adaptation to incredible feats of recognition. For influenza virus, the urgency of the situation may require an immediate germline response with no time to embark on the normal course of affinity maturation by somatic mutation before death ensues, as for H5N1 and 1918 influenza viruses. Whether these different modes of binding coupled to identification of the precise epitopes can be used in vaccine (or therapeutic48,49 or gene-transfer50) settings remains to be seen, but we believe it's worth exploring. VH1-69 recognition has also been observed elsewhere (in hepatitis C recognition51) and is likely to arise again. Finding antibody interactions that mimic previously identified drug-binding sites also seems likely to occur. Single chain–only recognition, perhaps for both light and heavy chains, will almost certainly be found in other systems, although most of these examples at present have been provided by selection of antibodies using phage display that can readily access large antibody repertoires.
In terms of utility, it seems that the particular mode of recognition may not be as important as what it shows about the underlying strategies for escaping immune detection by microbial pathogens. A tremendous amount has been learned from the study of these HIV-1 and influenza antibodies, and it is encouraging to note that broadly neutralizing antibodies are being found with increasing frequency as a consequence of enhanced focus as well as emerging new techniques52,53. Although the challenges presented by HIV-1, influenza virus and other evasive pathogens are daunting, the broadly neutralizing antibodies discussed above and the presence of high titers of neutralizing antibodies in subsets of infected people suggest that the armamentarium and repertoire of the human immune system are up to the challenge54. Whether modern vaccine design can appropriately harness this potential remains to be seen.
ACKNOWLEDGMENTS
We thank M. Elsliger, G. Nabel and L. Shapiro for comments on the manuscript; D. Ekiert for assistance with Figure 2; J. Stuckey for Figures 1–3; and members of the Structural Biology Section, Vaccine Research Center for comments on the manuscript. Supported by the National Institutes of Health (intramural program and grants) and by the International AIDS Vaccine Initiative.
Footnotes
Published online at http://www.nature.com/natureimmunology/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Contributor Information
Peter D Kwong, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA..
Ian A Wilson, Department of Molecular Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, California, USA..
References
- 1.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 2.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss RA, et al. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature. 1985;316:69–72. doi: 10.1038/316069a0. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhillon AK, et al. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 2007;81:6548–6562. doi: 10.1128/JVI.02749-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doria-Rose NA, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sather DN, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binley JM, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyatt R, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 10.Starcich BR, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 11.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 12.Labrijn AF, et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 14.Geijtenbeek TB, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 15.Korber B, et al. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 2001;58:19–42. doi: 10.1093/bmb/58.1.19. [DOI] [PubMed] [Google Scholar]
- 16.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447–52D. Structure. 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Bell CH, et al. Structure of antibody F425–B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J. Mol. Biol. 2008;375:969–978. doi: 10.1016/j.jmb.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan C, Lu H, Qi Z, Jiang S. Synergistic efficacy of combination of enfuvirtide and sifuvirtide, the first- and next-generation HIV-fusion inhibitors. AIDS. 2009;23:639–641. doi: 10.1097/QAD.0b013e328325a4cd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colman PM. New antivirals and drug resistance. Annu. Rev. Biochem. 2009 Mar 2; doi: 10.1146/annurev.biochem.78.082207.084029. published online, doi:10.1146/annurev.biochem.78.082207.084029. [DOI] [PubMed] [Google Scholar]
- 20.Luftig MA, et al. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat. Struct. Mol. Biol. 2006;13:740–747. doi: 10.1038/nsmb1127. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, et al. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 22.Decker JM, et al. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2005;201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang CC, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. USA. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farzan M, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 25.Choe H, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114:161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 26.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muster T, et al. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muster T, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alam SM, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorny MK, et al. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan-Hui P-Y, et al. Keystone Symposium on Prevention of HIV/AIDS, poster 119. Keystone; Colorado: 2009. Isolation of HIV-neutralizing human monoclonal antibodies from memory B cell repertoire using short term culture and high-throughput binding and neutralization screens. [Google Scholar]
- 33.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossmann MG. The canyon hypothesis. Viral Immunol. 1989;2:143–161. doi: 10.1089/vim.1989.2.143. [DOI] [PubMed] [Google Scholar]
- 35.Smith TJ, Chase ES, Schmidt TJ, Olson NH, Baker TS. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- 37.Saphire EO, et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 38.De Genst E, Saerens D, Muyldermans S, Conrath K. Antibody repertoire development in camelids. Dev. Comp. Immunol. 2006;30:187–198. doi: 10.1016/j.dci.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Dooley H, Flajnik MF. Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol. 2006;30:43–56. doi: 10.1016/j.dci.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 40.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kashyap AK, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. USA. 2008;105:5986–5991. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi M, Danev R, Nishiyama K, Sugawara K, Nagayama K. Zernike phase contrast electron microscopy of ice-embedded influenza A virus. J. Struct. Biol. 2008;162:271–276. doi: 10.1016/j.jsb.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Ksenofontov AL, Badun GA, Fedorova NV, Kordiukova LV. An approach the quantitative determination of the area of glycoprotein spikes at the surface of enveloped viruses. Mol. Biol. (Mosk.) 2008;42:1093–1096. [PubMed] [Google Scholar]
- 46.Dormitzer PR, Ulmer JB, Rappuoli R. Structure-based antigen design: a strategy for next generation vaccines. Trends Biotechnol. 2008;26:659–667. doi: 10.1016/j.tibtech.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida R, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W, Dimitrov DS. Human monoclonal antibodies and engineered antibody domains as HIV-1 entry inhibitors. Curr. Opin. HIV AIDS. 2009;4:112–117. doi: 10.1097/COH.0b013e328322f95e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez O, Tsibane T, Basler CF. Neutralizing anti-influenza virus monoclonal antibodies: therapeutics and tools for discovery. Int. Rev. Immunol. 2009;28:69–92. doi: 10.1080/08830180802593540. [DOI] [PubMed] [Google Scholar]
- 50.Johnson PR, et al. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. doi: 10.1038/nm.1967. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan CH, Hadlock KG, Foung SK, Levy SV. (H)1–69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood. 2001;97:1023–1026. doi: 10.1182/blood.v97.4.1023. [DOI] [PubMed] [Google Scholar]
- 52.Yu X, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 54.Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 56.Russell RJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. USA. 2008;105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantophlet R, Wilson IA, Burton DR. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 2003;77:5889–5901. doi: 10.1128/JVI.77.10.5889-5901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyatt R, et al. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu L, et al. Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored HIV-1 gp120 domain. J. Virol. 2009;83:5077–5086. doi: 10.1128/JVI.02600-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]