Abstract
SARS-CoV infection of human results in antigen-specific cellular and humoral immune responses. However, it is critical to determine whether SARS-CoV-specific memory T cells can persist for long periods of time. In this study, we analyzed the cellular immune response from 21 SARS-recovered individuals who had been diagnosed with SARS in 2003 by using ELISA, CBA, ELISpot and multiparameter flow cytometry assays. Our results demonstrated that low levels of specific memory T cell responses to SARS-CoV S, M, E and N peptides were detected in a proportion of SARS-recovered patients, and IFN-γ was the predominant cytokine produced by T cells after stimulation with peptides. Cytometry analysis indicated that the majority of memory CD8+ T cells produced IFN-γ, whereas memory CD4+ T cells produced IFN-γ, IL-2 or TNF-α. These results might provide valuable information on the cellular immune response in recovered SARS-CoV patients for the rational design of vaccines against SARS-CoV infection.
Keywords: Cellular Immune Response, Ionomycin, ELIspot Assay, Intracellular Cytokine Staining, Cytometric Bead Array
Introduction
SARS is a newly emerged infectious disease that has caused the deaths of hundreds of infected individuals. SARS-CoV is a positive-sense RNA virus that consists of three membrane proteins: spike (S), membrane (M) and envelope (E) and a nucleocapsid core (containing the structural component of helical nucleocapsid, N protein). It has been demonstrated that the viral surface proteins are associated with the embedding in the cell membrane of host cells, and N proteins play an important role in replication and RNA packaging [6, 10, 11, 17].
Previous studies by us and others have demonstrated that SARS-CoV spike, envelope, membrane and nucleocapsid proteins could elicit cellular immune responses, and the immune responses of SARS-CoV antigen-specific memory T cells were detectable more than 1 year after natural human infection. In addition, a proportion of immunodominant T-cell epitopes of SARS-CoV M, E and N peptides have been identified [12, 13, 19, 20]. Moreover, the existence of immunodominant B cell epitopes and neutralizing antibody responses have also been reported in both human and animals. More recently, Cao and his colleagues found that the levels of SARS-CoV-specific antibody appear to decrease 4 years after the burst of SARS [1, 7, 21]. Currently, however, there is no information available on the cellular immune response in SARS-CoV patients 4 years after recovery.
To investigate the persistence and quality of memory T cell responses to SARS-CoV in infected human, we analyzed the cellular immune response from SARS-recovered individuals who had been diagnosed as having SARS during the period of January–March in 2003.
Materials and methods
Subjects
Fully SARS-recovered individuals (n = 21; 2 males and 19 females, aged 22–44) were recruited in this study from the Second Affiliated Hospital of Sun Yat-sen University, Guangzhou, Guangdong, China. All participants had been diagnosed as having SARS patients based on clinical examination during the period of January–March 2003. The diagnostic criteria for SARS-CoV infection followed the World Health Organization’s definition of SARS, including a fever (temperature >38°C), a chest radiograph of the thorax showing evidence of new consolidation with respiratory symptoms (e.g. cough and shortness of breath) and a history of close contact with a person in whom SARS had been diagnosed. The diagnosis was further confirmed by serological detection of SARS-CoV-specific antibodies [8]. Ten normal subjects (2 males and 8 females, aged 24–52) without any contact history with SARS patients were used as negative controls. Our protocol was approved by the Clinical Research Ethics Committee of the Sun Yat-sen University, and informed consent was obtained from all participants.
Synthetic peptides
SARS-CoV S, M, E and N peptides were kindly provided by Drs. Koup and Bailer at the Vaccine Research Center (VRC), the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), USA. A total of 169 peptides spanning the entire SARS-CoV S protein, 30 peptides spanning the entire sequence of SARS-CoV M protein, 9 peptides spanning the entire sequence of SARS-CoV M protein and 57 peptides spanning the entire sequence of the SARS-CoV N protein (each peptide was 15–20 amino acids in length and overlapped by 10 amino acids) were used in these experiments.
Preparation of PBMCs
Venous blood was collected in a tube containing sodium heparin (15 U/ml, Shanghai Biochemistry Pharmaceuticals Company, Shanghai, China) and diluted 1:1 in Hank’s balanced salt solution. PBMCs were isolated by density centrifugation through a Histopaque 1.077 gradient (Shanghai Huajing, Shanghai, China) and washed twice in Hank’s solution. Cells were resuspended at a concentration of 2 × 106/ml in complete RPMI 1640 medium (GIBCO BRL, CA, USA) containing 10% heat-inactivated fetal bovine serum (Hyclone, UT, USA), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine and 1,000× 2-Mercaptoethahol (2-ME), all of which were purchased from GIBCO BRL.
