Abstract
Mouse has become an increasingly important organism for modeling human diseases and for determining gene function in a mammalian context. Unfortunately, transposon-tagged mutagenesis, one of the most valuable tools for functional genomics, still is not available in this organism. On the other hand, it has long been speculated that members of the Tc1/mariner-like elements may be less dependent on host factors and, hence, can be introduced into heterologous organisms. However, this prediction has not been realized in mice. We report here the chromosomal transposition of the Sleeping Beauty (SB) element in mouse embryonic stem cells, providing evidence that it can be used as an in vivo mutagen in mice.
Keywords: transposon
Tc1/mariner-like transposons are members of a superfamily of eukaryotic transposons that transpose in a so-called “cut-and-paste” manner via a DNA intermediate. Members of this superfamily have been identified in a wide variety of organisms, ranging from protozoa to vertebrates (1–4). The widespread phylogenetic distribution and indications for horizontal transmission of members of this family of transposons have led to the speculation that they may be less dependent on host-specific factors for transposition and therefore may be good candidates to be introduced into heterologous organisms (1, 5–7). Such a speculation has been supported by the recent report on transposition of the mariner element from Drosophila mauritiana in the parasitic flagellate protozoan Leishmania (8). More recently, it has been demonstrated that the Sleeping Beauty (SB) element, a reconstructed version of a Tc1/mariner-like transposon from fish, can transpose from transfected extrachromosomal DNA molecules (supercoiled plasmid DNA) into genomic loci in human cells (9). As part of our ongoing efforts to develop functional genomic tools in mice, we investigated whether the SB element can be used to establish transposon-tagged mutagenesis in mice.
MATERIALS AND METHODS
Establishment of the Tester Cell Lines.
The PGK-puro-bpA (10) expression cassette first was modified by oligoligation to introduce a KpnI site and an NheI site into the BglII site of the original construct to generate PGK-B2-puro-bpA. The SB element carrying a simian virus 40 promoter-driven neo gene then was excised from pT/neo (9) as a KpnI-XbaI fragment and cloned into the KpnI-NheI-digested PGK-B2-puro-bpA to obtain the excision testing construct. A 5-kb KpnI-XhoI fragment from IV6.8 (11) then was cloned into the excision construct to complete an insertional targeting vector for the mouse Hprt locus. The unique NheI site within the region of homology was used to linearize the vector for gene targeting, which has been described previously (33). Briefly, linearized targeting vector was electroporated into AB1 embryonic stem (ES) cells. Transfectants first were selected with 350 μg/ml G-418, taking advantage of the neo gene inside the SB element. Four days later, after the untransfected cells were killed, targeted clones were selected with 1.67 μg/ml of 6-thioguanine (6TG), because a targeting event will destroy the single copy of the Hprt gene on the X chromosome of the male AB1 cells. The fidelity of the targeting events was confirmed by Southern blot analysis (data not shown). Ninety-four of the 96 G-418/6TG-resistant clones analyzed by Southern blotting proved to be targeted correctly. Two clones were expanded for further analysis. To excise the SB element from the Hprt locus, targeted cell lines were expanded and 107 cells were electroporated with 50 μg supercoiled pSB10 (9), expressing the SB transposase gene from a human cytomegalovirus (CMV) enhancer/promoter. Immediately after electroporation, the cells were plated onto 3 × 90-mm puromycin-resistant feeder (SNLP) plates. Selection for excision events was initiated 72 hr after electroporation with medium containing 3 μg/ml of puromycin. Puromycin-resistant colonies were scored 12 days later, at which time colonies were picked and expanded for further analysis. The 17 clones described here were derived from three different electroporations.
Southern Blot Hybridization.
Genomic DNA samples were digested with BamHI and fractionated in 0.8% agarose gel. The membranes first were hybridized with H probe, which was prepared using 0.4-kb XmnI-BamHI of IV6.8 (11) as template. The same membranes then were stripped and hybridized with T probe, which was prepared using the 0.4-kb EcoRI fragment of pTneo (9) as template.
