Abstract
The POT1-TPP1 heterodimer, the major telomere-specific single-stranded DNA-binding protein in mammalian cells, protects chromosome ends and contributes to the regulation of telomerase. The recent discovery of telomeric RNA raises the question of how POT1 faithfully binds telomeric ssDNA and avoids illicit RNA binding that could result in its depletion from telomeres. Here we show through binding studies that a single deoxythymidine in a telomeric repeat dictates the DNA versus RNA discrimination by human POT1 and mouse POT1A. We solve the crystal structure of hPOT1 bound to DNA with a ribouridine in lieu of the critical deoxythymidine and show that this substitution results in burying the 2′-hydroxyl group in a hydrophobic region (Phe62) of POT1 in addition to eliminating favorable hydrogen-bonding interactions at the POT1–nucleic acid interface. At amino acid 62, Phe discriminates against RNA binding and Tyr allows RNA binding. We further show that TPP1 greatly augments POT1’s discrimination against RNA.
Keywords: crystal structure, DNA–protein interaction, POT1-TPP1, telomere
Telomeres are protein-DNA complexes that comprise the termini of linear chromosomes and help maintain the integrity of eukaryotic genomes (1). Telomeric DNA typically consists of a large number of short repeats of dsDNA ending with a single-stranded (ss) G-rich 3′ overhang (2–4). Specialized telomeric proteins bind to the ds and ss regions of telomeric DNA to prevent inappropriate degradation and fusion events at chromosome ends (5). One such protein, protection of telomeres 1 (POT1), binds specifically to the ss G-rich 3′ tail of chromosomes (6–11). Human POT1 (hPOT1) and hPOT1V2 (a splice variant of hPOT1 composed of its DNA-binding domain, used extensively to characterize POT1 structurally and biochemically) bind telomeric DNA with high affinity (KD ∼ 10 nM) and base specificity (10). POT1 is conserved among all mammals and has functional homologs in other species such as Oxytricha nova (12), Tetrahymena thermophila (13), and Schizosaccharomyces pombe (6, 14). A sequence-related protein in the plant Arabidopsis thaliana associates with the telomerase ribonucleoprotein rather than the telomeric DNA (15–17).
TPP1, another telomeric protein, binds POT1 and is critical for POT1 recruitment to telomeres (18–21). Although human TPP1 (hTPP1) does not bind telomeric DNA directly, it increases the affinity of hPOT1 for telomeric ssDNA (21). Additionally, the hPOT1-hTPP1 complex increases the processivity of telomerase, the unique reverse transcriptase that maintains telomere length by catalyzing the synthesis of telomeric DNA at 3′ ends of chromosomes (21). hTPP1-N, an N-terminal fragment of hTPP1 that includes an Oligonucleotide/oligosaccharide binding (OB) domain and the POT1-binding domain, fully recapitulates hTPP1’s POT1-ssDNA-binding-stimulation and telomerase processivity-enhancement functions (21).
TERRA is noncoding RNA-containing multiple G-rich telomeric repeats transcribed from chromosome ends (22–24). TERRA is found in mammals and budding yeast and implicated in the regulation of telomerase and in chromatin remodeling. TERRA in humans is 100 bases to 9 kilobases long (22, 24). In contrast, the 3′ ss G tails on the ends of human chromosomes are only 130–275 nucleotides long (2, 4). The pre-mRNA 3′ splice sites consensus sequence (Py)nNPyAG/G (Py = pyrimidine, n is an integer > 1, and “/” indicates the splice site) is essentially satisfied by the telomeric repeat sequence GTTAGG. Because 3′ splice sites are present on most pre-RNAs, their concentration in the nucleus is expected to exceed that of 3′ ss G-rich DNA tails.
In addition to TERRA, the number of RNA r(UUAGGGUUNG) sequences that match the POT1-binding site expected to occur randomly in the nucleus of a mammalian cell is ∼1,300–20,000 (25), which is greater than or equal to the number of d(TTAGGGTTAG) binding sites of hPOT1 (∼1,500) in a human cell (2, 4) (calculation in Supporting Information). Despite these potential RNA-binding alternatives, POT1 faithfully and strongly binds telomeric DNA showing no apparent affinity for any of these RNAs (6, 14, 23, 26). We wondered how mammalian POT1 displays such robust intolerance to RNA. Here, through binding experiments with mixed DNA-RNA oligonucleotides and a high-resolution crystal structure, we show that a single ribouridine in lieu of a deoxythymidine at the fourth position (rU4 instead of dT4) of a telomeric repeat sequence d(GGTTAGGGTTAG) is the primary determinant of RNA discrimination by hPOT1. We further show that POT1-TPP1 displays remarkably greater intolerance towards RNA than POT1 alone, uncovering a previously undescribed function for the TPP1 protein.
