Abstract
Increasing biochemical and genetic evidence indicates that the amyloid-β (Aβ) peptide derived from amyloid precursor protein (APP) plays a central role in Alzheimer’s disease (AD) pathogenesis. We previously reported that RanBP9 promotes Aβ generation by scaffolding APP/BACE1/LRP complexes together. Interestingly, the RanBP9-Δ1/N60 (residues 1–392) deletion mutant interacted much more strongly with APP/BACE1/LRP than full-length RanBP9. In this study, we found that RanBP9-N60, a processed form of RanBP9 virtually identical to the RanBP9-Δ1/N60 mutant, was strongly increased in AD brains compared with controls. To evaluate the potential pathogenic consequences of this phenotype, we studied the differential biological properties of full-length RanBP9 vs. RanBP9-Δ1/N60 in HEK293T and Neuro-2A cells. The RanBP9-Δ1/N60 fragment, which lacks a nuclear localization signal, displayed enhanced cytoplasmic vs. nuclear localization and >3-fold enhanced stability than full-length RanBP9. Importantly, RanBP9-Δ1/N60, which contains the LisH dimerization domain, retained the capacity to form self-interacting multimeric complexes and increased Aβ generation by ∼5-fold over vector controls, more potent than the ∼3-fold increase seen by full-length RanBP9. Taken together, these data indicate that RanBP9-N60 may further drive the amyloid cascade in AD and that the proteolytic processing of RanBP9 may be an attractive therapeutic target.—Lakshmana, M. K., Chung, J. Y., Wickramarachchi, S., Tak, E., Bianchi, E., Koo, E. H., Kang, D. E. A fragment of the scaffolding protein RanBP9 is increased in Alzheimer’s disease brains and strongly potentiates amyloid β peptide generation.
Keywords: APP, BACE1, LRP, LisH
Accumulation of the amyloid-β (Aβ) protein, a small peptide derived from β- and γ-secretase cleavages of the amyloid precursor protein (APP), is an indispensable pathogenic process in Alzheimer’s disease (AD). The vast majority of APP is constitutively cleaved in the middle of the Aβ sequence by α-secretase [a disintegrin and metalloprotease 10/17 (ADAM10/ADAM17)] in the nonamyloidogenic pathway, thereby abrogating the generation of an intact Aβ peptide. In the amyloidogenic pathway, a small proportion of APP is cleaved by β- and γ-secretases, known as β-site APP-cleaving enzyme 1 (BACE1) and presenilin, respectively (1), leading to the secretion of Aβ peptides. Such proteolytic processing requires the trafficking of APP such that APP and BACE1 are brought together in close proximity for the rate-limiting β-secretase cleavage to occur.
We and others have shown that the low-density lipoprotein receptor-related protein (LRP), a multifunctional endocytosis receptor (2), binds to APP and alters its trafficking to promote Aβ generation. The loss of LRP substantially reduces Aβ release, a phenotype that is reversed when full-length LRP (LRP-FL) or truncated LRP fragments containing its cytoplasmic tail are transfected (3,4,5,6). The proamyloidgenic activity of the LRP cytoplasmic tail is achieved at least in part by promoting APP/BACE1 interaction and enhancing APP localization to membrane rafts, where both β- and γ-secretase activities are highly enriched (5, 6). Recently, we narrowed down the region of LRP cytoplasmic tail capable of robustly enhancing Aβ production to the last 37 residues of LRP (LRP-C37) and identified 4 novel LRP-C37-interacting proteins (7). Among these, we discovered that the scaffolding protein RanBP9 not only interacts with LRP but also with APP and BACE1 and that it functions as a scaffold on which APP is brought together with BACE1 and LRP (8). Therefore, RanBP9 overexpression strongly increased and RanBP9 siRNA knockdown strongly decreased BACE1 cleavage of APP and Aβ generation (8). RanBP9 was also associated with membrane raft microdomains, consistent with its activity to promote APP localization to rafts (8).
At present, the normal physiological functions of RanBP9 are not well understood. However, RanBP9 contains 4 conserved domains, B30.2/SPRY (SPla and the ryanodine receptor), LisH (Lissencephaly type-1 like homology), CTLH (C-terminal to LisH), and CRA (CT11-RanBP9). The B30.2/SPRY domain is known to be involved in various protein-protein interactions, and the LisH domain has been implicated in protein dimerization or oligomerization (9, 10). While RanBP9 is ubiquitously expressed in all tissues, including brain, its subcellular localization appears to be variable and dependent on the cell type and differentiation state (11,12,13,14,15). For example, in rapidly dividing cells, RanBP9 appears to be mostly localized to the cytosol and nucleus, whereas in more differentiated cells (i.e., Madin-Darby canine kidney cells or muscle), the vast majority of RanBP9 is found discretely near the inner surface of the plasma membrane or associated with cytoskeletel elements (11,12,13,14,15). This type of subcellular localization is consistent with the postulated role of RanBP9 as scaffolding and signaling protein, bridging interactions between the cytoplasmic domains of a membrane receptors and intracellular signaling targets. These include MET receptor protein tyrosine kinase (16) and β2-integrin (12). Moreover, RanBP9 has also been shown to interact with plexin-A receptors and to strongly inhibit axonal outgrowth and branching (17).
During the course of our investigations in identifying the critical region within RanBP9 for mediating the interactions with LRP/APP/BACE1, we made an interesting observation that the RanBP9-Δ1/N60 mutant (expressing residues from 1 to 392), which retains the PRD, SPRY, and LisH domains but not CTLH and CRA domains, interacted more robustly with LRP/APP/BACE1 than the full-length RanBP9 or other deletion mutants (8). In this study, we show that RanBP9-N60, a processed form of RanBP9 virtually identical to the RanBP9-Δ1/N60 mutant, is significantly increased in AD brains and strongly potentiates Aβ generation. The latter phenotype is explained by the greatly increased cytoplasmic vs. nuclear localization and enhanced stability of the RanBP9-N60/Δ1 protein without losing the ability to form self-interacting complexes.