PCR Amplification and Sequencing of the Donor Sites.
The junction fragments were amplified by using a primer specific for the PGK promoter (PGKP: 5′-CTG TTC TCC TCT TCC TCA TC-3′) and a primer specific for the coding sequence of the puromycin-resistant gene (PURO.CDR: 5′-GAA GAG TTC TTG CAG CTC GGT-3′). This pair of primers should amplify a product of 259 bp if just the TATA is left behind at the site of excision. The PCR products amplified from individual clones were sequenced by using dye terminators and an ABI377 sequencer (Applied Biosystems).
Amplification of Novel Flanking Sequences by Inverse PCR.
The sequences flanking the reinserted SB elements were recovered by inverse PCR. The SB element belongs to the IR/DR subtype of the Tc1/mariner-like transposon, which contains a pair of direct repeats (DR) within the terminal inverted repeats (IR). Thus, three different sets of PCR primers were made for both the left end (IR/DR-L) and the right end (IR/DR-R) of the transposon (9) (Fig. 1). For the left end, the first set was: IPLF1, 5′-AAA CGA GTT TTA ATG ACT CCA AC-3′, and IPLF2, 5′-TCC CGG GAG CTT GGA TAT CCA T-3′; the second set was: IPLF1N, 5′-AAA-GAT CGA GTT TTA ATG ACT CCA ACT TAA-3′, and IPLF2N, GGG AGC TTG GAT ATC CAT TTT C-3′; and the third set was: IPLF1SN, 5′-TTT AAT GAC TCC AAC TTA AGT GT-3′, and IPLF2SN, GGG AGC TTG GAT ATC CAT TTT CGG-3′. For the right end, the first set was: IPRF1, 5′-AGA TTC CCT GTC TTA AGG TCA-3′, and IPRF2, AAT GTC AGG AAT TGT GAA AAA GTG-3′; the second set was: IPRF1N, 5′-ATT CCC TGT CTT AAG GTC AGT-3′, and IPRF2N, 5′-TTG ATC AGG AAT TGT GAA AAA GTG AG-3′; and the third set was: IPRF1SN, 5′-TGT GAA AAA GTG AGT TTA AAT GT-3′, and IPRF2SN, 5′-CCT GTC TTA AGG TCA GTT AG-3′. In both cases, the second set of primers are nested to the first set whereas the third set are nested to the second set. For inverse PCR, standard procedures were followed. Briefly, genomic DNA isolated from individual clones was digested with Sau3AI followed by ligation with T4 DNA ligase under conditions that favor intramolecular ligation. A 30-μl PCR first was performed with ligated genomic DNA and the first set of primers. Two microliters of this product was used in a second round of PCR using the second set of primers. The PCR product of these PCRs then was fractionated in a 3% agarose gel. Southern blot hybridization was performed by using an oligonucleotide probe specific for the very end of the transposon (IPIR, 5′-TGT AAA CTT CCG ACT TCA ACT-3′) to identify the specific products. The regions that hybridized to the probe then were excised from another gel run in parallel. The gel slices then were melted and used to set up PCRs to amplify the final products using the third set of primers. All sequencing reactions were performed by using dye terminators and an ABI377 sequencer.
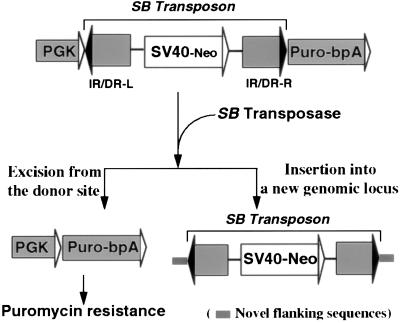
Figure 1.