IFN-γ secretion assays
PBMCs were seeded into 96-well culture plates (Becton Dickinson) (2 × 105 cells/well) in triplicate. S, M, E or N peptides and costimulatory mAb anti-CD28 (BD Biosciences Pharmingen, CA, USA) were each added to the wells at 1 μg/ml. In addition, PBMCs were stimulated with 20 ng/ml phorbol-12-myristate-13-acetate (PMA) (Sigma, St. Louis, MO, USA) and 1 μg/ml ionomycin (Sigma) as positive controls, and unstimulated PBMCs were used as negative controls. The plates were incubated for 72 h at 37°C in a humidified atmosphere containing 5% CO2. Supernatants were collected, and the level of IFN-γ was measured using an ELISA kit (BD Biosciences Pharmingen) according to the manufacturer’s instruction. The detection limit of the IFN-γ assay kit was 4.9 pg/ml.
Cytometric bead array analysis for cytokines
PBMCs from SARS-recovered donors (patients 1, 2, 5, 12 and 20) were seeded in 96-well plates with or without S, M, E or N peptides and anti-CD28 for 72 h. The culture supernatants were harvested, and Th1/Th2 cytokine production was measured with cytometric bead array (CBA) by flow cytometry (Becton Dickinson Immunocytometry Systems, CA, USA). Briefly, five bead populations with distinct fluorescence intensities were coated with capture antibodies specific for different cytokines including IFN-γ, IL-2, TNF-α, IL-4 and IL-10. After the beads were incubated with 50 μl of diluted supernatants, different cytokines in the sample were captured by their corresponding beads. The cytokine-captured beads were then mixed with phycoerythrin-conjugated detection antibodies to form sandwich complexes. Following incubation, fluorescence samples were washed, harvested and analyzed using CBA 6-bead analysis software (BD Biosciences Pharmingen). The assay sensitivity of these five cytokines was 20 pg/ml.
IFN-γ ELIspot assays
An ELIspot assay for IFN-γ (BD Biosciences Pharmingen) was performed. In brief, 96-well plates were coated with anti-IFN-γ mAb at 4°C overnight. The plates were washed twice before blocking with complete culture medium. Fresh PBMCs were plated in triplicate at 2 × 105 cells per well. S, M, E or N peptides and anti-CD28 were each added at 1 μg/ml. PBMCs were stimulated with 20 ng/ml PMA and 1 μg/ml ionomycin as positive controls, and unstimulated PBMCs were used as negative controls. After incubation for 20 h at 37°C in a humidified atmosphere containing 5% CO2, the cells were removed and incubated with biotinylated anti-human IFN-γ detection antibody for 2 h at room temperature. After washing, wells were developed for 1 h with streptavidin-HRP and incubated with substrate reagent according to the manufacturer’s protocol. Spot-forming cells (SFCs) were detected using the ELIspot image analysis system (Sage Creation, Beijing, China). The frequency of IFN-γ-producing cells was calculated as the number of spots/number of total PBMCs per well. The number of spots in negative control wells was in the range of 0–2 spots.
Cell-surface and intracellular cytokine staining and flow cytometric analysis
The following monoclonal antibodies (mAbs) were used in this study: anti-CD4 PerCP and anti-CD8 PerCP were purchased from BD Bioscience Immunocytometry Systems; anti-CD8 PE-Cy7, anti-IFN-γ APC, anti-IL-2 PE, anti-TNF-α FITC and isotype-matched control antibodies were obtained from BD Biosciences Pharmingen.
For cell-surface and intracellular cytokine staining and flow cytometric analysis, PBMCs were stimulated with or without S, M, E or N peptides in the presence of anti-CD28. After the first 1 h of incubation, brefeldin A (10 μg/ml Sigma-Aldrich) was added to the cultures to enable intracellular protein to accumulate in all stimulations. After incubation for a total of 5 h, the cells were washed twice in PBS and fixed for 8 min at room temperature in fixation buffer (PBS containing 4% paraformaldehyde). After an additional washing step, the cells were resuspended in permeabilization buffer (PBS containing 0.1% BSA, 0.05% sodium azide and 0.1% saponin) at 4°C for 2 h. The cells were then stained with anti-CD4, anti-CD8, anti-IFN-γ, anti-IL-2 and anti-TNF-α. After staining, all samples were washed and resuspended in FACS buffer (PBS containing 0.1% BSA, and 0.05% sodium azide), and more than 300,000 cells were acquired on a FACS Calibur flow cytometer. The data were analyzed by using FlowJo software (Tree Star Inc., OR, USA).