MATERIALS AND METHODS
Cell culture and generation of stable cells
Chinese hamster ovary (CHO), human embryonic kidney (HEK) 293FT, and Neuro-2A cells were grown in DMEME containing 10% FBS, 2 mM l-glutamine, 100 μg/ml penicillin, and 100 μg/ml streptomycin. CHO cells stably expressing APP751 + Flag-RanBP9-FL or Flag-Δ1/N60 deletion mutant cloned in pLHCX plasmid (Clontech, Palo Alto, CA, USA) have been described previously (8). One hundred percent confluency of CHO cells was defined as 106 cells/well of a 6-well plate (9.6 cm2 surface area).
Transient transfections
Transient transfections of HEK293FT and Neuro-2A cells with carried out using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and Opti-MEM I (Invitrogen). For the measurement of Aβ levels, 8 h following transient transfections, the medium was replaced with 1.0 ml of conditioned medium. After 48 h, the conditioned medium was collected, centrifuged to remove cell debris, and immunoprecipitated overnight using a monoclonal B436 antibody for Aβ.
DNA constructs
GFP-RanBP9 was a kind gift from Dr. Hideo Nishitani (Kyushu University, Fukuoka, Japan) (18). pcDNA-P3X-Flag-RanBP9 construct was a gift from Dr. Shim S-K (Yale University School of Medicine, New Haven, CT, USA). The pLHCX Flag-RanBP9-FL and pLHCX Flag-RanBP9-Δ1/N60 deletion mutant (expressing aa 1-392) were generated by restriction digestion followed by filling with plaque-forming unit polymerase in pcDNA-3X-Flag-RanBP9 vector. These cDNAs were sequenced and transferred to HindIII and ClaI sites of pLHCX vector (Clontech) for retrovirus production.
Chemicals and antibodies
The polyclonal antibody CT15, which reacts with the C-terminal 15-aa residues of APP, was used for the detection of full-length APP and CTFs and has been described previously (19, 20). Anti-Flag M2 monoclonal antibody was obtained from Sigma (St. Louis, MO, USA). RanBP9 monoclonal antibody was produced by immunizing mice with a peptide corresponding to 146–729 aa of RanBP9 (8, 12, 20). The 82E1 monoclonal antibody (IBL, Gunma, Japan) against 1–16 residues of human Aβ in combination with mouse monoclonal antibody, 6E10 (Abcam, Cambridge, MA, USA) raised against 1–17 of human Aβ was used for immunodetection of Aβ. Anti-GFP antibody was purchased from Invitrogen. All secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). The antibodies were diluted in 5% fat-free milk in TBS-T (Tris-buffered saline with 0.1% Tween-20) buffer. Cycloheximide [3-(2-(3–5-dimethyl-2-oxocyclohexyl)-2-hydroxyethyl) glutarimide] was obtained from Sigma. Hygromycin B in PBS, geneticin, and propidium iodide were purchased from Invitrogen.
Cell lysis, immunoprecipitation, and immunoblotting
For the immunoprecipitation of Aβ, HEK293FT cells, grown in 6-well plates, were transfected with 1 μg of pcDNA3-APP751 and 3 μg of either the pLHCX empty vector or pLHCX Flag-RanBP9 or Flag-RanBP9-Δ1/N60 deletion mutant. After 48 h, the conditioned medium was collected, centrifuged to remove cell debris, and immunoprecipitated overnight using a monoclonal B436 antibody for Aβ. To detect APP and Flag-RanBP9, the cells were lysed in cell lysis buffer (50 mM Tris-HCl, pH 8.0; 150 mM Nacl; 0.002% sodium azide; 400 nM Microcystin-LR; 0.5 mM sodium vanadate; and 1% Nonidet P-40) with complete protease inhibitor mix (Sigma). Pathologically confirmed human brain samples were obtained from the Shiley-Marcos University of California–San Diego (UCSD) Alzheimer’s Disease Research Center. Brain homogenates from AD and age-matched normal controls were prepared from frontal cortex in cell lysis buffer and a homogenizer (Kinematica AG, Luzern, Switzerland). Samples were subjected to SDS-PAGE, transferred, and immunoblotted with the indicated antibodies and detected by enhanced chemiluminescence (Pierce, Rockford, IL, USA). Gel loading was normalized to total protein concentration as measured by the microbicinchoninic acid (micro-BCA) method (Pierce).
Nuclear and cytoplasmic extract preparation
HEK293FT cells were transiently transfected with either Flag-RanBP9 or Flag-RanBP9-Δ1/N60, and after 48 h the cell lysates were prepared. For nuclear extract preparation, cells were incubated in 1 ml of buffer A (10 mM HEPES, pH 7.9; 10 mM NaCl; 0.1 mM EDTA; and 1 mM DTT) with fresh protease inhibitor cocktail (Sigma) for 15 min. Nonidet P-40 (Calbiochem) was added to 0.2%, and the samples were centrifuged for 5 min at 3000 rpm. The supernatants were used as cytoplasmic fraction. Pellets were vortexed for 5 min in 0.1 ml of buffer C (20 mM HEPES, pH 7.9; 400 mM NaCl; 1 mM EDTA; 1 mM DTT; and fresh protease inhibitor) at 4°C and sonicated. The samples were then centrifuged for 15 min at 14,000 rpm. Supernatants were diluted with buffer D (20 mM HEPES, pH 7.9; 1 mM EDTA; and fresh protease inhibitor) to the 200 mM final NaCl concentration and used as nuclear extracts.
Immunocytochemistry
For immunofluorescence staining, CHO-APP751 cells transiently transfected with Flag-RanBP9-FL or Flag-RanBP9-Δ1/N60 were grown on glass coverslips, and after 36 h cells were fixed in 4% paraformaldehyde in PBS and permeabilized with 0.4% Triton-X-100. After blocking with normal goat serum, cells were incubated with anti-Flag (M2) primary antibody followed by FITC-conjugated secondary antibody. Cells were also counterstained with propidium iodide to visualize nuclei. Images were acquired using the Olympus IX81 fluorescence microscope with attached CCD camera (Olympus, Tokyo, Japan).