A genetic-selection strategy for the detection of rare excision events. The puromycin-resistance gene driven by the PGK promoter is inactivated by the insertion of a nonautonomous SB element that separates the promoter from the coding sequence. Excision of the SB element mediated by SB transposase activates the puromycin-resistance gene. The SB element acquires novel flanking sequences when it reintegrates into the genome.
Linkage Analysis of the Reintegration Sites.
PCR-based mapping was performed by using six primer pairs specific for six novel SB integration sites: (i) 5NRF5, 5′-TAG TTG CCT CTG CCT CCC AGA-3′ and 3NRF5, 5′-ATC AGT GTA AAG CCA AGT ATG-3′; (ii) 5NLF6, 5′-GAT CAT GAA TAC ACC AGT GAA-3′ and 3NLF6, 5′-GAT CAA CCG AGT GAT TTC CAG-3′; (iii) 5NLF7, 5′-TAT GTA GAG GAC TAT GTC AGA C-3′ and 3NLF7, 5′-TGA TCA TAG GGT GCA CAG AA-3′; (iv) 5NLF11, 5′-CAG TTT GGC AGG GTG TGA GGT-3′ and 3NLF11, 5′-GAT CCT TTT TCC ATA GCA TAT C-3′; (v) 5NRF16, 5′-TAG CTT TTC AAT GAC CAC ACA GAC-3′ and 3NRF16, 5′-GAT CCC GAA TAC GAA AAA TAT AT-3′; (vi) 5NRF17, 5′-TAG CTT TTC AAT GAC CAC ACA-3′ and 3NRF17, ATA TTG ACG TTA CTA ATT CAT GC-3′. Genomic DNA from a monosomic hybrid cell line containing only the X chromosome from the mouse genome in a human genetic background was used as PCR template. Genomic DNA isolated from HeLa cells and from mouse ES cells also were included as templates in negative and positive controls, respectively. If PCR results were positive when using DNA both from the hybrid cell line and from the mouse ES cells but negative when using DNA from HeLa cells, the locus was scored as X-linked. If the PCR results were positive only when using DNA from mouse ES cells, the locus was excluded from the X-linked category.
RESULTS
A Selection System for Detecting Transposition in ES Cells.
A transposon must fulfill several criteria to be a useful in vivo mutagen in mice. First, it must be able to transpose into active genes and create mutations. Second, it must be able to transpose between chromosomal loci in the mouse genome, so that mutations can be continuously generated. Furthermore, it must transpose at a reasonably high frequency so that genetic screens are manageable.
To address these issues, a positive selection scheme was developed that allows the chromosomal transposition events of the SB element to be detected and the transposition frequency to be determined. In this strategy, a selectable excision testing cassette was constructed in which a nonautonomous SB transposon was cloned into a selectable marker cassette (the puromycin-resistance gene) to inactivate its function. This nonautonomous SB transposon contains only the cis-sequences required for transposition, the terminal inverted repeats (IR/DRs) (1, 9, 12), but lacks a transposase gene. Thus, it can transpose only when the transposase is provided in trans. The precise excision of the SB element as the first step of the transposition event would result in the reactivation of the puromycin-resistance gene, enabling positive selection for those cells in which an excision event had occurred. A single copy of this cassette then can be introduced into a defined locus in the mouse genome by gene targeting to facilitate both the analysis of the nature of the donor sites and the fates of the excised SB elements after the SB excision events (Fig. 1).
Transposase-Dependent Excision of SB Element.
The excision cassette was introduced into the Hprt locus on the mouse X chromosome by gene targeting (Fig. 2a). Targeted ES cell clones with this cassette were puromycin-sensitive and did not revert in the absence of transposase. Several independently targeted ES cell clones carrying the excision cassette were expanded, transiently transfected with a plasmid expressing the SB transposase, and placed under puromycin selection. Puromycin-resistant ES cell clones were obtained at a frequency of approximately 10−6 per transfected cell, providing the first indication that the SB transposase was able to excise the SB element from a defined chromosomal locus in mouse ES cells.
