Abstract
TAB2 and TAB3 activate the Jun N-terminal kinase and nuclear factor-κB pathways through the specific recognition of Lys 63-linked polyubiquitin chains by its Npl4 zinc-finger (NZF) domain. Here we report crystal structures of the TAB2 and TAB3 NZF domains in complex with Lys 63-linked diubiquitin at 1.18 and 1.40 Å resolutions, respectively. Both NZF domains bind to the distal ubiquitin through a conserved Thr-Phe dipeptide that has been shown to be important for the interaction of the NZF domain of Npl4 with monoubiquitin. In contrast, a surface specific to TAB2 and TAB3 binds the proximal ubiquitin. Both the distal and proximal binding sites of the TAB2 and TAB3 NZF domains recognize the Ile 44-centred hydrophobic patch on ubiquitin but do not interact with the Lys 63-linked isopeptide bond. Mutagenesis experiments show that both binding sites are required to enable binding of Lys 63-linked diubiquitin. We therefore propose a mechanism for the recognition of Lys 63-linked polyubiquitin chains by TAB2 and TAB3 NZF domains in which diubiquitin units are specifically recognized by a single NZF domain.
Keywords: crystallography, NF-κB, ubiquitin, zinc finger
Introduction
Post-translational modification of proteins with ubiquitin regulates a wide variety of biological processes (Hicke, 2001; Glickman and Ciechanover, 2002; Miranda and Sorkin, 2007; Hofmann, 2009; Wickliffe et al, 2009). The enzymatic cascade of ubiquitination includes three distinct classes of enzymes: E1 for ubiquitin activation, E2 for ubiquitin conjugation and E3 for ubiquitin transfer to substrate proteins. These three enzymes work together to conjugate the C-terminal group of ubiquitin with the Nɛ group of lysine residues of substrate proteins including ubiquitin itself. All seven lysine residues of ubiquitin (i.e. Lys 6, Lys 11, Lys 27, Lys, 29, Lys 33, Lys 48, Lys 63) can act as acceptors for further ubiquitination, producing polyubiquitin chains. Further, recent studies showed that the N-terminal group of ubiquitin can also act as an acceptor for ubiquitination to form linear polyubiquitin chains (Iwai and Tokunaga, 2009; Tokunaga et al, 2009). Among these eight types of polyubiquitin chains, the most abundant chain in vivo is Lys 48-linked polyubiquitin, which is the primary targeting signal for proteasomal degradation (Glickman and Ciechanover, 2002). Although the functions of the other types of polyubiquitin chains are not fully understood, recent proteomic studies suggest that polyubiquitin chains other than Lys 48-linked chains may also serve as targeting signals for proteasomal degradation and have potentially redundant functions (Meierhofer et al, 2008; Xu et al, 2009). Lys 63-linked polyubiquitin chains appear to function independent of proteasomal targeting and to instead have crucial functions in cellular processes such as DNA repair, membrane trafficking and inflammatory signalling (Miranda and Sorkin, 2007; Verstrepen et al, 2008; Chen and Sun, 2009; Hofmann, 2009; Wickliffe et al, 2009).
For instance, assembly of Lys 63-linked polyubiquitin chains is induced in the cytosol by stimulation of cells with cytokines such as tumour necrosis factor-α, interleukin-1β or Toll-like receptor ligands (Deng et al, 2000; Chiu et al, 2009; Skaug et al, 2009). Lys 63-linked chains recruit and activate a protein kinase complex containing transforming growth factor β-activated kinase 1 (TAK1), TAK1 binding protein 1 (TAB1) and TAB2 or its homologue TAB3. This TAK1 complex has a critical function in the activation of Jun N-terminal kinase (JNK) and nuclear factor-κB (NF-κB) signalling pathways (Ninomiya-Tsuji et al, 1999; Wang et al, 2001). TAK1 is a member of the MAPKKK family, whereas TAB1 is a co-activator of TAK1 (Shibuya et al, 1996; Sakurai et al, 2000). TAK1 directly phosphorylates both IκB kinase β and MKK6 to stimulate the NF-κB and JNK pathways, respectively (Wang et al, 2001). TAB2 and TAB3 have redundant functions in vivo and act as adaptors to recruit TAK1 and TAB1 to Lys 63-linked polyubiquitin chains for the activation (Ishitani et al, 2003; Cheung et al, 2004; Kanayama et al, 2004; Besse et al, 2007; Xia et al, 2009). The C-terminal Npl4 zinc-finger (NZF) domain of TAB2 and TAB3 (Supplementary Figure S1) specifically recognizes Lys 63-linked polyubiquitin chains unanchored or anchored to the substrate proteins such as RIP1 (Kanayama et al, 2004; Komander et al, 2009; Xia et al, 2009). This recognition is essential for the TAK1-induced activation of NF-κB and JNK pathway (Kanayama et al, 2004).