Statistical analysis
Data were analyzed by Instat3 software (GraphPad Software, San Diego, CA, USA) using either Student’s t test or 1-way analysis of variance followed by a Neuman-Keuls post hoc test. Data are expressed as means ± se. Differences were deemed significant when P < 0.05. The signal intensity from immunoblots was quantified using Image J software (U.S. National Institutes of Health, Bethesda, MD, USA).
RESULTS
RanBP9 is proteolytically processed to generate RanBP9-N60 in a cell-density-dependent manner
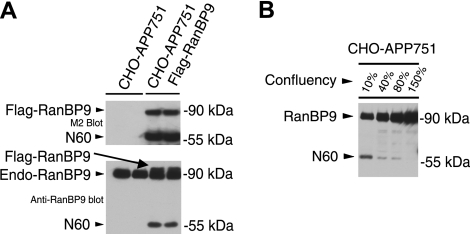
We previously reported that overexpression of RanBP9 either by transient or stable transfection resulted in robustly increased Aβ generation (8). In this study, we set out to characterize the potential regulation of RanBP9 proteolytic processing and its effects on Aβ generation. Lysates from confluent cultures of control CHO-APP751 cells and CHO-APP751 Flag-RanBP9 stably transfected cells were subjected to immunoblotting for Flag and RanBP9. The M2 antibody that detects the 3× Flag epitopes at the N terminus of exogenous RanBP9 not only detected the expected ∼90-kDa full-length band but also an apparent N-terminal proteolytic product migrating at ∼60 kDa (heretofore named N60; Fig. 1A, top panel). When lysates were probed with a monoclonal antibody recognizing RanBP9, the endogenous hamster RanBP9 could be detected together with the slightly slower migrating exogenous Flag-RanBP9 (Fig. 1A, bottom panel). In addition to the 90-kDa band expected of RanBP9-FL, N60 was also detected by the anti-RanBP9 antibody in RanBP9-transfected cells (Fig. 1A, bottom panel), demonstrating the identity of the N60 fragment using 2 different monoclonal antibodies. Previous studies (11,12,13,14) have shown that RanBP9 localization may depend on cell contact and/or growth state of cells. Because the endogenous N60 was not detected in contact-inhibited confluent cells, we next determined whether cell density has an effect on the proteolytic generation of N60 from endogenous RanBP9. Therefore, CHO-APP751 cells were cultured at 10, 40, 80, and 150% confluency, and equal volumes of lysates were immunoblotted with the RanBP9 monoclonal antibody. Surprisingly, cultures at 10% density demonstrated the highest levels of N60 generation, while increasing cell density gradually decreased N60 levels (Fig. 1B). At ∼150% cell density, no N60 product could be detected despite the ∼15-fold more protein than 10% density cultures (Fig. 1B). This clearly suggests that cell-to-cell contact normally inhibits the proteolytic cleavage of RanBP9 to form the N60 fragment. However, exogenously overexpressed RanBP9 appears to at least partially escape this tight physiological regulation.
Figure 1.
RanBP9 is proteolytically cleaved to generate RanBP9-N60 in CHO cells. A) CHO-APP751 parental cells and CHO-APP751-flag-RanBP9 stable cells were plated in duplicates, and confluent cultures were subjected to cell lysis and immunoblotting (equal protein amounts). Anti-Flag M2 antibody detected Flag-RanBP9 from stable cells but not from CHO-APP751 control cells (top panel, bottom band), demonstrating that stable cells express transfected RanBP9. In addition to the 90-kDa band expected of full-length RanBP9, an apparent proteolytic N-terminal 60-kDa fragment (RanBP9-N60) was also strongly detected with M2 antibody (top panel, bottom band). When lysates were probed with anti-RanBP9 antibody, endogenous hamster RanBP9 could be detected together with slightly slower migrating transfected human Flag-RanBP9 (bottom panel, arrow). N-terminal 60-kDa fragment was also detected with RanBP9 antibody, which was preferentially increased in RanBP9-transfected cells (bottom panel, bottom band). B) Regulation of RanBP9-N60 formation by cell density and microtubule dynamics. CHO-APP751 cells were plated at indicated densities and lysed in equal volumes, and equal volumes of lysates were probed with anti-RanBP9 monoclonal antibody. Note that N60 is formed only at low density and its formation is gradually reduced as cell density is increased, despite huge excess in protein in higher-density cultures.
N60 fragment of RanBP9 is significantly increased in AD brains
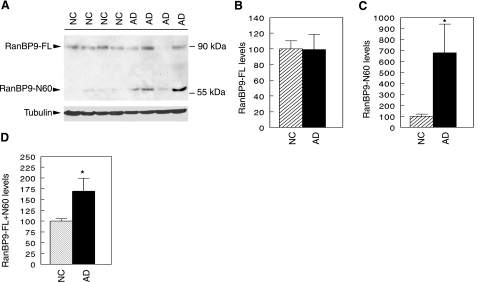
To determine whether RanBP9 protein expression and/or cleavage might be altered in AD, we prepared extracts from the frontal cortex of 8 AD brains and 10 age-matched normal control brains. Protein concentrations were determined by the micro-BCA method, and equal amounts of protein from each sample were subjected to immunoblotting for RanBP9 using a monoclonal antibody recognizing the N-terminal region of RanBP9. Consistent with our observations in cultured CHO-APP751 cells (Fig. 1), the anti-RanBP9 antibody detected 2 bands of ∼90 and 60 kDa in AD and normal control brains (Fig. 2A). However, the intensity of the N60 fragment appeared much stronger in most AD brains compared with normal controls (Fig. 2A). Quantitation of the 2 bands demonstrated that the level of the 90-kDa RanBP9-FL was comparable in AD and control brains (Fig. 2B). In contrast, the level of the N60 fragment was >6-fold higher on average in AD brains than in normal control brains (Fig. 2C), reaching statistical significance (P<0.02). Due to variations in postmortem interval and length of time in deep freeze of the human brain samples, we cannot be sure whether the detected N60 cleavage product occurred in the live brain or during postmortem time. Thus, it may be meaningful to measure the additive levels of both bands. When the full-length and N60 RanBP9 products were combined, there remained a significant ∼75% increase in RanBP9 in AD brains compared with age matched controls (P<0.02; Fig. 2D). Although the N60 fragment remains to be isolated and sequenced, both RanBP9-FL and N60 bands were specific to RanBP9, since depletion of the anti-RanBP9 antibody by recombinant GST-RanBP9 purified on glutathione Sepharose beads largely eliminated the immunoreactivity for both bands from human brain and CHO cells (Supplemental Fig. S1). As reduced cell density promoted N60 generation, these data suggest that increased RanBP9 cleavage in AD may emanate from compromised cell contacts in the neurodegenerative process.