Statistical analysis
The levels of cytokines and numbers of IFN-γ-producing cells were compared using Student’s t test between recovered SARS patients and healthy controls in the same condition. P < 0.05 was considered significant.
Results
Quantification of SARS-CoV S-, M-, E- and N-peptide-specific IFN-γ production 4 years after recovery
To determine whether memory-T cell-mediated immune responses were persistent in individuals who had completely recovered 4 years ago from SARS-CoV infections, PBMCs were prepared from peripheral blood of normal donors and SARS-CoV-recovered individuals. The cells were incubated in the presence or absence of a pool of SARS-CoV-derived peptides including S, M, E and N with or without anti-CD28 mAb. After stimulation for 3 days, the levels of IFN-γ in the cell cultures were assessed by ELISA. The results in Fig. 1a show that in the absence of stimulation, there was no detectable level of IFN-γ production. Remarkably, the stimulation of PBMCs from recovered individuals but not from normal donors with SARS-CoV S peptides resulted in significantly high levels of IFN-γ production (P < 0.01). Similar results were also obtained when the cells were stimulated with SARS-CoV N peptides (P < 0.05). Although the cells that were stimulated with M peptides produced IFN-γ, the level of IFN-γ production was not significantly different compared to using medium alone (P > 0.05). In addition, no IFN-γ was detected in SARS-CoV-recovered individuals after stimulation with SARS-CoV E peptides (P > 0.05). Furthermore, no marked difference in the production of IFN-γ by recovered individuals and healthy donors was observed when the PBMCs were stimulated with PMA plus ionomycin (P > 0.05). The statistical results showed that 66.70, 28.57, 14.29, and 38.10% of the SARS-CoV-recovered individuals tested in this study responded to stimulation by S, M, E, and N peptides, respectively, to produce IFN-γ (data not shown). Further analysis revealed that 9.52% of SARS-CoV-recovered persons produced IFN-γ in response to all four peptides, 14.29% to three, 19.05% to two, 28.57% to one, and 28.57% did not respond to any of four peptide antigens (Fig. 1b).
Fig. 1.

Production of IFN-γ by PBMCs from SARS-recovered individuals in response to SARS-CoV S, M, E and N peptides. a PBMCs from SARS-recovered individuals (n = 21) and normal control donors (n = 7) were stimulated with or without S, M, E and N peptides for 72 h. Cells stimulated with PMA plus inomycin were used as positive controls. The culture supernatants were collected and tested for the production of IFN-γ by ELISA. All assays were performed in triplicate. Each individual point represents one result from one donor. Bars indicate mean values. *P < 0.05, **P < 0.01, NS not significant (P > 0.05). b Percentage of donors responding to S, M, E and N peptides. ++++ percentage of donors responding to four all of S, M, E and N peptides; +++ percentage of donors responding to three of S, M, E and N peptides; ++ percentage of donors responding to two of S, M, E and N peptides; + percentage of donors responding to one of S, M, E and N peptides; − percentage of donors responding to none of S, M, E and N peptides
To confirm and extent the results obtained from ELISA, CBA was carried out to determine the production of Th1/Th2 cytokines in cell-free culture supernatants after cells from SARS-CoV-recovered individuals were stimulated with SARS-CoV peptides for 3 days. Consistent with the data obtained from ELISA, the results from CBA showed that IFN-γ, but not IL-2, TNF-α, IL-4 or IL-10, could be detected following stimulation with the peptides. The concentrations of IFN-γ were variable, depending on the types of SARS-CoV peptides. Among the four types of SARS-CoV peptides (Fig. 2a–d), S peptides induced the highest levels of IFN-γ production (Fig. 2a). These results suggested that IFN-γ-producing cells were persistent 4 years after recovery from SARS-CoV infection.
Fig. 2.