Zinc fingers are generally used as recognition modules that have divergent functions and zinc ligation topologies (Gamsjaeger et al, 2007). The NZF domain contains approximately 30 amino-acid residues that coordinate a single zinc ion with four conserved Cys residues (Wang et al, 2003; Alam et al, 2004). The NZF domain is frequently found in proteins involved in ubiquitin-dependent pathways, such as TAB2, TAB3 (Kanayama et al, 2004), Npl4 (Meyer et al, 2002), TRABID (Tran et al, 2008) and yeast Vps36 (Alam et al, 2004). Some of the isolated NZF domains bind with ubiquitin with Kd values of 100 μM or higher, whereas some others do not (Meyer et al, 2002; Wang et al, 2003; Alam et al, 2004). The ubiquitin-binding NZF domain comprises a highly conserved TF/Φ motif, where Φ represents a hydrophobic residue and 10 amino-acid residues are inserted between the -Thr-Phe- sequence and Φ (Wang et al, 2003). Most of the ubiquitin-binding NZF domains bind polyubiquitin chains without apparent specificity, whereas the NZF domains of TAB2 and TAB3 specifically bind Lys 63-linked polyubiquitin chains (Kanayama et al, 2004; Komander et al, 2009). Although structures of the NZF domain of Npl4 in isolation and in complex with ubiquitin have been reported (Wang et al, 2003; Alam et al, 2004), it has remained unclear how the NZF domains of TAB2 and TAB3 specifically recognize Lys 63-linked polyubiquitin chains. Here we report the crystal structures of the NZF domains of TAB2 and TAB3 in complex with Lys 63-linked diubiquitin (K63-Ub2) at 1.18 and 1.40 Å resolutions, respectively. The complex structures and structure-based mutagenesis analyses reveal the mechanism for specific binding of Lys 63-linked polyubiquitin chains by TAB2 and TAB3 NZF domains.
Results and discussion
Overall structures of the TAB2 and TAB3 NZF domains in complex with K63-Ub2
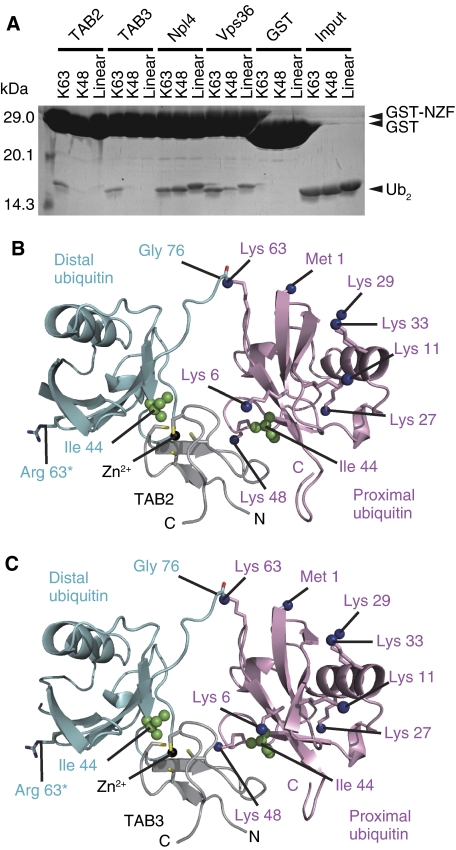
The NZF domains of mouse TAB2 (residues 665–693) and mouse TAB3 (residues 688–716) are sufficient for the specific binding with Lys 63-linkage polyubiquitin chains (Figure 1A). These regions of TAB2 and TAB3 are hereafter referred to as TAB2-NZF and TAB3-NZF, respectively. The crystal structure of the TAB2-NZF·K63-Ub2 complex was solved by the single-wavelength anomalous dispersion (SAD) method using the zinc edge (Supplementary Table S1) and the crystal structure of the TAB3-NZF·K63-Ub2 complex was solved by molecular replacement using the structure of the TAB2-NZF·K63-Ub2 complex as a search model. The TAB2- and TAB3-NZFs are 79.3% identical and, as expected, the TAB2-NZF·K63-Ub2 complex structure is nearly identical to the TAB3-NZF·K63-Ub2 complex structure (Figures 1B, C, 2 and Supplementary Figure S2). The Cα atoms of the TAB2-NZF·K63-Ub2 complex can be superposed onto those of the TAB3-NZF·K63-Ub2 complex with a root mean squared deviation (r.m.s.d.) value of 0.37 Å (177 residues in total, except for the structurally variable C-terminal tail of the proximal ubiquitin). We will therefore describe the structure of the TAB2-NZF·K63-Ub2 complex below and generally refer to Supplementary Data for discussion of the TAB3 complex. The TAB2-NZF consists of a pair of anti-parallel β-sheets and a following long loop. A zinc ion is coordinated by Cys 670, Cys 673, Cys 684 and Cys 687 of the TAB2-NZF, in a manner similar to those of the Npl4 NZF domain (Npl4-NZF) (Supplementary Figure S3; Wang et al, 2003; Alam et al, 2004). The Cα atoms of the TAB2-NZF can be superposed onto the Npl4-NZF with an r.m.s.d. value of 0.79 Å over 28 amino-acid residues. TAB2-NZF crystallized as a stoichiometric complex with K63-Ub2 (Figure 1B). The TAB2-NZF binds to the distal and the proximal ubiquitin moieties with buried surface areas of 333 and 302 Å2, respectively (Figures 1B and 2). Relative orientations between the distal and proximal ubiquitin moieties bound to TAB2- and TAB3-NZFs are considerably different from those of the free K63-Ub2 and Lys 63-linked tetraubiquitin chains (Komander et al, 2009; Weeks et al, 2009; Supplementary Figure S4). The TAB2-NZF interacts with the Ile 44-centred hydrophobic patches of both proximal and distal ubiquitin moieties, but does not directly recognize the Lys 63-linked isopeptide bond of K63-Ub2 (Supplementary Figure S3C).