Figure 2.
RanBP9-N60 is robustly increased in AD brains. A) Equal amounts of protein samples from AD brains (n=8) and age-matched normal controls (NC; n=10) were immunoblotted for RanBP9 using anti-RanBP9 antibody. Tubulin was detected with anti-tubulin as a loading control. A representative blot is shown. B) Relative level of full-length RanBP9 (RanBP9-FL) normalized to tubulin. There was no difference in the level of RanBP9-FL normalized to tubulin between AD brain and normal controls. C) Quantitative measurement of the 60-kDa fragment (RanBP9-N60) normalized to tubulin was ∼6-fold higher in AD brains than NC brains, reaching statistical significance. D) When signals from both RanBP9-FL and N60 were combined and normalized to tubulin, there remained ∼75% higher levels in AD brains than in NC brains, which is also highly significant. *P < 0.02.
Reduced nuclear localization and increased stability of RanBP9-Δ1/N60 lacking the CTLH and CRA domains
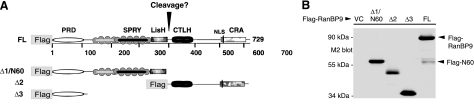
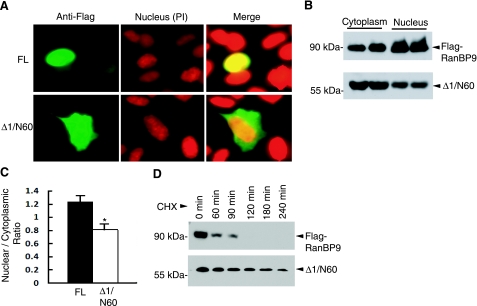
The observation that the N-terminal 60-kDa or N60 fragment of RanBP9 was elevated in AD prompted us to further investigate the potentially differential biological properties of this fragment vs. RanBP9-FL. The modular domain structure of RanBP9 is shown in Fig. 3A, and the potential function of each domain is outlined in Supplemental Table S1. We had previously generated several deletion mutants of RanBP9 (Δ1–Δ4; ref. 8). Remarkably, the Flag-RanBP9-Δ1/N60 mutant (residues 1–392), containing the PRD, SPRY, and LisH domains but lacking CTLH and CRA domains, migrated at ∼60 kDa on SDS-PAGE, very similar to the N60 fragment generated from transient transfection of full-length Flag-RanBP9 in 293T cells (Fig. 3B). Moreover, we had also previously observed that RanBP9-Δ1/N60 coimmunoprecipitated much more robustly with APP, LRP, and BACE1 than RanBP9-FL in transient cotransfection experiments (8). Therefore, we used the RanBP9-Δ1/N60 as a surrogate of the N60 fragment and initially asked whether the loss of the C-terminal region in RanBP9-Δ1/N60 influences its subcellular localization. Indeed, a bipartite nuclear localization signal sequence is located within the residues 635 and 649 of the CRA domain, suggesting that Δ1/N60 would be more readily retained in the cytoplasm. To test this, we transiently transfected CHO-APP751 cell with Flag-RanBP9 and Flag-RanBP9-Δ1/N60, and we stained them using the Flag M2 antibody. Previous studies (11, 12, 15, 21, 22) have shown that RanBP9 is present in both the nucleus and cytoplasm of different cell lines. In CHO-APP751 cells, RanBP9-FL was also found in both the nucleus and cytoplasm. However, a significant proportion of cells showed exclusive localization of RanBP9-FL in the nucleus as recognized by chromatin staining with propidium iodide (Fig. 4A). In contrast, the RanBP9-Δ1/N60 was mostly localized to the cytoplasm, although some nuclear staining was also detected (Fig. 4A). Representative pictures for each of RanBP9-FL and RanBP9-Δ1/N60 are shown in Fig. 4A. To confirm this finding in a different way, we next prepared nuclear and cytoplasmic extracts from HEK293FT cells transiently transfected with Flag-RanBP9-FL and Flag-RanBP9-Δ1/N60 and subjected equal amounts of protein to immunoblotting for Flag-RanBP9. Indeed, RanBP9-FL was enriched in the nucleus, whereas RanBP9-Δ1 was found in greater amounts in the cytoplasmic fraction (Fig. 4B). Taken as a ratio of cytoplasmic vs. nuclear distribution, the RanBP9-FL and RanBP9-Δ1/N60 significantly differed with each other (Fig. 4C; P<0.05).
Figure 3.
A) Schematic of domain organization of RanBP9 and Flag-tagged deletion constructs: Δ1/N60 (residues 1–392), Δ2, (residues 408–729), and Δ3 (residues 1–107). B) HEK293T cells were transiently transfected with vector control (VC), Flag-tagged RanBP9, Δ1, Δ2, or Δ3 deletion mutants. Equal protein amounts from lysates were immunoblotted for Flag (M2). Note that transient transfection of Flag-RanBP9 yields an N-terminal 60-kDa fragment (Flag-N60), which comigrates with Flag-RanBP9-Δ1/N60.
Figure 4.
