Abstract
The formation of heterochromatin at the centromeres in fission yeast depends on transcription of the outer repeats. These transcripts are processed into siRNAs that target homologous loci for heterochromatin formation. Here, high throughput sequencing of small RNA provides a comprehensive analysis of centromere-derived small RNAs. We found that the centromeric small RNAs are Dcr1 dependent, carry 5′-monophosphates and are associated with Ago1. The majority of centromeric small RNAs originate from two remarkably well-conserved sequences that are present in all centromeres. The high degree of similarity suggests that this non-coding sequence in itself may be of importance. Consistent with this, secondary structure-probing experiments indicate that this centromeric RNA is partially double-stranded and is processed by Dicer in vitro. We further demonstrate the existence of small centromeric RNA in rdp1Δ cells. Our data suggest a pathway for siRNA generation that is distinct from the well-documented model involving RITS/RDRC. We propose that primary transcripts fold into hairpin-like structures that may be processed by Dcr1 into siRNAs, and that these siRNAs may initiate heterochromatin formation independent of RDRC activity.
Keywords: centromeres, RNAi, small RNA, S. pombe
Introduction
Small RNA molecules are the effectors of RNA interference (RNAi), in which gene expression is regulated either on the transcriptional or the post-transcriptional level (Fire et al, 1998; Hamilton and Baulcombe, 1999; Volpe et al, 2002). RNAi typically involves the enzymes Dicer and Argonaute, and in some systems RNA-directed RNA polymerases. Animals and plants encode several isoforms of the Dicer and Argonaute enzymes, and small RNAs can be classified depending on their origin and the pathway in which they function (for review, see Djupedal and Ekwall (2009)). The three major classes are microRNA (miRNA), short interfering RNA (siRNA) and Piwi-interacting RNA (piRNA). In general, post-transcriptional silencing of genes is accomplished by miRNA, whereas siRNA can induce silencing of genes by recruitment of factors required for heterochromatin formation. The mechanism of transcriptional silencing has been extensively studied in the yeast Schizosaccharomyces pombe. This organism is a well-established model organism for the study of heterochromatin and has single genes coding for Dicer (dcr1+) and Argonaute (ago1+) (Volpe et al, 2002), which function in both transcriptional and post-transcriptional gene silencing (Sigova et al, 2004).
The current model for the RNAi-dependent formation of heterochromatin at the centromeres of S. pombe has been described as a self-reinforced feedback loop (Noma et al, 2004; Sugiyama et al, 2005) with siRNA, Ago1, Dcr1 and the RNA-directed RNA polymerase, Rdp1, as integral components (Volpe et al, 2002; Verdel et al, 2004). The RNA-induced initiation of transcriptional silencing (RITS) complex, which includes siRNA, Ago1, Tas3 and Chp1 (Verdel et al, 2004), is targeted to centromeres by dual mechanisms: through the recognition of nascent RNA transcripts by complementary siRNA and through Chp1 binding to the canonical heterochromatin mark, H3K9me. The RITS complex permits recruitment of the Clr4 complex (ClrC) (Motamedi et al, 2004; Zhang et al, 2008) that contains the histone methyl transferase (HMTase) Clr4KMT1, which is specific for lysine 9 of histone H3 (H3K9me) (Rea et al, 2000). ClrC creates new H3K9me marks that are specifically bound by chromodomain proteins, such as Swi6 and Chp2, homologues of metazoan heterochromatin protein 1 (Lorentz et al, 1994; Bannister et al, 2001), in addition to Chp1 (RITS) and Clr4 itself. The RITS complex also allows association of the RNA-directed RNA polymerase complex (RDRC) containing Rdp1, which can produce double-stranded (ds) RNA from nascent transcripts (Motamedi et al, 2004; Sugiyama et al, 2005). Dcr1 cleaves dsRNA into additional siRNAs, causing amplification of siRNAs and further increasing H3K9me levels. In this manner, both siRNAs and H3K9me are required for assembly of heterochromatin.
Heterochromatin, characterized by binding of chromodomain proteins to H3K9me and transcriptional silencing of embedded genes, is necessary at the centromeres of S. pombe for proper segregation of chromosomes during cell division (Ekwall et al, 1995; Bernard et al, 2001). The DNA sequences of the three centromeres consist of a central core domain (cnt) flanked by arrays of non-coding, inverted repeats of low GC content, which are interspersed with tRNA genes (Clarke et al, 1986; Nakaseko et al, 1986, 1987; Fishel et al, 1988; Chikashige et al, 1989; Takahashi et al, 1991, 1992; Wood et al, 2002). The repeats are divided into innermost repeats (imr) and outer repeats (otr), which are further subdivided into dg and dh elements. The smallest endogenous S. pombe centromere, cen I, has single copies of dg and dh elements on each chromosome arm, whereas cen III is estimated to have 12 copies of dg and dh elements in total (Wood et al, 2002). The copy number of dg and dh elements at the centromeres can vary in distinct S. pombe isolates as well as in different laboratory strains (Steiner et al, 1993). The use of plasmid-based constructs demonstrated that cnt together with a 2.1-kb fragment from the dg element is required to allow the formation of a mitotically functional centromere (Baum et al, 1994; Wood et al, 2002).
Analyses of RITS- and Ago1-associated siRNAs from S. pombe have revealed that a sizeable fraction of the siRNA originates from the dg and dh elements at the centromeres (Cam et al, 2005; Buhler et al, 2008). According to the current model for RNAi-dependent heterochromatin formation, RITS is guided by the encapsulated siRNAs to nascent RNA polymerase II transcripts from the centromeres. Transcription of the dg and dh elements has been shown to occur predominantly in the S-phase of the cell cycle when the non-permissive, heterochromatic structure is dispersed after DNA replication (Chen et al, 2008; Kloc et al, 2008). Hence, RNAi is involved in the maintenance of heterochromatin after DNA replication in each cell cycle. In unsynchronized, wild-type (WT) cells, transcripts derived from the ‘reverse' strand of the DNA are more easily detected (Volpe et al, 2002) and a strong promoter upstream of this transcript has been mapped (Djupedal et al, 2005). As a result of heterochromatin formation the expression of reporter genes inserted into the dg and dh elements is silenced in WT cells (Allshire et al, 1995). In strains defective for RNAi, such as ago1Δ and dcr1Δ, transcripts derived from both strands accumulate and reporter genes in dg or dh elements are not silenced (Provost et al, 2002; Volpe et al, 2002), probably due to the absence of heterochromatin at the centromeric region.