Figure 1.
Overall structures of the TAB2- and TAB3-NZFs in complex with K63-Ub2. (A) Pull-down assays using GST-fused TAB2-, TAB3-, Npl4- and Vps36-NZFs to assess their interaction with K63-, K48- or linear Ub2. The bound proteins were analysed by SDS–PAGE and stained with Coomassie Brilliant Blue. (B) The TAB2-NZF·K63-Ub2 complex. The NZF is coloured grey. The proximal and distal ubiquitin moieties are coloured pink and cyan, respectively. Ile 44 in each ubiquitin moiety is shown as green spheres. The Nɛ atoms of lysine residues and the nitrogen atom of the N-terminal Met in the proximal ubiquitin are shown as blue spheres. The K63R mutation in the distal ubiquitin is indicated as sticks. (C) The TAB3-NZF·K63-Ub2 complex. The drawing schemes are the same as in B.
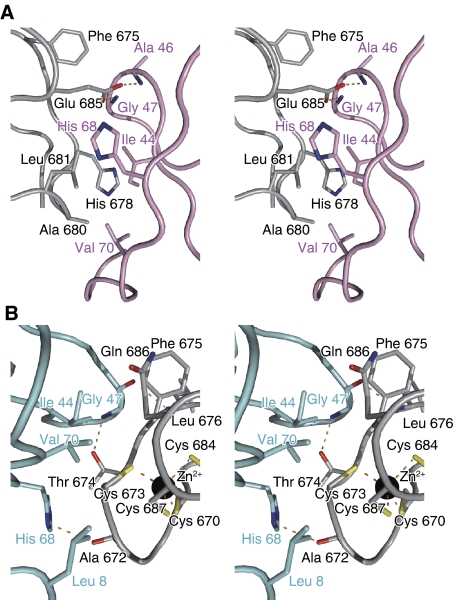
Figure 2.
Recognition of the distal and proximal ubiquitin moieties by TAB2. Stereo view of the detailed TAB2-NZF·K63-Ub2 binding interface. The colouring schemes are the same as in Figure 1. Hydrogen bonds are indicated as dashed orange lines. The labels of the NZF and the proximal and distal ubiquitin moieties are coloured black, pink and cyan, respectively. (A) The interface between the proximal ubiquitin and the TAB2-NZF. (B) The interface between the distal ubiquitin and the TAB2-NZF.
Proximal ubiquitin recognition
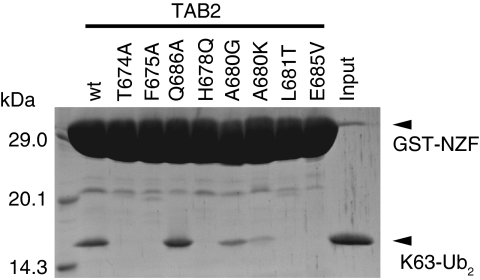
The TAB2-NZF domain recognizes the Ile 44-centred hydrophobic patch of the proximal ubiquitin moiety that is formed by the side chains of Ile 44, Ala 46 and Val 70 and aliphatic portions of the His 68 side chain. The side chains of Phe 675, Ala 680 and Leu 681 and aliphatic portions of the His 678 and Glu 685 side chains in the TAB2-NZF form a hydrophobic surface that interacts with the Ile 44-centred hydrophobic patch of the proximal ubiquitin (Figure 2A). Further, the two Oɛ atoms of Glu 685 in the TAB2-NZF hydrogen bond with the main-chain NH atoms of Ala 46 and Gly 47 in the proximal ubiquitin, respectively (Figure 2A). The functional importance of these interactions was confirmed by GST pull-down analyses using TAB2-NZF mutants (Figure 3). H678Q, L681T and E685V mutants completely lost the ability to bind K63-Ub2, whereas A680G and A680K mutants exhibit decreased binding to K63-Ub2. The proximal ubiquitin binding residues of TAB2 and TAB3 are well conserved among five organisms (Supplementary Figure S5), whereas those of the TAB2 and TAB3 are not fully conserved among the other ubiquitin-binding NZFs (Supplementary Figure S6). These facts suggest that the proximal binding residues of the TAB2- and TAB3-NZFs that we found in our complex structure are likely to have key functions in the specific recognition of Lys 63-linked polyubiquitin chains.
Figure 3.
Pull-down assays assessing the K63-Ub2 binding by proximal and distal ubiquitin binding site mutants of the TAB2-NZF. The bound K63-Ub2 molecules were analysed by SDS–PAGE and stained with Coomassie Brilliant Blue.