RanBP9-Δ1/N60 is relatively more stable and cytoplasmic than RanBP9-FL. A) CHO-APP751 cells were transiently transfected with either RanBP9-FL or RanBP9-Δ1/N60 mutant. After 24 h, cells were stained with M2 antibody, followed by nuclear counterstaining with propidium iodide (PI). While RanBP9-FL was found in both the nucleus and cytoplasm, a significant proportion of cells showed exclusive localization of RanBP9-FL in the nucleus. B) HEK293FT cells were transiently transfected with either Flag-RanBP9-FL or Flag-RanBP9-Δ1/N60, and nuclear and cytoplasmic extracts were analyzed by immunoblotting with the anti-Flag antibody (M2). C) Quantitative measurement of intensity of bands in the nucleus and cytoplasm by Image J software revealed significantly less Δ1/N60 in the nucleus relative to cytoplasm compared with RanBP9-FL (means±se; n=4). *P < 0.01; Student’s t test. D) CHO-APP751 cells stably expressing Flag-RanBP9 or Flag-RanBP9-Δ1/N60 were treated with cycloheximide (100 μg/ml) for the indicated time points. Immunoblot analysis using anti-Flag antibody (M2) showed that the half-life of RanBP9-FL was <1 h, while that of half-life of Δ1/N60 was >3 h.
We next asked whether RanBP9-Δ1/N60 alters the stability of the protein relative to RanBP9-FL. To test this, CHO-APP751 stable cells expressing Flag-RanBP9-FL or Flag-RanBP9-Δ1/N60 were treated with cycloheximide to stop protein synthesis and harvested at different time points. We found that RanBP9-FL is a short-lived protein with a half-life of <1 h (Fig. 4D). Within 2 h of cycloheximide treatment, Flag-RanBP9-FL was virtually undetectable (Fig. 4D). In contrast, Flag-RanBP9-Δ1/N60 was much more stable, with a half-life of >3 h (Fig. 4D). Thus, the loss of CTLH to CRA domains by proteolytic cleavage enhances the stability of RanBP9-Δ1/N60 in addition to reducing its nuclear localization. Therefore, we interpret these results to indicate that enhanced retention in the cytoplasm at the expense of nuclear translocation and increased stability of RanBP9-Δ1/N60 underlie the increased binding to APP, LRP, and BACE1 (8). Also, since RanBP9-Δ1/N60 has a much longer half-life than the RanBP9 holoprotein, even a small amount of N60 generation is predicted to result in its accumulation relative to RanBP9-FL, a phenotype seen in AD brains.
RanBP9-FL and RanBP9-Δ1/N60 are capable of forming complexes with each other
Previous studies (22, 23) have shown that RanBP9 forms large multimeric complexes with various LisH-domain-containing proteins, including muskelin. Because RanBP9 was capable of interacting with APP, LRP, and BACE1 and promoting APP complexes with LRP and BACE1 (8, 22), we hypothesized that RanBP9 itself might form dimeric or oligomeric complexes capable of scaffolding two or more proteins. Indeed, the LisH domain in Lis1 and other LisH-containing proteins has been implicated in dimerization and/or oligomerization (9, 10), but this has not been empirically demonstrated for RanBP9. To test whether RanBP9 can form self-interacting complexes, Neuro-2A cells were transiently cotransfected with increasing amounts of Flag-RanBP9 and GFP-RanBP9, and lysates were immunoprecipitated for either GFP or Flag tags. Indeed, GFP immune complexes contained Flag-RanBP9, and Flag immune complexes contained GFP-RanBP9 in amounts corresponding to the increasing amount of transfected Flag-RanBP9 (Fig. 5A, B). Therefore, RanBP9 is capable of forming complexes with itself, a capacity that may be important for its ability to enhance Aβ generation as a dimeric or multimeric scaffolding complex. Next, we tested whether Δ1/N60 retains the capacity to form complexes with RanBP9. In similar transient transfection experiments, Flag-Δ1/N60 and Flag-RanBP9 pulled down similar amounts of GFP-RanBP9 (Fig. 5C), demonstrating that the CTLH and CRA domains are not essential for this process. Taken together, these data demonstrate that RanBP9 forms dimeric or multimeric complexes and that this capacity is retained in RanBP9-Δ1/N60.
Figure 5.
Both RanBP9 and Δ1/N60 are capable of forming self-interacting complexes. A) Neuro-2A cells were transiently cotransfected with a constant amount of GFP-RanBP9 and increasing concentrations of Flag-RanBP9. After 48 h, lysates were immunoprecipitated with GFP antibody and then immunoblotted with anti-Flag (M2) antibody. B) In similar experiments as above, lysates were immunoprecipitated with anti-Flag M2 antibody and immunoblotted with anti-GFP antibody. Asterisk indicates IgG band. Note the concentration-dependent increase in the interactions between GFP-RanBP9 and Flag-RanBP9. C) Neuro-2A cells were cotransfected with GFP-RanBP9 along with Flag-RanBP9-FL, Flag-Δ1/N60, or empty vector. After immunoprecipitation with anti-Flag M2 antibody, immunoblot detection with GFP antibody showed that Flag-Δ1/N60 interacts with GFP-RanBP9 to a similar extent as full-length Flag-RanBP9.
