Abstract
In Drosophila, the PIWI proteins, Aubergine (Aub), AGO3, and Piwi are expressed in germlines and function in silencing transposons by associating with PIWI-interacting RNAs (piRNAs). Recent studies show that PIWI proteins contain symmetric dimethyl-arginines (sDMAs) and that dPRMT5/Capsuleen/DART5 is the modifying enzyme. Here, we show that Tudor (Tud), one of Tud domain-containing proteins, associates with Aub and AGO3, specifically through their sDMA modifications and that these three proteins form heteromeric complexes. piRNA precursor-like molecules are detected in these complexes. The expression levels of Aub and AGO3, along with their degree of sDMA modification, were not changed by tud mutations. However, the population of transposon-derived piRNAs associated with Aub and AGO3 was altered by tud mutations, whereas the total amounts of small RNAs on Aub and AGO3 was increased. Loss of dprmt5 did not change the stability of Aub, but impaired its association with Tud and lowered piRNA association with Aub. Thus, in germline cells, piRNAs are quality-controlled by dPRMT5 that modifies PIWI proteins, in tight association with Tud.
Keywords: Drosophila, piRNA, PIWI, sDMA, Tudor
Introduction
In RNA silencing, small RNAs of 20–30 nucleotides (nt) trigger gene silencing mechanisms by associating with Argonaute proteins, the core factors of the RNA silencing machinery (Carthew and Sontheimer, 2009; Siomi and Siomi, 2009). Each member of the Argonaute family of proteins contains two domains, the PAZ and PIWI domains, which confer two distinct activities to Argonaute: to associate with small RNAs and to catalytically interfere with target gene expression, respectively (Jinek and Doudna, 2009). Small RNAs guide Argonaute proteins to target transcripts and following this target recognition step, Argonaute proteins exhibit activities for gene silencing, either transcriptionally, or post-transcriptionally (Moazed, 2009; Siomi and Siomi, 2009). In the latter case, Argonaute proteins function as an endonuclease, or Slicer, to cleave the target transcripts, or as a translation inhibitor (Jinek and Doudna, 2009).
PIWI proteins are a subset of Argonaute proteins, primarily expressed in germlines (Siomi and Kuramochi-Miyagawa, 2009). In Drosophila, PIWI proteins consist of three members, Piwi, Aubergine (Aub), and AGO3. All PIWI members are associated with PIWI-interacting RNAs (piRNAs), a subset of germline-specific small RNAs that are mainly derived from transposons and other intergenic regions and which are found in clusters throughout the genome (Aravin et al, 2007; Siomi and Siomi, 2008). Earlier studies have shown that each PIWI protein loads a specific set of piRNAs with unique characteristics, such as piRNA origin and nucleotide and strand bias (Brennecke et al, 2007; Gunawardane et al, 2007). These bioinformatic observations led to the proposal of two models for piRNA production, the amplification loop model and the primary processing pathway (Aravin et al, 2007; Houwing et al, 2007). Dicer activities, which are necessary for processing small interfering RNA and microRNAs (Kim et al, 2009), are dispensable for piRNA production (Vagin et al, 2006). This suggests that the precursors of piRNAs may be single-stranded RNAs. In the amplification loop model, piRNAs induce reciprocal Slicer-dependent cleavage of sense and antisense transcripts of transposons, which is mediated by PIWI proteins. Aub-mediated silencing of sense transcripts generates sense piRNAs, which associate with AGO3 to direct the silencing of antisense transposon transcripts. The products of Slicer activity give rise to antisense piRNAs, which in turn bind to Aub and guide the slicing of sense transposon transcripts to generate sense piRNAs. Thus, in the amplification loop model, the interaction of AGO3 and Aub should occur at least transiently, in vivo. However, how they interact with each other and how the interaction is regulated remain unknown. Factors necessary for the primary processing pathway remain largely unidentified, except that loss of zucchini function leads to a marked reduction of processed piRNAs in ovaries and in ovarian somatic cells (Malone et al, 2009; Saito et al, 2009).