Here, we have performed high-throughput pyrosequencing of S. pombe size-selected small RNA to gain insight into the role of RNAi in this organism. We found that most centromeric small RNA are produced from two types of small RNA clusters that share a high degree of sequence identity, and we present a detailed description of these loci. Most small RNAs are found within regions of perfect sequence identity between repeats from all three centromeres, indicating functional conservation of these sequences. The predominant sequence class of small RNAs was validated by northern blots and has the characteristics of bona fide siRNAs. In addition, we have determined the in vitro secondary structure of a portion of the transcript from one of the small RNA clusters and have shown that it forms a partially double-stranded secondary structure that is processed by recombinant human Dicer. Furthermore, we demonstrate that a small fraction of centromeric small RNAs is synthesized independently of Rdp1, including small RNA corresponding to the experimentally determined hairpin. Therefore, we propose that the ability of nascent centromeric transcripts to fold into double-stranded ‘hairpin' structures may permit their Dcr1-dependent processing into siRNAs, which in turn contributes to the establishment of heterochromatin at the centromeres.
Results
Small RNA in S. pombe
With high-throughput 454 sequencing, 21 776 sequenced small RNAs were determined from WT S. pombe cells (Table I). In addition, sequences of small RNA were determined from rpb7G150D cells that carry a point mutation in the RNA polymerase II subunit Rpb7, which causes lower levels of transcription of centromeric siRNA precursors (Djupedal et al, 2005). A large percentage of sequences mapped to the transcribed region of tRNA or rRNA genes and were, together with reads without perfect match to the S. pombe genome, excluded from further analysis. Nearly half of the remaining 3552 WT sequences begin with a uracil (U), with lesser representation of A and C as a first base and strong selection against G (Figure 1A). Although this is a significant deviance from the genome average, Ago1-associated siRNAs (Buhler et al, 2008; subjected to the same matching and filtering as WT) show a strong bias for uracil in the first position (>85%; Figure 1A). Similarly, Ago1-associated siRNAs are primarily 22–23 nucleotides (nt) in length (Figure 1B; Buhler et al, 2008), whereas our WT small RNAs range in size from 20 to 30nt, with most being between 22 and 25nt (Figure 1B). These data indicate that Ago1 has a preference for 22–23-nt siRNAs that begin with U, and that it selects these siRNAs from a more diverse population generated in the cell. Interestingly, in the rpb7G150D mutant, there is a shift in the size of small RNAs with a larger proportion of short small RNAs than in the WT sample, combined with a lower preference for small RNAs that begin with U (Figure 1A and B).
Table 1.
Compilation of 454 deep sequencing analysis of small RNA from wild-type (WT) and the rpb7G150D mutant strain
| Total number of reads | Genome- matching readsa | Filtered matching readsb | Genome matches/ read | |
|---|---|---|---|---|
| 972 (WT) | 21776 | 16341 | 3531 | 4.9 |
| rpb7G150D | 51894 | 36990 | 11081 | 1.5 |
| Ago1-associated siRNAsc | 349251 | 257629 | 197170 | 6.3 |
| aReads with a perfect match in the Schizosaccharomyces pombe genome. | ||||
| bGenome-matching reads after removal of structural RNAs (tRNA, rRNA, snoRNA, and snRNA). | ||||
| cData are extracted from a study by Buhler et al (2008); GEO accession number: GSE12416. | ||||
Figure 1.
Characteristics of the small RNA populations of wild-type and rpb7G150D mutant cells in comparison to Ago1-associated siRNAs (Buhler et al, 2008). Small RNAs without a perfect match in the S. pombe genome or those matching tRNA and rRNA were removed. (A) Start nucleotide distribution of small RNA. The 5′ nucleotide on the x-axis and the relative abundance of small RNAs on the y-axis in wild-type and rpb7G150D mutant cells compared with Ago1-associated siRNAs. (B) Size distribution of small RNA, length in nucleotides on the x-axis and relative abundance on the y-axis in wild-type, rpb7G150D mutant cells and Ago1-associated siRNAs. (C) Percentages of genomic distributions of small RNA reads in wild-type cells, rpb7G150D mutant cells and Ago1-associated siRNAs.
Excluding sequences from transcribed structural RNAs, which are likely degradation products, the genomic regions that generate most small RNAs are centromeric repeats and protein-coding genes (Figure 1C). At the centromeres, hundreds of small RNAs cluster within the dg and the dh elements. In WT, 100 genes had three matching small RNA reads or more. Nearly all of these genes (98 genes) were also retrieved in the small RNA sample from the rpb7G150D mutant. The rpb7G150D sample, which generated nearly three times as many sequences as the WT sample in combination with a depletion of centromeric small RNAs, resulted in retrieval of over 600 genes with matching small RNA (cut-off: ⩾3 small RNAs per gene). All small RNAs towards genes were of the sense orientation, with the exception of the tlh1 gene (SPAC212.11) that has homology to the dh repeat and in which small RNAs of both orientations were abundant. The most likely explanation is that these sense-oriented small RNAs represent mRNA degradation products. However, Dcr1, Ago1, and Rdp1 were shown to mediate post-transcriptional silencing of an exogenous hairpin (Sigova et al, 2004). Furthermore, investigations of the proteome of dcr1Δ cells demonstrated increased protein levels of Hsp16, Pgk1, Tpx1, and Hsp104, whereas protein levels of Hxk2, Eno1, and Thi3 were decreased, with or without concomitant alterations of mRNA levels (Gobeil et al, 2008). We detected small RNAs homologous to six of these genes (Supplementary Figure S1). Thus, it is possible that these sense small RNAs somehow contribute to gene regulation. Although we did not carry out a systematic analysis of small RNAs in intergenic regions, we did notice 14 copies of a single small RNA in the intergenic downstream region of the convergent gene pair mei4+–act1+ (Supplementary Figure S1). This locus has been reported to be coated with heterochromatin factors (Cam et al, 2005) and these small RNAs are probably involved in controlling transcription termination in this intergenic region, as suggested by Gullerova and Proudfoot (2008).
Characterization of centromeric small RNA clusters
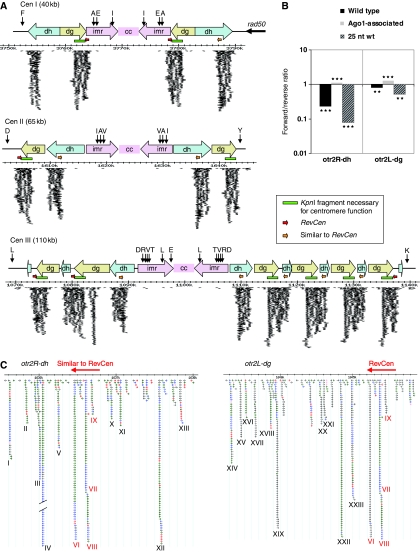
All clusters abundant for small RNA overlap the dg and dh elements that constitute the centromeric outer repeats (Figure 2A). These clusters are between 2000 and 3000 bp long with up to 1000 matching sequence reads. In general, there are two recurring sequences that produce small RNAs in the centromeres. The dg cluster overlaps the 3′-end of the dg repeat, including the fragment necessary for centromere function (Baum et al, 1994). The dh cluster is situated within the second half of the dh repeat. Alignment of these sequences from all centromeres reveals a high degree of conservation across both the dg and the dh clusters (Supplementary Figure S2A and B). The dg element has previously been reported to have 97% homology between different centromeres, whereas the homology of the dh element from centromere I, II, and III was reported to have 48% identity (Wood et al, 2002). If deletions of the dh elements are taken into account, the remaining dh sequences have up to 99% sequence identity between dhI and dhII (Nakaseko et al, 1987). The clusters are found at all centromeres. A 300-bp translocation from dh to dg (Chikashige et al, 1989) is common to both the dg and dh clusters.