Results
dT4 Dictates POT1 Specificity for DNA.
We observed that mouse POT1A, the essential POT1 paralog in mice (7), serves as an excellent source of recombinant mammalian POT1, because it can be obtained readily from overexpression in bacteria (unlike hPOT1, which is isolated after expression in baculovirus-infected insect cells). To examine the effects of ribonucleotide substitution within the telomeric sequence on mammalian POT1 binding, we carried out a parallel binding analysis of hPOT1 and mPOT1A with oligonucleotides that contain two telomeric repeats terminating in TAG-3′, which is optimal for mPOT1A and hPOT1 binding (27, 28).
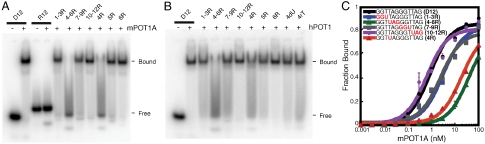
Like hPOT1 (6), mPOT1A failed to bind an all-RNA telomeric dodecamer R12 under conditions in which it formed a stable complex with D12 (Fig. 1A; oligonucleotide sequences in Table 1). To probe whether the DNA versus RNA specificity of POT1 is derived from determinants found throughout the ssDNA or from certain “hot spots” in the dodecameric sequence, we measured the affinity of hPOT1 and mPOT1A for variants of D12 that have stretches of three consecutive ribonucleotides in the telomeric sequence. Both hPOT1 and mPOT1A formed a discrete complex with 1–3R, 7–9R, and 10–12R but not with 4–6R (Fig. 1A and B). To evaluate whether the decreased binding seen with 4–6R arose from the presence of one particular ribonucleotide, we tested D12 variants with a single ribonucleotide at either the fourth (4R), fifth (5R), or sixth (6R) position from the 5′ end. Fig. 1A and B show that with hPOT1 and mPOT1A, 5R and 6R had binding profiles similar to that shown by D12. In contrast, the POT1-4R complex dissociated during gel electrophoresis much like POT1-4–6R. Hence, a single ribonucleotide (rU instead of dT) at position 4 interferes with POT1 binding.
Fig. 1.
Effect of ribonucleotide substitution on POT1-ssDNA binding. EMSA of mPOT1A (A) and hPOT1 (B) with 32P-labeled dodecameric ssDNA-RNA mixed oligonucleotides of telomeric sequence. Binding mixtures contained 1 nM 32P-labeled oligonucleotides and 1.5 nM mPOT1A (A) or 20 nM 32P-labeled oligonucleotides and 40 nM hPOT1 (B). The sequences of the oligonucleotides are detailed in Table 1. (C) Filter-binding experiments of mPOT1A with 32P-labeled ssDNA-RNA of the indicated sequence (ribonucleotides are depicted in red) were done in duplicate, and the mean of the fraction of mPOT1A-bound ssDNA-RNA was plotted against mPOT1A concentration. Error bars represent the standard deviation of the two measurements.
Table 1.
KD’s for hPOT1, hPOT1-hTPP1-N, mPOT1A, and mPOT1A-mTPP1-N with DNA-RNA mixed dodecameric oligonucleotides
Quantitation of the Deleterious Effects of Ribonucleotide Substitution on POT1-ssDNA Binding.
Using a filter-binding assay, we determined the KD of hPOT1-D12 to be 6.3 ± 3.5 nM and that for mPOT1A-D12 to be 0.9 ± 0.3 nM, values that agree well with previous reports (10, 28). To quantify the defect in binding for a particular dodecamer, we calculated the ratio of the KD for the complex formed by that dodecamer to the KD for the analogous complex formed by D12 (designated as “fold increase of KD” in Table 1). Consistent with our gel-shift analysis (Fig. 1A and B), 10–12R did not show a binding defect when compared to D12 (Fig. 1C and Table 1). hPOT1-1–3R and hPOT1-7–9R had 2-fold and 5-fold increases of KD, respectively. In agreement with the gel-shift analysis, the most significant binding defect was seen in the case of 4–6R (Table 1). With hPOT1, 4–6R binding was decreased by 15-fold. The deleterious effect of ribonucleotide substitutions in positions 4–6 was further magnified in the case of mPOT1A, where 47-fold reduced binding was observed.