RanBP9-Δ1/N60 robustly potentiates Aβ generation
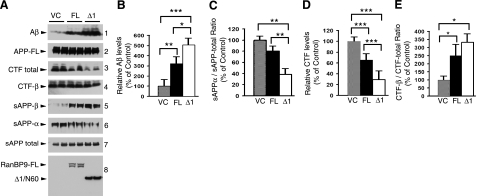
Given that RanBP9-Δ1/N60 was increased in AD, displayed increased stability, and interacted more robustly with LRP/APP/BACE1 than RanBP9-FL, we next compared the effects of RanBP9-FL vs. RanBP9-Δ1/N60 overexpression on APP processing and Aβ generation. HEK293FT cells were transiently cotransfected with APP751 and equal amounts of pLHCX-Flag-RanBP9, pLHCX-Flag-RanBP9-Δ1, or pLHCX empty vector for 8 h, and conditioned medium and cell lysates were harvested after 48 h. Under these conditions, full-length APP (APP-FL) levels were indistinguishable among all transfection conditions (Fig. 6A2), and N60 generation from RanBP9-FL was virtually undetectable in these very high density cultures (Fig. 6A8). As expected, both RanBP9 and RanBP9-Δ1/N60 increased Aβ generation over vector controls, but the effects of Δ1/N60 were noticeably stronger than those of RanBP9-FL for most measures of APP processing (Fig. 6). Specifically, quantitation from multiple experiments (n=4) showed that RanBP9-Δ1/N60 secreted ∼5-fold more Aβ than the vector control, whereas RanBP9-FL increased Aβ levels by ∼3-fold (Fig. 6A1, B). The increases in Aβ were mirrored by corresponding increases in the secretion of sAPP-β and decreases in sAPP-α, both of which were more robust with Δ1/N60 than RanBP9-FL (Fig. 6A5, 6). Thus, the ratio of sAPP-α relative to sAPP total was also more potently decreased by Δ1/N60 than by RanBP9-FL (Fig. 6C). Similar to our previous observations in CHO-APP751 cells (8), total APP-CTFs were decreased by RanBP9-FL, a phenotype that was further enhanced by Δ1/N60 (Fig. 6D). This CTF-decreasing phenotype is similar to that observed with overexpression of LRP-C37 (7). Despite the decrease in total APP CTFs, APP-CTF-β levels remained relatively unchanged. Therefore, Δ1/N60 further increased the CTF-β-to-APP-CTF total ratio beyond that of RanBP9-FL but without statistical significance (Fig. 6E). Taken together, these data demonstrate that RanBP9-Δ1/N60 is more potent than RanBP9-FL in enhancing β-secretase processing of APP and indicate that increased N60 generation in AD likely further contributes to amyloid pathology.
Figure 6.
RanBP9-Δ1/N60 is significantly more potent than RanBP9-FL in APP processing and Aβ generation. A) HEK293FT cells were transiently cotransfected with APP751 and equal amounts of VC, RanBP9-FL (FL), or RanBP9-Δ1/N60 (Δ1) plasmids, and conditioned media and lysates were harvested 48 h after transfection when cells were fully confluent. For detection of Aβ (1), the conditioned medium was immunoprecipitated with B436 monoclonal antibody and immunoblotted with a mixture of 6E10/82E1 antibody. Conditioned media were directly immunoblotted to detect sAPP total (63G; 7), sAPP-α (6E10; 6), and sAPP-β (18957, IBL; 5). In lysates, indicated antibodies were used to detect APP-FL (CT15; 2), APP-CTF total (CT15; 3), APP-CTF-β (6E10; 4), RanBP9 (M2; 8), and D1/N60 (M2; 8). B) Densitometric quantification of Aβ showed an average of ∼3-fold increase by RanBP9-FL compared with vector control. Increase by RanBP9-Δ1/N60 was ∼5-fold, which was also significantly higher than RanBP9-FL. C) Densitometric quantification of the ratio of sAPP-α relative to sAPP total revealed significant differences between VC and Δ1/N60 and also between FL and Δ1/N60 (P<0.01). D) Densitometric quantification of total CTF levels showed significant differences between VC and FL, VC and Δ1/N60, and FL and Δ1/N60. E) Densitometric quantification of the ratio of CTF-β relative to CTF total demonstrated significant differences between VC and FL and VC and Δ1/N60. Graphs show means ± se; n = 4. *P < 0.05, **P < 0.01, ***P < 0.001; 1-way ANOVA with Student-Newman-Keuls post hoc test.
DISCUSSION
The mechanisms by which APP and the secretases are brought together to generate Aβ are critical to discovering novel treatments for AD. We recently demonstrated that RanBP9 plays an important role in bringing APP together with LRP and BACE1 via a scaffolding mechanism (8). In this study, we discovered a novel N-terminal fragment of RanBP9 (N60) that is proteolytically generated in a cell density-dependent manner and significantly elevated in the brains of AD patients. The N60 fragment was virtually indistinguishable from RanBP9-Δ1/N60 (residues 1–392), a deletion variant that demonstrated far more robust interactions with APP/LRP/BACE1 than RanBP9-FL (8). The RanBP9-Δ1/N60 fragment, which lacks a nuclear localization signal, displayed enhanced cytoplasmic vs. nuclear localization and >3-fold enhanced stability than RanBP9-FL . Notably, RanBP9-Δ1/N60, which contains the LisH dimerization domain, retained the capacity to form self-interacting multimeric complexes and increased Aβ generation by 5-fold over vector controls, more potent than the ∼3-fold increase seen by RanBP9-FL. Taken together, these data indicate that RanBP9-N60 may further drive the amyloid cascade in AD and that the proteolytic processing of RanBP9 may be an attractive therapeutic target for AD.
The generation of the RanBP9-N60 fragment is intriguing in light of our observations that cell-to-cell contact appears to inhibit RanBP9 processing. This mode of physiological regulation is likely to have significant biological consequences. As cell density involves cell-to-cell contact and changes in cell morphology, regulated formation of N60 suggests a role for RanBP9 in these processes. Indeed, RanBP9 has recently been shown to regulate both cell adhesion and cell morphology (15, 24). More specifically, knockdown of RanBP9 in adherent cells led to protrusive phenotypes (15), and overexpression resulted in altered cell migration (15, 16, 25). These observations suggest that RanBP9 plays a vital role in regulating these events by linking the extracellular signals via transmembrane receptors (i.e., plexin-A, Met, and integrin LFA-1) with the intracellular signal transduction machinery (12, 16, 17). RanBP9 is highly conserved across species, and in particular, the SPRY, LisH, and CTLH domains are identical between mice and humans. Although the search for the protease responsible for the formation of N60 was not addressed in this study, we found that nocodazole, a microtubule-disrupting agent, prevented the formation of RanBP9-N60 (not shown). Therefore, the results from this study, taken together with the earlier demonstrations that RanBP9 regulates cell morphology, suggest a mechanistic link between RanBP9 processing, microtubule dynamics, and the regulation of cell morphology and migration.