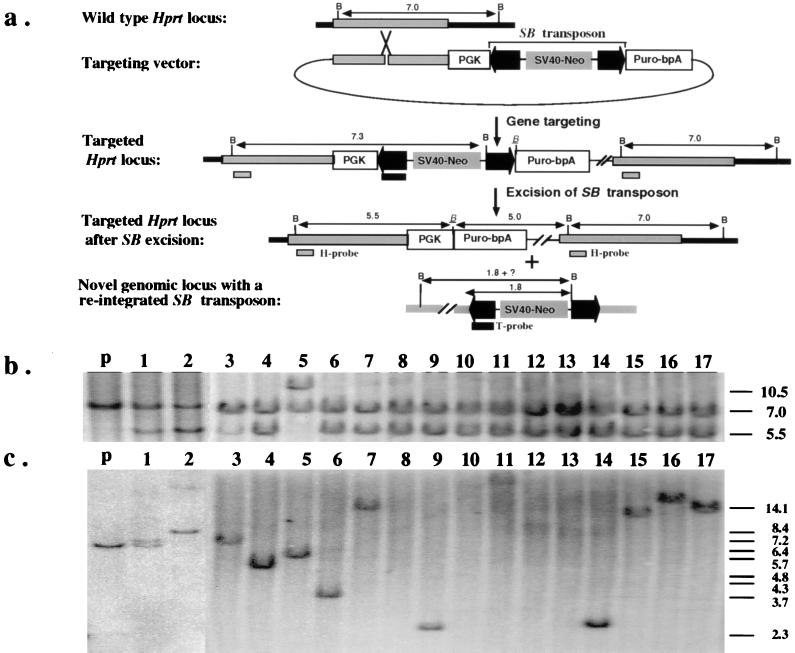
Figure 2.
(a) Introduction of a single copy of the excision testing construct into the Hprt locus by gene targeting. The region of homology for the insertional-type targeting vector is indicated as gray bars. The PGK promoter and the coding sequence of the puromycin-resistance gene are shown as open boxes. The IR/DR sequences of the SB transposon are highlighted as dark arrowheads. A complete, nonautonomous SB element also is indicated in the targeting vector. The positions of the Hprt- (H) and transposon-specific (T) probes that detect excision and reinsertion events, respectively, also are indicated. All relevant BamHI sites are shown as “B” except the one that is adjacent to the excision site, which is italicized and underlined. Notice that the SB element contains a unique BamHI site approximately 1.8 kb from the end that hybridizes to the T probe. (b) Southern hybridization analysis with the H probe confirms the excision of the SB element from the testing construct. The lower intensity of the 5.5-kb fragment compared with the 7.0-kb fragment in clone 1 and clone 3 is a result of contamination with parental cells as the result of cross-feeding. The parental line has a predicted doublet consisting of a 7.0-kb and a 7.3-kb fragment. (c) Southern hybridization analysis with the T probe confirms reintegration of the excised SB element. The sizes of the bands in lanes 1, 2, 3, 4, 5, 6, 9, 11, 14, 15, 16, and 17, are variable but are all longer than 2.3 kb. No hybridization was detected in lanes 8, 10, 12, and 13. The parental fragment detected in clones 1 and 3 is due to parental cell contamination. The size marker is BstEII-digested λ-phage DNA.
Molecular Characterization of the Donor Sites.
To confirm that authentic transposition had occurred, these puromycin-resistant clones were characterized at the molecular level. First, we investigated whether the SB transposons had been excised. Given that many DNA transposons exhibit precise excision, we predicted the postexcision structure of the Hprt locus in the puromycin-resistant clones (Fig. 2a). Indeed, 17 of 18 clones analyzed had the predicted 5.5-kb BamHI restriction fragment when a Hprt-specific probe was used in a Southern hybridization experiment (Fig. 2 a and b), consistent with excision of the SB transposon from the puromycin-resistance gene. A single clone had a 10.5-kb fragment (Fig. 2b, lane 5). This is likely to be the result of imprecise excision of the transposon accompanied by a small deletion that destroyed the BamHI site close to the insertion site (Fig. 2a). This subsequently was confirmed by sequence analysis.