Single ribonucleotide substitutions in the context of D12 at positions 4, 5, and 6 (4R, 5R, and 6R) were assessed to quantify the contributions of each of these positions to DNA specificity. Again, in agreement with the binding data shown in Fig. 1A and B, the greatest defect in binding is seen for 4R (4-fold for hPOT1 and 22-fold for mPOT1A; Table 1). Because rU differs from dT by having a 2′-OH and by lacking a 5-methyl group on the base, we tested oligonucleotides containing an rT and a dU at position 4. Compared to D12, both 4rT and 4dU showed modest binding defects with mPOT1A (4rT showing a 3-fold defect and 4dU showing a 2-fold defect). With hPOT1, no significant binding defect was observed with 4rT or 4dU. We conclude that the effects of the 5-methyl and 2′-H groups on dT4 are not additive in terms of binding energy (or not multiplicative in terms of KD) but rather synergistic in giving rise to DNA specificity for POT1.
hPOT1 binds the all-RNA oligonucleotide R12 with KD ∼ 1,200 nM, whereas it binds D12 with KD = 6.3 nM. Given that dT4 makes the largest single contribution to DNA specificity for hPOT1, we asked whether introducing dT4 in the context of R12 (4D, Table 1) could rescue POT1 binding. Indeed, 4D bound POT1 with 4 times greater affinity than R12 (KD ∼ 300 nM). It seemed possible that reinforcing the DNA-like 2′-endo sugar pucker of dT4 in 4D by introducing an adjacent deoxyribonucleotide could lead to further POT1-binding recovery. This was the case, because hPOT1 bound 4–5D (Table 1) with a KD of ∼110 nM. However, binding remained ∼17-fold weaker than that of hPOT1-D12, providing additional evidence that the difference between RNA and DNA binding is not completely localized to a single position.
Structural Basis for DNA versus RNA Specificity of POT1.
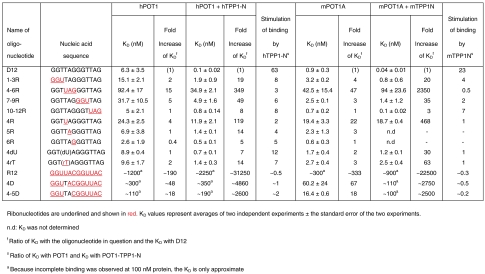
The binding studies reported here use oligonucleotides that include two 5′ G nucleotides not present in the hPOT1V2-d(TTAGGGTTAG) structure [Protein Data Bank (PDB): 1XJV] (10). To ask whether the presence of the additional G nucleotides alters the POT1-ssDNA interface, we crystallized hPOT1V2 in complex with D12 and solved the structure at 1.8 Å (See Supporting Table 1 and Material and Methods). Structural comparison showed that the protein backbone and side chains of 1XJV and hPOT1V2-D12 (rmsd = 0.2 Å over 291 Cα atoms) superimpose well, as do the 10 nucleotides common to the two structures (Fig. S1A). The eleventh nucleotide (dG2) in our hPOT1V2-D12 structure stacks against dT3 and makes no other contacts to the DNA or protein (Fig. S1B). No electron density was observed for the twelfth nucleotide, dG1. We conclude that D12 and d(TTAGGGTTAG) have similar modes of binding to POT1, and hence ribonucleotide substitutions at the 10 conserved positions of D12 and d(TTAGGGTTAG) are expected to have similar effects on POT1 binding. Indeed, hPOT1 binds d(TTAGGGTTAG) with 10-fold greater affinity than it binds dTrUd(AGGGTTAG) (Fig. S2).
To analyze the structural basis for the binding defect seen in oligonucleotides containing the 4rU substitution, we crystallized the hPOT1V2-dTrUd(AGGGTTAG) complex and solved the structure at 1.8 Å resolution (Supporting Table 1). The final refined model had an Rcrystal/Rfree = 0.228/0.238. A 2Fo - Fc map (Fig. 2A) and a simulated annealing composite omit map (Fig. S3A) generated using the CNS program (29) unambiguously confirmed the position of the O2′ of rU4 (Fig. 2A). There were no significant changes to the protein backbone (rmsd = 0.2 Å over 289 Cα atoms), although subtle changes to the nucleic acid backbone were observed when compared to 1XJV (Fig. 2D, E, and F). The potential role of nucleic acid backbone geometry in RNA discrimination by POT1 and by POT1-TPP1 is detailed in Discussion.