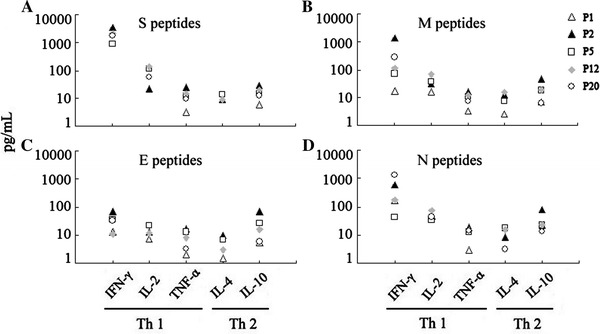
Cytometric bead array (CBA) analysis of Th1/Th2 cytokine production by PBMCs from SARS-recovered individuals in response to S, M, E and N peptides. PBMCs from five SARS-recovered donors (patient 1, 2, 5, 12 and 20) were seeded in 96-well plates with or without S, M, E and N peptides for 72 h. The culture supernatants were collected, and Th1/Th2 cytokine production was measured serially by CBA using a FACSCalibur flow cytometer. The data were generated in graphical format using the BD CBA software. The assay sensitivity with these five cytokines was 20 pg/ml. a S peptides, b M peptides, c E peptides, d N peptides
Frequency of IFN-γ-producing cells from SARS-CoV-recovered individuals following stimulation with SARS-CoV S, M, E and N peptides 4 years after recovery
To evaluate the frequency of antigen-specific IFN-γ-producing cells, PBMCs from SARS-CoV-recovered individuals and normal donors were stimulated with SARS peptides, and IFN-γ-producing cells were counted using the ELIspot assay. The results in Fig. 3a, from five representative individuals (P1, P2, P5, P12 and P20), indicated that no spots were observed when PBMCs were cultured in the absence of stimulation. Patient 1 only responded to N peptides to produce IFN-γ, patient 2 and patient 12 responded to S, M and N but not E peptides, patient 5 responded to S and N peptides, and patient 20 responded to all four peptides. The statistical results in Fig. 3b show that the majority of SARS-recovered individuals responded significantly to S and N peptides compared to medium (P < 0.01). Few of them responded to M and E peptides. Among SARS-CoV-recovered individuals tested in this study, the statistical results showed that 61.90% of them responded to S peptides, 28.57% responded to M, 9.52% responded to E and 33.33% responded to N peptides (data not shown). Further analysis, shown in Fig. 3c, indicated that 4.76% of individuals responded to all four peptides, 9.52% responded to three peptides, 33.33% responded to two peptides, 19.05% responded to one peptide and 33.33% did not respond to any of the four peptides. These results demonstrate that memory T cells from SARS-CoV-recovered individuals in response to SARS-CoV stimulation are heterogeneous.
Fig. 3.

IFN-γ-producing cells in PBMCs from SARS-recovered individuals in response to S, M, E and N peptides. a Representative images of IFN-γ ELISPOT results from five donors, patients 1, 2, 5, 12 and 20. b Mean spot numbers in PBMCs from 21 SARS-recovered donors and 7 healthy control donors in response to S, M, E and N peptides. The number of spots in the medium controls ranged from 0 to 2. All assays were performed in triplicate. Bars indicate mean values. *P < 0.05, **P < 0.01, NS: P > 0.05. # P2 in response to S, M and N; △P15 in response to S and N. c Percentage of donors responding to S, M, E and N peptides. ++++ percentage of donors responding to all four of S, M, E and N peptides; +++ percentage of donors responding to three of S, M, E and N peptides; ++ percentage of donors responding to two of S, M, E and N peptides; + percentage of donors responding to one of S, M, E and N peptides; − percentage of donors responding to none of S, M, E and N peptides
Characterization of SARS-CoV S, M, E and N peptide-specific memory T cells
To determine the subpopulations of cytokine-producing cells, PBMCs from recovered individuals with SARS-CoV infection and healthy unexposed donors were stimulated with SARS-CoV S, M, E and N peptides, respectively. After incubation for 5–6 h, cells were harvested, washed and stained with antibodies to CD4 and CD8 together with the antibodies to cytokines IFN-γ, IL-2 and TNF-α. After incubation, cells were washed, acquired by FACS Calibur and analyzed using FlowJo software. The statistical results in Fig. 4a indicate that after stimulation with SARS-CoV S, M, E or N peptides, both CD4+ and CD8+ T cells expressed IFN-γ. Among these, the S peptides induced a markedly higher percentage of IFN-γ-producing cells in the recovered patients than in normal donors (P < 0.05), and the percentage of IFN-γ-producing CD8+ T cells was higher than that of CD4+ T cells when stimulated with all four peptides. In addition, we detected the frequency of IL-2 and TNF-α-producing memory T cells in SARS-CoV-recovered individuals, and the results in Fig. 4a–d indicate that cytokine-producing CD4+ and CD8+ T memory cells (including both IL-2 and TNF-α-producing cells) were very rare, and there was no significant difference compared to the healthy donors (P > 0.05). In the analysis of multiple functional CD4+ and CD8+ T memory cells, we found that the majority of SARS-CoV S, M, E or N peptide-responding CD8+ T memory cells were IFN-γ-producing cells, and CD4+ T memory cells could produce IFN-γ, IL-2 or TNF-α, except those from patient 12, after stimulation with S peptides (Fig. 4e). These data indicate that IFN-γ production was the main property of SARS-CoV-responding CD8+ T cells, whereas CD4+ T cells produced different cytokines but only expressed one of the Th1 cytokines.
