Abstract
Cullin-based E3 ubiquitin ligases are activated through covalent modification of the cullin subunit by the ubiquitin-like protein Nedd8. Cullin neddylation dissociates the ligase assembly inhibitor Cand1, and promotes E2 recruitment and ubiquitin transfer by inducing a conformational change. Here, we have identified and characterized Lag2 as a likely Saccharomyces cerevisiae orthologue of mammalian Cand1. Similar to Cand1, Lag2 directly interacts with non-neddylated yeast cullin Cdc53 and prevents its neddylation in vivo and in vitro. Binding occurs through a conserved C-terminal β-hairpin structure that inserts into the Skp1-binding pocket on the cullin, and an N-terminal motif that covers the neddylation lysine. Interestingly, Lag2 is itself neddylated in vivo on a lysine adjacent to this N-terminal-binding site. Overexpression of Lag2 inhibits Cdc53 activity in strains defective for Skp1 or neddylation functions, implying that these activities are important to counteract Lag2 in vivo. Our results favour a model in which binding of substrate-specific adaptors triggers release of Cand1/Lag2, whereas subsequent neddylation of the cullin facilitates the removal and prevents re-association of Lag2/Cand1.
Keywords: Cand1, cell cycle, cullin, Nedd8, ubiquitin
Introduction
Ubiquitination regulates many cellular processes by targeting proteins for degradation through the 26S-proteasome (Hershko and Ciechanover, 1998). During this process, the small protein ubiquitin is attached to substrate proteins in three steps. First, ubiquitin is activated by an E1 activating enzyme, which results in the formation of a thioester bond between the C-terminus of ubiquitin and the active site cysteine of the E1. From the E1, ubiquitin is transferred to the active site cysteine of E2 ubiquitin-conjugating enzymes, which subsequently interact with E3 ubiquitin ligases. E3 enzymes recognize the substrate and promote ubiquitin transfer from the E2 onto substrate proteins (Pickart, 2001). Substrate ubiquitination is achieved by the formation of an isopeptide linkage between the C-terminus of ubiquitin and a lysine residue of the substrate protein. Multiple rounds of ubiquitination extend a ubiquitin chain on the first ubiquitin, which depending on the ubiquitin lysine used in chain formation can lead to recognition by 26S-proteasomes, which subsequently degrade the substrate.
The largest class of E3 ubiquitin ligases is represented by the multi-subunit cullin–RING E3s (CRLs). Cullins function as scaffolds within the CRL complex, which bind through their N-terminus to variable substrate-specific modules and their C-terminus to the small RING-finger protein Rbx1 (Hrt1 in budding yeast). The cullin/Rbx1 heterodimer acts as the catalytic core by recruiting ubiquitin-charged E2 to the complex. The composition of the substrate-specific module varies depending on the cullin and the substrate (Sumara et al, 2008). The best characterized CRL, the Saccharomyces cerevisiae SCF (Skp1–Cdc53/cullin1-F-box) complex, uses Skp1 to interact with one of several F-box proteins that in turn directly bind targets. For example, the F-box protein Cdc4 promotes cell-cycle progression by mediating degradation of the cyclin-dependent kinase inhibitor Sic1 at the G1/S transition (Schwob et al, 1994; Verma et al, 1997).
Owing to their critical role in substrate selection, the activity of E3 ubiquitin ligases is highly regulated. One mode of CRL regulation is the modification of the cullin subunit with the ubiquitin-like protein Nedd8 (Rub1 in S. cerevisiae: related to ubiquitin 1; Lammer et al, 1998; Liakopoulos et al, 1999). Nedd8 is similar to ubiquitin in sequence and structure and is also covalently linked to target proteins. Nedd8 substrates are generally mono-neddylated, and though there are countless substrates for ubiquitination, only very few neddylated proteins are known to date. The best characterized Nedd8 substrates are cullin proteins. Mono-neddylation of cullins at a specific C-terminal lysine residue results in the activation of CRL complexes by triggering structural changes that allow for efficient ubiquitin transfer to the substrate (Duda et al, 2008; Fang et al, 2008; Saha and Deshaies, 2008) and by increasing the affinity of ubiquitin-charged E2 enzyme to the ligase (Kawakami et al, 2001).
In addition, Nedd8 counteracts the association of a cullin ligase assembly inhibitor called Cand1 (cullin-associated and neddylation-dissociated 1; Liu et al, 2002; Zheng et al, 2002b). Cand1 preferentially associates with unneddylated cullin and prevents binding of substrate-specific factors, thus inhibiting the formation of an active ligase complex. The X-ray crystal structure of the human Cand–Cul1 complex showed that Cand1 interacts with both the cullin C- and N-termini (Goldenberg et al, 2004). At the N-terminus, Cand1 inserts a β-hairpin loop into the Skp1-binding pocket, sterically preventing association of the substrate-specific module with the ligase (Zheng J et al, 2002; Zheng N et al, 2002). At the cullin C-terminus, Cand1 covers the lysine residue that becomes neddylated in the active complex, providing an explanation for why cullin neddylation and Cand1 binding appear mutually exclusive. However, it remains unclear whether neddylation is required to remove Cand1 and as a result allows the association of substrate-specific factors, or whether the presence of substrate-specific modules counteracts Cand1 and neddylation subsequently activates the complex.
Using bioinformatic analysis, we have identified budding yeast Cand1 as a gene described earlier to be involved in longevity assurance, called Lag2 (longevity assurance gene 2). As expected, Lag2 directly interacts with non-neddylated yeast cullin Cdc53 (cullin1), and prevents cullin neddylation in vivo and in vitro. Interestingly, Lag2 is only released from Cdc53 in the presence of substrate-specific adaptors, which subsequently allows neddylation of the cullin, thus establishing a plausible order of molecular events required for CRL activation. Finally, we found that Lag2 is itself a substrate for neddylation and thus the first non-cullin neddylation target identified in yeast.
Results
Identification of Lag2 as the Cand1 orthologue in S. cerevisiae
To determine whether S. cerevisiae carries a Cand1-like activity, we performed a bioinformatic analysis of the yeast genome searching for open reading frames that contained two functionally important regions of human Cand1: the C-terminal β-hairpin structure that inserts into the Skp1-binding pocket on cullins, as well as an N-terminal motif that binds to conserved surfaces in the cullin C-terminus (Figure 1A; Supplementary Figure 1; Zheng J et al, 2002; Zheng N et al, 2002). Using this approach (MAFFT program, L-INS-I algorithm; Katoh et al, 2005), we identified Lag2 as the only gene in the budding yeast genome that carries significant homology in both regions (Figure 1A, alignment). Lag2 homologues were also present in other species that did not carry an obvious Cand1 gene, whereas Lag2 was absent from all examined species with an identifiable Cand1 (Supplementary Table I). This observation further correlates with the notion that Lag2 may act as Cand1 orthologue in these organisms.
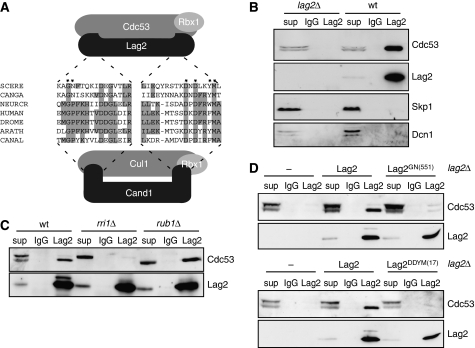
Figure 1.