Distal ubiquitin recognition
The TAB2-NZF also recognizes the Ile 44-centred hydrophobic patch of the distal ubiquitin moiety that is formed by the side chains of Leu 8, Ile 44 and Val 70 and aliphatic portions of the His 68 side chain. The side chain of Phe 675 and aliphatic portions of the Cys 673, Thr 674, Gln 686 and Cys 687 side chains in the TAB2-NZF form a hydrophobic surface to interact with the Ile 44-centred hydrophobic patch of the distal ubiquitin (Figure 2B). Thr 674 and Phe 675 of the TAB2-NZF are included in the aforementioned TF/Φ motif for the ubiquitin binding, whereas Cys 673 and Cys 687 are also involved in the Zn2+ coordination. Further, the main-chain CO of Ala 672, the main-chain NH of Leu 676 and the Oγ atom of Thr 674 in the TAB2-NZF hydrogen bond with the Nδ atom of His 68, the main-chain CO and the main-chain NH of Gly 47 in the distal ubiquitin, respectively (Figure 2B). We sought to confirm the functional importance of these interactions by GST pull-down analyses using TAB2-NZF mutants (Figure 3). As expected from our structure, T674A and F675A mutants completely lost their binding activities. However, the Q686A mutant can bind to K63-Ub2 as efficiently as the wild type, indicating that Gln 686 of the TAB2-NZF contributes little to the binding affinity with the distal ubiquitin. It is important to note that Thr 674 and Phe 675 of the TAB2-NZF are completely conserved among five organisms, whereas Gln 686 of the TAB2-NZF is replaced by Ala or Glu in fruit fly or zebra fish, respectively (Supplementary Figure S5A). Intriguingly, Gln 686 of the TAB2-NZF is located in the position corresponding to the hydrophobic residue (Φ) in the TF/Φ motif. The role of this Φ–Gln replacement will be discussed below, in the context of the linkage specificity.
Amino-acid sequence alignment of the ubiquitin-binding NZFs of TAB2, TAB3, Npl4, TRABID and Vps36 shows that the amino-acid residues that are required for the distal ubiquitin binding by TAB2 are almost completely conserved (Supplementary Figure S6A). Only Phe 675 of the TAB2-NZF is replaced by Tyr in TRABID. The near-perfect sequence conservation of the distal ubiquitin binding residues suggests that all ubiquitin-binding NZFs share a common strategy for the ubiquitin binding. In fact, the physical basis for distal ubiquitin binding by the TAB2- and TAB3-NZFs is similar to that for monoubiquitin binding by the Npl4-NZF (Supplementary Figure S3). The Cα atoms of the TAB2-NZF and the distal ubiquitin moiety (100 residues in total) in the TAB2-NZF·K63-Ub2 complex can be superposed onto those of the Npl4-NZF·ubiquitin complex with an r.m.s.d. value of 1.63 Å (Alam et al, 2004). Their ubiquitin binding modes are essentially identical. However, the relative orientations between ubiquitin and NZF are slightly different and the Thr rotamers in the TF/Φ motif are different between the Npl4-NZF and TAB2-NZF. The dihedral angle around the Cα–Cβ bond of Thr 558 in the Npl4 rotates 120 deg, relative to that of Thr 674 in the TAB2-NZF, lacking a hydrogen bond with ubiquitin. Thr 674 of the TAB2-NZF in our high-resolution structure is highly ordered with low temperature factors (∼11 Å2) and clear electron density. Further, the proximal ubiquitin does not interact with Thr 674 of the TAB2-NZF, and thus is unlikely to affect the side-chain rotamer of Thr 674 in the TAB2-NZF. Therefore, we suggest that the Thr rotamer in the TF/Φ motif of the Npl4-NZF should likely be the same as that of the TAB2-NZF observed in our structures, consistent with the perfect sequence conservation of Thr in the TF/Φ motif among the ubiquitin-binding NZFs.
Linkage specificity
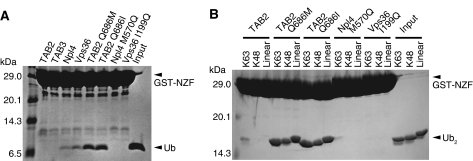
For linkage-specific ubiquitin chain recognition, binding proteins must exclusively bind to ubiquitin chains but not to monoubiquitin, as we observe for the TAB2- and TAB3-NZFs (Figures 1A and 4A). In contrast, the Npl4-NZF and the second NZF domain of Saccharomyces cerevisiae Vps36 (Vps36-NZF) bind to both monoubiquitin and all diubiquitins tested (Figures 1A and 4A).
Figure 4.
Pull-down assays using NZF variants containing point mutations in the Φ position of the TF/Φ motif to assess their mono- or diubiquitin binding. (A) Pull-down assays using the GST-fused TAB2-, Npl4- and Vps36-NZF mutants to assess their monoubiquitin binding. The bound monoubiquitin molecules were analysed by tricine SDS–PAGE and stained with Coomassie Brilliant Blue. (B) Pull-down assays using GST-fused TAB2-, Npl4- and Vps36-NZF mutants to assess their binding with K63-Ub2, K48-Ub2 or linear Ub2. The bound diubiquitin molecules were analysed by SDS–PAGE and stained with Coomassie Brilliant Blue.
Chain-selective binding of the TAB2- and TAB3-NZFs seems to be related to the hydrophilic Gln that is located in the position of Φ in the TF/Φ motif for ubiquitin binding. The Npl4- and Vps36-NZF domains contain the complete TF/Φ motif (Φ is Met 570 in the Npl4-NZF or Ile 199 in the Vps36-NZF) where Phe and Φ form a hydrophobic surface to interact with the Ile 44-centred hydrophobic patch of ubiquitin. In contrast, Φ is replaced by the hydrophilic Gln 686/709 in TAB2/TAB3 (Supplementary Figure S5A), which contributes little to the distal ubiquitin binding, as mentioned above (Figure 3).
