Abstract
Maternal choline availability is essential for fetal neurogenesis. Choline deprivation (CD) causes hypomethylation of specific CpG islands in genes controlling cell cycling in fetal hippocampus. We now report that, in C57BL/6 mice, CD during gestational days 12–17 also altered methylation of the histone H3 in E17 fetal hippocampi. In the ventricular and subventricular zones, monomethyl-lysine 9 of H3 (H3K9me1) was decreased by 25% (P<0.01), and in the pyramidal layer, dimethyl-lysine 9 of H3 (H3K9me2) was decreased by 37% (P<0.05). These changes were region specific and were not observed in whole-brain preparations. Also, the same effects of CD on H3 methylation were observed in E14 neural progenitor cells (NPCs) in culture. Changes in G9a histone methyltransferase might mediate altered H3K9me2,1. Gene expression of G9a was decreased by 80% in CD NPCs (P<0.001). In CD, H3 was hypomethylated upstream of the RE1 binding site in the calbindin 1 promoter, and 1 CpG site within the calbindin1 promoter was hypermethylated. REST binding to RE1 (recruits G9a) was decreased by 45% (P<0.01) in CD. These changes resulted in increased expression of calbindin 1 in CD (260%; P<0.05). Thus, CD modulates histone methylation in NPCs, and this could underlie the observed changes in neurogenesis.—Mehedint, M. G., Niculescu, M. D., Craciunescu, C. N., Zeisel, S. H. Choline deficiency alters global histone methylation and epigenetic marking at the Re1 site of the calbindin 1 gene.
Keywords: G9a, hippocampus, REST/NRSF, progenitors
During gestation, maternal dietary choline deficiency (CD) increases neural tube closure defects in rodent and human fetuses (1,2,3,4). Later in gestation, maternal CD alters the development of fetal hippocampus by decreasing neural progenitor cell (NPC) proliferation and by increasing apoptosis and expression of markers of differentiation (5,6,7). We suggested that these effects were mediated by changes in gene-specific DNA methylation (8), and we now suggest that they also are mediated by histone methylation.
Brain neurogenesis is modulated by epigenetic mechanisms that involve histone methylation and chromatin remodeling (9,10,11). Functional interactions between histones and DNA are modulated by their methylation, monomethyl and dimethyl H3K9 (H3K9me1,2) silence specific sites within euchromatic regions (12, 13), whereas dimethyl and trimethyl lysine 4 H3 (H3K4me2,3) (14,15,16) are enriched in areas with transcriptional active chromatin (17). Other modifications of histone residues, such as acetylation of lysines on histone 3 (H3K9Ac), also alter chromatin architecture (18) and inhibit DNA methylation at promoter regions resulting in transcriptionally permissive chromatin (19).
The effects of CD on histone methylation are probably mediated by interactions between multiple enzyme systems. Monomethyl and dimethyl H3K9 are formed by G9a histone methylase (20, 21), while H3K9me3 is formed by SUV39 (22). G9a HMTase has 10- to 20-fold higher activity than does SUV39 HMTase in fetal brain (20). Mice, in which G9a was deleted, exhibit growth retardation, embryonic lethality, and diminished levels of H3K9me2,1 (20). Transcriptional repressor neuron restrictive silencing factor (REST) binding mediates the inhibition of expression of numerous neuronal genes (23). REST recruits a corepressor complex to repressor element 1 (RE1), which includes histone deacetylase (HDAC), methyl CpG binding protein 2 (MeCP2), (24), and G9a (25). Demethylases also modulate chromatin permissivity; methylated H3K4 and H3K9 are substrates for several histone demethylases such as the LSD1 and JHDM family (26,27,28).
In this study, we examined whether choline availability during pregnancy altered the “histone code” in NPCs of fetal mouse hippocampus, whether these changes were associated with altered cell proliferation or apoptosis, and whether the underlying mechanisms involved altered gene expression and protein levels for G9a, SUV39 as well as REST binding to RE1 of Calb1.
MATERIALS AND METHODS
Animals
Timed-pregnant C57BL/6 mice were ordered from Jackson Laboratory, Inc. (Bar Harbor, ME, USA) at gestation day 7 and used in all experiments according to a protocol described elsewhere (5). The animals were kept in a temperature-controlled environment at 24°C and exposed to a 12-h light and dark cycle. The mice consumed an AIN-76A pelleted diet with the standard 1.1 g/kg (7.8 mmol/kg) choline chloride (Dyets, Inc., Bethlehem, PA, USA) and water ad libitum until the end of embryonic day 11 (E11) when they were randomly assigned to 1 of the 2 feeding groups: a choline-deficient (CD) group with 0.0 g/kg choline chloride AIN-76A (Dyets), and control group (CT) with 1.1 g/kg choline chloride AIN-76A (Dyets) as described previously (5).
Fetal brain collection
On E17, the fetal brains were collected as described previously (5). In two male pups from each litter, skulls were opened and immersed in fixation buffer containing 4% formaldehyde and 0.2% glutaraldehyde (Polysciences, Inc., Warrington, PA, USA) in 0.1 M PBS. The remaining brains from the litter were collected individually, frozen in liquid nitrogen, and stored at −80°C for further determinations of mRNA levels. For the overnight postfixation of the brains, the heads were stored in perfusion fixative for 24 h and then kept in 70% ethanol at 4°C until processing for paraffin embedding. All samples were cut in 5-μm coronal sections, and anatomically equivalent areas containing hippocampus and septum were selected according to a standard atlas of the developing brain (29) and used for further immunohistochemical analysis.
Fetal mouse NPCs in culture
According to previously published data, NPCs in culture behave in a manner similar to that observed in fetal hippocampal NPCs from the hippocampal ventricular (VZ) and subventricular (SVZ) in terms of chromatin architecture, remodeling, histone modifications, and gene expression (7, 8, 10, 30, 31). In addition, previous studies (7) using gene array analysis of CD-treated NPCs showed expression changes for genes that modulate cell cycle, apoptosis, neuronal differentiation, methyl metabolism, and calcium-binding proteins in a pattern similar to that seen in fetal hippocampal NPCs when maternal dietary choline was varied. NPCs from E14 C57BL/6 mice were obtained from Lonza (Walkersville, MD, USA) and plated according to the manufacturer’s protocol using Neurobasal medium (Invitrogen, Carlsbad, CA, USA). The medium was supplemented with 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin-streptomycin (Invitrogen), 2% B27 supplement without vitamin A (Invitrogen), 20 ng/ml murine epidermal growth factor (Invitrogen), 20 ng/ml human βFGF, and 2 mg/ml heparin. The cells were plated at a density of 105 in 10-cm untreated Petri dishes (Fisher Scientific, Pittsburgh, PA, USA) and incubated for 5 d at 37°C, 5% CO2. One-third of the medium volume, along with the cellular debris, was changed every 3 d, and fresh growing factors were added. After 5 d in culture, the floating neurospheres were collected in sterile 15 ml tubes and pelleted by centrifugation at 120 g for 5 min at 37°C. The neurospheres were dissociated using Accutase (Innovative Cell Technologies, San Diego, CA, USA), passaged, and reseeded as suspension. The cycle was repeated until small tertiary neurospheres were generated. The cells were assigned to two different treatment groups (up to n=7 plates/group), passaged to new uncoated plates, and suspended in custom Neurobasal medium (D700SA, Atlanta Biologicals, Lawrenceville, GA, USA) containing 5 μM choline chloride (CD) or 70 μM choline chloride (CT) with the ingredients described above. The optimal exposure time, 72 h, was determined after the assessment of proliferation and apoptosis changes as described below. NPC colonies were pelleted by centrifugation and immediately processed for total protein extraction, genomic DNA extraction, total mRNA isolation, or chromatin precipitation. Modifications in the proliferation of NPCs were detected after the first 48 h in low-choline medium, whereas changes in apoptosis only became obvious after 96 h exposure; for subsequent experiments, we chose the 72-h period so as to minimize artifacts due to cell death. This timeframe allowed the NPCs to complete several cell cycles at an expansion rate of 10- to 15-fold/7 d (32).