Lag2 interacts with Cdc53 in a neddylation-dependent manner in vivo. (A) Schematic representation of Cul1/Cand1 and Lag2/Cdc53 complexes. Depicted with dashed lines are regions important for Cand1 inhibitory function (C-terminal β-hairpin and N-terminal patch). The sequence conservation in these regions between Lag2 and Cand1 homologues from different species is shown; identical residues are shaded. The asterisks mark residues that have been mutated to alanine to probe for their functional importance. (B) Extracts prepared from wild type or lag2Δ cells were immunoprecipitated with either control IgG or Lag2-specific antibodies, and bound proteins were immunoblotted as indicated with Cdc53, Lag2, Skp1 or Dcn1 antibodies. One percent of total extracts were loaded as input (sup). (C) Extracts from wild type (wt), deneddylation (rri1Δ) and neddylation-deficient (rub1Δ) cells were subjected to immunoprecipitation assay with either control IgG or Lag2-specific antibodies. Bound proteins were visualized by immunoblotting with Cdc53 (upper panel) and Lag2 (lower panel) antibodies. (D) Extracts from lag2Δ cells expressing wild-type Lag2, Lag2GN(551) (upper panel) or Lag2DDYM(17) (lower panel) were immunoprecipitated with control IgG or Lag2-specific antibodies. Bound proteins were visualized by immunoblotting with Cdc53 and Lag2 antibodies.
Lag2 interacts in vivo with cullins in a neddylation state-dependent manner
To test whether Lag2 is indeed a budding yeast Cand1, we first determined whether Lag2 interacts with Cdc53. An antibody to Lag2 specifically co-immunoprecipitated Cdc53 from yeast extract (Figure 1B), but not Skp1 or Dcn1, and Lag2 and Cdc53 readily interacted in yeast two-hybrid assays (Supplementary Figure 2A). Conversely, Cdc53 but not Lag2 was found in Skp1 immunoprecipitates (Supplementary Figure 2B). Importantly, Lag2 preferentially co-immunoprecipitated non-neddylated Cdc53, even though the majority of Cdc53 in cells is neddylated (Figure 1C). This observation suggests that similar to Cand1, Lag2 may sequester non-neddylated cullin core complexes. Consistent with this hypothesis, deletion of Rub1/Nedd8, which prevents cullin neddylation, resulted in a slightly stronger interaction between Lag2 and Cdc53 (Figure 1C).
Cullin neddylation can be reversed by a specific isopeptidase activity of the COP9–signalosome (CSN) complex (Lyapina et al, 2001). Yeast cells lacking CSN activity are viable, but Cdc53 accumulates in its neddylated form. As predicted, Lag2 no longer co-immunoprecipitated Cdc53 from cells deleted for the CSN core component Csn5/Rri1 (Figure 1C), strongly indicating that neddylation of Cdc53 prevents its interaction with Lag2.
The interaction between Lag2 and Cdc53 not only depends on the cullin neddylation state, but also requires the sequences conserved between Lag2 and Cand1. Mutational disruption of either the Lag2 β-hairpin (G551A, N552A, hereafter referred to as Lag2GN(551)) or the N-terminal cullin interaction site (D17A, D19A, Y22A, M23A, hereafter referred to as Lag2DDYM(17)) strongly diminished or abolished the interaction of Lag2 and Cdc53 by co-immunoprecipitation (Figure 1D). Together, these results show that Lag2 resembles Cand1 with respect to cullin binding, and suggest that Lag2 may function as an orthologue of Cand1 in budding yeast.
Lag2 inhibits the SCF complex in vivo in a neddylation-dependent manner
Association of Cand1 with CRL core complexes prevents their activation and association with substrate-specific adaptors, and as such is predicted to act as an inhibitor of ligase function. To determine whether Lag2 indeed inhibits cullin complexes in vivo, we examined the effect of Lag2 deletions and overexpression on the function of the SCF complex. Interestingly, neither deletion nor overexpression of Lag2 had any discernable effect on viability of wild-type cells (data not shown, Figure 2A). However, Lag2 overexpression was lethal in cells deleted for the yeast Nedd8 homologue Rub1 or the yeast Nedd8 E3 ligase Dcn1 (Kurz et al, 2005, 2008; Figure 2A), and this effect was not additive on simultaneous deletion of rub1 and dcn1. In contrast, Lag2 overexpression had no effect on viability of cells lacking the de-neddylating enzyme Rri1/Csn5 (Figure 2A), supporting the notion that cullin neddylation counteracts the interaction and thus function of Lag2. Indeed, overexpression of the Lag2GN(551) and Lag2DDYM(17) mutants was not detrimental to neddylation-deficient cells (Figure 2B), showing that the conserved stretches between Lag2 and Cand1 and the interaction with cullins are important for Lag2 function.
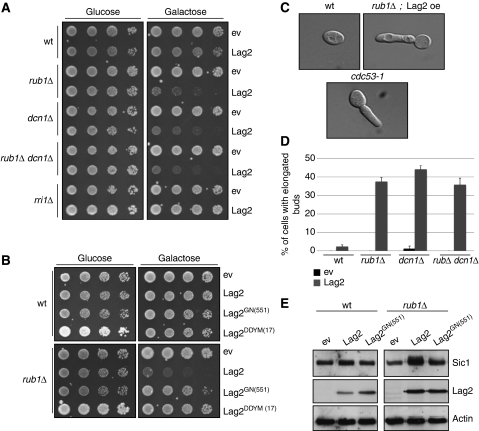
Figure 2.
Lag2 inhibits SCF function in vivo. (A) Five-fold serial dilution of an equal number of wild type, rub1Δ, rri1Δ, dcn1Δ and rub1Δ dcn1Δ cells transformed with an empty control plasmid (ev) or a plasmid allowing overexpression of Lag2 from the regulatable GAL1,10-promoter, were spotted on media containing glucose (left, GAL-promoter off) or galactose (right, GAL-promoter on). The plates were photographed after 3 days at 30°C. (B) Five-fold serial dilutions of an equal number of wild-type (wt) (upper plates) or rub1Δ (lower plates) cells transformed with an empty control plasmid (ev) or plasmids allowing as indicated overexpression of wild-type Lag2, Lag2GN(551) or Lag2DDYM(17) from the regulatable GAL1,10-promoter were spotted on media containing glucose (GAL-promoter off) or galactose (GAL-promoter on). The plates were photographed after 3 days at 30°C. (C, D) The morphology of wild type (wt) or rub1Δ cells overexpressing Lag2 from the GAL-1,10 promoter was examined by DIC microscopy 20 h after induction with 2% galactose (C, upper pictures). For control, the morphology of temperature-sensitive cdc53-1 cells shifted to the restrictive temperature for 7 h was included below (C, lower picture). The accumulation of wt, rub1Δ, dcn1Δ and rub1Δ dcn1Δ cells cells with elongated buds was quantified (D), and plotted as percentage of the total number of cells with standard deviations (n=500 in each case). (E) Sic1 levels were analysed by immunoblotting of extracts prepared from wild type (wt) or rub1Δ cells transformed with an empty control plasmid (ev) or plasmids allowing overexpression of wild type Lag2 or Lag2GN(551) from the regulatable GAL1,10-promoter. Immunoblotting with antibodies against Lag2 and actin control for equal Lag2 expression and gel loading, respectively.