Fig. 4.
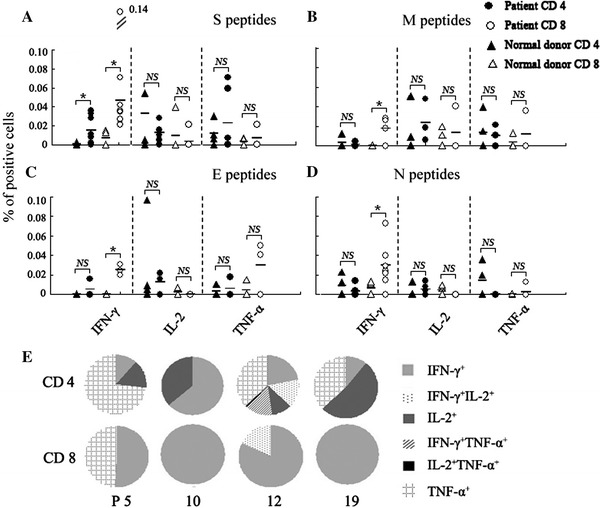
Characterization of SARS-CoV S-, M-, E- and N-antigen-specific cytokine-producing cells in PBMCs. PBMCs from SARS-recovered donors and healthy unexposed individuals were incubated with S, M, E or N peptides for 6 h. Cell-surface and intracellular cytokine staining for IFN-γ, IL-2 and TNF-α were performed. More than 300,000 cells were acquired on a FACSCalibur flow cytometer. The cells were first gated on CD4+ T or CD8+ T cells and subsequently analyzed for cytokine expression. The results shown are the percentage of cytokine-producing cells in CD4+ or CD8+ T cells from different patients after stimulation and subtracting the isotype control. Each individual point represents one result from one donor. Bars indicate mean values. *P < 0.05, NS: P > 0.05. a S peptides, b M peptides, c E peptides, d N peptides. e Multiple cytokine analysis of CD4+ and CD8+ T cells following S peptide stimulation
Discussion
In the current study, we analyzed the cellular immune responses in individuals who had recovered from SARS after stimulation with SARS-CoV peptides ex vivo. Our results demonstrated that very low responses of SARS-CoV specific memory T cells were detected in a proportion of individuals 4 years after natural infection. In line with our observations, it has recently been shown that the antibodies to SARS-CoV waned over the time [1].
Memory T cells consist of both CD4+ and CD8+ T cells that can rapidly acquire effector functions to kill and eliminate infected cells and secrete cytokines to inhibit replication of pathogens and to regulate immune responses. After stimulation with antigen, memory CD4+ T cells differentiate into effector cells. According to the cytokine production, CD4+ T cells can be divided into Th1 and Th2 cells. Th1 cells secrete IFN-γ, IL-2 and TNF-α and participate in the cellular immune response. On the other hand, Th2 cells secrete IL-4, IL-5, IL-10 and IL-13 and enhance humoral immune response [9, 14–16]. Our previous studies on immune response of SARS-recovered individuals have demonstrated that after natural infection, both CD4+ and CD8+ T cells are involved in immune responses to SARS-CoV S, M, E and N antigens. Memory CD8+ T cells display an effector memory cell phenotype expressing CD45RO− CCR7− CD62L−. In contrast, the majority of IFN-γ+ CD4+ T cells are central memory cells expressing CD45RO+ CCR7+ CD62L− [12, 13, 19, 20]. The existence of memory T cells in response to SARS-CoV antigens after immunization of mice with DNA vaccine has been described. T cell proliferation, IFN-γ production, DTH responses and cytotoxic T cell activities as well as the induction of high levels of specific antibodies were also demonstrated in our studies and those of other [3–5].