Figure 2.
Distribution of small RNA of wild-type cells at the centromeres. (A) Schematic on-scale representation of the centromeres of chromosome I, II and III based on the S. pombe GeneDB. Vertical black arrows represent tRNAs with standard one letter abbreviations representing amino-acid specificity. The green bars represent the KpnI restriction fragment that was shown to be necessary for centromere function (Baum et al, 1994). Red horizontal arrows represent RevCen; orange horizontal arrows represent the region within the dh element with a translocation similar to RevCen. A 700-bp region is deleted from the dh cluster at the left arm of centromere I and the sequence in between dg and dh siRNA clusters is deleted in centromere III. Each arrow represents a sequenced, small RNA match; thicker arrows indicate multiple sequenced small RNAs. Owing to the high degree of sequence similarity, most sequenced centromeric small RNA have perfect match to several clusters within the centromeres. These sequences have been plotted at each perfect match, that is, more than once per sequence. (B) Histogram of small RNA strand distribution at the otr2R-dh and otrR2-dg siRNA clusters. The x-axis depicts ratio of forward to reverse strand siRNAs (log10 scale). All bars are significantly different from 1; ***P<0.001; **P<0.01. (C) Representation of typical centromeric siRNA clusters, here exemplified by the clusters at otr2R-dh and otr2L-dg. Matching sequenced small RNAs are represented by arrows according to their position, orientation and length; pink=14–19 bases, red=20–21 bases, green=22–23 bases, blue=24–25 bases, and grey >25 bases. Hotspots of siRNAs have been named in roman numerals, the most abundant siRNA, ‘IV', was sequenced 400 times and has been cropped. The red horizontal arrow depicts the location of the RevCen sequence and roman numerals in red indicate the siRNA hotspots that match to both dg and dh siRNA clusters.
Although small RNAs match both strands of the centromeric clusters, there is a preference for small RNAs from the reverse strand of the repeat, with stronger bias at the dh cluster than at the dg cluster (Figure 2B). No strand bias is expected according to the current models for RNAi-dependent heterochromatin formation at the centromeres in S. pombe because cleavage of Rdp1-derived dsRNA will produce equal amounts of plus- and minus-stranded siRNAs. Interestingly, a direct comparison with Ago1-associated siRNAs (Buhler et al, 2008) from these clusters showed no minus strand bias. Instead there was a small but significant enrichment for the positive strand. Consistent with this, 25nt small RNAs from WT, which are unlikely to associate with Ago1 due to the size, show the strongest minus strand bias (>10-fold).
The accumulation of small RNAs is not equal across the clusters, resulting in several hotspots of small RNAs. The 26 most abundant small RNA hotspots were named with roman numerals (Figure 2C). The most abundant centromeric small RNA hotspot, small RNA IV, was sequenced 400 times, with the length of the small RNA varying by a few nucleotides. The GC content of the small RNAs is significantly higher than in the surrounding sequence (Supplementary Figure S3). As the small RNAs are homologous to regions in which the sequence is identical in most or all copies of the dg and dh elements, it is not possible to determine from which centromeric repeat copy these small RNAs originate.
Hotspot small RNAs are Dcr1 dependent, carry a 5′-monophosphate, and are associated with Ago1
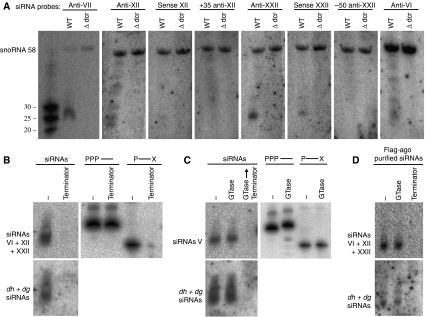
To verify the occurrence of small RNA hotspots, single oligonucleotide probes corresponding to four of the most abundant sequenced small RNAs were synthesized and used as probes on northern blots of small RNA preparations. In addition, control sense oligonucleotides or oligonucleotides homologous to neighbouring sequence with few matching small RNAs were used. In accordance with the small RNA distribution determined by sequencing, the four hotspot small RNA probes (anti-VII, anti-XII, anti-XXII, and anti-VI) readily detected small RNAs by northern analyses, whereas little or no signal was detected with sense probes (sense XII and sense XXII) or nearby control probes (+35-anti-XII and 50-anti-XXII). No signals were detected in small RNA preparations from dcr1Δ cells (Figure 3A).
Figure 3.
Validation and analyses of siRNAs by northern blots. (A) Single oligonucleotide probes, antisense, sense, or nearby to siRNAs IV, XII, XXII and VI were used for detection of small RNAs from wild-type or dcr1Δ cells. SnoRNA 58 was used as loading control. (B) and (C) Analyses of 5′-termini of small RNAs by enzymatic reactions followed by northern blots. (B) Terminator exonuclease digests monophosphorylated 5′-termini and (C) guanylyltransferase (GTase) caps di- or triphosphorylated 5′-termini. The control oligonucleotides are 5′-triphosphorylated RNA oligonucleotide (PPP—) and 5′-monophosphorylated RNA oligonucleotide with a blocked 3′-end (P—X) (Ule et al, 2005). The blots were probed sequentially with specific oligonucleotide probes and with a random-primed probe spanning the whole siRNA cluster (dh +dg siRNAs). (D) Analysis of Ago1-purified siRNA 5′-ends by enzymatic reactions followed by northern blot as described above.
Sequencing of Ago1-associated siRNAs in S. pombe indicates that 5′-monophosphate siRNAs are present (Buhler et al, 2008). Our analysis corroborated this as we prepared small RNA libraries in a 5′-monophosphate-dependent manner. However, in Caenorhabditis elegans, 5′-monophosphate siRNAs are a minority. The majority of siRNAs have been reported to have 5′-triphosphates in accordance with being products of an RNA-directed RNA polymerase (Pak and Fire, 2007; Sijen et al, 2007). To determine if 5′-triphosphate siRNAs were also present in S. pombe, we treated small RNAs with Terminator exonuclease, a 5′-exonuclease that digests RNA with 5′-monophosphates. Small RNAs completely disappear from the centromere after treatment with Terminator exonuclease, whereas a 5′-triphosphate control oligonucleotide was unaffected (Figure 3B). Furthermore, small RNAs were not capped by guanylyltransferase (GTase) that caps 5′-di- or triphosphorylated RNA and produces approximately two-nucleotide slower gel migration. In addition, GTase-treated small RNAs were digested with Terminator exonuclease, which cannot digest if a 5′-cap is present (Figure 3C). Finally, in RNA samples purified from Ago1–FLAG (Buhler et al, 2007), single oligonucleotide probes, as well as random-primed probes that detect all small RNAs, detected small RNA with 5′-monophosphates (Figure 3D). These data indicate that sequenced S. pombe centromeric small RNAs reported here are Dcr1 dependent, possess 5′-monophosphates, and associate with Ago1 in vivo and thus seem to be true siRNAs.