Recently, PIWI proteins in various species, including Xenopus, Drosophila and mice, were shown to have a post-translational modification, called symmetric dimethyl-arginine (sDMA), in their N-terminal regions (Kirino et al, 2009; Reuter et al, 2009; Vagin et al, 2009). sDMA is a methyl-group modification that occurs on specific arginine residues in protein molecules (Bedford and Clarke, 2009). Like other methyl-group modifications found on arginines, such as mono methyl-arginine and asymmetric dimethyl-arginine (aDMA), sDMA is known to modify the ability of a protein to interact with other factors and to modulate its biological activity. A good example is Sm splicing factors (Yong et al, 2004; Neuenkirchen et al, 2008). Nascent Sm proteins, such as B/B′, D1, and D3, have been shown to contain sDMAs in their arginine- and glycine-rich (RG) domains. Further studies showed that a complex, termed the methylosome and which contains protein arginine methyltransferase 5 (PRMT5), a member of the PRMT family of proteins, catalyses sDMA modification in eukaryotes. After such dimethylation at specific arginines in the RG domains, Sm proteins associate though their sDMAs, with a Tudor (Tud) domain-containing protein, survival motor neuron (SMN) protein. This Sm-SMN association concomitantly recruits U snRNA. Finally, by this sequential mechanism, U snRNP is efficiently assembled. Recent studies have shown that the PRMT5-SMN-system serves as a chaperone system that prevents the mis-assembly of Sm proteins to non-target RNA and also prevents Sm protein aggregation (Pellizzoni et al, 2002; Chari et al, 2008). Many other Tud-domain proteins have been shown to interact with particular proteins, and this binding requires methylated arginine residues in the target proteins (Côté and Richard, 2005). These observations in mammalian cells led us to infer that PIWI proteins with sDMA modification could be associated with Tud domain-containing proteins and that such sDMA-specific interaction may regulate piRNA association with PIWI proteins in germline cells. In fact, a fly homolog of PRMT5, dPRMT5 (also known as DART5, or Capsuleen (Csul)) (Gonsalvez et al, 2006; Anne et al, 2007) was shown to be responsible for the sDMA modification of PIWI proteins in Drosophila (Kirino et al, 2009).
In this study, we found that in Drosophila, a Tud domain-containing protein, Tud, associates with Aub and AGO3 in an sDMA-dependent manner. The fly homolog of mouse Tud domain-containing protein-1 (Tdrd1), dTdrd1, and Spindle-E (Spn-E), both of which belong to the fly Tudor domain-containing protein family, did not show sDMA-dependent interaction with Aub or AGO3. It seems that Tud, Aub, and AGO3 are able to form various heteromeric complexes in different combinations. Of these, particular complexes that contain both Tud and Aub (and/or AGO3) contained pre-piRNA-like molecules. The expression levels of Aub and AGO3, along with their degree of sDMA modification, were not changed by tud mutations. However, the association of transposon-derived piRNAs with Aub and AGO3 was altered by the loss of tud, whereas the total amounts of small RNAs on Aub and AGO3 were increased. Loss of dprmt5 did not change the stability of Aub, but did impair its association with Tud and lowered piRNA association with Aub. Taken together, these results suggest that, in germline cells, piRNAs are quality-controlled by two factors: dPRMT5, which sDMA-modifies PIWI proteins, and Tud, which associates with PIWI proteins specifically through such sDMA modification.
Results
LC-MS/MS analysis of Aub-sDMA modification
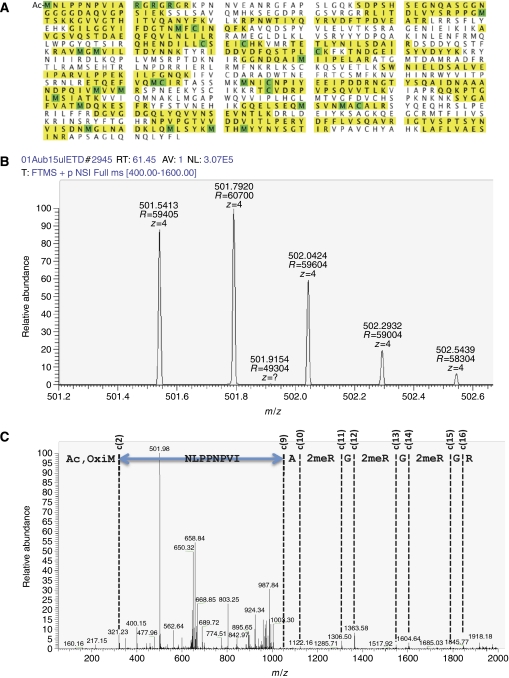
To examine which arginines in Aub are sDMA modified, LC-MS/MS analysis was performed on Aub immunopurified from ovaries using anti-Aub antibodies (Gunawardane et al, 2007; Nishida et al, 2007). The isolated protein was first trypsinized and analysed with LC-MS. A database search confirmed the protein as Aub with 54% sequence coverage (Figure 1A). The m/z=501.5413 peak of the MS spectrum (Figure 1B) matched to the N-terminal end of Aub, spanning from methionine1 to arginine17 (M1–R17). N-terminal acetylation, oxidation on M1, and three sDMAs on arginines, R11, R13, and R15 (Figure 1C) were observed with a parent mass error of 0.4 p.p.m. and a Mascot ion score of 38. No other peptides corresponding to M1–R17, with or without partial modifications, were detected.
Figure 1.