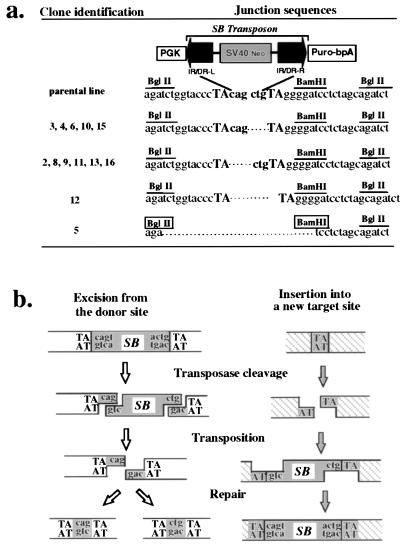
One molecular signature of Tc1/mariner-like transposons is that they often leave unique footprints at their excision sites that are composed of a few base pairs from either ends of the transposons flanked by a pair of TA dinucleotide repeats. These footprints are left behind as a consequence of a staggered double-strand DNA break introduced by the transposase and the repair of this break by the DNA repair machinery of the host cell. Typical footprints of Tc1 and Tc3 in Caenorhabditis elegans consist of the last 2 nt of the transposons (13, 14), whereas the mariner element in Drosophila tends to generate 3-bp footprints (15). To examine the molecular characteristics of the SB transposase-mediated excision events in more detail, for each puromycin-resistant clone, a fragment of the locus from which the SB transposons had been excised was amplified by the PCR and sequenced. In 11 of the 13 clones, the footprints consisted of the terminal 3 nt from either end of the SB transposon flanked by a pair of TA dinucleotides (Fig. 3a). Of the two atypical clones, #12 had a pair of TA dinucleotides with no intervening sequence whereas #5 had a small deletion of 22 bp (Fig. 3a). The majority of footprints observed are consistent with the “cut-and-paste” model of transposition proposed by van Luenen et al. for Tc1 and Tc3 transposition in C. elegans (13), except that the SB transposase appears to generate a 3-bp overhang similar to those of mariner in Drosophila (15) and Tc3 in zebrafish (17) (Fig. 3b). Thus, it appears that the SB transposon is excised from a defined chromosomal locus in mouse ES cells through a mechanism similar to that found in other Tc1/mariner-like transposons.
Figure 3.
(a) DNA sequence analysis of the donor sites after excision of the SB element from the testing construct. The four different classes of excision products are shown together with the original parental sequence. The TA dinucleotide sequence flanking the site of excision is shown in uppercase letters while the three nucleotide sequences from either ends of the SB element are shown in lowercase. The BamHI site adjacent to the site of excision and the pair of BglII sites are indicated. Both the BamHI and the BglII sites that were destroyed by the 22-bp deletion in clone #5 are boxed. (b) Schematic illustration of SB element transposition. The transposition of SB element is illustrated as a series of sequential events with events for the excision locus on the left and those for the insertion locus on the right. First, the transposase cleaves both ends of the SB element to create a 3-base 5′ overhang at the excision locus and also cleaves the TA dinucleotide at the insertion locus to create a gap with a 3′ TA overhang at both ends. After that, the excised SB element is transferred from the excision site to the new insertion site and reintegrates back into the genome. Finally, the broken ends at both the excision locus and the insertion locus are rejoined and the gaps are repaired to complete the entire transposition event.
Characterization of the Reintegration Sites.