The 5′-end of the transcript from the dg cluster forms a partially double-stranded RNA structure and is processed by human recombinant Dicer in vitro
The high degree of sequence identity at the centromeric dg and dh elements could be caused by frequent events of homologous recombination. Alternatively, functional conservation could maintain important features of the sequence, such as the ability to form secondary structures. Within the transcripts from the dg and dh clusters, siRNAs are derived from several hotspots with intervening cold spots. One possible explanation is that the transcript itself folds into a secondary structure that provides a substrate for siRNA generation. The hotspots would correspond to double-stranded regions whereas the cold spots would represent unstructured regions, and the preservation of such a structure would presumably be selected for in all copies of these repeats. We decided to investigate the hypothesis that transcripts from the centromeres can form double-stranded structures, which could be processed by Dcr1, and thereby initiate siRNA production at the centromeres. To test this, we initially used M-fold (calculates secondary structures with minimal free energy) to predict the presence of structured RNA (Zuker, 2003). These analyses encouraged us to directly test for secondary structure and we selected the 5′-end of the transcript originating from the dg cluster for secondary structure probing. The in vitro transcribed fragment, which we call RevCen, is 432-nt long and starts immediately downstream of a strong promoter that has been previously characterized (Djupedal et al, 2005). High-throughput sequencing data from the rpb7G150D mutant, in which transcription from the promoter is severely compromised, reveal a sharp reduction in the number of centromeric siRNAs, including the downstream dg cluster and the RevCen fragment (Figure 4A and B). The promoter was originally mapped to the left arm of centromere I, within the imrIL element. However, the promoter region is perfectly conserved, present and probably functional at all outer repeats except from the otr2R-dg (Figure 4C). The RevCen fragment traverses a 300-bp translocation (Chikashige et al, 1989) from within the dh cluster (Figure 4D), and siRNAs originating from this extremely well-conserved sequence are complementary to 10 out of 12 dg and dh clusters (Figure 4D). The WT levels of transcription of the reverse strand of the dg cluster, therefore, seem to be necessary for accumulation of siRNAs from both strands of the DNA.
Figure 4.
The RevCen fragment. (A) Representation of siRNA from wild type (WT) and rpb7G150D cells at otr2L-dg according to their position and orientation. (B) Histogram of siRNA size distributions from WT and rpb7G150D cells, siRNA length in nucleotides on x-axis, and percentage of number of siRNA on y-axis. (C) Sequence alignment of the RevCen promoter region characterized in Djupedal et al. (2005) in the dg repeats from all centromeres. Coordinates are: otr1L-dg from 3752406, otr1R-dg from 3778228, otr2L-dg from 1604242, otr3L-dg from 1073094, and otr3R-dg from 1137620. The region is not present in otr2R-dg. The arrow indicates the direction of transcription. (D) Sequence alignment showing the 340-nt translocation present in all dh and dg elements from the three centromeres, one dh and dg repeat per chromosome arm is shown as they are identical. The siRNAs VII and VIII are shown in boxes with orientation. Coordinates are: otr1R-dh from 3781697, otr2L-dh from 1611493, otr2R-dh from 1636391, otr3L-dh from 1088967, otr3R-dh from 1109980, otr1L-dg from 3763484, otr1R-dg from 3784652, otr2L-dg from 1604792, otr3L-dg from 1082192, and otr3R-dg from 1116691. The region is not found in otr1L-dh and otr2R-dh.
The secondary structure of the 432-nt long RevCen RNA was assessed by enzymatic and chemical probing. The ribonucleases T1, T2, and V1 cleave after unpaired guanosines, unpaired adenosines and paired/stacked nucleotides, respectively. Lead (II) acetate cleaves the RNA backbone in unstructured regions without sequence preference. Partial cleavages were obtained using end-labelled RevCen (Figure 5A). The method gave structural information covering the 5′-most 350 nucleotides, consistent with a partially double-stranded hairpin-like structure (Figure 5B). The siRNA hotspots VIII and VII, of which siRNA VII has been validated by northern analysis in vivo (Figure 3A), map within the double-stranded regions of RevCen, and are indicated in Figure 5B. Less abundantly sequenced siRNAs (<5 reads per sequence) map along the basic stem loop. The siRNAs VI and IX correspond to the opposite strand of the DNA and are not labelled in the figure. Next, we wanted to test whether this sequence could be a substrate for Dcr1-mediated cleavage. Indeed, incubation of internally labelled, in vitro transcribed RevCen with recombinant Dicer leads to the appearance of small RNA species of the expected sizes (Figure 5C). Hence, the 5′ region of the siRNA precursor from the dg cluster has the ability to fold into a partially double-stranded secondary structure that is recognized and processed by human recombinant Dicer in vitro.
Figure 5.
Secondary structure determination of RevCen. (A) Secondary structure probing of in vitro transcribed, [γ-32P]ATP 5′-end-labelled RevCen RNA analysed on polyacrylamide gels. Partial RNA cleavages were performed as described in Materials and methods section. An OH ladder and a T1 ladder were used to assess cleavage positions. The positions of G residues are marked on the right. (B) Refined prediction of the RevCen secondary structure using the Mfold software using constraints from structural probing data in (A). The siRNAs VII and VIII are indicated in light and dark orange. The grey shade indicates the part of RevCen, which could not be resolved. Nucleotides are highlighted with different colours to show probe-dependent cleavage, as indicated in the colour key. (C) Cleavage of RevCen by human Dicer in vitro. RNA fragments ranging from 21–33 nt were formed when treating RevCen with recombinant human Dicer. RNA was labeled after incubation. The increasing dose of Dicer is indicated. Incubation times were as follows: 5 min for lanes 1 and 4; 30 min for lanes 2 and 5; 2 h for lanes 3, 6 and 7. Concentrations of Dicer: lane 7: 0 units; lanes 1–3: 0.1 units and lanes 4–6: 1.0 units.