Fig. 2.
Structural basis for RNA discrimination by hPOT1. (A) A stereo view showing the electron density (2Fo - Fc) around rU4 contoured at 1σ obtained from rigid body refinement of hPOT1V2-d(TTAGGGTTAG) (PDB: 1XJV) against crystallographic data of hPOT1V2-dTrUd(AGGGTTAG) at 1.8 Å. The final refined model is superimposed on the map as a stick model in Corey–Pauling–Koltun (CPK) atomic coloring. Density for the O2′ is clearly defined in the electron density. The polar and stacking interactions involving dT3, rU4/dT4, dA5, and dG6 in the hPOT1V2-dTrUd(AGGGTTAG) (B) and the hPOT1V2-d(TTAGGGTTAG) (PDB: 1XJV) (C) structures are shown. The two-headed red arrow in (B) indicates a van der Waals contact between the O2′ of rU4 and the Phe62 of hPOT1. (D–F) hPOT1V2-dTrUd(AGGGTTAG) (red) and hPOT1V2-d(TTAGGGTTAG) (green) structures were superposed to highlight differences in the nucleic acid backbone along nucleotides dT3-dG6 (D), dG7-dT9 (E), and dT10-dG12 (F). The double-headed black arrows indicate instances of significant DNA-C2′–protein proximity (< 4.5 Å). Note that the numbering scheme for the nucleic acid in this paper (and associated PDBs) is G(1)G(2)T(3)T(4)A(5)G(6)G(7)G(8)T(9)T(10)A(11)G(12). We have numbered the DNA strand for hPOT1V2-d(TTAGGGTTAG) according to this scheme, although previous references, including PDB: 1XJV, adopt the following numbering scheme: [T(1)T(2)A(3)G(4)G(5)G(6)T(7)T(8)A(9)G(10)].
The O2′ of rU4 in the hPOT1V2-dTrUd(AGGGTTAG) structure (Fig. 2B) is located at a distance of 3.3 Å from the ε-C of the aromatic side chain of Phe62, which is involved in a π-sandwich with the bases of rU4 and dT3. We suggest that positioning of the O2′ in close proximity to Phe62, which is part of a hydrophobic patch of hPOT1 (Fig. 3A), precludes solvation of the 2′OH group. The conformation adopted by the rU4 ribose sugar, which is very different from that of the dT4 sugar, perturbs the conformation of the phosphate backbone link to dT3 (compare Fig. 2B and C, and see superposition in Fig. 2D). This perturbation results in the rearrangement of the base of dT3 in the rU4-containing complex, such that the hydrogen bond between N3 of dT3 and the γ-O of Thr41 (seen in 1XJV) is lost and the Thr41 side chain adopts an alternative rotameric conformation. Additionally, a possible hydrogen-bonding interaction between the carboxylate of Asp42 and the O4 of dT3 in 1XJV (requiring protonation of either the δ-O of Asp42 or the O4 of dT3) is no longer observed in the rU4-containing complex. In addition to the lost direct protein–nucleic acid interactions, two water-mediated interactions that are present in 1XJV are absent in the hPOT1V2-dTrUd(AGGGTTAG) structure (Fig. S3B and C).
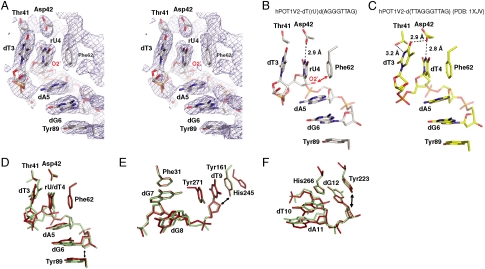
Fig. 3.