In addition, the heterogeneity of Th1 cells indicated that T cells specific for SARS-CoV antigen could be divided into three subsets based on IL-2 and IFN-γ expression, including IFN-γ+ cells, IL-2+ and IFN-γ+ cells and IL-2+ cells [18]. It has been confirmed recently that Th1 cells producing multiple cytokines are more efficient as effector cells. Single CD4+ IFN-γ-producing cells are more limited in mediating optimal and sustained protection. The factors controlling the quality of Th1 responses seem to be the amount and duration of antigen, the type of antigen-presenting cell and the innate cytokine milieu [2]. In our study, we found that in the absence of antigen, memory T cells can still persist for 4 years in a proportion of SARS-CoV-recovered individuals, but most of the memory Th1 cells and memory CD8+ T cells are single-cytokine-producing cells. This might imply that the memory T cells will ultimately become unresponsive or die. The poor Th1 response could be one of the reasons for the diminished antigen-specific neutralizing antibody production. Therefore, the naturally acquired immune response does not persist for a long period of time and might not provide a protection upon re-exposure to SARS-CoV infection.
In conclusion, our study indicates that both memory CD4+ T cells and CD8+ T cells specific for SARS-CoV in SARS-recovered individuals could be maintained for 3–4 years and gradually decreased in the absence of antigen. These results may have important implications in developing effective SARS vaccines.
Acknowledgments
This study was supported by grants from National “863” Project (No. 2007AA02Z415),Scientific Technology Program of Guangdong Province (No. 2006B36005005) and the Science and Technology Program of Guangzhou (No. 2008J1-C141-3). We sincerely thank the individuals who donated their blood for this study.
Conflict of interest statement
All authors have read and approved the content and have contributed significantly to the work, and none of the authors has any potential financial conflict of interest related to this manuscript.
References
- 1.Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 2.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 3.Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, Ma R, Wu C. Immunization with SARS-CoV S DNA vaccine generates memory CD4+ and CD8+ T cell immune responses. Vaccine. 2006;24:4905–4913. doi: 10.1016/j.vaccine.2006.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin H, Xiao C, Chen Z, Kang Y, Ma Y, Zhu K, Xie Q, Tu Y, Yu Y, Wang B. Induction of Th1 type response by DNA vaccinations with N, M, and E genes against SARS-CoV in mice. Biochem Biophys Res Commun. 2005;328:979–986. doi: 10.1016/j.bbrc.2005.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai MM. SARS virus: the beginning of the unraveling of a new coronavirus. J Biomed Sci. 2003;10:664–675. doi: 10.1007/BF02256318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, Fontanet A, Zhang PH, Zhan L, Xin ZT, Baril L, Tang F, Lv H, Cao WC. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J Infect Dis. 2006;193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X, Duan C, Wang H, Jiang C, He C, Liu X, Zeng H. Significance of detection of Serum SARS-Ab in medical workers exposed to SARS. Chin Trop Med. 2007;7:782–783. [Google Scholar]
- 9.Okada R, Kondo T, Matsuki F, Takata H, Takiguchi M. Phenotypic classification of human CD4+ T cell subsets and their differentiation. Int Immunol. 2008;20:1189–1199. doi: 10.1093/intimm/dxn075. [DOI] [PubMed] [Google Scholar]
- 10.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:588–597. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng H, Yang L, Li J, Lu Z, Wang L, Koup R, Bailer R, Wu C. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 2006;8:2424–2431. doi: 10.1016/j.micinf.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng H, Yang L, Wang L, Li J, Huang J, Lu Z, Koup R, Bailer R, Wu C. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351:466–475. doi: 10.1016/j.virol.2006.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sallusto F, Geinat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 15.Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. 2003;9:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 16.Sprent J, Surh CD. T cell memory. Annu Rev Immunol. 2002;20:551–579. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu CY, Kirman JR, Rotte MJ, Davey DF, Perfetto SP, Rhee EG, Freidag BL, Hill BJ, Douek DC, Seder RA. Distinct lineages of TH1 cells have differential capacities for memory cell generation in vivo. Nat Immunol. 2002;3:852–858. doi: 10.1038/ni832. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Peng H, Zhu Z, Li G, Huang Z, Zhao Z, Koup R, Bailer R, Wu C. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120:171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Peng H, Zhu Z, Li G, Huang Z, Zhao Z, Koup RA, Bailer RT, Wu C. Persistent memory CD4 + and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J Gen Virol. 2007;88:2740–2748. doi: 10.1099/vir.0.82839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, Kong W, Huang Y, Roberts A, Murphy B, Subbarao K, Nabel G. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]