To complete a cycle of transposition, the excised SB transposons need to reintegrate back into the genome. To address this, Southern hybridization was performed on genomic DNA samples from the puromycin-resistant cell clones by using a probe specific for the left IR/DR sequence of the transposon. In 13 of 17 clones examined, a novel fragment was detected indicating that the excised SB transposons had reintegrated into mouse chromosomes (Fig. 2c). For the remaining 4 clones (Fig. 2c, lanes 8, 10, 12, and 13), the SB transposons apparently were lost after the excision events. These data indicate that in the majority of cases the excised transposons reintegrated back into the mouse genome to complete a full cycle of a typical transposition event.
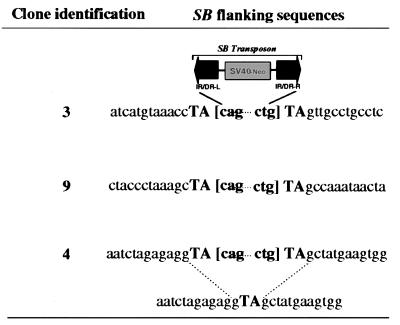
It has been shown previously that Tc1/mariner-like transposons integrate preferentially into TA dinucleotides and lead to duplication of this TA dinucleotide sequence upon their integration into the target sites (13, 14, 18). The flanking sequences of the reintegrated SB transposons were cloned from several of the puromycin-resistant clones by inverse PCR. This analysis revealed that in every case an intact end of the SB transposon was flanked by a TA dinucleotide sequence followed by a novel sequence (Fig. 4 and data not shown). To verify that the TA dinucleotide sequences at both ends of the transposons were the result of the duplication of an otherwise single TA unit at the site of insertion, the corresponding wild-type sequence of a single insertion site was amplified from mouse DNA. The sequence confirmed that there was only a single TA unit in the insertion site in the wild-type locus (Fig. 4). These results are consistent with recent observations for SB transposition in human cells (9). Taken together, our results indicate the precision of SB transposition in terms of both the excision and the reintegration reactions in mouse ES cells through the classical cut-and-paste mechanism.
Figure 4.
Novel flanking sequences of the reintegrated SB element. The flanking sequences (14 nt from each side) of the reintegrated SB elements in clones #3, #9, and #4 are shown. The TA dinucleotides adjacent to the SB elements are shown in uppercase text while the SB elements are indicated with the last 3 nt from each end of the element in brackets. The sequence corresponding to the wild-type locus before SB insertion in clone #4 also is shown below the sequences flanking the SB element for comparison.
The Frequency of SB Transposition in ES Cells.
Transient expression of the transposase in the targeted cell lines generated excision clones at a frequency of 10−6 per transfected cell. This result indicated that the SB element excises at low frequency from this particular locus with the particular transposase expression cassette. To obtain an estimate for the excision frequency under conditions of continued expression of the SB transposase, we carried out a Luria–Delbrük fluctuation analysis (19). In this experiment, the pSB10 (9) transposase construct was cotransfected with a PGK-hprt minigene (20) into a targeted ES cell line carrying a single copy of testing construct at the Hprt locus (Fig. 2a). A total of 48 HAT-resistant lines were established, each was expanded, and a defined number of cells from each clone were plated and subjected to puromycin selection. As expected, the number of puromycin-resistant colonies recovered per million cells plated varied, ranging from 0 to as many as 485, which reflects primarily the different timing of the first excision event in different clones during the expansion. Luria–Delbrück fluctuation analysis determined the frequency of the SB excision to be 3.5 × 10−6 events/cell per generation under these conditions.
The frequency of excision from a particular donor site can vary depending on host background, the chromosomal location of the transposon, as well as particular sequence constituents of the transposons (7, 21). Keeping the transposon sequence constant, we addressed the question of whether the chromosomal context of preintegrated SB transposons had an effect on the efficiency of subsequent excision events. Stable lines carrying the testing construct at random chromosomal locations were generated and tested for their excision frequencies by transiently expressing the transposase. Six of seven lines displayed detectable excision frequencies, ranging from 0.2 × 10−6 to 5.1 × 10−6 per treated cell. The line that displayed the highest frequency of excision contained a single copy of the testing construct (data not shown), and the frequency of excision in this line was about 10-fold higher than that observed in the lines with a single copy of the testing construct targeted to the Hprt locus. Thus, the frequency of SB excision in ES cells appears to be affected by its chromosomal location. In addition, these data also indicate that the frequency of SB transposition can be as high as 3.5 × 10−5 events/cell per generation.