We next examined the phenotype of these cells and determined whether the lethality associated with Lag2 overexpression is indeed a result of SCF inhibition. The cyclin-dependent kinase inhibitor Sic1 is one of the major substrates of the Cdc53Cdc4 complex, and a failure to degrade Sic1 results in a G1/S cell-cycle arrest accompanied by abnormal, elongated growth of the forming bud. Consistent with Lag2 counteracting SCF function, overexpression of Lag2 in neddylation-deficient cells resulted in the formation of elongated buds (Figure 2C), similar to the morphology of cdc53-1 temperature-sensitive mutants (Willems et al, 1996). The fraction of cells displaying this defect was lower in rub1Δ, dcn1Δ or rub1Δ dcn1Δ cells overexpressing Lag2 (approx. 40%) compared with cdc53-1 mutants at the restrictive temperature (∼60%), suggesting that a failure to degrade Sic1 may not be the only cause of Lag2 overexpression-induced lethality in neddylation-deficient cells (Figure 2D). To verify that Sic1 degradation is indeed impaired, we determined total Sic1 protein levels using western blot analysis in wild type and neddylation-deficient cells overexpressing Lag2. Although overexpression of Lag2 in wild-type cells only marginally increased Sic1 levels (Figure 2E), Sic1 protein abundance was approximately three-fold higher in neddylation-deficient cells overexpressing Lag2 (Figure 2E). Consistent with the phenotypic analysis, this increase was not observed on overexpression of the Lag2GN(551) (Figure 2E), implying that Sic1 accumulation requires the ability of Lag2 to interact with Cdc53.
Lag2 prevents Cdc53 neddylation in vitro by forming a heterotrimeric complex with Cdc53 and Hrt1
To investigate the molecular mechanism of Lag2 function in vitro, we tested whether purified Cdc53/Hrt1 forms a heterotrimeric complex similar to Cul1/Rbx1/Cand1. We expressed GST-tagged Hrt1, 6xHis-tagged Cdc53 and untagged Lag2 from a poly-cistronic bacterial expression vector in Escherichia coli, and purified complexes using a two-step affinity purification against the 6xHis and GST-tags. Subsequent gel filtration over a Superose 6 column showed the existence of a stable complex with an apparent size of 670 kDa (Figure 3A), suggesting that Cdc53/Hrt1 and Lag2 form a complex approximately three times the expected size of a single heterotrimeric unit. This oligomerization may be facilitated through dimerization of the GST-tag on Hrt1 and/or the previously reported dimerization of cullins.
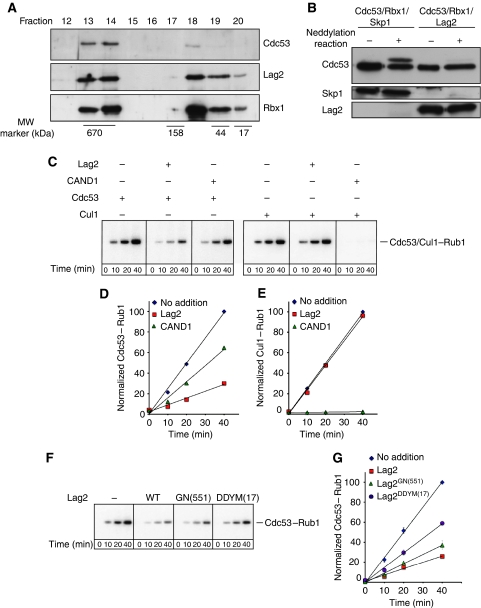
Figure 3.
Lag2 prevents Cdc53 neddylation in vitro. (A) Extracts prepared from E. coli expressing 6xHis-Cdc53, GST-Hrt1 and untagged Lag2 were purified by sequential affinity purification against the 6xHis and GST-tags, and the eluate separated on a Superose 6 column. The fractions were analysed for the presence of Cdc53, Lag2 and Hrt1 by immunoblotting. The positions of marker proteins with known molecular weight (kDa) are indicated below. (B) Cdc53/Rbx1/Skp1 and Cdc53/Rbx1/Lag2 complexes expressed in E. coli and purified using the two-step affinity purification protocol were subjected to in vitro neddylation reactions as described in ‘Materials and methods'. The neddylation state of Cdc53 was visualized by immunoblotting with antibody against Cdc53 (upper panel). The presence of Skp1 (middle panel) or Lag2 (lower panel) was controlled with specific antibodies. Note that the presence of Lag2 but not Skp1 prevents Cdc53 neddylation in vitro. (C–E) Purified Cdc53–Hrt1 or Cul1–Rbx1 complexes were subjected to neddylation reactions as described in ‘Materials and methods' in the presence of [32P]-Rub1, and analysed by autoradiography after the times indicated. Where indicated (+), the complexes were pre-incubated for 30 min with purified Lag2 or CAND1. The neddylation efficiency of Cdc53 (D) or Cul1 (E) was normalized by phosphorimager analysis from at least three experiments and plotted against time (min). Diamonds: no addition; squares: pre-incubation with Lag2; triangles: pre-incubation with CAND1. (F, G): The ability of purified wild-type (wt) Lag2 (squares), Lag2GN(551) (triangles) and Lag2DDYM(17) (circles) to inhibit neddylation of Cdc53–Hrt1 complexes was examined as described above. Pre-incubation with buffer alone (‘−' in F; diamonds in G) controls for the efficiency of Cdc53 neddylation.
To test whether Lag2 prevents cullin neddylation in vitro, we subjected purified 6xHis-Cdc53/GST-Hrt1/Lag2 complexes to neddylation reactions, and monitored Cdc53 modification by immunoblotting. For control, we included 6xHis-Cdc53/GST-Hrt1/Skp1 complexes, which we purified like the Lag2 complex after expression in E. coli from a poly-cistronic vector. Importantly, though the Cdc53/Hrt1/Skp1 complex was readily neddylated in vitro (Figure 3B), the Cdc53/Hrt1/Lag2 complex was refractory to this modification. To corroborate these results, we purified hCul1/hRbx1 and Cdc53/Hrt1 from baculovirus-infected insect cells (Supplementary Figure 3A), and subsequently monitored Cdc53/hCul1 neddylation using radioactive yeast Rub1 to visualize the conjugates by autoradiography. Titration of purified Lag2 to Cdc53/Hrt1 complexes showed that Lag2 directly binds to Cdc53 in nearly stoichiometric amounts (Supplementary Figure 3B). Although both hCul1/hRbx1 and Cdc53/Hrt1 complexes were efficiently modified by Rub1 (Figure 3C), pre-incubation of Cdc53/Hrt1 with recombinant Lag2 inhibited neddylation of Cdc53 in vitro (Figure 3C). Likewise, pre-incubation of hCul1/hRbx1 with hCand1 almost entirely prevented neddylation of hCul1. When quantified, the presence of Lag2 reduced Cdc53 neddylation to ∼ 20% of the amount detected in the absence of Lag2 (Figure 3D), whereas Cand1 almost entirely abolished neddylation of Cul1 (2%, Figure 3E). The molecular basis for this difference remains unclear, but it is possible that Lag2 requires additional not yet identified factors to fully inhibit cullin neddylation. Despite the large difference of Lag2 and Cand1 on primary sequence level, hCand1 was able to partially inhibit neddylation of ScCdc53 (40%). As expected, purified Lag2GN(551) and in particular Lag2DDYM(17) were defective to block Cdc53 neddylation compared to wild-type controls (Figure 3F and G), implying that efficient binding of Lag2 to the Cdc53/Hrt1 complex is necessary to prevent cullin neddylation in vitro.