To clarify the relationship between the monoubiquitin binding and Φ in the TF/Φ motif, we mutated Met 570 of the Npl4-NZF and Ile 199 of the Vps36-NZF to Gln, and also mutated Gln 686 of the TAB2-NZF to Ile or Met for GST pull-down analyses. Strikingly, the Q686M and Q686I mutations enabled the TAB2-NZF to bind to monoubiquitin and resulted in linkage-independent binding to K63-, K48- and linear diubiquitins (Figure 4). In contrast, M570Q and I199Q mutations disrupt the abilities of the Npl4- and Vps36-NZF domains to bind to monoubiquitin, respectively. These results clearly show that Φ in the TF/Φ motif is a key residue that regulates the ability of the NZF domain to bind to monoubiquitin. The presence of a non-hydrophobic residue in the Φ position of the TF/Φ motif of the TAB2- and TAB3-NZFs diminishes binding to monoubiquitin and enables linkage-specific binding when combined with a proximal ubiquitin binding surface.
The Φ–Gln replacement in the TF/Φ motif of the TAB2- and TAB3-NZFs is essential but not sufficient for Lys 63-linkage-specific polyubiquitin binding. Because mutations in both the distal and proximal ubiquitin binding sites of the TAB2-NZF disrupt binding of K63-Ub2, simultaneous binding of the distal and proximal ubiquitin moieties to the TAB2-NZF is essential for Lys 63-linkage-specific binding (Figure 3). The distal and proximal ubiquitin binding sites of the TAB2-NZF are organized to optimize simultaneous binding to each ubiquitin moiety in a configuration characteristic to Lys 63-linked ubiquitin chains. Among all the lysine residues and the N-terminal methionine residue of the proximal ubiquitin, Lys 63 is the closest to the C-terminal tail of the distal ubiquitin in the present complex structure (Figure 1 and Supplementary Figure S7A). Because of this proximity of Lys 63 of the proximal ubiquitin to the C-terminal tail of the distal ubiquitin, the distal and proximal ubiquitin moieties of K63-Ub2 bind simultaneously to the TAB2-NZF. In contrast, in the orientations of the distal and proximal ubiquitin moieties bound to TAB2- and TAB3-NZFs, the C-terminal tail of the distal ubiquitin cannot reach either the Nɛ group of Lys 48 or the N-terminal group in the proximal ubiquitin, even if considering the structurally flexible C-terminal tail of the distal ubiquitin. One of the most elongated forms of the ubiquitin C-terminal tail has been observed in the complex with the de-ubiquitinating enzyme (DUB) AMSH-LP (Sato et al, 2008), where the distance between the Cα atom of Leu 71 (positioned in the end of the last β-sheet) and that of Gly 76 in the distal ubiquitin is 19 Å (Supplementary Figure S7B). This is shorter than the distance between the Cα atom of Leu 71 in the distal ubiquitin and the Lys 48 Nɛ group (23 Å) or the N-terminal group in the proximal ubiquitin (24 Å) in the TAB2-NZF·K63-Ub2 complex (Supplementary Figure S7A). Unexpectedly, the distance between the Cα atom of Leu 71 and the Nɛ group of Lys 6 is 14 Å, and the ubiquitin C-terminal tail conformation observed in the Npl4-NZF·ubiquitin complex appears likely to enable the bonding between Gly 76 in the distal ubiquitin and Lys 6 in the proximal ubiquitin (Supplementary Figures S3 and S7A). From a structural point of view, we cannot exclude the possibility that TAB2- and TAB3-NZFs might be able to bind to Lys 6-linked chains.
Strategies for the linkage specificity
Our crystal structures of the TAB2- and TAB3-NZFs in complex with K63-Ub2 and structure-based mutagenesis analyses revealed that a single zinc-finger domain contains both distal and proximal ubiquitin binding sites that enable Lys 63-linked chains to simultaneously approach both sites in the specific geometry required for binding. The TAB2- and TAB3-NZFs effectively measure the length between Ile 44-centred hydrophobic patches and Lys 63-linked chains have the correct spacing to bind both the proximal and distal ubiquitin binding sites simultaneously. It is important to note that direct recognition of the isopeptide linkage is not required for this mode of linkage-specific binding. This strategy seems to be typical for linkage-specific ubiquitin chain binding by ubiquitin binding domains (UBDs), as illustrated by crystal structures of the RAP80 tandem UIMs (ubiquitin-interacting motifs) in complex with K63-Ub2 (Sato et al, 2009) and the UBAN (ubiquitin binding domains of ABINs and NEMO) of NEMO in complex with linear Ub2 (Rahighi et al, 2009) and the NMR-based structural model of the second UBA (ubiquitin-associated motif) of hHR23A in complex with K48-Ub2 (Varadan et al, 2005).