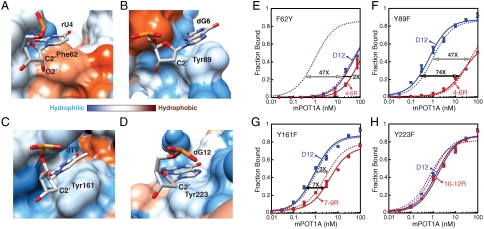
Hydrophobicity of Phe62 is critical for RNA discrimination. The crystal structure of hPOT1V2-dTrUd(AGGGTTAG) is shown as a surface representation of the protein and a stick representation of rU4 (A), dG6 (B), dT9 (C), or dG12 (D). The protein is colored according to hydrophobicity of the amino acids [according to the Kyte–Doolittle scale (36)] such that blue to white to dark orange represents increase in hydrophobicity. For clarity, only the pertinent nucleotide (in CPK coloring convention) of the nucleic acid of hPOT1V2-dTrUd(AGGGTTAG) is shown in each panel. The appropriate O2′, C2′, and aromatic stacking residues of hPOT1 are indicated. (E–H) Binding data and curve fits for mPOT1A-F62Y with D12 and 4–6R (E), mPOT1A-Y89F with D12 and 4–6R (F), mPOT1A-Y161F with D12 and 7–9R (G), and mPOT1A-Y223F with D12 and 10–12R (H). Binding curves of D12 with mPOT1A mutants are shown as solid blue lines and those with wild-type mPOT1A as dotted blue lines. Binding curves of indicated ribonucleotide-substituted oligonucleotides with mPOT1A mutants are shown as solid red lines and those with wild-type mPOT1A as dotted red lines. Error bars represent the standard deviation of two independent measurements. The ratio of the KD with a particular oligonucleotide and KD with D12 for mPOT1A mutants is indicated in black and that for wild-type is indicated in gray.
With the exception of Thr41, no significant differences in the protein side-chain conformations between 1XJV and the rU4-containing structure are seen at the protein–nucleic acid interface. Superposition of the nucleotides from the two structures shows that all bases involved in base stacking (rU/dT4, dG6, dG7, dT9, dG12) with aromatic POT1 side chains overlay precisely on each other with the exception of rU4/dT4 (Fig. 2D, E, and F). For nucleotides not involved in base stacking directly with aromatic amino acids, the superposition is less perfect.
To gain insight as to why POT1 is specifically sensitive to RNA substitution at position 4 and not at other positions in the sequence, we inspected the vicinity of the C2′ (the atom that bears the 2′OH of RNA) for each nucleotide in the hPOT1V2-dTrUd(AGGGTTAG) structure (Fig. 3). Four nucleotides (rU4, dG6, dT9, and dG12) involved in base stacking with POT1 have their C2′ atoms within 4.5 Å of their cognate aromatic protein side chains (Fig. 2D, E, and F). We observe that three of these (dG6, dT9, and dG12), which have their bases stacking against tyrosine side chains of POT1 (Fig. 2D, E, and F), are in predominantly hydrophilic environments (Fig. 3B, C, and D). The fourth, rU4 that stacks against Phe62, is in a hydrophobic environment (Fig. 3A), as discussed above. It is possible that ribonucleotide substitutions at positions dG6, dT9, and dG12 of a telomeric oligonucleotide do not adversely affect hPOT1 binding because the polar O2′ (covalently bonded to C2′) is well accommodated in the hydrophilic environment provided by hPOT1 (Fig. 3B, C, and D). In contrast, the dT4 to rU4 change that introduces an O2′ and removes a 5-methyl group (both changes reduce hydrophobicity) places rU4 in a predominantly hydrophobic environment (Fig. 3A), possibly contributing to the energetic destabilization reflected in the binding defect of hPOT1 with 4R and 4–6R (Fig. 1 and Table 1).
We hypothesized that, in the context of the POT1 structure, the hydrophobic environment created by phenylalanine discriminates against ribonucleotide binding, whereas the more hydrophilic environment created by tyrosine allows ribonucleotide binding. To test this hypothesis, we expressed and purified mPOT1A-F62Y, mPOT1A-Y89F, mPOT1A-Y161F, and mPOT1A-Y223F mutants and compared their binding affinities for ribonucleotide-containing oligonucleotides versus D12 (Fig. 3E–H). Interestingly, the F62Y mutant showed only a 2-fold binding defect with 4–6R compared to the 47-fold defect seen with wild-type protein (Fig. 3E and Table 1). In further support of our hypothesis, Y89F and Y161F showed modest increases in binding defects with 4–6R (70-fold versus 47-fold for wild-type; Fig. 3F) and 7–9R, respectively (7-fold versus 3-fold for wild-type; Fig. 3G and Table 1). Although Y223F behaved like a wild-type protein, showing no binding defect with 10–12R (Fig. 3H), it is possible that further increase in hydrophobicity around Tyr223 is required to observe an increase in RNA discrimination.