Proliferation and apoptosis assessment of NPCs
NPCs were exposed to CD and CT conditions and harvested at different time points: 24, 48, 72, 96, and 120 h. Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate (dUTP)-digoxigenin anti-digoxigenin HRP conjugate antibody nick end labeling (TUNEL) was used as a marker of DNA fragmentation. NPC nuclei with single- and double-stranded DNA breaks were detected using the ApopTag Plus Peroxidase In Situ Apoptosis Detection Kit (Chemicon International, Temecula, CA, USA), according to the manufacturer’s protocol.
Cell proliferation was assessed using 5-bromo-2-deoxyuridine (BrdU) incorporation at 48 and 72 h. Before being harvested, 10 μM BrdU was added to the culture medium, and cells were incubated for 2 h. Cells were fixed with 4% paraformaldehyde and subsequently processed using a BrdU Immunohistochemistry Kit (Chemicon International), according to the manufacturer’s protocol. Diaminobenzoic acid was utilized for color resolution, and methyl-green was used for counterstaining. BrdU-positive (nuclei that were in S phase) and TUNEL-positive cells (with small nuclei/nuclear fragments, undergoing pyknosis, or karyorrhexis) were identified using transmitted light microscopy and expressed as a percentage of the total number of cells. At least 500 cells/sample were counted.
Immunohistochemistry in paraffin-embedded sections
Paraffin-embedded hippocampal coronal sections were selected from anatomically identical regions to minimize differences due to the anterior-posterior gradient in neurogenesis (33). The slides were deparaffinized with xylene and then rehydrated using ethanol in decreasing concentrations. The tissue autofluorescence was minimized by incubating the slides at room temperature for 1 h in 0.25% NH4OH (Sigma, St. Louis, MO, USA) in ethanol 70%. Antigen retrieval for nuclear antigens was performed using citric acid buffer, pH 6, at 100°C for 10 min, followed by incubation in Proteinase K 20 μg/ml (Sigma). As a reducing agent, sodium borohydride (Sigma, 10 mg/ml in distilled water) was used for 40 min at room temperature. An incubation in blocking buffer containing 10% goat serum in PBS + 0.1% Tween 20 (PBST) was performed overnight at 4°C. The slides, except for the negative controls, were labeled for H3K9me1 (ab9045; Abcam Inc., Cambridge, MA, USA) and H3K9me2 (ab1220; Abcam), according to the manufacturer’s protocol. For each staining, 7 different fetal brains per group (CD and CT, respectively) were used, each from a different dam. An additional slide per group was placed into the blocking buffer, without the first antibody, and used as a negative control. The slides were incubated for 48 h at 4°C, then rinsed in PBST, and placed in the same blocking buffer as described above, and a secondary antibody was added: goat polyclonal anti-rabbit, CY3-conjugated (AP132, Chemicon) at a 1:500 dilution of 1 mg/ml reconstituted antibody, for 2 h at room temperature. DAPI (0.1 μg/ml for 20 min, Sigma) was used for staining the nuclear DNA.
Western blotting
E17 whole fetal brains or cultured NPCs were used for the assessment of methylated H3K9 levels. The samples were homogenized in lysis buffer (1% sodium dodecil sulfate, 10 mM sodium-orto-vanadate, and 10 mM Tris) supplemented with 10% protease inhibitor cocktail (P2714; Sigma). Total protein concentration for all samples was determined using the Lowry method (34). Equal amounts of proteins were separated by electrophoresis using polyacrylamide Tris-HCL gels (4–15%; Bio-Rad Laboratories, Hercules, CA, USA) and then blotted on nitrocellulose membranes (0.2-μm pore size; Invitrogen). The equal transfer to the membrane was verified by Poinceau staining. An overnight incubation was performed at 4°C, in blocking buffer containing 2% normal goat serum (Chemicon) in PBST. Immunolabeling was accomplished using chromatin immunoprecipitation (ChIP)-grade polyclonal antibodies for H3K9me1 (ab9045; Abcam), H3K9me2 (ab1220; Abcam), H3K9me3 (ab8898; Abcam), total H3 (ab1791; Abcam), β-actin (ab8227; Abcam), calbindin 1 (ab25085; Abcam), and G9a (07-551; Upstate-Millipore, Billerica, MA, USA). The secondary antibody utilized was a polyclonal goat anti-rabbit, HRP-conjugated (ab6721; Abcam), diluted 1:7000 in blocking buffer. The labeling was performed at room temperature for 40 min on a rocker platform. The blot was incubated in primary antibodies at 1:5000 dilution of anti-G9a IgG and 1:7000 dilution of anti-Calb1 IgG in 2% goat serum PBST. After the membranes were incubated in Western blotting luminol reagent (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 3 min at room temperature, bands were captured on a BioMax Film (Eastman Kodak Company, Rochester, NY, USA) and the molecular imaging system Kodak Image Station 4000MM Pro (Carestream Health, Inc., Rochester, NY, USA) at different exposure times.
Image analysis
Fluorescent images (×4, ×10, and ×20) for H3K9me1-, H3K9me2-, and H3K9me3-stained sections were collected with a Nikon FXA microscope (Nikon, Garden City, NY, USA) equipped with an Optronics TEC-470 CCD Video Camera System (Optronics Engineering, Goleta, CA, USA). The images, acquired from the same field with different fluorescent probes, were then merged. The labeling level and background fluorescence level for selected areas were measured as optical density (OD) using the ScionImage software (Scion Corp., Frederick, MD, USA). Epitope staining level for each slide was obtained by subtracting the background OD and normalizing to the total number of nuclei in the selected area. As a confirmation, specific for VZ, SVZ, and the pyramidal layer of hippocampus, a second image acquisition was performed using a state-of-the-art Zeiss LSM5 Pascal inverted confocal laser scanning microscope (Carl Zeiss, Inc., Thornwood, NY, USA), equipped for epifluorescence imaging with a multiline argon laser and a He-Ne laser, an objective PlanNeoFluar ×40/1.3 DIC3 with oil immersion, a filter set LP560, and a LSM 5 Pascal 3.2 SP2. Using the range indicator function of the software, the maximum red fluorescence of the erythrocyte and the minimum background fluorescence of the acellular areas were utilized as an internal control. Sequential image acquisition was performed with z-stack function at an interval of 0.42 μm, with a resolution of 0.11 μm/pixels, with an average 10 images/stack. The mean optical density (MOD) for the images in each z stack was measured using the ImageJ 1.37v free software from the National Institutes of Health (http://rsb.info.nih.gov/ij/) with the stack function.
Western blot and endpoint PCR analyses were performed by assessing the MOD of protein/PCR bands, and the values were normalized to total H3 for each sample (for Western blot) and to input DNA for endpoint PCR.
ChIP
Cell pellets, CD and CT (72 h exposure), were generated from NPCs as described above (n=7 plates/group). Shearing and precipitation were performed using the ChIP-IT Express Enzymatic Kit (Active Motif, Carlsbad, CA, USA), and optimization was performed according to the manufacturer’s protocol. The antibodies used in this procedure were all ChIP-grade rabbit polyclonal against H3K4me2 (ab7766; Abcam), H3K9me1 (ab9045; Abcam), and H3K9me2 (ab7312; Abcam) and anti-REST (sc-15118, Santa Cruz). Their concentrations, relative to the chromatin amount, were adjusted as indicated in the manufacturer’s protocol. Throughout the procedure, the negative controls negative IgG (mock immunoprecipitation; mock IP) and negative primer pair and the positive controls IgG anti RNA pol II and EF1-alpha primers were used as part of the Chip-IT Control Kit-Mouse (cat. no. 53011, Active Motif). Before immunoprecipitation, a small amount of chromatin was saved, purified, and later used as calibrator (input DNA) for quantitative PCR measurements. A total number of 4 samples/treatment were generated. The resulting precipitated chromatin and the input DNA were used as templates for quantitative real-time PCR analysis. The chromatin sequence around the RE1 binding site of calbindin 1 (Calb1) gene was analyzed. The primer sets used in this assay were previously described by Ballas et al. (24): sense 5′-GAGAACTCCGGAGGACGCCCGAAC-3′ and antisense 5′-GATGGTTGGGATGAACAAGCACAGTC-3′. In addition, we designed 6 primer pairs covering 1.8 kb of overlapping amplicons around the RE1 site: pair 1, sense 5′-GGCTGCTACGGAGTCA-3′ and antisense 5′-TTGCGCTAATGGAGA-3′; pair 2, sense 5′-CTTAAACAGCCACGTGA-3′ and antisense 5′-GGAGGCTTTCACTCCTGA-3′; pair 3, sense 5′-GAGTGAAAGCCTCCCTGA-3′ and antisense 5′-TCCTGCCTCCTTTTCCTA-3′; pair 5, sense 5′-CCTGCTCTTCCCTTCGA-3′ and antisense 5′-CGCATAAAGGGCATAGGA-3′; pair 6, sense 5′-AAACACCATCCCCAGCA-3′ and antisense 5′-TTGAAGGGTCCTGCTCA-3′; and pair 7, sense 5′-AGAGGTTTTGTGGGAGA-3′ and antisense 5′-TGGTGGAGGTGGGGAA-3′. For the assessment of H3K9me1, H3K9me2, and H3K4me2 levels interacting with RE1, real-time PCR amplification was performed using the Takara SYBR Premix Ex Taq (Takara Bio Inc., Shiga, Japan) according to the manufacturer’s protocol. The real-time PCR machine used was the ICycler IQ Multicolor Real-Time PCR Detection System with Optical System Software Version 3.1 (Bio-Rad). For the assessment of REST binding to RE1, endpoint PCR was used within the linear increase range, using an Eppendorf Mastercycler EP gradient machine (Eppendorf, Westbury, NY, USA). For both real-time and endpoint amplifications, the conditions were 95°C for 10 min and 40 cycles of 95°C for 20 s, 56°C for 30 s, and 72°C for 50 s.