In the crystal structure of the RAP80 tandem UIMs, UIM1-UIM2, in complex with K63-Ub2, the RAP80 UIM1 and UIM2 recognize the Ile 44-centred hydrophobic patch on the proximal and the distal ubiquitin moieties, respectively, without any interactions with the Lys 63-linked isopeptide bond (Sato et al, 2009). The affinity between each UIM of RAP80 and monoubiquitin is too weak (Kd value is ∼500 μM) to form a stable complex (Sims and Cohen, 2009). Therefore, a robust interaction between RAP80 and Lys 63-linked ubiquitin chains is only achieved by simultaneous interactions between multiple UIMs and multiple ubiquitin moieties in polyubiquitin chains, termed avidity. The key determinant of the Lys 63-linkage-specific avidity is the length of the inter-UIM region (Sato et al, 2009; Sims and Cohen, 2009). The appropriate length of the inter-UIM region organizes the UIM1 and UIM2 to optimize the spatial relationship between UIM1 and UIM2 for simultaneous binding to both ubiquitin moieties of K63-Ub2. The inter-UIM region therefore measures the inter-molecular distance between the proximal and distal ubiquitin moieties of K63-Ub2. Similarly, the structures of the hHR23A·K48-Ub2 and NEMO·linear Ub2 complexes show that hHR23A and NEMOoptimize the spatial relationship between multiple ubiquitin binding sites to enable simultaneous binding to the multiple ubiquitin moieties of specific polyubiquitin chains (Varadan et al, 2005; Rahighi et al, 2009). Intriguingly, this strategy is also observed in anti-Lys 63-linked ubiquitin chain antibodies in complex with K63-Ub2 (Newton et al, 2008).
In contrast, the strategy for the linkage specificity of UBDs differs from that of the linkage-specific DUBs. To date, the crystal structure of the AMSH-LP DUB domain in complex with K63-Ub2 is the only reported structure of a linkage-specific DUB in complex with an isopeptide-linked ubiquitin chain (Sato et al, 2008). AMSH-LP is a zinc-dependent DUB that specifically cleaves Lys 63-linked polyubiquitin chains. A single AMSH-LP DUB domain interacts with both proximal and distal ubiquitin moieties. Compared with the extensive binding site for distal ubiquitin, the binding site for proximal ubiquitin makes few contacts with AMSH-LP and contributes little to the binding affinity. However, AMSH-LP directly interacts with the isopeptide-linked Lys 63 and neighbouring residues in the proximal ubiquitin. This interaction facilitates the correct orientation and accurate positioning of the isopeptide bond for efficient linkage-specific de-ubiquitination. A similar mechanism has been proposed for other linkage-specific DUBs, CYLD (Komander et al, 2008) and OTU1 (Wang et al, 2009). Therefore, linkage specificity of DUBs seems to be predominantly achieved in the hydrolysis step rather than in the binding step. Further, the linkage-specific DUBs directly recognize the (iso)peptide linkage in striking contrast to the linkage-specific UBDs.
Triple ubiquitin-binding NZF domains of TRABID
TRABID contains three NZFs (TRABID-NZF1–3), which specifically bind to Lys 63-linked and linear polyubiquitin chains (Tran et al, 2008; Komander et al, 2009). Individually, each TRABID-NZF domain fails to bind monoubiquitin by GST pull-down (data not shown). Chain-selective binding by the TRABID-NZF1–3 can be partially understood on the basis of its amino-acid sequence and our complex structure. In the TF/Φ motifs of the TRABID-NZF1–3, Φ in the TRABID-NZF2 is replaced by the hydrophilic Gln, similar to the TAB2- and TAB3-NZFs (Supplementary Figure S6A). In addition, as mentioned above, the phenylalanine residue of each TRABID-NZF1–3 is replaced by Tyr, which may also diminish binding to monoubiquitin. However, the TRABID-NZF1–3 domains seem to lack proximal ubiquitin interacting sites, suggesting that the structural basis for linkage-specific recognition by TRABID-NZF1–3 differs from that of the TAB2- and TAB3-NZFs. The most plausible mechanism for the linkage specificity of the TRABID-NZF1–3 is that the multiple NZF domains bind a single ubiquitin chain and linkage-specific binding is accomplished by an avidity-based mechanism (Komander et al, 2009; Sims and Cohen, 2009). Because Lys 63-linked and linear polyubiquitin chains have similar distances between Ile 44 centred hydrophobic patches, it is possible that the multiple ubiquitin moieties of Lys 63-linked and linear polyubiquitin chains can simultaneously bind to the multiple NZFs of TRABID whereas those of Lys 48-linked chains cannot. Elucidation of the basis for linkage-specific binding by the TRABID-NZF1–3 awaits the structural analysis of it in complex with a Lys 63-linked or linear ubiquitin chain.
In summary, linkage-independent ubiquitin-binding NZFs (i.e. Npl4 and Vps36) contain a complete TF/Φ motif and bind tightly to monoubiquitin, whereas linkage-specific ubiquitin-binding NZFs (i.e. TAB2, TAB3 and TRABID) contain an incomplete TF/Φ motif (i.e. TF/Q for TAB2 and TAB3, and TY/Φ or Q for TRABID) and bind weakly to monoubiquitin. Linkage-specific binding further requires simultaneous interaction with both the proximal and distal ubiquitin moieties for a specific and robust interaction, which is achieved by the additional, proximal binding site for the TAB2 and TAB3 or likely by an avidity-based mechanism involving binding by the multiple NZFs for TRABID. These variations enable NZF domains to show a wide range of affinities and specificities that allow them to perform their specific functions in ubiquitin-dependent signalling pathways.