Lag2 is itself a substrate for neddylation
The in vivo function of Lag2 requires an intact neddylation machinery, and the responsible neddylated substrate underlying this effect is thought to be the cullin. Interestingly, however, we noticed that a fraction of Lag2 migrated about 10 kDa slower on SDS–PAGE gels (Figure 4A), which is indicative of a covalent modification by ubiquitin or a ubiquitin-like protein (UBL). To determine whether Lag2 is indeed modified by a UBL, we examined Lag2 modification in yeast cells deleted for all non-essential UBLs. Interestingly, deletion of the yeast Nedd8 homologue Rub1 resulted in the loss of modified Lag2 (Figure 4A). Moreover, expression of an N-terminally HA-tagged version of Rub1 (HA-Rub1) in rub1Δ cells restored the appearance of modified Lag2 (Figure 4A), which as expected migrated slightly slower compared to untagged Rub1. Finally, in contrast to vector controls, modified Lag2 readily co-immunoprecipitated with HA-Rub1 (Figure 4B). Taken together, these data show that a fraction of Lag2 is neddylated in vivo.
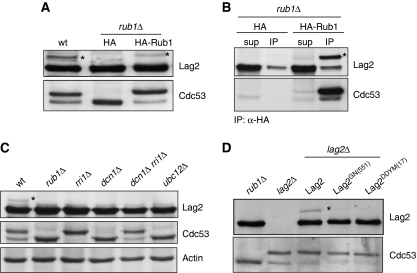
Figure 4.
Lag2 bound to Cdc53 is neddylated in vivo. (A) Total cell extract prepared from wild type (wt) or rub1Δ cells transformed with either an empty HA-vector (HA) or a plasmid expressing HA-tagged Rub1 was analysed by immunoblotting with antibodies against Lag2 (upper panel) and Cdc53 (lower panel). The asterisk marks the position of a slower migrating Lag2 species, which is absent in rub1Δ cells and slightly upshifted in rub1Δ cells expressing HA-Rub1. (B) HA-immunoprecipitates (IP) from extracts prepared from rub1Δ cells transformed with either an empty HA-vector (HA) or a plasmid expressing HA-tagged Rub1 were immunoblotted for the presence of Lag2 (upper panel) or Cdc53 (lower panel) with specific antibodies. An aliquot of the extracts was loaded as input (sup). The asterisk marks the position of Lag2 modified by HA-Rub1. (C) Lag2 (upper panel) and Cdc53 (middle panel) neddylation were examined by immunoblotting of cell extracts prepared from cells lacking the indicated components of the neddylation pathway. An antibody specific for actin controls for equal loading (lower panel). The asterisk marks the position of a neddylated Lag2. (D) Extracts prepared from rub1Δ, lag2Δ or lag2Δ cells with integrated wild-type Lag2, Lag2GN(551) or Lag2DDYM(17) were immunoblotted with antibodies against Lag2 (upper panel) or Cdc53 (lower panel). The asterisk marks the position of neddylated Lag2.
Similar to cullin neddylation, Lag2 neddylation required all known components of the Nedd8 conjugation machinery. Deletion of either the Nedd8 E2 Ubc12 or its E3 ligase Dcn1 resulted in a loss of both, Lag2 and Cdc53 neddylation (Figure 4C). However, though deletion of the de-neddylase Csn5/Rri1 shifted all of Cdc53 to its neddylated form, it prevented neddylation of Lag2 (Figure 4C). As Lag2 no longer binds to neddylated Cdc53, the opposite outcomes of Csn5/Rri1 deletion on the neddylation state of Cdc53 and Lag2 could be explained if Lag2 required prior binding to Cdc53 for neddylation. Indeed, Lag2 mutants (Lag2GN(551) and Lag2DDYM(17)) that lost the ability to bind to non-neddylated Cdc53 were no longer neddylated (Figure 4D), indicating that the interaction between Lag2 and Cdc53 is a pre-requisite for Lag2 neddylation.
Antagonizing Lag2 function in vivo requires neddylation of Cdc53 but not Lag2
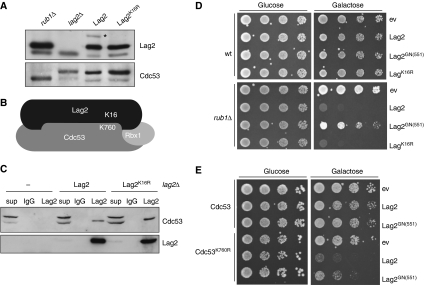
To determine the functional significance of Lag2 neddylation in vivo, we next sought to identify the neddylated lysine on Lag2. To achieve this goal, we systematically mutated all lysine residues to arginine, integrated the mutants into the yeast genome and subsequently determined the neddylation state of the mutated Lag2 by western blot analysis. Using this approach, we identified a single lysine, K16, which when mutated to arginine resulted in a loss of Lag2 neddylation (Figure 5A). Interestingly, K16 is located in close proximity to the site where Lag2, in analogy to Cand1, would interact with the cullin C-terminus (Figure 5B). The Lag2K16R mutant immunoprepitated Cdc53 as efficiently as wild-type controls (Figure 5C), excluding the possibility that the observed loss of Lag2 neddylation is simply caused by a defect in Cdc53 binding.
Figure 5.
Neddylation of Cdc53 but not Lag2 is necessary to overcome the inhibitory effects of Lag2 on SCF activity. (A) Extracts prepared from rub1Δ, lag2Δ or lag2Δ cells with integrated wild-type Lag2 or Lag2 with lysine 16 mutated to arginine (Lag2K16R) were immunoblotted with antibodies against Lag2 (upper panel) or Cdc53 (lower panel). The asterisk marks the position of neddylated Lag2. (B) Schematic representation of the Cdc53/Lag2 complex with the position of the lysine residues modified by Rub1 indicated (K16 in Lag2 and K760 in Cdc53). (C) Extracts from lag2Δ cells expressing wild-type Lag2 or Lag2K16R were immunoprecipitated with control IgG or Lag2-specific antibodies. Bound proteins were visualized by immunoblotting with Cdc53 (upper panel) and Lag2 (lower panel) antibodies. (D) Five-fold serial dilution of an equal number of wild type (wt) (upper plates) or rub1Δ (lower plates) cells transformed with an empty control plasmid (ev) or plasmids allowing overexpression of wild-type Lag2 (wt), Lag2GN(551) or Lag2K16R from the regulatable GAL1,10-promoter was spotted on media containing glucose (left, GAL-promoter off) or galactose (right, GAL-promoter on). The plates were photographed after 3 days at 30°C. (E) Five-fold serial dilution of an equal number of cdc53Δ cells complemented with wild-type Cdc53 or the neddylation-deficient Cdc53-K760R mutant transformed with an empty control plasmid (ev) or plasmids allowing overexpression of wild-type Lag2 or Lag2GN(551) from the regulatable GAL1,10-promoter was spotted on media containing glucose (left, GAL-promoter off) or galactose (right, GAL-promoter on). The plates were photographed after 3 days at 30°C. Note that neddylation of Cdc53 is important for cells to cope with increased levels of Lag2.