Real-time RT-PCR
RNA was extracted from cultured NPCs and whole E17 fetal brain tissue using the RNeasy Mini Kit (Qiagen, Valencia, CA). After the concentration and quality were determined, the resulting total mRNA was stored at −80°C. A 2-step real-time PCR method was used for the assessment of gene expression. First, a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) containing random primers was used as described in the manufacturer’s protocol to generate single stranded DNA template. After transcription, a volume of sample that did not exceed 2 ng/μl concentration of the cDNA template per PCR reaction was amplified in the second step. Amplification conditions were adjusted using the standard curve method for an optimal slope, 100% PCR efficiency and no nonspecific products. Almost identical efficiencies were achieved for the gene of interest and the TBP housekeeping gene. The following primers were designed for the G9a (euchromatic histone lysine N-methyltransferase 2, EHMT2): forward 5′-GAGTTCATAGCTCTTTGGGGGACA-3′ and reverse 5′-ATCGCCGACGGTGACAGTGA-3′ (accession no. NM_123456); calbindin D28K (Calb1): forward 5′-GCTGCAGAACTTGATCCAGGA-3′ and reverse 5′-TCCGGTGATAGCTCCAATCC-3′ [2]; and TBP (TATA box binding protein): forward 5′-ACCCACACAGGGTGCCTCA-3′ and reverse 5′-TGGCAGGAGTGATAGGGGTCA-3′ (NT_123456) genes. The primers were designed using the Gene Fisher interactive PCR primer design software support (http://bibiserv.techfak. uni-bielefeld.de/genefisher/; ref. 35). SUV39 gene expression was measured using a primer set (PPM25519A; SuperArray, Frederick, MD, USA). The primer pairs were tested by RT-PCR for dimer formation. To ensure against nonspecific priming, nonredundant BLAST searches were performed for all primers (http://www.ncbi.nlm.nih.gov/BLAST/).
A 25-μl reaction mixture containing 450 nM primers, 2 ng cDNA, and the Real-Time PCR Master Mix IQ SYBR Green Supermix Bio-Rad) was used for all reactions. The conditions for the amplification reactions were 95°C for 10 min and 40 cycles of 94°C for 15 s, 56°C for 30 s, and 72°C for 50 s, followed by a melting curve to ensure single amplicon generation. The relative quantification method (2−ΔΔCT) was used to analyze gene expression changes as described elsewhere (36).
Calb1 CpG methylation
DNA from NPCs was extracted using a FlexiGene DNA Kit (Qiagen) according to the manufacturer’s protocol. One microgram of genomic DNA from each sample was bisulfite treated using an EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA), according to the manufacturer’s protocol. The genomic sequence, including the promoter and exon 1 of the Calb1 gene, was retrieved using the NT_039258.6 locus (as retrieved from the NCBI genome database, last accessed May, 31, 2007). CpG island identification was performed using the EMBOSS on-line software (ref. 37; available at http://www.ebi.ac.uk/emboss/cpgplot/). The DNA sequence selected for methylation sequencing included 9 CpG sites, where sites 7–9 are located within the 21-bp RE-1 regulatory site of exon 1, as described elsewhere (24). Four microliters of bisulfite-treated DNA from each sample served as template for PCR amplification using the following primers: forward 5′-GATAAATATAGAGAATTGGGTG-3′ and reverse 5′-biotinilated TAACTACAAATAAAATTCTACCAT-3′; the amplification was performed on an Eppendorf Mastercycler EPgradient (Eppendorf), under the following conditions: 94°C for 5 min, followed by 45 cycles (94°C for 1 min, 60°C for 30 s, and 72°C for 25 s), and 72°C for 1 min final extension. The PCR products were sequenced using a PyroMark MD machine (Biotage AB, Uppsala, Sweden). CpG methylation analysis was performed using the Pyro Q-CpG 1.0.9 software (Biotage AB), and the results were expressed as percent methylated cytosine for any CpG site analyzed. The PCR amplification primers and the sequencing primer (5′-GATAAATATAGAGAATTGGG-3′) were designed using the PSQ Assay Design software (Biotage AB) and ordered from Operon Biotechnologies (Huntsville, AL, USA). PCR was performed using the Apex Taq Master Mix (Genesee Scientific, San Diego, CA, USA). Two independent experiments were performed, each consisting of 4 CT samples and 4 CD samples.
Statistical analysis
The optical density values of protein bands, normalized to total H3, were analyzed using a Student’s t test, with differences considered significant at P < 0.05. For the ChIP assay and for gene expression, analysis was performed using the Relative Expression Software Tool–Multiple Condition Solver (REST-MCS) version 2 (free from the website http://www. gene-quantification.de/rest.html), which uses a pairwise fixed reallocation randomization test. Statistical significance was defined as P < 0.05. For Calb1 CpG methylation assessment, Epi Info 3.4.1 (Centers for Disease Control and Prevention, Atlanta, GA, USA; available at http://www.cdc.gov/epiinfo/) was used. Homogeneity of variances between treatments and for each CpG site was determined by Bartlett’s χ2 test (P=0.5716). Because multiple measurements (2 treatment groups, each with 9 CpG sites) were performed simultaneously, Bonferroni correction was applied and an α value of 0.00555 was established for hypothesis testing. The statistical significance was tested using a t test for each CpG site (n=4 independent samples for each group, CT and CD, respectively), and statistical significance was defined as P < 0.00555.