Materials and methods
Preparation of the TAB2K63-Ub2 and TAB3K63-Ub2 complex
The genes encoding NZF domains of mouse TAB2 (residues 665–693) and TAB3 (residues 688–716) were PCR amplified from a mouse cDNA library. The amplified genes were cloned into the pCold-GST expression vector (Hayashi and Kojima, 2008) with NdeI and XhoI sites to produce N-terminal GST fusion protein and were confirmed by DNA sequencing. Escherichia coli strain RosettaTM (DE3) cells (Invitrogen) were transformed with the expression vector, and were cultured in LB containing 100 mg/l ampicillin at 37°C. When the optical density at 600 nm of the culture reached ∼0.5, the culture was incubated for 30 min at 15°C. Thereafter, isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 0.3 mM to induce protein expression for 24 h at 15°C. The cells were collected by centrifugation at 8000 g for 15 min and were disrupted by sonication in phosphate-buffered saline (PBS), containing 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulphonyl fluoride and 0.5% Triton X-100. The lysates were centrifuged at 30 000 g for 60 min and the supernatants were then loaded onto a Glutathione Sepharose FF column (GE Healthcare) that had been pre-equilibrated with PBS containing 1 mM DTT and 0.5% Triton X-100. The column was washed with PBS containing 1 mM DTT and 0.5% Triton X-100 and then with PBS containing 1 mM DTT. GST fusion proteins were eluted with 50 mM Tris–HCl buffer (pH 8.0), containing 200 mM NaCl, 1 mM DTT and 15 mM reduced glutathione. The GST tags were cleaved by Turbo3C (HRV3C) protease (Accelagen), and the samples were dialysed against 50 mM Tris–HCl buffer (pH 8.0), containing 40 mM NaCl and 1 mM DTT. TAB2 was loaded onto a ResourceQ anion exchange column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl buffer (pH 8.0) containing 40 mM NaCl and 1 mM DTT, and were eluted with a linear gradient of 0–1 M NaCl. TAB3 was passed over a ResourceQ anion exchange column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl buffer (pH 8.0) containing 40 mM NaCl and 1 mM DTT. The samples were loaded onto a Superdex 75 16/60 (prep grade) column (GE Healthcare) pre-equilibrate with 10 mM Tris–HCl buffer (pH 7.2) containing 50 mM NaCl and 5 mM β-mercaptoethanol. The fractions rich in the purified proteins were collected for crystallization.
Crystallization and data collection
K63-Ub2 was prepared as described previously (Sato et al, 2008). The purified mouse TAB2 (665–693) and TAB3 (688–716) were concentrated to 10 g/l by using Amicon Ultra-15 3000 MWCO filter (Millipore), following the manufacturer's instructions. The TAB2 (665–694) or TAB3 (688–716) was mixed with K63-Ub2 in a 2:1 molar ratio for crystallization. Initial crystallization screening was performed using the sitting drop vapour diffusion method at 20°C, with a Mosquito® liquid-handling robot (TTP Lab Tech). We tested about 500 conditions, using crystallization reagent kits supplied by Hampton Research, and initial hits were further optimized. The best crystals of the TAB2·K63-Ub2 complex were obtained at 20°C with the sitting drop vapour diffusion method by mixing 0.5 μl of protein solution with an equal amount of precipitant solution containing 90 mM Bis–Tris–HCl buffer (pH 6.5), 180 mM ammonium acetate, 21% PEG3350 and 4% pentaerythritol ethoxylate, and equilibrating against 500 μl of reservoir solution containing 100 mM Bis–Tris–HCl buffer (pH 6.5), 200 mM ammonium acetate and 27% PEG3350. The best crystals of the TAB3·K63-Ub2 complex were obtained at 20°C with the sitting drop vapour diffusion method by mixing 0.5 μl of protein solution with an equal amount of precipitant solution containing 100 mM Bis–Tris–HCl buffer (pH 6.5) and 21% PEG3350, and equilibrating against 500 μl of reservoir solution containing 100 mM Bis–Tris–HCl buffer (pH 6.5) and 21% PEG3350. The crystal of the TAB2K63-Ub2 complex belonged to space group P212121, with unit cell dimensions a=30.1 Å, b=71.5 Å and c=72.0 Å. The crystal of the TAB3·K63-Ub2 complex belonged to space group P212121, with unit cell dimensions a=29.9 Å, b=71.0 Å and c=71.6 Å.
Structural determination and refinement
Diffraction data sets were collected at the beamline BL41XU in SPring-8 (Hyogo, Japan), and were processed with the program HKL2000 (Otwinowski and Minor, 1997) and the CCP4 program suite (Collaborative Computational Project, 1994). To solve the structure of the TAB2·K63-Ub2 complex from the SAD data set, we used SHELX97 (George and Schneider, 1997), SHARP (Fortelle and Bricogne, 1997), SOLOMON/DM (Cowtan and Main, 1993; Abrahams and Leslie, 1996) and ARP/warp (Morris et al, 2003) for heavy-atom site search, phase calculation, density improvement and automatic model-building, respectively. These programs were controlled by autoSHARP (Vonrhein et al, 2007). MOLREP (Vagin and Teplyakov, 1997) was used to solve the structure of the TAB3·K63-Ub2 complex by molecular replacement. The crystal structure of the TAB2·K63-Ub2 complex was used as a search model. Atomic models were corrected by using COOT (Emsley and Cowtan, 2004) with careful inspection. Refinement was carried out by using Refmac5 (Murshudov et al, 1997) with iterative correction and refinement of the atomic models. The final models have excellent stereochemistry and Rfree values of 0.191 for the TAB2·K63-Ub2 complex and 0.218 for the TAB3·K63-Ub2 complex at 1.18 and 1.40 Å resolution, respectively. Data collection, phasing and refinement statistics are shown in Supplementary Table S1. All molecular graphics were prepared with PyMOL (DeLano Scientific; http://www.pymol.org).