To distinguish whether Lag2 and/or Cdc53 neddylation is important to regulate Lag2 function, we first overexpressed the non-neddylatable Lag2K16R mutant in wild type and rub1Δ cells. Similar to wild-type Lag2, Lag2K16R overexpression had no effect on wild-type cells, whereas it interfered with viability in rub1Δ cells (Figure 5D), implying that Lag2 neddylation is not required for Lag2 or Cdc53 activity under these conditions. Moreover, purified Lag2K16R efficiently interfered with Cdc53 neddylation in vitro (Supplementary Figure 4), and overexpression of wild type, but not the binding-defective Lag2GN(551) mutant, was toxic in cells expressing the non-neddylatable Cdc53K760R mutant as their only source of Cdc53 (Figure 5E). Taken together, these results suggest that neddylation of Cdc53 but not Lag2 is functionally important to inhibit Lag2 activity in vivo.
Skp1 in complex with substrate-specific adaptors counteract Lag2 function in vivo and in vitro
Although neddylation of Cdc53 antagonizes Lag2 function in vivo, bound Lag2/Cand1 efficiently prevents neddylation by blocking access of the neddylation machinery to the specific lysine residue. Thus, additional factors must have a role in regulating the interaction of these two proteins. A plausible candidate may be Dcn1, which interacts with the cullin close to its neddylation site (Kurz et al, 2008) and is required for Cdc53 neddylation in vivo (Kurz et al, 2005). Although purified Dcn1 may bind Cdc53/Hrt1/Lag2 complexes in vitro (Supplementary Figure 5A), the addition of Dcn1 was not sufficient to promote cullin neddylation in the presence of Lag2 (Figure 3C; Supplementary Figure 5B). Furthermore, if the only role of Dcn1 was to remove Lag2/Cand1 from the cullin, one may expect that Dcn1 would be dispensable for neddylation in the absence of Lag2/Cand1. However, Cdc53 remained unneddylated in dcn1Δ cells, even if Lag2 was deleted (Supplementary Figure 5C). Taken together, we conclude that the neddylation machinery is not sufficient to remove Cand1/Lag2 from cullins and that other factors must be required to activate the assembly of cullin ligases.
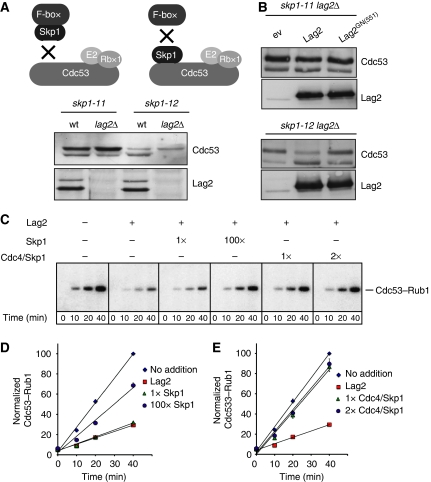
Previous reports implicated that substrate-specific adaptors may counteract Cand1 function (Bornstein et al, 2006) and based on the X-ray crystal structure, binding of Cand1 or Skp1 to Cul1 appears mutually exclusive (Goldenberg et al, 2004). To examine whether Skp1 is required to release Lag2 from Cdc53 in vivo, we first analysed the neddylation state of Cdc53 in two temperature-sensitive genetic alleles of Skp1, skp1-11 and skp1-12. The skp1-11 allele carries two mutations (G160E and R167K) in its C-terminal domain that interfere with binding to the F-box adaptors, but not to Cdc53, whereas skp1-12 carries a single mutation (L8G) that disrupts binding to Cdc53 (Figure 6A) (Bai et al, 1996). Although we detected no obvious difference in skp1-11 cells, Cdc53 neddylation was markedly reduced in skp1-12 cells, suggesting that association of Skp1 with Cdc53 is required for efficient neddylation in vivo (Figure 6A, Lammer et al, 1998). Moreover, total Cdc53 levels were significantly lower in skp1-12 mutants compared to wild type or skp1-11 mutants, indicating that association of Skp1 may help to stabilize the cullin. To test whether the lower Cdc53 neddylation level is due to ectopic Lag2 activity, we next deleted Lag2 in these strains. Indeed, Cdc53 neddylation was restored in skp1-12 lag2Δ double mutants (Figure 6A), showing that Skp1 counteracts Lag2 function in vivo. This Lag2-mediated neddylation defect required binding of Lag2 to Cdc53, as the Lag2GN(551) mutant was not able to prevent Cdc53 neddylation in skp1-12 cells (Figure 6B). Consistent with these findings, overexpression of wild-type Lag2 in skp1-12 mutants strongly reduced viability at semi-permissive temperature presumably by sequestering Cdc53/Hrt1 core complexes, whereas this effect was much less severe with the Lag2GN(551) mutant (Supplementary Figure 6). On the basis of these results we conclude that Skp1 is required to trigger Lag2 dissociation from cullins in vivo, which is a prerequisite for subsequent cullin neddylation.
Figure 6.
Skp1 in complex with substrate-specific adaptors counteracts Lag2 activity in vivo and in vitro. (A) Schematic illustration of the molecular defects attributed to the temperature-sensitive skp1 mutants (upper panel): Skp1-11 carries two mutations (G160E and R167K) in its C-terminal domain that allows binding to Cdc53 but not F-box adaptors (drawing on the left), whereas Skp1-12 carries a single mutation (L8G) that disrupts binding to Cdc53 (drawing on the right). The neddylation state of Cdc53 was analysed by immunoblotting of total extracts prepared as indicated from skp1-11, skp1-12, skp1-11 lag2Δ or skp1-12 lag2Δ cells grown at the semi-permissive temperature of 30°C. (B) Extracts from skp1-11 lag2Δ (upper panel) or skp1-11 lag2Δ (lower panel) cells harbouring an empty control plasmid (ev) or expressing wild-type Lag2 or Lag2GN(551) were immunoblotted with specific antibodies for Cdc53 (upper blots) and Lag2 (lower blots). (C) Purified Cdc53–Hrt1 complexes pre-incubated with (+) or without (−) Lag2 were subjected to neddylation reactions as described in ‘Materials and methods' in the presence of [32P]-Rub1, and the neddylation of Cdc53 was analysed by autoradiography after the times indicated. Where indicated, different stoichiometric amounts of purified Skp1 or Skp1–Cdc4 complexes were added at time 0. (D, E) The neddylation efficiency of Cdc53/Hrt1/Lag2 complexes incubated with purified Skp1 (D) or Skp1–Cdc4 complexes (E) as described above was normalized by phosphorimager analysis from at least three experiments and plotted against time (min. Diamonds: Cdc53/Hrt1 complexes without Lag2 (no addition); squares: Cdc53/Hrt1/Lag2 complexes; triangles and circles: Cdc53/Hrt1/Lag2 complexes with Skp1 or Cdc4/Skp1 at the indicated stoichiometric ratios.