RESULTS
CD decreased H3K9me1 and H3K9me2 in the fetal hippocampus and in cultured NPCs
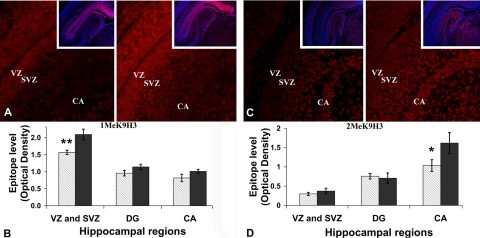
Dietary availability of choline during pregnancy (38) or alteration in the endogenous biosynthesis of choline (39) influenced global levels of S-adenosylmethionine and altered methylation in the developing fetal brain, changing both global and gene-specific DNA methylation (8). Using semiquantitative measurements, immunohistochemistry, and Western blot analysis we determined whether dietary choline altered global methylation levels of histones, around which DNA is coiled. The 5 major classes of histones include the core histones H2A, H2B, H3, and H4, and the linker histone H1, and they can be methylated or acetylated on specific amino acid residues. In this study, we examined lysine 9 of histone H3 (H3K9me1 and H3K9me2; see Fig. 4) in the fetal hippocampus because epigenetic marks on this amino acid are associated with gene silencing in the brain and other tissues (25). At E17, maternal CD decreased both H3K9me1 and H3K9me2 in specific fetal hippocampal layers containing NPCs and pyramidal neurons (Fig. 1). The decrease in H3K9me1 was confined to the proliferation area of the hippocampus (VZ and SVZ), with a CD/CT methylation ratio of 0.74 ± 0.03 (P<0.01; values are means ± se; Fig. 1A, C). The decrease in H3K9me2 was localized exclusively to the pyramidal cell layer of cornus ammoni (CA), with a CD/CT methylation ratio of 0.64 ± 0.1 (P<0.05; Fig. 1C, D). Z-stack confocal image analysis at high magnification and oil immersion was used to further validate the region-specific variations in total H3K9 methylation levels between the different diets. The VZ and SVZ images corresponding to the CA2 region, containing H3K9me1 labeled nuclei of the NPCs, showed differences in H3K9me1, with a CD/CT ratio of 0.7 ± 0.1 (P<0.01). Also, there were differences in H3K9me2-labeled nuclei from the pyramidal layer of CA region 2, with a CD/CT ratio of 0.65 ± 0.10 (P<0.01). These changes were not seen in whole-brain preparations, as there were no changes in the total levels of H3K9me1 and H3K9me2 within the whole-brain histone pool as assessed by Western blotting (n=7 independent samples of E17 fetal brains per treatment; immunofluorescent labeling or Western blotting). No significant changes were found in the OD of H3K9me1 and H3K9me2 immunoreactive bands relative to the total H3 levels (H3K9me1: CD 0.75±0.19 vs. CT 0.52±0.09; H3K9me2: CD 0.40±0.11vs. CT 0.39±0.12) by Student’s t test. The original images are available online (http://www.unc.edu/zeisel_lab/). Values are given as relative MOD levels (see Materials and Methods).
Figure 1.
CD decreases H3K9me1 and H3K9me2 levels in specific areas of the fetal hippocampus. Pregnant mice were fed a CD diet or a CT diet from E12-E17. E17 fetal brains were used to assess the relative levels of H3K9me1 and H3K9me2 in the hippocampus and whole brains (n=7 independent samples/treatment; immunofluorescent labeling and Western blotting, respectively). A) Representative fluorescent images for both treatments (CD and CT); red staining indicates H3K9me1. Insets: nuclear staining. B) H3K9me1 levels are decreased in CD hippocampi as compared with CT (1.56±0.06 vs. 2.09±0.15), specifically in VZ and SVZ of hippocampus, which encompass the main proliferation area. CA, cornus ammoni; DG, dentate gyrus. C) Representative fluorescent images for both treatments; red staining indicates H3K9me2. Insets: nuclear staining. D) H3K9me2 levels were also decreased in pyramidal layer of CA (CD 1.03±0.15 vs. CT 1.61±0.27). Values are given as relative MOD levels (see Materials and Methods). Gray bars, CD; black bars, CT. Error bars = se. *P < 0.05, **P < 0.01; Student’s t test.
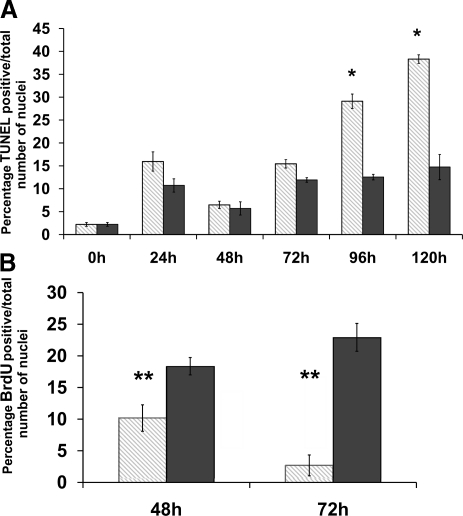
Figure 2.
CD changes the proliferation and apoptosis index in E14 mouse neural progenitors. Mouse neural progenitors were exposed to either a CD medium (5 μM choline, CD) or a control medium (70 μm choline, CT) for 0, 24, 48, 72, 96, and 120 h. TUNEL and BrdU immunolabeling was performed as described in Materials and Methods. Number of labeled nuclei was reported as percentage from total number of cells counted (n=5 independent samples/treatment). A) CD increases number of TUNEL-positive nuclei at 96 h (CD 29.12±1.57 vs. CT 12.55±0.59%) and 120 h (CD 38.33±0.94 vs. CT 14.74±2.75%) but not at 72 h. B) CD decreases proliferation at 48 h (CD 10.16±2.08 vs. CT 18.36±1.37%) and 72 h (CD 2.68±1.64 vs. CT 22.9±2.19%). Gray bars, CD; black bars, CT. Error bars = se. *P < 0.05, **P < 0.01; Student’s t test.
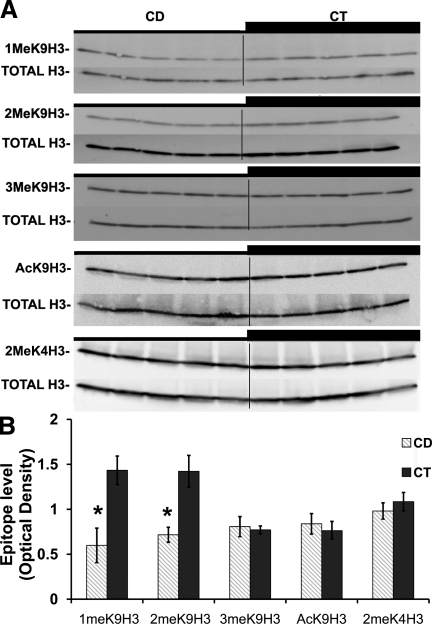
Figure 3.
CD decreases H3K9me1 and H3K9me2 but has no influence on H3K9me3, H3K9Ac, and H3K4me2 levels in the E14 NPC histone extracts. Mouse neural progenitors were exposed to either a CD medium (5 μM choline, CD) or a control medium (70 μm choline, CT) for 72 h. Immunolabeling was performed as described in Materials and Methods, and OD was measured (n=5 independent samples/treatment). A) Representative Western blot images of H3K9me1, H3K9me2, H3K9me3 (CD 0.81±0.11 vs. CT 0.77±0.043), H3K9Ac (CD 0.83±0.09 vs. CT 0.76±0.1), and H3K4me2 (CD 0.98±0.11 vs. CT 1.08±0.09). B) Values for each sample were normalized to the OD of total histone H3. CD decreased the H3K9me1 and H3K9me2 protein levels (H3K9me1: CD 0.59±0.19 vs. CT 1.43±0.15; H3K9me2: CD 0.71±0.08 vs. CT 1.42±0.17). Values are given as relative MOD levels (see Materieals and Methods). Gray bars, CD; black bars, CT. Error bars = se. *P < 0.05, **P < 0.01; Student’s t test.
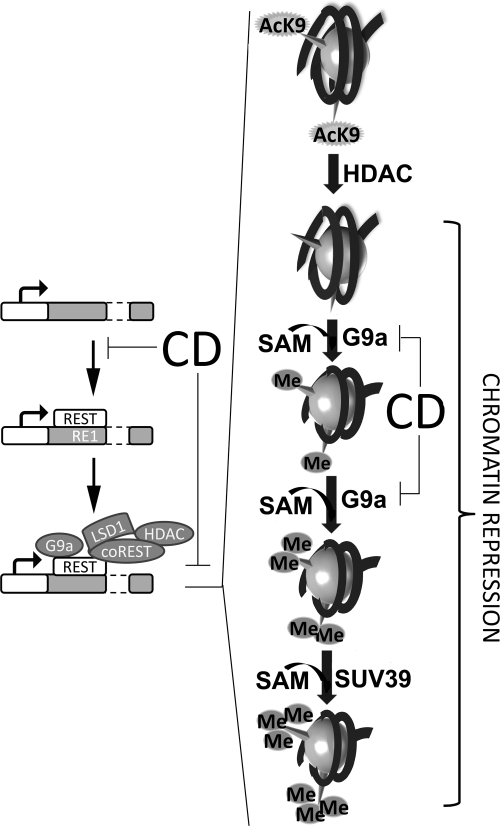
Figure 4.
Regulation of chromatin compaction by histone methylation and deacetylation via REST mechanism. Chromatin repression is achieved following REST binding to the RE1 site, by recruiting histone modifying enzymes (G9a, SUV39, and HDAC). First HDAC performs deacetylation of H3K9 residues. After this event, G9a utilizes S-adenosylmethionine (SAM) as the methyl donor and methylates the available residue, decreasing chromatin availability. In a separate set of events, G9a and DNMT1 (which maintains the DNA methylation patterns established during development) are recruited by PCNA at the replication fork during DNA synthesis. They transmit the methylation status of both histones and DNA to the newly formed nucleosomes.