LC-MS/MS analysis of Aub modification. (A) Aub peptide identified by database searching. Letters with yellow background are search hits. Those with green background indicate amino acids with modifications; of those, M, R, and C indicate methionine with oxidation, arginine with sDMA, and cysteine with carbamidomethylation, respectively. Ac on the first methionine indicates that the amino acid is N-terminal acetylated. (B) MS spectrum of the Aub peptide, from M1 to R17 (M1–R17; m/z=501.5413, theoretical value=501.5415). (C) ETD MS/MS spectrum of the Aub peptide, M1–R17. Ion masses in the fragmentation well match those of C ions from the Aub peptide, spanning from alanine10 to glycine16 (A10–G16) A-2meR-G-2meR-G-2meR-G-R, where ‘2meR' indicates an arginine that is sDMA modified. Fragmentation peaks of the region, asparagine2 to isoleucine10 (N2–L9), are not assigned.
Tud associates with Aub in an sDMA-dependent manner
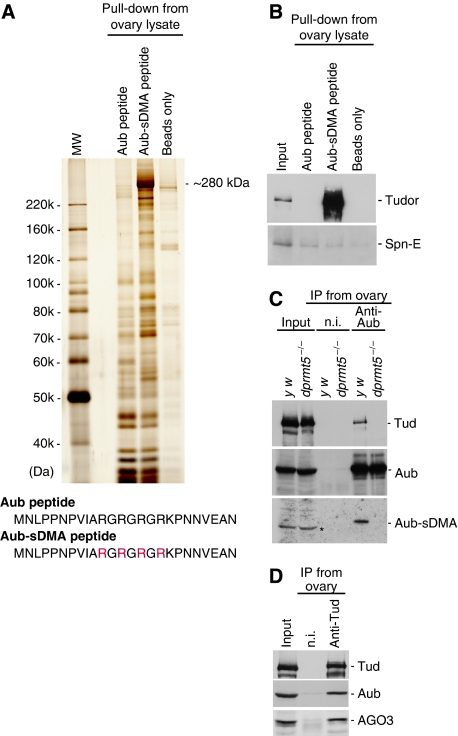
To identify proteins that associate with Aub in a manner dependant on sDMA modification, pull-down assays were performed using a peptide corresponding to the first 25 amino acids of Aub (the sequence is shown in Figure 2A). The peptide was synthesized with and without sDMAs according to the LC-MS/MS results (Figure 1). We could not entirely exclude the possibility of an sDMA modification on R17; therefore, we made the Aub-sDMA peptide with sDMA on R17. A prominent, ∼280 kDa protein was shown to be specifically associated with the sDMA-containing Aub (Aub-sDMA) peptide (Figure 2A). MS analysis identified this protein as Tud, one of the Tudor domain-containing proteins expressed in ovaries. Mutants lacking Tud and Aub show very similar phenotypes; for example, both fail to make germplasm, which is a specialized, maternally provided cytoplasm that is sequestered at the posterior pole of the oocyte during oogenesis and that contains the determinants of the germline (Snee and Macdonald, 2004; Thomson and Lasko, 2004; Arkov et al, 2006). Thus, the identification of Tud in this experiment as a candidate Aub-binding partner is consistent with the findings of earlier genetic analyses.
Figure 2.
Tud is specifically associated with sDMA-modified Aub. (A) Pull-down assays from ovary lysates were performed using Aub peptides corresponding to methionine1 to asparagine25 (M1–N25). The sequences of the peptides are shown beneath the figure. ‘Aub peptide' and ‘Aub-sDMA peptide' were made without and with sDMA modification, respectively. Rs in red indicates Rs that were sDMA modified. A prominent protein of ∼280 kDa associated specifically with the Aub-sDMA peptide. MS analysis revealed that the protein corresponds to Tud. (B) Western blot analysis with anti-Tud antibodies of the protein pools associated with Aub peptides in (A) confirmed that the ∼280 kDa band is Tud. Spn-E was not detected in either protein pool. (C) Immunoprecipitation experiments were performed from ovary lysates of wt and dprmt5 mutants using anti-Aub antibodies. Non-immune IgG (n.i.) was used as a negative control. The immunopurified complexes were then probed with anti-Tud, anti-Aub, and SYM11, an antibody specifically recognizing sDMA-modified proteins. Tud was observed only in the Aub complex obtained from wt ovaries. Aub in dprmt5 mutants was not detected with SYM11, as has been reported earlier (Kirino et al, 2009). The signals marked with an asterisk in ‘Input' lanes were background. (D) The anti-Tud immunopurified complexes from wt ovary lysates were probed with anti-Tud, anti-Aub, and anti-AGO3 antibodies. Both Aub and AGO3 were detected in the complexes, suggesting the sDMA-dependent association of Tud with AGO3.