TPP1 Binding Greatly Enhances Discrimination against RNA by POT1.
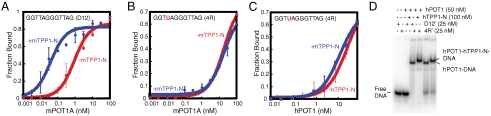
TPP1 is the in vivo protein-binding partner of POT1. In vitro binding experiments have shown that, although TPP1 by itself does not bind ssDNA, the POT1-TPP1 complex exhibits an ∼10-fold greater affinity for telomeric DNA than POT1 alone (21). We performed filter-binding experiments of hPOT1 and mPOT1A with the various dodecamers listed in Table 1 in the presence of hTPP1-N (21) and mTPP1-N, respectively (see Materials and Methods). As expected, the addition of TPP1-N increases the affinity of POT1 for D12 (compare KD for D12 with hPOT1 and with hPOT1 + TPP1-N in Table 1 and similarly for D12 with mPOT1A and with mPOT1A + mTPP1-N; also see Fig. 4A). Surprisingly, substitution with a single ribonucleotide (rU4) alters the TPP1 dependence profile entirely. The binding curves for mPOT1A-4R in the presence and absence of mTPP1-N are virtually superimposable as are the curves for the corresponding human proteins (Fig. 4B and C). Hence, a single ribonucleotide in lieu of a deoxyribonucleotide at the fourth position of the telomeric ssDNA dodecamer abrogates the ability of TPP1 to increase the affinity of POT1 for this oligonucleotide.
Fig. 4.
A single ribonucleotide substitution abrogates TPP1’s ability to stimulate POT1-ssDNA binding. Binding curves for mPOT1A-D12 (A), mPOT1A-4R (B), and hPOT1-4R (C) complexes in the presence (blue) and absence (red) of cognate TPP1-N proteins. Error bars represent the standard deviation of two independent measurements. (D) EMSA showing that hTPP1-N binds hPOT1-D12′ and hPOT1-4R′ complexes to give distinct bands corresponding to hPOT1-hTPP1-N-D12′ and hPOT1-hTPP1-N-4R′, respectively. Note that D12′ and 4R′ contain eight deoxythymidines introduced at the 5′ end of D12 and 4R sequences, respectively, to resolve the POT1-ssDNA versus POT1-TPP1-N-ssDNA complexes.
The lack of stimulation of hPOT1-4R binding in the presence of TPP1 might arise from prevention of POT1-TPP1-N complex formation by the rU4 substitution. To ascertain whether TPP1-N forms a complex with POT1-4R, we carried out an EMSA of hPOT1-D12′ and hPOT1-4R′. D12′ and 4R′ differ from D12 and 4R by the presence of 8 dTs at their 5′ ends, which enhances the electrophoretic separation of the POT1-only and POT1-TPP1 complexes. Fig. 4D shows that addition of hTPP1-N to POT1-D12′ and POT1-4R′ led to the formation of stable ternary complexes, as evidenced by the appearance of a slower migrating band. Thus, TPP1-N remains bound to POT1 in the presence of the rU4 substitution but has lost its capacity to stabilize POT1 binding to the 4R oligonucleotide. Interestingly, hTPP1 shows no stimulation of POT1-oligonucleotide binding with 4D or 4–5D (Table 1). Put together, these data indicate that, although a dT at position 4 is important for high affinity of POT1-oligonucleotide binding, it is not sufficient to explain the mechanism by which TPP1 further enhances this affinity. We next probed the effect of ribonucleotide substitutions at other positions on the stimulation of POT1-ssDNA binding by TPP1-N. We computed the ratio of the KD of POT1-ssDNA-RNA complexes in the absence and presence of TPP1-N [see “stimulation of binding by hTPP1-N” (or mTPP1-N) in Table 1]. For D12, this ratio is 63 with hPOT1 and 23 with mPOT1A. Upon inspection of Table 1, it becomes immediately evident that stimulation of oligonucleotide binding by POT1 in the presence of TPP1-N is weaker for all ribonucleotide-substituted oligonucleotides, although maximal interference is seen in cases where there is a 4dT to 4rU change (4–6R and 4R in Table 1). Hence, the stimulation of POT1-ssDNA binding by TPP1-N is extremely sensitive to the presence of ribonucleotides throughout the telomeric sequence.