CD decreased mitosis and increases apoptosis in NPCs in culture
Rodent dams fed CD diets during late pregnancy have offspring with diminished progenitor cell proliferation and increased apoptosis in fetal hippocampus VZ and SVZ (5, 40, 41). To further investigate the in vivo findings, we used an in vitro model of fetal mouse NPCs. According to previously published data, this model closely resembles the in vivo NPCs from the VZ and SVZ. The cultured NPCs and the in vivo cells behave similarly in terms of chromatin architecture, remodeling, and histone modifications (7, 8, 10, 30, 31). In addition, previous studies (7) using gene array analysis of CD-treated NPCs showed expression changes for genes that modulate cell cycle, apoptosis, neuronal differentiation, methyl metabolism, and calcium-binding proteins. Choline availability during the essential stages of fetal development changes hippocampal neurogenesis. The number of dividing cells populating the VZ and SVZ of the hippocampus decreases under low maternal choline diet (5). Conversely, activated caspase 3 as a marker of apoptosis is increased in the hippocampal area on 5 d exposure in utero (5). A similar pattern of effects was found when NPCs were grown in tissue culture media that varied in choline content. CD resulted in decreased NPC proliferation as measured by BrdU incorporation into newly formed DNA strands during the S phase (after 48 h CD 10.16±2.08 vs. CT 18.36±1.37% BrdU-positive cells, P<0.01; after 72 h CD 2.68±1.64 vs. CT 22.9±2.19%; P<0.01; data are means ± se; Fig. 2B). Also, CD resulted in increased rates of apoptosis in NPCs after 96 h (CD 29.12±1.57 vs. CT 12.55±0.59% TUNEL-positive cells; P<0.05) and 120 h (CD 38.33±0.94 vs. CT 14.74±2.75%; P<0.05; Fig. 2A). These results suggest that CD induces effects on proliferation and apoptosis in vitro that are similar to those observed in vivo. Interestingly, modifications in the proliferation of NPCs were detected after the first 48 h, whereas changes in apoptosis phenomenon only become obvious after the 96 h exposure. To eliminate a possible influence of this variable on the histone modification measurements, we chose the 72-h time point.
CD decreased histone methylation in NPCs in culture
H3K9me1 and H3K9me2, both markers of repressed chromatin, were decreased in NPCs grown in low choline medium at 72 h (based on relative OD for H3K9me1: CD 0.59±0.19 vs. CT 1.43±0.15, P<0.05; and for H3K9me2: CD 0.71±0.08 vs. CT 1.42±0.17, P<0.05; data are means ± se; Fig. 3). In most cases, methylation changes in H3K9 residue are accompanied by opposite changes in its acetylation level as part of the histone code (Fig. 4; refs. 42, 43). However, as compared with total H3, no changes were detected in the levels of H3K9me3 (heterochromatin hallmark; Fig. 3) nor in the levels of acetyl-lysine 9 of histone H3 (H3K9Ac) and H3K4me2 (euchromatin markers; Fig. 3).
CD decreases G9a methylase, but not SUV39 histone methylase, gene expression and protein levels
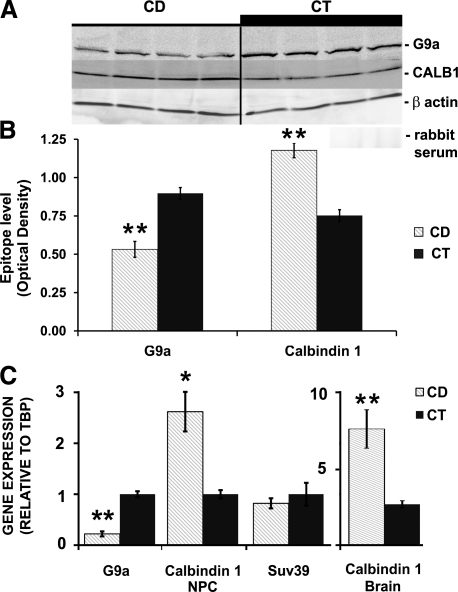
Epigenetic marks on histones are the result of the activities of specific enzymes. G9a histone methyl transferase (HMTase) is responsible for covalently modifying H3K9 to form H3K9me1,2 (Fig. 4), using S-adenosylmethionine as the methyl donor (44, 45). Changes in the activity of G9a HMTase in NPCs could explain the reduced methylation of H3K9 that we observed. Possible transcriptional changes in the mature RNA levels of G9a were evaluated by quantitative RT-PCR analysis of NPCs cultured in low or normal choline medium. CD decreased G9a HMTase RNA (CD/CT RNA ratio 0.22± 0.02, P<0.001) but had no effect on SUV39 HMTase RNA (CD/CT RNA ratio 0.82±0.24; Fig. 5C). G9a protein levels measured by Western blot analysis mirrored the gene expression changes (OD of 140-kDa immunoreactive bands attributed to G9a protein relative to β-actin were CD 0.53±0.05 vs. CT 0.9±0.04; P<0.01; Fig. 5A, B).
Figure 5.
CD alters G9a and Calb1 gene expression and protein levels but not SUV39. After 72 h exposure to a CD medium (CD) or a control medium (CT), gene expression of G9a histone methylase (in NPCs) and Calb1 (in NPCs and whole brain) was determined using quantitative real time RT-PCR (see Materials and Methods). A, B) No changes were detected in the SUV39 transcripts level in NPCs. G9a protein levels (A) were decreased by CD 0.53 ± 0.05 vs. CT 0.9 ± 0.04 in NPCs (B); increased immunoreactive bands of Calb1 (A) were detected under CD 1.18 ± 0.05 vs. CT 0.75 ± 0.04 in NPCs (B). Negative control blots were probed with reimmune serum (A). C) CD decreased G9a expression (CD/CT ratio 0.22±0.02, P<0.001; n=4/group), while Calb1 expression was increased (CD/CT ratio 2.62±0.05in NPCs and 2.84±0.8 in brain). Gray bars, CD; black bars, CT. Error bars = se. *P < 0.05, **P < 0.01; randomization test.
CD increased Calb1 gene expression and calbindin 1 protein levels
We further investigated whether the changes in epigenetic marks on histones altered the expression of specific genes involved in neurogenesis. Previously, we (5,6,7, 46) reported that CD increased the expression of several genes encoding calcium binding proteins such as calretinin, and we now report that CD alters formation of calbindin 1, a marker of mature GABAergic neurons. To determine whether changes in histone methylation correlated with alterations in Calb1 gene expression, real-time RT-PCR was performed using total RNA from cultured NPCs and E17 whole fetal brain tissue. At 72 h, CD induced an increase in Calb1 gene expression (CD/CT RNA ratio 2.62±0.05, P<0.05; Fig. 5C), which resulted in increased calbindin protein in NPCs (ratio of OD of Calb1/OD of β-actin for CD 1.18 ±0.05 vs. CT 0.75±0.04 OD, P<0.01; Fig. 5B). Similarly, the mRNA levels of Calb1 were increased in the fetal brain tissue by the CD diet (CD/CT ratio 2.84±0.8, P<0.01; Fig. 5C).