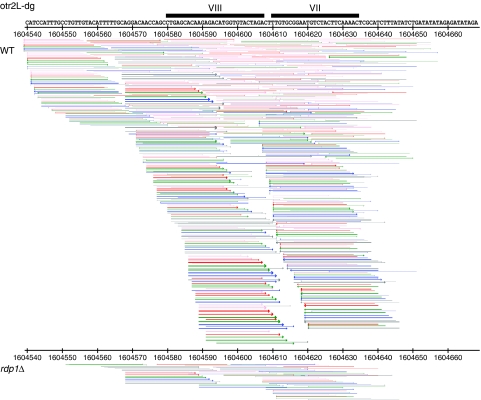
If Dcr1 processing of primary transcripts contributes to centromeric silencing in vivo, small RNAs should be synthesized independently of RDRC activity. Deep sequencing of small RNAs in rdp1Δ cells with an Illumina Genome Analyzer (F Simmer et al, unpublished data), reveals the presence of small centromeric RNAs (Figure 6). The total number of centromeric small RNAs are drastically reduced as compared with WT (Supplementary Figure S4), which reflects the degree of amplification of the siRNA signal maintained by Rdp1 in WT cells. A fraction of centromeric small RNAs matching both strands of the DNA are synthesized independently of Rdp1, of which a few correspond to the experimentally determined hairpin in RevCen. These siRNAs show base composition and size distribution similar to WT and rpb7G150D (Supplementary Figure S5). Furthermore, the distribution pattern over the centromeric clusters shows peaks and deserts similar to WT but with lower amplitude (compare Supplementary Figure S6 and S7). Thus, small RNA produced in rdp1Δ cells resembles those of WT cells.
Figure 6.
Reduction of centromeric siRNAs in rdp1Δ cells compared with wild type (WT), here displayed at otr2L-dg. Representation of the RevCen stem loop, in sense orientation, containing the siRNAs VII and VIII. Matching sequenced siRNAs in wild type and in rdp1Δ are represented by arrows according to their position, orientation, and length; pink=14–19 bases, red=20–21 bases, green=22–23 bases, blue=24–25 bases, and grey >25 bases. Each arrow represents a region with a matching siRNA; thicker arrows indicate regions with multiple siRNAs on a log scale.
Production of small RNA from centromeric clusters is independent of heterochromatin
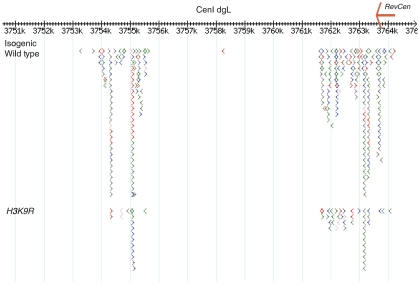
Small RNA production from centromeres has been suggested to result from targeting of RITS/RDRC through interactions between the chromo-domain of the RITS component Chp1 and H3K9me (Petrie et al, 2005). As Rdp1 is not required for low levels of siRNA production, we tested whether H3K9me was also dispensable for this process. We used a strain carrying a point mutation in the histone H3 gene (H3K9R), which abolishes methylation of lysine 9 (Mellone et al, 2003). This strain and isogenic WT were subjected to high-throughput 454 sequencing and the centromeric regions were examined (Figure 7). The siRNAs could be detected using this method but there was a clear relative reduction versus WT similar to that observed for rpb7G150D. Hence siRNAs are produced even in the absence of heterochromatin.
Figure 7.
Detection of centromeric siRNAs in H3K9R cells as compared with isogenic wild type at cen I otr1L-dg. Each arrow represents a sequenced RNA, colour code as in Figure 2C. Position within chromosome on x-axis is given in kb. Location of RevCen is marked with red arrow.
Discussion
Small RNAs mediate complex networks of gene regulation in plants and animals. The yeast S. pombe has one of the most basic RNAi systems, which function in both post-transcriptional and transcriptional gene silencing. S. pombe is an established model organism for the study of the latter. Here we present a deep sequence analysis of small RNA in S. pombe. As in previous analyses of Ago1- or RITS-associated siRNAs (Cam et al, 2005; Buhler et al, 2008), large fractions of WT small RNAs match the repetitive regions of the centromeres and protein-coding genes. The deep sequence analysis of small RNA from the rpb7G150D mutant strain resulted in a larger number of sequences and relatively few small RNA from the repetitive regions of the centromeres, and thus more genes with matching small RNA were found. Nearly all protein-coding genes with small RNA matches in the WT sample also match small RNAs in the mutant sample, which supports the existence of these molecules in vivo. It has been shown previously that most siRNA-matching genes are sense to gene direction (Cam et al, 2005; Buhler et al, 2008). In this study, all small RNAs that matched genes were sense to gene direction, indicating that they may be processed from mRNA transcripts. It has been reported that siRNAs have an additional role in the regulation of transcriptional termination at convergent transcribed genes (Gullerova and Proudfoot, 2008). Double-stranded RNA was detected and transient RNAi-dependent heterochromatin was shown to form in the intergenic region of three convergently transcribed gene pairs. Under the growth conditions used here, both genes are actively transcribed at only one of these gene pairs and we detected small RNAs at this gene pair (Supplementary Figure S1). However, more specific conditions may be required to detect more small RNAs at such regions, for example, before, during, and after the S-phase. The deep sequence analysis of small RNA from S. pombe presented here is not likely to be saturated. A higher number of sequenced small RNA from WT cells should reveal rare small RNAs that are not covered by this analysis. Furthermore, RNA samples from different growth conditions or from cells that are synchronized in different phases of the cell cycle would probably generate different small RNA profiles as a reflection of the active regulatory mechanisms. Being a genetically tractable organism, S. pombe may also serve as an excellent model for understanding the basis of post-transcriptional RNAi.
The repetitive, heterochromatic centromeres of S. pombe were previously shown to be transcribed and abundant for siRNAs (Volpe et al, 2002; Cam et al, 2005; Sugiyama et al, 2005; Buhler et al, 2008). We provide a detailed description of these centromeric sequences. Two types of small RNA clusters, 2.1- and 2.3-kb long and overlapping the dg and dh elements, are reiterated two or more times at each centromere and account for most of the small RNAs in WT cells. A promoter upstream of the dg cluster has been characterized and northern blots probed for dg or dh reveal centromeric transcripts to be up to 2.4 kb in size (Volpe et al, 2002; Djupedal et al, 2005). Therefore, these clusters seem to correspond to transcription units, whose transcripts are processed into siRNAs. Furthermore, the dg cluster traverses the 2.1-kb fragment that was demonstrated to be required for centromere function (Baum et al, 1994). In conclusion, only specific regions of the dg and dh elements are transcribed and subsequently processed into siRNAs, and at least one such region is required for centromere function.