To investigate whether Skp1 may antagonize Lag2 function in vitro, we incubated purified Cdc53/Hrt1/Lag2 complexes with neddylation enzymes in the presence or absence of recombinant Skp1. Although addition of a 1:1 molar ratio of Skp1 to the Cdc53/Lag2 neddylation reactions had no discernable effect on the inhibition of Cdc53 neddylation by Lag2 (Figure 6D and E), a 100 × molar excess of Skp1 significantly restored neddylation, suggesting that Skp1 counteracts Lag2, presumably by replacing it from Cdc53. The large amount of Skp1 required for Lag2 inhibition suggests, however, that there are additional cellular factors that facilitate Skp1-dependent removal of Lag2 from Cdc53. Obvious candidates for this role are the F-box proteins, as the functional E3 ligase requires both, Skp1 and the F-box adaptor for function. We thus tested whether simultaneous addition of Skp1 and the F-box protein Cdc4 would facilitate in vitro Cdc53 neddylation. Indeed, addition of equimolar amounts of Skp1/Cdc4 to Cdc53/Lag2 neddylation reactions resulted in a strong increase of Cdc53 neddylation, whereas a two-fold molar excess restored neddylation to the levels measured in the absence of Lag2 (Figure 6D and E). Taken together, these data show that a functional substrate-specific adaptor module counteracts Lag2/Cand1 to allow cullin neddylation.
Discussion
The regulation of CRL activity occurs at multiple levels and involves a fine-tuned interplay of specific inhibitors and post-translational modifications such as neddylation. Here, we report the identification and characterization of Lag2 in S. cerevisiae, which functions like its mammalian counterpart Cand1 as an inhibitor of CRL assembly in vitro and in vivo. Our functional analyses indicate an ordered pathway of Cdc53 activation, which is initiated by binding of a substrate-specific adaptor module to the Cdc53/Lag2 complex, which in turn favours dissociation of Lag2 and allows subsequent neddylation of the cullin subunit.
S. cerevisiae Lag2 functions as an orthologue of mammalian Cand1
Several lines of evidence suggest that S. cerevisiae Lag2 functions like mammalian Cand1 with respect to CRL regulation. First, Lag2 directly binds to Cdc53 and sequesters non-neddylated cullin core complexes. Lag2 also interacts with the yeast Cul4-type cullin Rtt101 (Supplementary Figure 7), suggesting that it may regulate several if not all cullin subfamilies. Second, Lag2 prevents Cdc53 neddylation in vivo and in vitro, and finally overexpression of Lag2 inhibits Cdc53 function in a neddylation-dependent manner. These results exclude the notion that cullin neddylation in budding yeast may not be essential because of the lack of a CAND1-like activity. However, despite the functional resemblance of Lag2 and CAND1, the only similarity between Lag2 and CAND1 lies within short N- and C-terminal stretches that in the Cand1/Cul1 crystal structures have been shown to contact the cullin. These motifs are also of functional importance: the C-terminal β-hairpin structure inserts into the Skp1-binding pocket on the cullin and the N-terminal sequence patch binds to the cullin C-terminus to cover the neddylation lysine. Accordingly, mutation of these Lag2 sequences strongly reduces the interaction with Cdc53 and abolishes the inhibitory effect of Lag2 on CRLs. Apart from these two short sequence stretches, Lag2 and Cand1 seem to be surprisingly different proteins. Human Cand1 is a large 120 kDa protein made up of 1159 amino acids, whereas Lag2 is only 75 kDa in size (660 amino acids). The Cand1 crystal structure showed that the protein forms a horseshoe-like clamp that binds to the cullin at its N- and C-termini. Owing to its smaller size, it is unlikely that Lag2 takes up a similar shape. In fact, Cdc53 is itself ∼90 kDa in size and forms an elongated rod-like structure. Thus, in order for Lag2 to bind both, the Cdc53 N- and C-termini, it will most likely have to reach directly across the protein, rather than forming a horseshoe. It is interesting to note in this context that the function of the A. nidulans Cand1 protein is split into two separate proteins that represent the mammalian N- and C-terminal domains (Pick and Pintard, 2009). This raises the question about the functional role of the central part of Cand1. If it is important for cullin regulation, these functions must be fundamentally different in yeast and higher organisms. Alternatively, it is possible that this area of the protein is dispensable for cullin regulation, but serves an additional, non-cullin-related function. Not much is known about the role of Cand1 or Lag2 outside of cullin regulation, and in the future it will be interesting to investigate if they exist and how they differ from each other.
Regulation of the Cullin–Cand1/Lag2 cycle
Cullin neddylation and substrate-specific adaptor availability are major regulators of the Cullin–Cand1/Lag2 cycle (Figure 7). However, in contrast to some previous models (Liu et al, 2002; Zheng et al, 2002a), neddylation enzymes are not sufficient to trigger the release of Lag2/Cand1, and bound Lag2/Cand1 prevents access of the neddylation machinery to the specific lysine residue on cullins, even in the presence of Dcn1. Nevertheless, neddylation may help to prevent re-association of the Lag2/Cand1 inhibitor. Indeed, overexpression of Lag2 inhibits Cdc53 activity only when expressed in cells defective for components of the neddylation pathway. Interestingly, both Lag2 and Cdc53 are direct substrates for neddylation in vivo. Surprisingly, we were unable to reconstitute Lag2 neddylation in vitro, suggesting that some additional components may be missing in our reconstituted system. Although our results suggest that the critical target to counteract Lag2 function seems to be neddylation of the cullin, it is possible that Lag2 neddylation contributes to its inactivation. Indeed, the site of Lag2 neddylation is located in close proximity to the cullin interaction site, and may thus regulate its binding affinity for Cdc53. These results favour a model whereby Lag2 is neddylated on release from cullins and that like cullin neddylation, Lag2 neddylation may help to prevent re-association of the two proteins. Alternatively, Lag2 neddylation may regulate aspects of Lag2 function that are unrelated to cullin binding. As cullin binding is a prerequisite for Lag2 neddylation, this modification marks a specific pool of Lag2, which may thus carry a signalling function or target the protein to novel binding partners or subcellular compartments. It remains to be investigated if higher eukaryotic Cand1 is also a substrate for neddylation. Although the neddylated lysine on Lag2, K16, is not conserved in hCand1, there are multiple conserved lysines in the vicinity that could substitute.
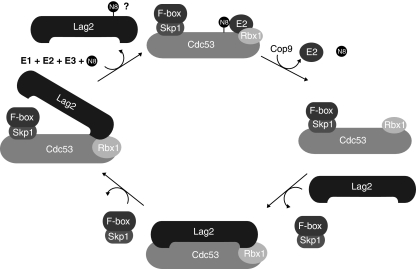
Figure 7.
Proposed model of SCF regulation by Lag2/CAND1. Non-neddylated Cdc53 in complex with Hrt1/Rbx1 is sequestered in an inactive, non-assembled form by Lag2/CAND1. SCF/CRL1 activation is initiated by binding of Skp1 in complex with an appropriate F-box protein, which promotes dissociation of Lag2/CAND1 by competing for the amino-terminal binding site on Cdc53. This initial step may be further facilitated by neddylation of Lag2. Skp1/F-box binding now allows access of the neddylation machinery to modify lysine 760 on Cdc53, which counteracts Lag2/CAND1 binding and triggers a conformational change that allows recruiting activated E2. De-neddylation by the CSN/signalosome may subsequently contribute to SCF/CRL1 inactivation and allow reformation of a stable Lag2/Cand1–cullin-Hrt1/Rbx1 complex.