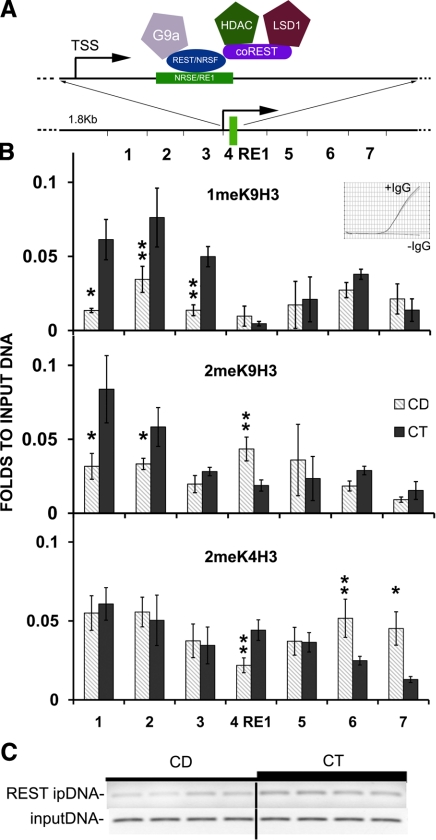
CD decreases REST binding to the RE1 site of Calb1 and altered methylation of H3K9me1, H3K9me2, and H3K4me2 in the proximity of this RE1 site
The presence of REST at the RE1 site is associated with a specific repressive histone pattern and, consequently, with inhibition of neuronal gene expression (47). Calb1 gene expression is regulated by REST repressor complex binding to the RE1 element in the gene promoter, and this binding alters both H3K9, H3K4 (Fig. 4) and DNA methylation in proximity to the RE1 site (24, 48, 49). In cultured NPCs, using ChIP, we observed decreased binding of REST to the RE1 site within the Calb1 promoter (CD 0.2±0.02 vs. CT 0.36±0.01 OD, P<0.01, relative to input DNA; Fig. 6C). DNA-histone interactions were analyzed by ChIP assay; an immunoprecipitation step was carried out utilizing antibodies against H3K9me1, H3K9me2, or H3K4me2. The DNA was then analyzed by quantitative PCR, measuring the enrichment of each form of methylated histone in proximity to the RE1 sequence of Calb1 (Fig. 6A). After 72 h of CD, the combined levels of H3K9me1 residue over this 1.8-kb sequence were decreased (to 51.8±3.7%, P<0.01 vs. CT). Similarly the combined levels of H3K9me2 were decreased (to 74±4.1%, P<0.01 vs. CT), while the combined levels of H3K4me2 were increased (to 115±3.4%; P<0.05 vs. CT; Fig. 6A, B).
Figure 6.
CD changes the DNA-histone 3 interaction at the RE1 site of Calb1. After 72 h exposure to a CD medium (CD) or a control medium (CT), ChIP was used to assess changes in the levels of H3K9 and H3K4 methylation at the RE1 locus (B) and abundance of REST at the Calb1 site (C; Materials and Methods). A) Schematic representation of the RE1 site of Calb1 gene. RE1/NRSE (neuron-restrictive silencer element) site is within exon 1 of Calb1 and is in proximity to the transcription starting site. REST/NRSF (neuron-restrictive silencer factor) and coREST repressive complex (G9a, G9a HMTase; LSD1, lysine-specific demethylase 1) interacts with the RE1/NRSE site [adapted from Ooi et al. (49)]. B) CD decreased H3K9me1 and H3K9me2 levels upstream of RE1 in Calb1 decreased. Conversely, H3K4me2 was increased in proximity to the Re1 site. Inset indicates no amplification for negative controls (negative IgG and negative primer set) by qRT-PCR. *P < 0.05, **P < 0.01 vs. CT; randomization test. C) CD decreased the binding of REST to the RE1 site of Calb1 (CD 0.2±0.02 vs. CT 0.36±0.01, normalized to input DNA, P<0.01 by t test). Gray bars, CD; black bars, CT. Error bars = se. ipDNA, immunorecipitated DNA against REST antibody.
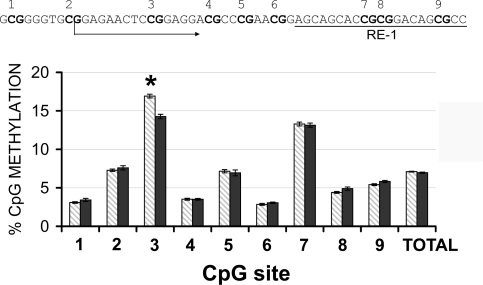
CpG methylation of Calb1 promoter
The ability of chromatin to bind transcription factors at a specific site involves covalent changes of both histone tails and DNA. Hypermethylation of DNA at specific promoter regions or transcription binding sites inhibits gene expression (50) and allows for modifications of the transcriptome without mutations of the DNA code. In different model systems, H3K9 methylation and DNA methylation are tightly linked (51). Thus, we assessed DNA methylation changes in NPCs in culture. The first 9 CpG sites within the CpG island of the Calb1 promoter were studied by pyrosequencing; CD increased methylation of CpG site 3 (CD 16.9±0.2 vs. CT 14.2±0.2% methylated, P<0.005; Fig. 7).
Figure 7.
CD induces hypermethylation of a specific CpG site within Calb1 gene. DNA pyrosequencing after bisulfite treatment indicates that CD induces specific hypermethylation of the CpG site 3 within the CpG island of Calb1 promoter (CD 16.9±0.2 vs. CT 14.2±0.2%). CpG sites 7–9 are located within the RE1 binding site (RE1 sequence underscored in inset). Gray bars, CD; black bars, CT. Error bars = se. *P < 0.00555. Each CpG site is marked with bold letters. Arrow indicates transcription start site for Calb1.
DISCUSSION
For the first time, we observed that, in mice, maternal CD during E12–17 altered methylation of H3 in E17 fetal NPCs in the areas of the hippocampus where neurogenesis was occurring (SVZ and VZ). Also, we report the same effects of CD on H3 methylation in mouse E14 NPCs in culture. Gene expression of G9a HMTase was decreased by CD and likely contributed to the changes in histone methylation. SUV39 HMTase gene expression was unchanged. There was diminished REST binding to RE1, and this decreased recruitment of G9a (25). This resulted in decreased H3K9me1,2 residues in proximity to the RE1 binding site in the Calb1 promoter sequence in CD, while H3K4me2 residues were increased. In addition, 1 CpG site within the Calb1 promoter was hypermethylated in CD. These changes resulted in increased expression of Calb1 in CD. Thus, the availability of choline modulates histone methylation in fetal NPCs, and we suggest that these epigenetic changes contribute to choline-mediated changes in hippocampal development.
Choline is an essential nutrient needed for membrane formation, methylation, and acetylcholine biosynthesis (52). More choline (∼4× dietary levels) in maternal rodent diet during d 11–17 of gestation increased proliferation and decreased apoptosis in hippocampal NPCs (5, 40, 41) and enhanced memory in the adult animals throughout their lifetimes (53,54,55,56,57,58,59). Conversely, CD during pregnancy had the opposite effects (57). In the rodent models, dietary CD decreased S-adenosylmethionine concentrations in tissues (38, 60), affecting methylation reactions. Neurogenesis is regulated, in part, by epigenetic mechanisms that involve methylation (8, 10, 11, 25, 38, 61, 62). Indeed, previously we reported that choline availability changed DNA methylation in mouse fetal hippocampus (8, 39). Now, we report that maternal dietary CD alters methylation of histones in fetal hippocampus.
Human diets vary greatly in choline content, and some pregnant women eat less than half of the recommended adequate intake (4, 63). In our study, we examined the effects on the brain when pregnant mice were fed a diet devoid of choline vs. a control diet for 5 d. The control diet is associated with normal concentrations of choline in maternal liver and plasma as well as in fetal brains (5), while the low choline diet depletes these choline pools (5). Although pregnant women likely never eat a choline-devoid diet, they consume low-choline diets for months, not days, and they deplete tissue stores of choline (64). We found that in cell culture 70 μM choline in the medium resulted in intracellular concentrations of choline and metabolites that were similar to those measured in the whole brain from control fetuses, while 5 μM choline in the medium resulted in intracellular concentrations of choline and metabolites that were similar to those measured in the whole brain from CD fetuses (65). We suggest that these experimental conditions can be used to estimate the possible effects of a low-choline diet in pregnant mothers.
We assessed changes in histone methylation using whole brain and using an in vitro model in which NPCs proliferate and differentiate. The NPCs in culture undergo a complex pattern of neurogenesis that replicates the patterns seen in vivo, and mitosis and apoptosis in these cells are changed by exposure to low choline in a manner that is similar to what we report in vivo.
DNA methylation and H3K9 methylation mechanisms exhibit cross-talk, making it difficult to determine which modification occurs first for specific genes (42, 48, 49, 51, 66,67,68). We report that the H3 methylation, notably H3K9, is not altered within the whole fetal brain (Supplemental Data). However, the specific decrease in H3K9me1 in the VZ and H3K9me2 in the pyramidal layer (Fig. 1), along with the timing of hippocampal development, suggests that there is a preference for maintaining the levels of different methylated residues in specific cellular phenotypes with unique epigenomes. Ergo, H3K9me1 is altered in the NPC layer, and after a second wave of epigenomic changes occurs in the migrating/differentiating cells, H3K9me2 is altered in daughter cells derived from VZ (pyramidal neurons in CA area; Fig. 1). H3K9me3 levels were not changed in the whole brain or in NPCs (Fig. 3).