Local Transposition of the SB Element.
Many DNA transposons exhibit preferential integration into linked loci. This can be a particularly useful feature in the use of these elements as mutagens in complex genomes. Because targeted tester cells contain a single copy of the SB element on the X chromosome, we examined whether any of the novel integration sites recovered by inverse PCR were X-linked. For this purpose, we used a human–mouse monochromosomal hybrid cell line that contains the major portion of the mouse X-chromosome in a human background (22). Because the tester ES cell line was derived originally from a male embryo, if the reintegration events are random, 1 of 40 of the novel integration sites would be X-linked. PCR analysis showed that half (three of six) of the novel integration sites were X-linked (Table 1), indicating that SB element undergoes preferential local transposition in ES cells.
Table 1.
Chromosomal localization of novel integration sites
| Novel insertion sites | Genomic DNA sources for PCR
|
Chromosomal locations* | ||
|---|---|---|---|---|
| Human | Hybrid 8.0 | Mouse | ||
| IPRF-3 | − | + | + | X |
| IPLF-4 | − | − | + | A or Y |
| IPRF-5 | − | − | + | A or Y |
| IPLF-6 | − | − | + | A or Y |
| IPRF-16 | − | + | + | X |
| IPRF-17 | − | + | + | X |
X, X chromosome; A or Y, autosomes or Y chromosome.
DISCUSSION
Mutant mice have became the dominant means of obtaining information on gene function in a mammalian context. Although gene-driven approaches, such as overexpression and loss of function mutations, are useful for understanding gene function, it is likely that phenotype-driven genetic screens will be necessary for large-scale functional genomic studies given the complexity of mammalian genomes. Large-scale mutagenesis schemes are possible in mice by using agents such as N-ethyl-N-nitrosourea; however, the identification of the causative point mutation is often difficult and time-consuming (23, 24). Transposon-tagged mutagenesis has proven to be one of the most effective means in establishing a genotype–phenotype relationship in model organisms such as C. elegans (25), Drosophila (26, 27), and in a few special situations, in mice (28–32). The limited application of this approach in mice has been caused by the lack of an appropriate transposon, namely a classical cut-and-paste-type transposon. The chromosomal transposition of the SB element in the ES cells reported here provides direct evidence that this element may be used to establish a general transposon-tagged mutagenesis scheme in mice. Although the frequency of transposition is quite low (3.5 × 10−5 events/cell per generation), it is conceivable that it can be improved. Because the SB transposon/transposase system was reconstructed from inactive elements so that the element can transpose in mammalian cells, neither the transposon nor the transposase may represent the SB transposon/transposase system in the original host genome and, therefore, are likely to be suboptimal. Both the transposase and the cis-acting components could be subjected to random in vitro mutagenesis to generate more effective versions of this transposon/transposase system. In addition, the finding that the SB element transposes preferentially into linked loci is particularly advantageous in performing regional, specific mutagenesis in mice with deletions. The combination of segmental hemizygosity (10, 33) and mutagenesis by using the SB transposon could greatly improve the efficacy of genetic screens in mice. Such a scheme could be applied somatically to identify tumor-suppressor genes or applied in the germ line as a general screen for recessive mutations.
Acknowledgments
We thank Hugo Bellen, Richard Behringer, Alea Mills, Pentao Liu, and Binhai Zhang for comments on this manuscript, and S. Perez and Y.-C. Cheah for technical assistance. We also thank Dr. P. Hackett and R. Plasterk for valuable discussions. A.B. is an Investigator and G.L. is a Research Associate with the Howard Hughes Medical Institute. A.B. acknowledges support from the National Institutes of Health.