GST pull-down assays
For GST pull-down assays, GST-fused NZFs of TAB2, TAB3, Npl4 and Vps36 were overproduced in E. coli strain Rosetta™ (DE3) (Invitrogen), and purified by Glutathione Sepharose FF and Resource Q anion exchange columns (GE Healthcare). Mutations were generated by PCR. 100 μg of the GST-fused wild-type and mutants of NZFs were immobilized on Glutathione Sepharose FF beads (GE healthcare), and then incubated with 55 μg of K63-Ub2, K48-Ub2, linear Ub2 or monoubiquitin for 15 min at 4°C in 25 mM Tris–HCl buffer (pH 7.2) containing 20 μM zinc chloride, 1 mM DTT and 0.1% Triton X-100. The beads were washed with the same buffer three times. The diubiquitin or monoubiquitin molecules bound to the beads were released by boiling in SDS loading buffer and analysed by SDS–PAGE or tricine SDS–PAGE, respectively. The gels were stained with Coomassie Brilliant Blue.
Accession codes
The coordinates and structure factors of the TAB2- and TAB3-NZFs in complex with K63-Ub2 have been deposited in the Protein Data Bank with accession codes and , respectively.
Supplementary Material
Supplementary Figure S1–S7
Supplementary Table S1
Supplementary Information
Review Process File
Acknowledgments
We thank C Toyoshima for support of this research. We thank S Kaiser for critical reading and improvement of this paper. We are grateful to K Iwai for providing us with reagents for the overexpression of E1, E2-25K and linear Ub2. We are grateful to M Komada for providing us with the overexpression vectors of ubiquitin mutants for K63-Ub2 and K48-Ub2 synthesis. We thank the beam-line staffs at NW12A and BL5A of Photon Factory (Tsukuba, Japan) and BL41XU of SPring8 (Hyogo, Japan) for technical help during data collection. This work was supported by grants from MEXT to SF and AYa, and YS and MY are supported by JSPS research fellowships for young scientists.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abrahams JP, Leslie AG (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr 52: 30–42 [DOI] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI (2004) Ubiquitin interactions of NZF zinc fingers. EMBO J 23: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse A, Lamothe B, Campos AD, Webster WK, Maddineni U, Lin SC, Wu H, Darnay BG (2007) TAK1-dependent signaling requires functional interaction with TAB2/TAB3. J Biol Chem 282: 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33: 275–286 [DOI] [PubMed] [Google Scholar]
- Cheung PC, Nebreda AR, Cohen P (2004) TAB3, a new binding partner of the protein kinase TAK1. Biochem J 378: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Zhao M, Chen ZJ (2009) Ubiquitin in NF-κB signaling. Chem Rev 109: 1549–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cowtan KD, Main P (1993) Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr D Biol Crystallogr 49: 148–157 [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fortelle EDL, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276: 472–494 [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP (2007) Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 32: 63–70 [DOI] [PubMed] [Google Scholar]
- George G, Schneider T (1997) SHELXL: high resolution refinement. Methods Enzymol 277: 319–343 [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kojima C (2008) pCold-GST vector: a novel cold-shock vector containing GST tag for soluble protein production. Protein Expr Purif 62: 120–127 [DOI] [PubMed] [Google Scholar]
- Hicke L (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195–201 [DOI] [PubMed] [Google Scholar]
- Hofmann K (2009) Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 8: 544–556 [DOI] [PubMed] [Google Scholar]
- Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K (2003) Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J 22: 6277–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F (2009) Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep 10: 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548 [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B Box module. Mol Cell 29: 451–464 [DOI] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D (2009) Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10: 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhofer D, Wang X, Huang L, Kaiser P (2008) Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res 7: 4566–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J 21: 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Sorkin A (2007) Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol Interv 7: 157–167 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Perrakis A, Lamzin VS (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol 374: 229–244 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134: 668–678 [DOI] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K (1999) The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398: 252–256 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Sakurai H, Miyoshi H, Mizukami J, Sugita T (2000) Phosphorylation-dependent activation of TAK1 mitogen-activated protein kinase kinase kinase by TAB1. FEBS Lett 474: 141–145 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S (2009) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455: 358–362 [DOI] [PubMed] [Google Scholar]
- Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K (1996) TAB1: an activator of the TAK1 MAPKKK in TGF-β signal transduction. Science 272: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell 33: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ (2009) The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem 78: 769–796 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Tran H, Hamada F, Schwarz-Romond T, Bienz M (2008) Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev 22: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D (2005) Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell 18: 687–698 [DOI] [PubMed] [Google Scholar]
- Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R (2008) TLR-4, IL-1R and TNF-R signaling to NF-κB: variations on a common theme. Cell Mol Life Sci 65: 2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Wang B, Alam SL, Meyer HH, Payne M, Stemmler TL, Davis DR, Sundquist WI (2003) Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J Biol Chem 278: 20225–20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C (2009) Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol 386: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SD, Grasty KC, Hernandez-Cuebas L, Loll PJ (2009) Crystal structures of Lys-63-linked tri- and di-ubiquitin reveal a highly extended chain architecture. Proteins 77: 753–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe K, Williamson A, Jin L, Rape M (2009) The multiple layers of ubiquitin-dependent cell cycle control. Chem Rev 109: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461: 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S7
Supplementary Table S1
Supplementary Information
Review Process File