A 300-bp translocation is common to both small RNA clusters and most siRNAs are found in regions of perfect, or near perfect, sequence identity in between dg and dh elements from the centromeres. It would therefore be possible for one such siRNA to recruit RITS in trans to all peri-centromeric heterochromatin loci in S. pombe. Consequently, siRNA production at one cluster could suffice to direct formation of heterochromatin to all centromeres. Trans-activity of siRNAs could be facilitated by the clustering of the centromeres throughout the cell cycle (Funabiki et al, 1993; Appelgren et al, 2003), causing accumulation and high local concentrations of centromere-specific siRNAs at these compartments. If trans-activity is an important function for centromeric siRNAs in S. pombe, high degrees of sequence identity between dg and dh elements at the centromeres would be selected and maintained.
How are centromeric transcripts processed into siRNA? Each type of cluster has a distinct pattern of siRNAs. There is a non-random distribution of siRNAs within the clusters and siRNAs have significantly higher GC content than the surrounding sequence (Supplementary Figure 3). This may indicate that the stability of base pairs formed is relevant for the selection or accumulation of siRNAs. Detailed analysis of the deep sequencing data reveals accumulation of small RNAs with the same basic sequence, but of variable length due to different start and/or stop nucleotides. This indicates that S. pombe Dcr1 does not cleave at specific nucleotides. The dcr1 gene lacks the PAZ domain, a typical feature of Dicer proteins. The distance between the PAZ and the RNase III domains determines the length of the siRNAs (Lingel et al, 2003; Macrae et al, 2006). The size specificity of Dcr1 was, therefore, suggested to be determined by additional factors in vivo (Colmenares et al, 2007). We have observed a few examples of sequential Dcr1 cleavage, in which the 3′-end of one abundant siRNA is adjacent to the 5′-end of another siRNA. Similarly, we have observed examples of siRNAs of opposite orientation with the typical two-nucleotide 3′ overhang that is expected of Dcr1 cleaving double-stranded siRNA precursors. However, there does not seem to be a general defined sequential cleavage of siRNA precursors by Dcr1, as there is very little accumulation of phased siRNAs or siRNA passenger strands from the centromeres. Instead, we observe a few specific siRNAs from within the clusters that accumulate in high number. In conclusion, highly abundant siRNAs neighbour regions with no or very few siRNAs.
Are there different classes of small RNAs in S. pombe? Unlike the two previous studies, which reported RITS-associated small RNA to be 20–22 nt long (Cam et al, 2005) and Ago1-associated small RNA to be 21–23 nt long (Buhler et al, 2008), we found the size distribution of total, WT small RNA to be between 21 and 25 nucleotides (Figure 1A). The size distribution of small RNAs from the rpb7G150D sample has a statistically significant increase in small RNAs ⩽21 nt versus small RNAs ⩾22 nt as compared with the WT sample. This could, in part, be due to the depletion of centromeric siRNAs that are predominantly 22–23 or 25 nt in length in the WT (Figure 4B). The difference in size between the WT small RNAs from our study and siRNAs reported to associate with Ago1 could be due to different experimental techniques. However, Ago1-associated siRNAs were also reported to have a much stronger bias towards uridine as the 5′-starting nucleotide (Buhler et al, 2008). The smaller size and 5′-uridine may be preferred features for Ago1 loading and/or retention. We have, however, validated the most frequently sequenced centromeric small RNAs by northern blots and also detected these in association with Ago1. As described above, small RNAs of different lengths are produced from the same sequence and northern analyses of centromeric siRNAs from several studies show siRNA signals of up to 25 nt in S. pombe (Motamedi et al, 2004; Li et al, 2005; Buhler et al, 2006, 2007). If 21–23 nt length and 5′-uridines are preferred by Ago1, we conclude that small RNAs of variable size and start/stop nucleotide are synthesized and accumulate in WT S. pombe cells.
In C. elegans, two classes of siRNAs have been reported: rare primary siRNAs that are generated by Dicer cleavage and abundant secondary siRNAs that are short products of an RNA-directed polymerase, and as such have triphosphorylated 5′-termini (Pak and Fire, 2007; Sijen et al, 2007). As in C. elegans, the RNA-dependent polymerase Rdp1 is necessary for detectable siRNA accumulation and transcriptional silencing in S. pombe (Volpe et al, 2002; Motamedi et al, 2004; Sugiyama et al, 2005). To determine whether S. pombe siRNAs are synthesized in the same pathways as in C. elegans, we investigated the structure of the 5′-termini of small RNAs and found the majority of small RNAs to have 5′-monophosphate groups. Therefore, most small RNAs in S. pombe seem to be products of Dcr1 cleavage of double-stranded siRNA precursors. We conclude that unlike the RNA-directed RNA polymerases in C. elegans, which generate several short transcripts along the length of the RNA template, Rdp1 of S. pombe predominantly generates full-length double-stranded RNA that is subsequently cleaved into siRNAs by Dcr1. If there are primary and secondary siRNAs in S. pombe, these have routes of synthesis different from those in C. elegans.
The reduction in size of small RNAs in the sample from the rpb7G150D mutant may, however, indicate that there are different classes of small RNAs in S. pombe, the ‘longer' small RNAs, 24–25 nt in length, which are produced through transcription of centromeric repeats, and shorter small RNAs that are produced through a distinct pathway. We suggest that primary siRNAs could be produced by direct Dcr1-mediated cleavage of nascent transcripts that are folded into hairpin-like structures, whereas secondary siRNAs originate from Dcr1-mediated processing of Rdp1-generated double-stranded RNA. This would be similar to the production of trans-acting (ta-)siRNAs in Arabidopsis thaliana, whereby miRNA cleavage triggers production of dsRNA and ta-siRNAs (Chapman and Carrington, 2007). Unlike miRNAs, which cleave the ta-siRNA precursor transcript in trans, primary siRNAs in S. pombe could function both in cis and in trans. For example, siRNA VII and VIII could represent primary siRNAs whereas siRNA VI and IX, which are of the opposite orientation, could be secondary siRNA products (Figure 2A). In WT, there was a bias for centromeric siRNAs from the more highly transcribed reverse strand, whereas siRNAs associated with Ago1 (Buhler et al, 2008) were slightly biased for the opposite strand. The shift in strand bias could be due to differences in routes of synthesis whereby Dcr1-mediated cleavage of nascent transcripts to generate minus-strand-biased primary siRNAs might occur before Ago1 localization to these loci and subsequent production of secondary siRNAs from both strands. Alternatively, production of primary siRNAs by cleavage of the nascent transcript may occur at some distance from the chromatin and be unavailable for association with Ago1. The slight bias for the positive strand among Ago1-associated siRNAs is logical if the reverse strand, which accumulates more highly, is the primary target for Ago1-mediated transcript cleavage.