What is the consequence of the lack of stimulation of POT1-oligonucleotide binding by TPP1-N on RNA discrimination? Inspection of the “fold increase of KD” columns in Table 1 reveals that discrimination against a particular RNA-containing oligonucleotide by hPOT1 (and mPOT1A) is significantly larger in the presence of hTPP1-N (and mTPP1-N) than in its absence. Most notably, hPOT1-4R shows only a 4-fold increase of KD due to ribo-substitution, whereas hPOT1-TPP1-N-4R shows an increase in KD of 119-fold. These data imply that the discrimination against RNA of telomeric sequence is much greater for POT1 when it is bound to TPP1 and that the mechanism of RNA discrimination contributed by TPP1 binding is distinct from that exhibited by POT1 alone.
Discussion
Here we have shown through qualitative and quantitative POT1-ssDNA-RNA-binding experiments that a single thymidine (dT4) in the telomeric repeat contributes the most to hPOT1’s specificity for DNA. A previous study of RNA binding by S. pombe POT1 revealed that the same dT4 residue as well as the preceding dT3 in the pombe telomeric sequence GGTTAC were most responsible for the specificity for DNA over RNA (14). Although the absense of a 5-methyl is responsible for reduced binding of rU4-containing oligonucleotides to S. pombe POT1, we find here that both the 2′OH and the lack of a 5-methyl group on rU4 interfere synergistically with mammalian POT1 binding. The protein used in the previous study consisted of only the first OB fold of S. pombe POT1, and its nucleic acid binding characteristics differ from that of the entire two-OB-fold DNA-binding domain (30, 31). However, we note that the TTA trinucleotide, which is common to the two telomeric sequences, is bound very similarly by hPOT1 and the first OB fold of S. pombe POT1, with the same kind of dT-dT-Phe-stacking interactions and similar amino acid side chains involved in hydrogen bonding, so it seems likely that this part of the S. pombe POT1–OB–nucleic acid interaction is correct. We conclude that the basis for POT1 discrimination against RNA binding may be substantially conserved between fission yeast and mammals.
We solved the structure of POT1 in complex with an oligonucleotide with the rU4 substitution to provide a physical picture of the deleterious effect of this substitution on POT1 binding. The structure shows that the presence of rU4 causes a change in the DNA conformation such that two hydrogen-bonding interactions of dT3 with POT1 are lost. In addition, the position of rU4 in the structure renders its polar O2′ in close proximity to the hydrophobic region around Phe62 (Fig. 3A). It therefore seems possible that burying the 2′OH in a hydrophobic environment in the rU4-containing complex leads to desolvation of this functional group, resulting in energy destabilization (32). Intriguingly, the only nucleotide where the C2′ (and O2′) is in a strictly hydrophobic environment is rU4 (Fig. 3A). To test the idea that, in the context of POT1, having a more hydrophobic Phe near a 2′OH is less favorable than having a more hydrophilic Tyr, we probed the effects of F62Y, Y89F, Y161F, and Y223F mutations on mPOT1A binding to ribonucleotide-substituted oligonucleotides. mPOT1A-F62Y showed merely a 2-fold binding defect with 4–6R as opposed to the 47-fold defect shown by the wild type. Y89F and Y161F showed modest increases in binding defects for 4–6R and 7–9R, respectively, also consistent with the hypothesis that Phe stacking discriminates against RNA binding and Tyr stacking permits RNA binding. The Phe-Tyr switch dictating DNA versus RNA specificity seen here has also been noted in T7 RNA polymerase, where the Y639F mutant relaxes RNA specificity by allowing incorporation of dNTPs (33).
Another intriguing result that emerged from our binding analysis is the fact that TPP1 binding greatly increases the DNA versus RNA discrimination of POT1. In the presence of TPP1-N, the sensitivity of POT1-ssDNA binding to any ribonucleotide substitution in the binding sequence is greatly increased, suggesting that TPP1 provides a layer of RNA discrimination above that presented by POT1 alone. Although the structure of the POT1-TPP1 complex has not been solved, there is a high-resolution crystal structure of ssDNA bound to the structurally analogous O. nova TEBPα-β dimer (PDB: 1OTC) (12). In this structure, the α and β subunits form a groove that engulfs the ssDNA. If POT1-TPP1 sandwiches ssDNA in an analogous manner, the altered conformation of the sugar-phosphate backbone caused by ribo-substitution (Fig. 2D, E, and F) may result in the loss of shape complementarity between the nucleic acid-POT1 surface and TPP1. Alternatively, the 2′OH groups might directly perturb the TPP1–nucleic acid interface.