ABBREVIATIONS
- ES
embryonic stem
- IR
inverted repeat
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Ivics Z, Izsvák Z, Minter A, Hackett P B. Proc Natl Acad Sci USA. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robertson R H T. Nature (London) 1993;311:485–486. [Google Scholar]
- 3.Robertson H M, MacLeod E G. Insect Mol Biol. 1993;2:25–139. doi: 10.1111/j.1365-2583.1993.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 4.Radice A R, Bugaj B, Fitch D H A, Emmons S W. Mol Gen Genet. 1994;244:606–612. doi: 10.1007/BF00282750. [DOI] [PubMed] [Google Scholar]
- 5.Kidwell M G. Nature (London) 1993;362:202. doi: 10.1038/362202a0. [DOI] [PubMed] [Google Scholar]
- 6.Lidholm D A, Lohe A R, Hartl D L. Genetics. 1993;134:859–868. doi: 10.1093/genetics/134.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doak T G, Doerder F P, Jahn C L, Herrick G. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerios-Filho F J, Beverly S M. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- 9.Ivics Z, Hackett P B, Plasterk R H, Izsvák Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 10.Ramirez-Solis R, Pentao L, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Hasty P, Bradley A. Mol Cell Biol. 1994;14:2404–2410. doi: 10.1128/mcb.14.4.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izsvák Z, Ivics Z, Hackett P B. Mol Gen Genet. 1995;247:312–322. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- 13.van Luenen H G A M, Colloms S D, Plasterk R H. Cell. 1994;79:293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 14.Vos J C, De Baere I, Plasterk R H. Genes Dev. 1996;10:755–761. doi: 10.1101/gad.10.6.755. [DOI] [PubMed] [Google Scholar]
- 15.Bryant G, Garza D, Hartl D. Genetics. 1990;125:103–114. doi: 10.1093/genetics/125.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plasterk R H A. EMBO J. 1991;10:1919–1925. doi: 10.1002/j.1460-2075.1991.tb07718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raz E, van Luenen H G A M, Schaerringer B, Plasterk R H, Driever W. Curr Biol. 1998;2:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- 18.Lampe D J, Churchill M E A, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 19.Luria S E, Delbrück M. Genetics. 1943;28:491–510. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matzuk M M, Finegold M J, Su J-G, Hseuh A J W, Bradley A. Nature (London) 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 21.Lohe A R, Lidholm D-A, Hartl D L. Genetics. 1995;140:183–192. doi: 10.1093/genetics/140.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman G E, Berry M, Craig I W, Levy E R. Genomics. 1991;10:961–970. doi: 10.1016/0888-7543(91)90186-i. [DOI] [PubMed] [Google Scholar]
- 23.Rinchik E M. Trends Genet. 1991;7:15–21. doi: 10.1016/0168-9525(91)90016-j. [DOI] [PubMed] [Google Scholar]
- 24.Bedell M A, Jenkins N A, Copeland N G. Genes Dev. 1997;11:1–10. doi: 10.1101/gad.11.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Plasterk R H. Methods in Cell Biology. New York: Academic; 1995. pp. 59–80. [DOI] [PubMed] [Google Scholar]
- 26.Bellen H J, O’Kane C J, Wilson C, Grossniklaus U, Pearson R K, Gehring W J. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 27.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnieke A, Harbers K, Jaenisch R. Nature (London) 1981;304:315–320. doi: 10.1038/304315a0. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Nature (London) 1918;293:370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- 30.Nusse R, Varmus H E. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 31.Varmus H E. Prog Clin Biol Res. 1983;119:23–35. [PubMed] [Google Scholar]
- 32.Allen J D, Berns A. Semin Cancer Biol. 1996;7:299–306. doi: 10.1006/scbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- 33.You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, Ticknor C, Jaenisch R, Schimenti J. Nat Genet. 1997;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]