At present, the manner in which RanBP9 stability is controlled is unknown. Our observation that RanBP9-Δ1/N60 is far more stable than RanBP9-FL indicates that the C-terminal region containing the CTLH and CRA domains is critical for the rapid turnover of RanBP9. Some of its turnover is mediated by endoproteolysis to generate the N60 fragment, while another component may be mediated by the ubiquitin-proteasome system. The localization of RanBP9 is also an important factor for its activity. In polarized MDCK cells, RanBP9 is mainly found associated with proteins in the inner leaflet of the plasma membrane and is directed to the basolateral surface, like APP and LRP (12, 26, 27). RanBP9 is also localized to synaptic sites in neurons and cytoskeletal elements in muscle (11, 14). In other undifferentiated cell lines, RanBP9 is distributed throughout the cytoplasm as well as the nucleus (13, 22). Within the nucleus, it has been shown that RanBP9 can act as a transcriptional coactivator of various hormone receptors, including the androgen receptor, glucocorticoid receptor, and thyroid hormone receptor (13, 21). Given that a putative bipartite nuclear localization signal is located in the C-terminal CRA domain, the proteolytic cleavage of RanBP9 would be predicted to functionally alter its different activities in the nucleus and cytoplasm.
We found that the RanBP9-N60 fragment was significantly elevated in AD brains compared with age-matched normal controls. Based on the size of the N60 fragment in comparison with RanBP9-Δ1/N60, N60 is expected to contain the PRD, SPRY, and LisH domains and retain the functionality of Δ1/N60. We propose a model in which unhealthy conditions such as loss of synaptic contacts or neuronal insults (i.e., Aβ) could promote the cleavage of RanBP9 to generate N60. Due to its increased stability and cytoplasmic localization, N60 would form more stable complexes with LRP/APP/BACE1. Due to its ability to form dimeric or multimeric complexes via its LisH dimerization domain, N60 would retain the capacity to simultaneously scaffold APP with LRP or BACE1, thereby enhancing APP association with rafts, β-secretase cleavage, and Aβ generation. This positive feedback model is analogous to the induction of BACE1 under stressful conditions both at the mRNA and protein levels (28,29,30). In this model, RanBP9 normally promotes Aβ generation, but proteolytic conversion to the N60 fragment renders it more potent than its precursor in promoting all aspects of APP processing, as empirically demonstrated in this study. In stable CHO-APP751 cell lines expressing RanBP9-FL or RanBP9-Δ1/N60, both cell lines similarly enhanced Aβ generation (8). However, the difference was likely masked due to large amounts of constitutive N60 generation in the CHO-APP751-RanBP9-FL cell line. In the transient transfection model using high-density HEK293FT cells in this study, we did not detect appreciable amounts of N60 generation from RanBP9-FL.
The cellular and biochemical mechanisms by which Aβ is generated are critical for designing therapeutic strategies for AD. Although directly inhibiting β- or γ-secretase activities are obvious therapeutic strategies, γ-secretase inhibitors have unintended side effects, such as inhibition of Notch cleavage, an important activity for neurogenesis and maintenance of multiple stem-cell populations (31). Highly specific β-secretase inhibitors are not yet available for therapeutic purposes, and the potential side effects are uncertain, especially in light of new observations that BACE1 controls the myelination of central nervous system and peripheral axons (32). The results of this study indicate that RanBP9 represents an alternative therapeutic target for inhibiting contact between APP and BACE1. Furthermore, inhibition of RanBP9-N60 generation may also be a viable therapeutic strategy.
Supplementary Material
Acknowledgments
This work was supported in part by the American Health Assistance Foundation (A2007-05 to D.E.K.), the Alzheimer’s Association (NIRG-07-60584, M.K.L.), the National Institute of Aging (AG 005131-24S1 to D.E.K.), and a World Class University grant from the Korea Science and Engineering Foundation (to D.E.K.).
References
- De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113:1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- Herz J, Strickland D K. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzik C U, Busse T, Merriam D E, Weggen S, Koo E H. The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 2002;21:5691–5700. doi: 10.1093/emboj/cdf568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulery P G, Beers J, Mikhailenko I, Tanzi R E, Rebeck G W, Hyman B T, Strickland D K. Modulation of beta-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer’s disease. J Biol Chem. 2000;275:7410–7415. doi: 10.1074/jbc.275.10.7410. [DOI] [PubMed] [Google Scholar]
- Yoon I S, Pietrzik C U, Kang D E, Koo E H. Sequences from the low density lipoprotein receptor-related protein (LRP) cytoplasmic domain enhance amyloid beta protein production via the beta-secretase pathway without altering amyloid precursor protein/LRP nuclear signaling. J Biol Chem. 2005;280:20140–20147. doi: 10.1074/jbc.M413729200. [DOI] [PubMed] [Google Scholar]
- Yoon I S, Chen E, Busse T, Repetto E, Lakshmana M K, Koo E H, Kang D E. Low-density lipoprotein receptor-related protein promotes amyloid precursor protein trafficking to lipid rafts in the endocytic pathway. FASEB J. 2007;21:2742–2752. doi: 10.1096/fj.07-8114com. [DOI] [PubMed] [Google Scholar]
- Lakshmana M K, Chen E, Yoon I S, Kang D E. C-terminal 37 residues of LRP promote the amyloidogenic processing of APP independent of FE65. J Cell Mol Med. 2008;12:2665–2674. doi: 10.1111/j.1582-4934.2008.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmana M K, Yoon I S, Chen E, Bianchi E, Koo E H, Kang D E. Novel role of RanBP9 in BACE1 processing of amyloid precursor protein and amyloid beta peptide generation. J Biol Chem. 2009;284:11863–11872. doi: 10.1074/jbc.M807345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G, Darhin E, Giorgio G, Franco B, Reiner O. Novel functional features of the Lis-H domain: role in protein dimerization, half-life and cellular localization. Cell Cycle. 2005;4:1632–1640. doi: 10.4161/cc.4.11.2151. [DOI] [PubMed] [Google Scholar]