Finally, the issue of ssDNA versus RNA discrimination also occurs with single-stranded DNA binding protein (SSB) and replication protein A (RPA), proteins that bind single-stranded DNA at replication forks and strongly discriminate against ssRNA (34, 35). It will be interesting to investigate how these proteins execute RNA discrimination.
Materials and Methods
Protein Expression and Purification.
hPOT1, hPOT1V2, hTPP1-N (aa 91–334), mPOT1A (wild-type and mutants), and mTPP1-N (aa 1–246) proteins were expressed in bacteria and purified as detailed in SI Materials and Methods.
Electrophoretic Mobility Shift Assays.
Binding mixtures (10 µL) containing specified concentrations of 5′ 32P-labeled oligonucleotides and POT1/TPP1 proteins were incubated at 4 °C and were then analyzed by electrophoresis through a nondenaturing 4–20% gradient polyacrylamide gel (Invitrogen). Further details are provided in SI Materials and Methods.
Filter-Binding Assays for KD Determination.
Filter-binding experiments were performed in a 96-well dot blot apparatus. The protein-DNA mixtures were incubated on ice and were then filtered through a precooled (at 4 °C) membrane sandwich containing a nitrocellulose membrane (BA85, Whatman), a positively charged nylon membrane (Hybond N+, GE), and a filter paper (Whatman). The membranes were air-dried and quantified by using a PhosphorImager. Further details are provided in SI Materials and Methods.
Structure Determination of hPOT1V2-dTrUd(AGGGTTAG) and hPOT1V2-D12.
X-ray crystallography methods and solution of structures by molecular replacement using PDB:1XJV as a starting model were performed as detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Corie Ralston from beam line 8.2.1 at the Advanced Light Source, Berkeley, for help with x-ray diffraction data collection. We thank Dr. Sandy Chang (University of Texas, M.D. Anderson Cancer Center) for providing a plasmid containing the cDNA sequence of mPOT1A. J.N. is a Howard Hughes Medical Institute fellow of the Helen Hay Whitney Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3KJO and 3KJP).
This article contains supporting information online at www.pnas.org/cgi/content/full/0911099107/DCSupplemental.
References
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 3.Rhodes D, Fairall L, Simonsson T, Court R, Chapman L. Telomere architecture. EMBO Rep. 2002;3:1139–1145. doi: 10.1093/embo-reports/kvf246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 6.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 7.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 8.Baumann P, Podell E, Cech TR. Human Pot1 (protection of telomeres) protein: Cytolocalization, gene structure, and alternative splicing. Mol Cell Biol. 2002;22:8079–8087. doi: 10.1128/MCB.22.22.8079-8087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 10.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 11.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 12.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 13.Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007;27:1592–1601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 15.Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol Cell Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tani A, Murata M. Alternative splicing of Pot1 (protection of telomere)-like genes in Arabidopsis thaliana. Genes Genet Syst. 2005;80:41–48. doi: 10.1266/ggs.80.41. [DOI] [PubMed] [Google Scholar]
- 17.Surovtseva YV, et al. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye JZ, et al. POT1-interacting protein PIP1: A telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 20.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 21.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 22.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 23.Luke B, Lingner J. TERRA: Telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luke B, et al. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson M. A rapid and convenient method for isolation of nuclear, cytoplasmic and total cellular RNA. Nucleic Acids Res. 1988;16:10934. doi: 10.1093/nar/16.22.10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa F, Matunis MJ, Dreyfuss G, Cech TR. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He H, et al. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr, Sect D: Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: The telomeric single-stranded DNA-binding activity of Pot11-389. J Mol Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Trujillo KM, Bunch JT, Baumann P. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of heterogeneous fission yeast telomeres. J Biol Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- 32.Hansch C, Coats E. Alpha-chymotrypsin: A case study of substituent constants and regression analysis in enzymic structure-activity relationships. J Pharm Sci. 1970;59:731–743. doi: 10.1002/jps.2600590602. [DOI] [PubMed] [Google Scholar]
- 33.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–2711. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim C, Snyder RO, Wold MS. Binding properties of replication protein A from human and yeast cells. Mol Cell Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.