Rather than cullin neddylation, our data show that the presence of Skp1 bound to substrate-specific adaptors is required to counteract the inhibitory association of Cand1/Lag2 with Cdc53 (Figure 7). Indeed, in contrast to Cdc53, Skp1 did not co-immunoprecipitate with Lag2. Moreover, overexpression of Lag2 is lethal in Skp1 mutants, and Skp1 mutants that are impaired in cullin binding show a dramatic reduction in Cdc53 neddylation because of enhanced association of Lag2. Structural work showed that binding of Skp1 and Cand1 is mutually exclusive, because Cand1 inserts a β-hairpin loop into the Skp1-binding pocket on hCul1 (Zheng et al, 2002b; Goldenberg et al, 2004). As this β-hairpin loop is conserved and important for Lag2 function in vivo, Skp1 may generally dissociate Lag2/Cand1 by competing for the same binding site. However, Skp1 alone is a rather poor competitor in vitro, and requires the presence of an associated F-box protein. This dependence on substrate-adaptors seems less pronounced in vivo, where in contrast to the cullin non-binding mutant of Skp1, skp1-12, the F-box-binding mutant skp1-11 did not significantly affect cullin neddylation. The reason for this difference is not clear, but may possibly be due to the temperature-sensitive nature of the skp1-11 mutant. Furthermore, it is likely that binding of F-box proteins triggers structural changes or facilitates the folding of Skp1 and thus its association with cullins.
On the basis of these results, we propose the following model for cullin activation and inactivation by Lag2/Cand1 (Figure 7). Lag2/Cand1 tightly binds to the cullin/Hrt1 heterodimer, and thereby prevents the assembly of an active CRL complex. Skp1 in association with a substrate-adaptor (and possibly bound substrate) facilitates dissociation of Lag2/Cand1, which allows the neddylation machinery to gain access to the neddylation lysine on Cdc53/Cul1. Freed from Lag2/Cand1 and neddylated, the cullin can undergo a conformational change that allows recruitment of the activated E2 and efficient ubiquitin transfer to the bound substrate. Cullin and possibly Lag2 neddylation also prevent re-association of Lag2/Cand1 and thereby contribute to sustained CRL activity. De-neddylation by the CSN/signalosome may then be required for CRL inactivation and reformation of a stable Lag2/Cand1–cullin complex. Although this cycle is now well substantiated for SCF/CRL1 complexes, it remains to be examined whether substrate-adaptor modules specific for other cullin subfamilies such as BTB-proteins or Ddb1/DCAF heterodimers may function in a similar manner to displace Lag2/Cand1.
A physiological role of Lag2/Cand1 in CRL regulation?
Although it is now well established that Cand1 sequesters cullin–RING core complexes, the functional importance of this regulatory mechanism remains unclear. Cells lacking Lag2 are viable and exhibit no obvious growth defect associated with CRL function. However, under nutrient-rich conditions, the majority of Cdc53 is neddylated, and as Lag2 only binds to un-neddylated Cdc53, there is more unbound cullin than Lag2-bound cullin present at any given time (Supplementary Figure 8). Thus, though Lag2/Cand1 may be dispensable for rapidly growing cells, it may become important to regulate CRL activity under specialized physiological conditions. It is noteworthy in this context that Lag2 is preferentially expressed in young cells and was originally described as a protein involved in determination of longevity. Although the connection of this function with respect to its role in CRL regulation has not been explored, it is intriguing that Lag2/Cand1 may regulate some aspects of aging.
It has been proposed that Cand1 keeps the cullin complex inactive to prevent auto-ubiquitination of the ligase in the absence of substrate. If this were the case, one would expect a dramatic reduction in cullin ligase levels in the absence of Lag2/Cand1. However, deletion of Lag2 has no discernable effect on total cullin levels, at least under nutrient-rich conditions. Nevertheless, Cdc53 levels are significantly reduced in skp1-12 cells, which are defective for the Skp1–cullin interaction. This effect is even more pronounced by deletion of Lag2, suggesting that Cdc53 is unstable in the absence of subunits interacting with its amino-terminal domain. Although it is unlikely therefore that the major role for Lag2/Cand1 is to protect CRLs from auto-ubiquitination, Skp1 and Lag2/Cand1 may cooperate to stabilize Cdc53/Cul1 in vivo.
Alternatively, one could imagine a role of Lag2/Cand1 in maintaining some cullin core complexes in an inactive reservoir that can be quickly activated. Such a mechanism may be particularly important to ensure that rare substrate-adaptors are able to compete for available cullins and rapidly mount an appropriate CRL balance when needed. For example, it is possible that specific pools of Cullin–Cand1/Lag2 complexes may be locally activated by the presence of substrate-specific adaptor and/or substrate. Such rare or discretely localized substrate-adaptors may exhibit a higher affinity to displace Lag2/Cand1 to efficiently compete with their more abundant counterparts. With the identification of Lag2 in yeast, we will be able to harness the powerful genetic and biochemical techniques established in this model organism to investigate these speculative models in the future.
Materials and methods
Plasmids and yeast strains
All plasmids are described in Supplementary Table II. Lag2 with its the endogenous promoter (500 base pair upstream region) was amplified from genomic DNA with primers 5′-ATCTGACACCGGCGGAATATACCACTTCGTTATAAG -3′ and 5′-GGTCATCTCGAGCTAGTCATGAGGGAGATAAGG-3′ and ligated into NotI–XhoI cut vectors generating pES63. The Lag2GN(551) and Lag2K16R mutants were generated by QuikChange mutagenesis (Stratagene) with following primers: Lag2GN(551) 5′-GATACATTAAAAGCAGCTAATGCTACCCAAAAAATTGATG-3′and 5′-CATCAATTTTTTGGGTAGCATTAGCTGCTTTTAATGTATC-3′; Lag2K16R 5′-CAATATCGTTCCACTAGGGACAATCACCTTA-3′ and 5′-TAAGGTCATTGCCCTAGTGGAACGATATTG-3′.
The poly-cistronic E. coli expression vectors were constructed using the pETr3a/pST39 system (Tan, 2001). In short, GST-TEV-Hrt1 was cloned in position 1 of the poly-cistronic expression cassette using Xba1/BamH1, Lag2 or Skp1 were cloned in position 3 using Sac1/Kpn1 and 6xHis-Cdc53 (non-cleavable) was cloned into position 4 using BspE1/Mlu1.
Yeast were transformed and cultured by standard methods (Amberg et al, 2005). All carbon sources were used at 2%. pGAL1-10 expression constructs were grown to (OD600) of 0.5–0.8 in raffinose on galactose induction. The Lag2 mutants were generated by homologous recombination methods. All the yeast strains are listed in Supplementary Table III.
Purification of Lag2 and CRL complexes
To express the 6xHis-Cdc53/GST-HRT1/Lag2 and 6xHis-Cdc53/GST-HRT1/Skp1 complexes, the plasmids were transformed into E. coli Rosetta cells. A starter culture in LB medium was grown over night. Subsequently, 2 l of auto-induction medium (Studier, 2005) were inoculated at OD600:0.05 and grown at 37°C until an OD600 of 0.8. The cultures were subsequently cooled to 16°C on ice and grown overnight in a 16°C shaker to induce expression. The cells were collected and the pellet resuspended in 100 ml of NiNTA buffer. After two French Press passages the lysate was cleared for 30 min at 48 000 g in an SS34 rotor and the complexes subsequently purified over NiNTA resin (Qiagen). The eluted complexes were then purified a second time over GSH resin (GE Healthcare) to specifically isolate 6xHis-Cdc53/GST-HRT1 complexes. The presence of either Lag2 or Skp1 in the complexes was determined by Coomassie staining of SDS–PAGE gels, or western blotting with specific antibodies. The purified complexes were separated on a Superose 6 10/300GL gel filtration column (GE Healthcare) and their relative size determined against the mobility of gel filtration chromatography standards from Bio-Rad.