- Mateja A, Cierpicki T, Paduch M, Derewenda Z S, Otlewski J. The dimerization mechanism of LIS1 and its implication for proteins containing the LisH motif. J Mol Biol. 2006;357:621–631. doi: 10.1016/j.jmb.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Bowman A L, Catino D H, Strong J C, Randall W R, Kontrogianni-Konstantopoulos A, Bloch R J. The rho-guanine nucleotide exchange factor domain of obscurin regulates assembly of titin at the Z-disk through interactions with Ran binding protein 9. Mol Biol Cell. 2008;19:3782–3792. doi: 10.1091/mbc.E08-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti S, Sirri A, Cheli A, Rogge L, Innamorati G, Putignano S, Fabbri M, Pardi R, Bianchi E. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem. 2004;279:13027–13034. doi: 10.1074/jbc.M313515200. [DOI] [PubMed] [Google Scholar]
- Rao M A, Cheng H, Quayle A N, Nishitani H, Nelson C C, Rennie P S. RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. J Biol Chem. 2002;277:48020–48027. doi: 10.1074/jbc.M209741200. [DOI] [PubMed] [Google Scholar]
- Seebahn A, Rose M, Enz R. RanBPM is expressed in synaptic layers of the mammalian retina and binds to metabotropic glutamate receptors. FEBS Lett. 2008;582:2453–2457. doi: 10.1016/j.febslet.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Valiyaveettil M, Bentley A A, Gursahaney P, Hussien R, Chakravarti R, Kureishy N, Prag S, Adams J C. Novel role of the muskelin-RanBP9 complex as a nucleocytoplasmic mediator of cell morphology regulation. J Cell Biol. 2008;182:727–739. doi: 10.1083/jcb.200801133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Li Z, Messing E M, Wu G. Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. J Biol Chem. 2002;277:36216–36222. doi: 10.1074/jbc.M205111200. [DOI] [PubMed] [Google Scholar]
- Togashi H, Schmidt E F, Strittmatter S M. RanBPM contributes to Semaphorin3A signaling through plexin-A receptors. J Neurosci. 2006;26:4961–4969. doi: 10.1523/JNEUROSCI.0704-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Hirose E, Uchimura Y, Nakamura M, Umeda M, Nishii K, Mori N, Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- Kang D E, Yoon I S, Repetto E, Busse T, Yermian N, Ie L, Koo E H. Presenilins mediate phosphatidylinositol 3-kinase/AKT and ERK activation via select signaling receptors. Selectivity of PS2 in platelet-derived growth factor signaling. J Biol Chem. 2005;280:31537–31547. doi: 10.1074/jbc.M500833200. [DOI] [PubMed] [Google Scholar]
- Repetto E, Yoon I S, Zheng H, Kang D E. Presenilin 1 regulates epidermal growth factor receptor turnover and signaling in the endosomal-lysosomal pathway. J Biol Chem. 2007;282:31504–31516. doi: 10.1074/jbc.M704273200. [DOI] [PubMed] [Google Scholar]
- Poirier M B, Laflamme L, Langlois M F. Identification and characterization of RanBPM, a novel coactivator of thyroid hormone receptors. J Mol Endocrinol. 2006;36:313–325. doi: 10.1677/jme.1.01891. [DOI] [PubMed] [Google Scholar]
- Umeda M, Nishitani H, Nishimoto T. A novel nuclear protein, Twa1, and Muskelin comprise a complex with RanBPM. Gene. 2003;303:47–54. doi: 10.1016/s0378-1119(02)01153-8. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Yang J, Ueda A, Suzuki T, Tomaru K, Takeno M, Okuda K, Ishigatsubo Y. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236–247. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Dansereau D A, Lasko P. RanBPM regulates cell shape, arrangement, and capacity of the female germline stem cell niche in Drosophila melanogaster. J Cell Biol. 2008;182:963–977. doi: 10.1083/jcb.200711046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Lim S, Lee K, Deng X, Friedman E. Serine/threonine kinase Mirk/Dyrk1B is an inhibitor of epithelial cell migration and is negatively regulated by the Met adaptor Ran-binding protein M. J Biol Chem. 2003;278:49573–49581. doi: 10.1074/jbc.M307556200. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Craessaerts K, Dewachter I, Moechars D, Greenberg B, Van Leuven F, Van den B H. Basolateral secretion of amyloid precursor protein in Madin-Darby canine kidney cells is disturbed by alterations of intracellular pH and by introducing a mutation associated with familial Alzheimer’s disease. J Biol Chem. 1995;270:4058–4065. doi: 10.1074/jbc.270.8.4058. [DOI] [PubMed] [Google Scholar]
- Takeda T, Yamazaki H, Farquhar M G. Identification of an apical sorting determinant in the cytoplasmic tail of megalin. Am J Physiol Cell Physiol. 2003;284:C1105–C1113. doi: 10.1152/ajpcell.00514.2002. [DOI] [PubMed] [Google Scholar]
- Bourne K Z, Ferrari D C, Lange-Dohna C, Rossner S, Wood T G, Perez-Polo J R. Differential regulation of BACE1 promoter activity by nuclear factor-kappaB in neurons and glia upon exposure to beta-amyloid peptides. J Neurosci Res. 2007;85:1194–1204. doi: 10.1002/jnr.21252. [DOI] [PubMed] [Google Scholar]
- Tamagno E, Bardini P, Obbili A, Vitali A, Borghi R, Zaccheo D, Pronzato M A, Danni O, Smith M A, Perry G, Tabaton M. Oxidative stress increases expression and activity of BACE in NT2 neurons. Neurobiol Dis. 2002;10:279–288. doi: 10.1006/nbdi.2002.0515. [DOI] [PubMed] [Google Scholar]
- Tesco G, Koh Y H, Kang E L, Cameron A N, Das S, Sena-Esteves M, Hiltunen M, Yang S H, Zhong Z, Shen Y, Simpkins J W, Tanzi R E. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J A, Yuan J S, Tan J B, Visan I, Guidos C J. Notch signaling in development and disease. Clin Genet. 2003;64:461–472. doi: 10.1046/j.1399-0004.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- Hu X, Hicks C W, He W, Wong P, Macklin W B, Trapp B D, Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.