Do transcripts from centromeric small RNA clusters form double-stranded secondary structures? In plants and animals, miRNAs are processed from transcripts that fold back on themselves to form double-stranded hairpin structures. In S. pombe long inverted repeats form hairpin structures that are processed into siRNAs, which mediate post-transcriptional (Raponi and Arndt, 2003), as well as transcriptional gene silencing (Iida et al, 2008). By using the M-fold algorithm on the first 500 nucleotides from the dg cluster, a hairpin-like structure was predicted and we therefore decided to experimentally determine the secondary structure of the transcript. Owing to the difficulty of experimentally determining the full length transcript of >2 kb, we selected for structural probing a 432-nt sequence immediately downstream of the characterized promoter that traverses the translocation common to both types of small RNA clusters. The tentative secondary structure of the 350-nt region that was resolved was partially double-stranded and reminiscent of the structures of the pre- and pri-miRNA substrates of Dicer and Drosha. This sequence was recognized by human recombinant Dicer and processed into small RNA in vitro. Furthermore, small RNAs from within the double-stranded structure are present in rdp1Δ cells. This demonstrates a route of synthesis that does not involve secondary strand production by this RNA-directed polymerase, but rather by Dcr1 cleavage of primary transcripts. These primary transcripts may be folded into hairpins or, alternatively, primary transcripts from opposite strands may be paired. We suggest the former alternative, which is in accordance with the reported strand bias of centromeric small RNAs. This model is consistent with the finding that siRNA can be produced at low levels even in the absence of heterochromatin, as RITS/RDRC targeting by H3K9me would not be required. All that would be necessary for siRNA production is transcription of centromeres by RNA pol II, RNA folding, and Dicer processing.
Finally, there is strong experimental support for the current model of a positive feedback loop (Noma et al, 2004; Sugiyama et al, 2005), in which siRNAs are produced from Rdp1-generated double-stranded siRNA precursors in cis to transcription. We propose that, in addition to the current model, information in the primary sequence of the DNA may help de novo formation of heterochromatin in situations in which both H3K9me and centromere-specific siRNA are lost. The half-life of siRNAs in S. pombe is not known, and if these are only produced in a short window of the cell cycle after replication of the DNA (Chen et al, 2008; Kloc et al, 2008), it is plausible that there may be depletion of siRNAs when cells have been in stationary phase for long periods of time or when spores are re-entering the cell cycle after quiescence. It remains to be proven, however, that the secondary structure formed in vitro has a biological role in heterochromatin formation in vivo.
Materials and methods
Yeast strains and medium
Yeast strains, listed in Supplementary Table S1, were grown in YEA medium and were collected in log phase.
Small RNA library preparation. Total nucleic acid was extracted from cells in log phase, and purified according to the method given by White and Kaper (1989). The siRNAs (15–30 nt), from 500 μg of total nucleic acid, were excised from a 15% denaturing acrylamide gel (0.5 × TBE, 0.42 g/ml urea, 15% 19:1 acrylimide:bisacrylimide). After elution from the gel (twice incubated with rotation at 4°C in 0.3 M NaCl for several hours), small RNAs were precipitated with glycogen, sodium acetate and ethanol. A 5′-adapter (TGGGAATTCCTCACTaaa, lowercase bases are RNA) was ligated to the small RNA (20 μM adapter, 15% DMSO, 50 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 10 mM 2-ME, 0.2 mM ATP, 0.1 mg/ml BSA, 1 U/μl T4 RNA ligase and 1 U/μl RNase inhibitor) before gel extraction (30- to 50-nt fragments) and elution as above. After precipitation, a 3′ adapter (uuuCAATCCATGGACTGT) was ligated in the same manner, gel extracted (50–70 nt), eluted and precipitated. The adapter-ligated small RNA was subjected to reverse transcription using standard protocols and SuperScript RT II (Invitrogen). Reverse transcription reactions were stopped by addition of 150 mM KOH, 20 mM Tris base and neutralized with HCl (final volume approximately 180 μl). 50 μl of this was used as template for PCR (5′-GCCTCCCTCGCGCCATCAGTGGGAATTCCTCACT-3′ and 5′-GCCTTGCCAGCCCGCTCAGACAGTCCATGGATTG-3′). PCR products were gel extracted on a native polyacrylamide gel (0.5 × TBE, 10% 19:1 acrylimide:bisacrylamide). The 90–105-nt products were excised and eluted. Purified amplicons were sent to 454 Life Sciences for pyrosequencing.
Sequence analysis. Raw sequences were searched for adapter motifs. Where both adapters were identified, the sequence in between was extracted and positioned on the S. pombe genome (GeneDB) using a local alignment script. Only small RNAs with a perfect genomic match were included in further analyses and small RNAs matching the transcribed strand of tRNA, rRNA, or other structural RNAs were removed from further analyses. Genome annotation was collected from geneDB, http://www.sanger.ac.uk/Projects/S_pombe. Analyses of small RNA size and strand bias were performed using Perl scripts (available on request). Sequence alignments were performed using DNAMAN 4.13.
Northern blots
The RNA extraction was carried out as described above. Small RNAs were concentrated by precipitating away large RNAs with 7% PEG 8000, 0.5 M NaCl, and then by ethanol precipitation.
For Ago1-associated small RNA preparation S. pombe cultures (3 × Flag-ago1:KanMX6) were grown to a cell density of 108 cells per ml in 4 × concentrated YES medium. For each sample 10 g of cells were lysed in solid phase in the presence of liquid nitrogen using a mortar grinder (Retsch) for 30 min. Extracts were prepared by dilution of the crushed cells in 20 ml lysis buffer (50 mM HEPES-NaOH (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 0.1% NP-40, 5 mM DTT, yeast protease inhibitors (Sigma), Superasin (Ambion), and 0.2 mM PMSF), and filtered through GD/X 1.6 μm (Whatman). Immunoprecipitations were performed using 20 μg of M2 anti-Flag antibody (Sigma) coupled to 4 μl protein G Dynabeads resin (Invitrogen) for 15 min. The RNA samples bound to Dynabeads were washed with lysis buffer, treated with 200 ng/ml proteinase K (Sigma) for 2 h in TENS/2 buffer (25 mM Tris–HCl (pH 7.5), 5 mM EDTA, 50 mM NaCl, and 0.5% SDS) at 37°C, extracted with phenol/chloroform (5:1; pH 4.7), and ethanol precipitated.
Enzymatic reactions: the small RNAs were further purified on 12% polyacrylamide, 8 M urea gel in 1 × TBE, eluted, precipitated and desalted. Enzymatic reactions were performed according to the manufacturer's instruction: guanylyltransferase (Epicentre), Terminator exonuclease (Epicentre) and T4 RNA ligase (New England Biolabs).
Northern blot: the RNAs were run on denaturing polyacrylamide gels as above, electrotransferred onto Hybond-NX membranes (Amersham) and UV-cross-linked. Membranes were hybridized overnight at 42°C in 0.5 M NaPO4 (pH 7.2), 7% SDS, and 1 mM EDTA. Probes were either radioactively phosphorylated oligos or random-primed dh or dg PCR fragments (see Supplementary Table S2 and S3). Membranes were washed twice at 42°C in 2 × SSC, 0.2% SDS.