Purification of human Cul1–Rbx1 and CAND1 has been described (Zheng et al, 2002b; Goldenberg et al, 2004; Duda et al, 2008). S.cerevisiae Ubc12, Dcn1, Rub1 (harbouring a Protein Kinase A phosphorylation site), Skp1, Cdc4–Skp1 and Uba3-Ula1 were expressed in BL21 (DE3) GOLD E. coli, and Cdc53–Hrt1 in insect cells and initially purified by glutathione affinity chromatography, and treated with either TEV or thrombin protease to cleave off the GST-tags. Dcn1, Skp1, Uba3-Ula1 and Cdc53–Hrt1 were further purified by ion exchange. Rub1 and Ubc12 were subjected to dialysis, and GST was removed by glutathione affinity chromatography. All proteins were polished by gel filtration chromatography. A contaminant in Cdc4–Skp1 was removed by nickel affinity chromatography, and subsequently passed over a buffer exchange column in 25 mM HEPES pH 7.0, 200 mM NaCl, 1 mM DTT.
Antibodies
Antibody against full-length GST-Lag2 were raised in rabbits (Eurogentec, Seraing, Belgium) and affinity purified on GST-Lag2. The following commercial antibodies were used: Cdc53 (yN-18, Santa Cruz), Skp1 (yC-20, Santa Cruz), HA11 (MMS-101R, Covance), actin (MAB1501R, Chemicon International).
Preparation of yeast extracts, immunoblotting and imunoprecipitation experiments
Total yeast cell extracts were prepared as follows: 2 ml of cells corresponding to OD600 0.8–1.2 were precipitated by addition of 1.85 M NaOH and 50% trichloroacetic acid and pelleted at 13 000 g for 2 min at 4°C. The pellets were washed in acetone and resuspended in urea loading buffer (120 mM Tris–HCl pH 6.8, 5% glycerol, 8 M urea, 143 mM 2-mercaptoethanol, 8% SDS). Samples were separated by SDS–PAGE followed by western blotting with desired antibodies.
For immunoprecipitation experiments, yeast cells were lysed by FreezerMill (SPEX) in lysis buffer (10 mM Hepes pH7.9, 10 mM KCl, 1.5 mM Mg2Cl, 0.5 mM DTT, 0.5 mM PMSF and Roche protein inhibitor cocktail) and cleared by centrifugation for 30 min at 4°C. The extracts were incubated with 25 μg of Lag2-specific antibody or rabbit IgG and 10 μl proteinA beads (Amersham Bioscience) overnight at 4°C. Next, beads were washed five times with washing buffer (10 mM Hepes pH7.9, 300 mM KCl, 2 mM CHAPS, 0.5 mM DTT, 0.5 mM PMSF and Roche protein inhibitor cocktail) and bound proteins were denatured in 30 μl sample buffer for 5 min at 95°C.
In vitro neddylation assays
Bacterially expressed complexes (100 ng) were neddylated in reaction buffer (50 mM Tris pH7.5, 1 mM DTT, 10 mM MgCl2, 5 mM ATP) in the presence of 1 mg of 6xHis-Nedd8, 400 ng Nedd8 E1 and 400 ng Nedd8 E2. The reactions were started by the addition of Nedd8 and incubated at 30°C for 30 min. After addition of 3 × Laemmli buffer to stop the reactions, neddylation of Cdc53 was analysed by western blot with anti-His or anti-Cdc53 antibodies.
In vitro rubylation of Cdc53 and Cul1 purified from insect cells was performed with Rub1 phosphorylated on a Protein Kinase A consensus site, as described earlier for NEDD8 (Walden et al, 2003). Cdc53 and Cul1 rubylation was performed with 80 nM Cdc53–Hrt1 or Cul1–Rbx1, 80 nM Dcn1, 8 nM Uba3-Ula1, 80 nM Ubc12, ∼1 μM [32P]-Rub1, and 160 nM Lag2 in 50 mM Tris pH 7.6, 55 mM NaCl, 5 mM MgCl2, 4 mM ATP and 1 mM DTT. To allow Lag2 binding, reactions were incubated at 30°C for 5 min in the absence of Rub1. Reactions were initiated by the addition of either Rub1 or Rub1+Skp1–Cdc4 at the indicated ratios relative to Cdc53–Hrt1. Serial dilutions of [32-P]-Rub1 were separated on SDS–PAGE and exposed simultaneously with reactions on a phosphoimager cassette. The amount of Cdc53–Rub1 formed at each timepoint was calculated from a [32-P]-Rub1 standard curve. Reactions were normalized by setting the amount of Cdc53–Rub1 formed at 40 min in the absence of Lag2 to 100%.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank DT Huang for the pGEX-Uba3-Ula1 plasmid, Y-C Chou and F Sicheri for the GST-TEV-HRT1 clone and for assistance with the poly-cistronic expression vectors, M Schneider and F Rudolf for initial work on Lag2, C Rupp and C Zbinden for technical assistance and members of the Schulman and Peter laboratories for helpful discussions. This work was supported by funding from the NIH (R01GM069530) and ALSAC (American Lebanese Syrian Associated Charities) to BAS, and the ETHZ, Oncoswiss and the Swiss National Science Foundation to MP. BAS is an Investigator of the Howard Hughes Medical Institute, and TK a recipient of a Marie-Curie Intra-European fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amberg DC, Burke D, Strathern JN (2005) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 2005 edn. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 [DOI] [PubMed] [Google Scholar]
- Bornstein G, Ganoth D, Hershko A (2006) Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci USA 103: 11515–11520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Wang X, Yamoah K, Chen PL, Pan ZQ, Huang L (2008) Characterization of the human COP9 signalosome complex using affinity purification and mass spectrometry. J Proteome Res 7: 4914–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119: 517–528 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, Omata M, Tanaka K (2001) NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J 20: 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F (2008) Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell 29: 23–35 [DOI] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O'Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M (2005) The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435: 1257–1261 [DOI] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M (1998) Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev 12: 914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A (1999) Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci USA 96: 5510–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 10: 1511–1518 [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385 [DOI] [PubMed] [Google Scholar]
- Pick E, Pintard L (2009) In the land of the rising sun with the COP9 signalosome and related Zomes. Symposium on the COP9 signalosome, Proteasome and eIF3. EMBO Rep 10: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244 [DOI] [PubMed] [Google Scholar]
- Studier FW (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Sumara I, Maerki S, Peter M (2008) E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol 18: 84–94 [DOI] [PubMed] [Google Scholar]
- Tan S (2001) A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif 21: 224–234 [DOI] [PubMed] [Google Scholar]
- Verma R, Feldman RM, Deshaies RJ (1997) SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell 8: 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL Jr, Holton JM, Schulman BA (2003) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell 12: 1427–1437 [DOI] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M (1996) Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86: 453–463 [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H (2002a) CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell 10: 1519–1526 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002b) Structure of the Cul1-Rbx1-Skp1-F box Skp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File