Although it remains possible that low S-adenosylmethionine concentrations associated with CD (38) caused H3 hypomethylation, other mechanisms are tenable. It is possible that HDAC-catalyzed deacetylation of euchromatin leads to subsequent changes in histone methylation (69, 70). We observed no changes in H3K9Ac associated with CD in NPCs (Fig. 3). Choline modified the activity of histone methylases; we report here that G9a HMTase gene expression and protein levels were diminished by CD (Fig. 5). The observed changes in G9a may be mediated by decreased binding of the REST repressor complex (Fig. 6), which activates G9a (25). It is interesting that relatively high levels of H3K9me3 were maintained despite CD (Fig. 3). Since H3K9me1 and 2 are formed by G9a histone methylase, and H3K9me3 is formed via SUV39 (22), we suggest that G9a HMTase activity is preferentially affected by CD (we observed no CD effect on SUV39; Fig. 5). G9a has 10- to 20-fold higher activity than does SUV39 HMTase (20, 44, 45). Since G9a HMTase, along with EZH2 HMTase (part of the PCR2 complex) (71), also are responsible for methylating H3K27 (44), future studies should compare the methylation status of H3K27 and H3K9 to discriminate between lower histone methylation due to reduced G9a HMTase vs. increased demethylation of H3K9 via increased expression/activity of the demethylases. In addition, since methylated H3K9 is a substrate for several histone demethylases such as the JHDM family of enzymes, and H3K4 is specifically demethylated by the LSD1 (26,27,28, 72), further studies are needed to determine whether other HMTase activities contribute to the altered H3 K9 and H3 K4 methylation. Indeed, decreased activity of JHDM3A could preserve H3K9me3 at the expense of H3K9me2.
Low H3K9me1 and 2 levels induced by CD, coupled with normal H3K9me3, should be associated with reallocation of heterochromatin protein 1 (HP1) toward pericentric heterochromatin (20), recruitment of G9a, DNMT1, and HDAC (73), or DNMT3a (22) maintaining a repressive status of pericentric heterochromatin. A secondary mechanism of HP1 recruitment involves automethylation of G9a HMTase at the ARKT motif (74), and decreased S-adenosylmethionine concentrations in CD could impair automethylation.
We report that REST complex binding was altered in CD (Fig. 6). Not only does REST binding to RE1 activate G9a (25), it also is associated with a repressive histone pattern and with inhibition of neuronal gene expression (47). REST is expressed in the VZ and SVZ of the hippocampal neuroepithelium (48). Isolated NPCs and less-differentiated neurons from the hippocampus contain REST protein (48), which diminishes as neurons differentiate (24). The loss of REST binding to the RE1 site induces activation of the Calb1 promoter and, subsequently, Calb1 expression (47). Once the chromatin architecture has been established, other chromatin binding proteins (coREST and MeCp2) preserve its epigenetic status (24, 49).
The levels of H3K9 methylation across 1.8 kb of chromatin surrounding the RE1 site indicate that choline-mediated histone tail changes are not confined to the RE1 site but rather extend around RE1 and the transcriptional starting site of Calb1 gene. This supports previous findings (48). Overall, the levels of H3K9me1 and 2 (robust markers of repression) in CD indicate a permissive chromatin state of the promoter area, with important changes upstream from the RE1 site (Fig. 6). CD increases H3K4me2 (indicative of active loci) downstream from the RE1 site. The overall chromatin state around the RE1 site under CD can be explained by the low REST binding to its RE1 site (Fig. 6C) and leads to a high Calb1 expression (Fig. 5). However, in the proximity (200 bp) of the RE1 site, histone methylation at H3K9me2 was increased and H3K4me2 was decreased; this observation is interesting and needs further exploration. The activity of the REST suppressor on the RE1 site is mediated in the adult hippocampus by small noncoding dsRNAs (49, 75). Our current data do not exclude a possible decrease in the level of this modulatory RNA as well as in the REST protein under CD.
CD also induced specific DNA hypermethylation of 1 CpG site, situated upstream of the RE1 site within the Calb1 sequence, without changing the total methylation levels of the entire CpG island (Fig. 7). This confirms previous findings (76) in other rodent models that CD during pregnancy can induce DNA hypermethylation at specific CpG sites. In addition, CD induces hypomethylation of a CpG island within the DNMT1 gene, leading to overexpression of DNMT1 (76). Decreased G9a and DNMT1 could impair the protein-protein interaction within the PCNA-G9a-DNMT1 complex (67), leading to alterations in both histone and DNA methylation.
Future studies should better characterize the modifications in DNA-histone interactions induced by CD and the role played by histones in altering gene-specific DNA methylation. We suggest that maternal dietary choline influences histone methylation and thereby modulates gene expression and ultimately alters neurogenesis and apoptosis in fetal brain.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (AG-09525, DK-55865). Support for this work was also provided by grants from the National Institutes of Health to the Clinical Nutrition Research Unit (DK-56350). We thank Dr. Robert Bagnell, Jr., Professor and Director of the Microscopy Service Laboratory, UNC Department of Pathology, for assistance.
References
- Fisher M C, Zeisel S H, Mar M H, Sadler T W. Inhibitors of choline uptake and metabolism cause developmental abnormalities in neurulating mouse embryos. Teratology. 2001;64:114–122. doi: 10.1002/tera.1053. [DOI] [PubMed] [Google Scholar]
- Fisher M C, Zeisel S H, Mar M H, Sadler T W. Perturbations in choline metabolism cause neural tube defects in mouse embryos in vitro. FASEB J. 2002;16:619–621. doi: 10.1096/fj.01-0564fje. [DOI] [PubMed] [Google Scholar]
- Blom H J, Shaw G M, den Heijer M, Finnell R H. Neural tube defects and folate: case far from closed. Nat Rev Neurosci. 2006;7:724–731. doi: 10.1038/nrn1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G M, Carmichael S L, Yang W, Selvin S, Schaffer D M. Periconceptional dietary intake of choline and betaine and neural tube defects in offspring. Am J Epidemiol. 2004;160:102–109. doi: 10.1093/aje/kwh187. [DOI] [PubMed] [Google Scholar]
- Craciunescu C N, Albright C D, Mar M H, Song J, Zeisel S H. Choline availability during embryonic development alters progenitor cell mitosis in developing mouse hippocampus. J Nutr. 2003;133:3614–3618. doi: 10.1093/jn/133.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright C D, Siwek D F, Craciunescu C N, Mar M H, Kowall N W, Williams C L, Zeisel S H. Choline availability during embryonic development alters the localization of calretinin in developing and aging mouse hippocampus. Nutr Neurosci. 2003;6:129–134. doi: 10.1080/1028415031000084418. [DOI] [PubMed] [Google Scholar]
- Niculescu M D, Craciunescu C N, Zeisel S H. Gene expression profiling of choline-deprived neural precursor cells isolated from mouse brain. Brain Res Mol Brain Res. 2005;134:309–322. doi: 10.1016/j.molbrainres.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Niculescu M D, Craciunescu C N, Zeisel S H. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20:43–49. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim J H, Miniaci M C, Schwartz J H, Thanos D, Kandel E R. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Gage F H. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev. 2004;14:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Levenson J M, Sweatt J D. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Peters A H, Kubicek S, Mechtler K, O'Sullivan R J, Derijck A A, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens J H, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Rice J C, Briggs S D, Ueberheide B, Barber C M, Shabanowitz J, Hunt D F, Shinkai Y, Allis C D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Bernstein B E, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey D K, Huebert D J, McMahon S, Karlsson E K, Kulbokas E J, 3rd, Gingeras T R, Schreiber S L, Lander E S. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ng H H, Robert F, Young R A, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister A J, Sherriff J, Bernstein B E, Emre N C, Schreiber S L, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Mutskov V J, Farrell C M, Wade P A, Wolffe A P, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson J R, Walker P A, Vasisht N, Kelly G, Howell S, Taylor I A, Blackburn G M, Gamblin S J. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck A A, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters A H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu D D, Speh J C, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley T J, Morrison J F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine J R, Cole P A, Casero R A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren M E, Borchers C H, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Ma D K, Chiang C H, Ponnusamy K, Ming G L, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobowitz D M, Abbott L C, editors. Boca Raton, FL, USA: CRC Press; Chemoarchitectonic Atlas of the Developing Mouse Brain. 1998 [Google Scholar]