Structural probing
Genomic DNA was used as template for amplification of the RevCen sequence (432 bp) from chromosome I using High Fidelity Expand Polymerase (Roche Applied Science) and the forward primer containing a T7 RNA polymerase promoter (underlined); RevCenF (5′-GAAATTAATACGACTCACTATAGCGGTTTTCATTGTGTATCATCTTCCTGG-3′). Reverse primer: RevCenR (5′-ATGGTACCAAAGCTCGAACATAGAAAGAAATCC-3′). The PCR product was purified using a Qiagen gel purification kit.
The PCR product carrying the T7 promoter was in vitro transcribed using T7 RNA polymerase (Ambion). Removal of the DNA template was performed using DNase I Amp Grade (Invitrogen) and stopped with 2.5 mM EDTA. The RevCen RNA was purified on an 8% polyacrylamide, 7 M urea, 1 × TBE gel. The RNA was excised and eluted from the gel in a shake incubator overnight at 4°C. The eluted RNA was precipitated and re-suspended in 50 μl RNase-free water. About 10 pmol of RevCen RNA was dephosphorylated by 1 U of shrimp alkaline phosphatase (Fermentas), phenol-extracted and precipitated. About 10 pmol dephosphorylated RevCen RNA were incubated in 10 U T4 polynucleotide kinase (PNK) (Fermentas) and 20 μCi [γ-32P]ATP (Perkin Elmer) for 15 min at 37°C. The labelled RNA was purified from an 8% denaturing polyacrylimide gel as described above. Secondary structure probing was performed using 5′-end-labelled RevCen RNA. For each reaction, 0.1 pmol RNA was denatured before addition of 1 μg yeast tRNA (Ambion) and TMN buffer (to a concentration 20 mM Tris–Cl (pH 7.6), 100 mM Na acetate, 5 mM Mg acetate) to a final volume of 10 μl. The reaction mixture was incubated for 5 min at 30°C before addition of ribonuclease T1, V1 (Ambion), or T2 (Invitrogen), or lead (II) acetate (Merck). Ribonucleases were used at concentrations of 2 × 10−3 and/or 4 × 10−3 U/μl for 5 min at 30°C. Lead (II) acetate, prepared fresh, was added in a final concentration of 2 mM and incubated for 30 s, 1 min and 5 min at 30°C. All reactions were stopped by the addition of 50 mM EDTA and 1 volume of denaturing loading dye. T1 ladders (G-specific cleavages) used as markers were obtained under denaturing conditions (Brunel and Romby, 2000), and alkaline hydrolysis ladders were obtained according to the manufacturer's protocol (Ambion). Samples were analysed on 6, 8, or 15% denaturing polyacrylamide gels. Gels were dried and exposed to PhosphoImager screens, and analysis was performed using ImageQuaNT software.
Sequence analysis and RNA secondary structure prediction
RNA secondary structure predictions were performed using Mfold version 2.3 with constraints based on the results from structural probing.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table I
Supplementary Table II
Supplementary Table III
Review Process File
Acknowledgments
The KE laboratory acknowledges financial support from the Swedish Cancer Fund and Swedish Research Council, VR. RCA was supported by MRC Strategic Grant G0301153, Wellcome Trust Programme Grant 065061/Z. RCA is a Wellcome Trust Principal Research Fellow. KE, RCA, and DCB are members of the EC P6 Network ‘The Epigenome' LSHG-CT-2004–503433. We thank Sander Grannemenn and Femke Simmer for reagents and help; Alastair Kerr for his help with supporting Supplementary Figure S3. We thank Jenna Persson for critical reading of the paper. DCB is funded as a Royal Society Research Professor and RAM acknowledges support from the National Science Foundation, USA. DCB thanks the Gatsby Charitable Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 9: 218–233 [DOI] [PubMed] [Google Scholar]
- Appelgren H, Kniola B, Ekwall K (2003) Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J Cell Sci 116 (Pt 19): 4035–4042 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Baum M, Ngan VK, Clarke L (1994) The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell 5: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Brunel C, Romby P (2000) Probing RNA structure and RNA-ligand complexes with chemical probes. Methods Enzymol 318: 3–21 [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D (2008) TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D (2006) Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886 [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8: 884–896 [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI (2008) Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451: 734–737 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M (1989) Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell 57: 739–751 [DOI] [PubMed] [Google Scholar]
- Clarke L, Amstutz H, Fishel B, Carbon J (1986) Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA 83: 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D (2007) Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Djupedal I, Ekwall K (2009) Epigenetics: heterochromatin meets RNAi. Cell Res 19: 282–295 [DOI] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K (2005) RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev 19: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269: 1429–1431 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L (1988) Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 8: 754–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121: 961–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil LA, Plante P, Rohani M, Ouellette M, Provost P (2008) Involvement of Dcr1 in post-transcriptional regulation of gene expression in Schizosaccharomyces pombe. Front Biosci 13: 2203–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, Proudfoot NJ (2008) Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132: 983–995 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Iida T, Nakayama J, Moazed D (2008) siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell 31: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R (2008) RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 18: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ (2005) Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol 15: 1448–1457 [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M (2003) Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469 [DOI] [PubMed] [Google Scholar]
- Lorentz A, Ostermann K, Fleck O, Schmidt H (1994) Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene 143: 139–143 [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA (2006) Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198 [DOI] [PubMed] [Google Scholar]
- Mellone B, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC (2003) Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol 13: 1748–1757 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M (1986) Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J 5: 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Kinoshita N, Yanagida M (1987) A novel sequence common to the centromere regions of Schizosaccharomyces pombe chromosomes. Nucleic Acids Res 15: 4705–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI (2004) RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36: 1174–1180 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF (2005) RNA interference (RNAi) dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol 25: 2331–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Silverstein RA, Dishart D, Walfridsson J, Djupedal I, Kniola B, Wright A, Samuelsson B, Radmark O, Ekwall K (2002) Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc Natl Acad Sci USA 99: 16648–16653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M, Arndt GM (2003) Double-stranded RNA-mediated gene silencing in fission yeast. Nucleic Acids Res 31: 4481–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Sigova A, Rhind N, Zamore PD (2004) A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev 18: 2359–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH (2007) Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Steiner NC, Hahnenberger KM, Clarke L (1993) Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol Cell Biol 13: 4578–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI (2005) RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA 102: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Niwa O, Yanagida M (1991) A large number of tRNA genes are symmetrically located in fission yeast centromeres. J Mol Biol 218: 13–17 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB (2005) CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37: 376–386 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- White JL, Kaper JM (1989) A simple method for detection of viral satellite RNAs in small plant tissue samples. J Virol Methods 23: 83–93 [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880 [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI (2008) Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Table I
Supplementary Table II
Supplementary Table III
Review Process File