- Cremisi F, Philpott A, Ohnuma S. Cell cycle and cell fate interactions in neural development. Curr Opin Neurobiol. 2003;13:26–33. doi: 10.1016/s0959-4388(03)00005-9. [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu Y, Chesnut J D, Rao M S. Isolation of neural stem and precursor cells from rodent tissue. Methods Mol Biol. 2008;438:39–53. doi: 10.1007/978-1-59745-133-8_5. [DOI] [PubMed] [Google Scholar]
- Wachs F P, Couillard-Despres S, Engelhardt M, Wilhelm D, Ploetz S, Vroemen M, Kaesbauer J, Uyanik G, Klucken J, Karl C, Tebbing J, Svendsen C, Weidner N, Kuhn H G, Winkler J, Aigner L. High efficacy of clonal growth and expansion of adult neural stem cells. Lab Invest. 2003;83:949–962. doi: 10.1097/01.lab.0000075556.74231.a5. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer S A. Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J Comp Neurol. 1990;301:325–342. doi: 10.1002/cne.903010302. [DOI] [PubMed] [Google Scholar]
- Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Giegerich R, Meyer F, Schleiermacher C. GeneFisher–software support for the detection of postulated genes. Proc Int Conf Intell Syst Mol Biol. 1996;4:68–77. [PubMed] [Google Scholar]
- Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Zeisel S H, Zola T, daCosta K A, Pomfret E A. Effect of choline deficiency on S-adenosylmethionine and methionine concentrations in rat liver. Biochem J. 1989;259:725–729. doi: 10.1042/bj2590725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Mar M H, Song J, Zeisel S H. Deletion of the Pemt gene increases progenitor cell mitosis, DNA and protein methylation and decreases calretinin expression in embryonic day 17 mouse hippocampus. Brain Res Dev Brain Res. 2004;149:121–129. doi: 10.1016/j.devbrainres.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Albright C D, Tsai A Y, Friedrich C B, Mar M H, Zeisel S H. Choline availability alters embryonic development of the hippocampus and septum in the rat. Brain Res Dev Brain Res. 1999;113:13–20. doi: 10.1016/s0165-3806(98)00183-7. [DOI] [PubMed] [Google Scholar]
- Albright C D, Friedrich C B, Brown E C, Mar M H, Zeisel S H. Maternal dietary choline availability alters mitosis, apoptosis and the localization of TOAD-64 protein in the developing fetal rat septum. Brain Res Dev Brain Res. 1999;115:123–129. doi: 10.1016/s0165-3806(99)00057-7. [DOI] [PubMed] [Google Scholar]
- Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Turner B M. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright C D, Mar M H, Craciunescu C N, Song J, Zeisel S H. Maternal dietary choline availability alters the balance of netrin-1 and DCC neuronal migration proteins in fetal mouse brain hippocampus. Brain Res Dev Brain Res. 2005;159:149–154. doi: 10.1016/j.devbrainres.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Greenway D J, Street M, Jeffries A, Buckley N J. RE1 silencing transcription factor maintains a repressive chromatin environment in embryonic hippocampal neural stem cells. Stem Cells. 2007;25:354–363. doi: 10.1634/stemcells.2006-0207. [DOI] [PubMed] [Google Scholar]
- Ooi L, Wood I C. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zeisel S H. Choline: critical role during fetal development and dietary requirements in adults. Annu Rev Nutr. 2006;26:229–250. doi: 10.1146/annurev.nutr.26.061505.111156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meck W, Williams C. Perinatal choline supplementation increases the threshold for chunking in spatial memory. Neuroreport. 1997;8:3053–3059. doi: 10.1097/00001756-199709290-00010. [DOI] [PubMed] [Google Scholar]
- Meck W, Williams C. Characterization of the facilitative effects of perinatal choline supplementation on timing and temporal memory. Neuroreport. 1997;8:2831–2835. doi: 10.1097/00001756-199709080-00005. [DOI] [PubMed] [Google Scholar]
- Meck W, Williams C. Simultaneous temporal processing is sensitive to prenatal choline availability in mature and aged rats. Neuroreport. 1997;8:3045–3051. doi: 10.1097/00001756-199709290-00009. [DOI] [PubMed] [Google Scholar]
- Meck W H, Smith R A, Williams C L. Pre- and postnatal choline supplementation produces long-term facilitation of spatial memory. Dev Psychobiol. 1988;21:339–353. doi: 10.1002/dev.420210405. [DOI] [PubMed] [Google Scholar]
- Meck W H, Williams C L. Choline supplementation during prenatal development reduces proactive interference in spatial memory. Brain Res Dev Brain Res. 1999;118:51–59. doi: 10.1016/s0165-3806(99)00105-4. [DOI] [PubMed] [Google Scholar]
- Meck W H, Williams C L. Metabolic imprinting of choline by its availability during gestation: implications for memory and attentional processing across the lifespan. Neurosci Biobehav Rev. 2003;27:385–399. doi: 10.1016/s0149-7634(03)00069-1. [DOI] [PubMed] [Google Scholar]
- Williams C L, Meck W H, Heyer D D, Loy R. Hypertrophy of basal forebrain neurons and enhanced visuospatial memory in perinatally choline-supplemented rats. Brain Res. 1998;794:225–238. doi: 10.1016/s0006-8993(98)00229-7. [DOI] [PubMed] [Google Scholar]
- Shivapurkar N, Poirier L A. Tissue levels of S-adenosylmethionine and S-adenosylhomocysteine in rats fed methyl-deficient, amino acid-defined diets for one to five weeks. Carcinogenesis. 1983;4:1051–1057. doi: 10.1093/carcin/4.8.1051. [DOI] [PubMed] [Google Scholar]
- Levenson J M, O'Riordan K J, Brown K D, Trinh M A, Molfese D L, Sweatt J D. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Park J J, Baum M J, Paredes R G, Tobet S A. Neurogenesis and cell migration into the sexually dimorphic preoptic area/anterior hypothalamus of the fetal ferret. J Neurobiol. 1996;30:315–328. doi: 10.1002/(SICI)1097-4695(199607)30:3<315::AID-NEU1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Jensen H H, Batres-Marquez S P, Carriquiry A, Schalinske K L. Choline in the diets of the U.S. population: NHANES, 2003–2004. FASEB J. 2007;21:lb219. [Google Scholar]
- Ozarda Ilcol Y, Uncu G, Ulus I H. Free and phospholipid-bound choline concentrations in serum during pregnancy, after delivery and in newborns. Arch Physiol Biochem. 2002;110:393–399. doi: 10.1076/apab.110.5.393.11832. [DOI] [PubMed] [Google Scholar]
- Albright C D, Liu R, Bethea T C, Da Costa K A, Salganik R I, Zeisel S H. Choline deficiency induces apoptosis in SV40-immortalized CWSV-1 rat hepatocytes in culture. FASEB J. 1996;10:510–516. doi: 10.1096/fasebj.10.4.8647350. [DOI] [PubMed] [Google Scholar]
- Bernstein B E, Humphrey E L, Erlich R L, Schneider R, Bouman P, Liu J S, Kouzarides T, Schreiber S L. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P O, Chin H G, Smallwood A, Feehery G R, Gangisetty O, Karpf A R, Carey M F, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y H, Zhang W, Chen X, George J, Ng H H. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J C, Allis C D. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken A P, Jensen M R, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R J, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers W A, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Chin H G, Esteve P O, Pradhan M, Benner J, Patnaik D, Carey M F, Pradhan S. Automethylation of G9a and its implication in wider substrate specificity and HP1 binding. Nucleic Acids Res. 2007;35:7313–7323. doi: 10.1093/nar/gkm726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage F H. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- Kovacheva V P, Mellott T J, Davison J M, Wagner N, Lopez-Coviella I, Schnitzler A C, Blusztajn J K. Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J Biol Chem. 2007;282:31777–31788. doi: 10.1074/jbc.M705539200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.