Western blot analysis using anti-Tud antibodies confirmed that Tud is abundant in the protein pool obtained from the Aub-sDMA peptide pull-down (Figure 2B). An earlier study showed that Aub and Tud co-precipitate; however, no evidence was provided for their direct interaction or for the requirement of the sDMA modification (Thomson et al, 2008). Another Tud domain-containing protein, Spn-E (also known as Homeless) (Gillespie and Berg, 1995; Ponting, 1997), was not detected with either of the Aub peptides, regardless of sDMA modification (Figure 2B). The specificity of the anti-Spn-E antibodies raised in our laboratory is shown in Supplementary Figure 1A and B. In mice, Tdrd1 associates with Mili, Miwi, and Miwi2 (PIWI proteins in mice) in an sDMA-dependent manner throughout spermatogenesis (Reuter et al, 2009; Vagin et al, 2009); however, the closest homolog of Tdrd1 in flies, dTdrd1 (CG14303), was only weakly associated with the Aub peptides, independent of sDMA modification (Supplementary Figure 1C). Thus, dTdrd1 does not seem to be the functional counterpart of mouse Tdrd1. Mouse Tdrd6 is a putative orthologue of Drosophila Tud, based on their peptide sequence similarities (Hosokawa et al, 2007), and Tdrd6 was shown to bind Miwi and Mili (Vagin et al, 2009; Vasileva et al, 2009). However, sDMA-dependency of such interactions remains undetermined.
dprmt5 deletion mutant flies, csulRM50/Df(2R)Jp7, showed a strong impairment in Aub-sDMA modification (Kirino et al, 2009). Using another dprmt5 mutant strain, csule00797/csule00797, we also verified that dPRMT5 is required for Aub-sDMA modification, (Figure 2C). In the dprmt5 mutant ovaries, the association of Tud with Aub was no longer detectable (Figure 2C). These data correlate well with the observation that Tud recognizes the sDMA-modified Aub peptide as a binding partner (Figure 2A and B). Immunoprecipitation from wild-type (wt) ovary lysates using anti-Tud antibodies confirmed the association of Aub with Tud (Figure 2D). Like Aub, AGO3 is also sDMA modified in ovaries (Kirino et al, 2009). Thus, we also performed western blot analysis on the anti-Tud immunoprecipitates with anti-AGO3 antibodies and AGO3 was also detected in the complexes (Figure 2D). We inferred from these data that Tud might also recognize AGO3 in an sDMA-dependent manner.
Tud also associates with AGO3 in an sDMA-dependent manner
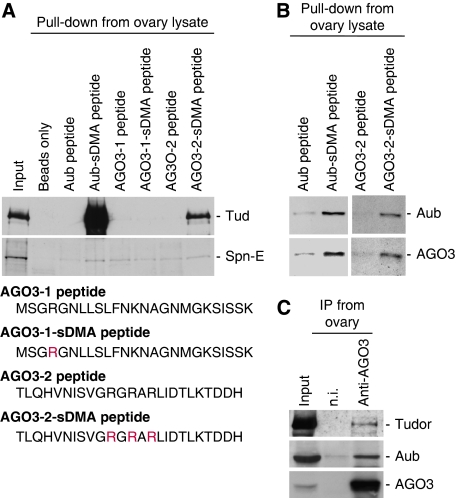
To determine whether Tud indeed associates with AGO3 in an sDMA-dependent manner, we produced AGO3 peptides, AGO3-1 and AGO3-2, which correspond to the far N-terminal end and to the region from threonine58 to histidine82 (T58–H82) of AGO3, respectively. Both peptides were synthesized with and without sDMAs (Figure 3A) according to the results of the LC-MS/MS analysis, which revealed that R4, R68, and R70 of AGO3 have sDMAs (Supplementary Figure 2). The possibility that R72 is also sDMA modified was not entirely clarified; thus, AGO3-2 peptide was synthesized with the R72-sDMA modification (Figure 3A). Pull-down experiments from ovary lysates revealed that Tud was able to associate with AGO3, through the T58–H82 peptide only, in an sDMA-dependent manner (Figure 3A). Spn-E was again undetected with all the peptides used (Figure 3A). We also confirmed that the AGO3-2-sDMA peptide even without R72-sDMA interacted with Tud (Supplementary Figure 3A). To examine whether Tud, when associated with Aub (or AGO3), co-associated with AGO3 (or Aub), the protein pools pulled down with the peptides were probed with anti-Aub and anti-AGO3 antibodies. Although the signals were not so strong, Aub and AGO3 were both specifically detected with the Aub-sDMA and AGO3-2-sDMA peptides (Figure 3B). In addition, the anti-AGO3 immunoprecipitates contained both Tud and Aub (Figure 3C). Linear density gradient experiments also showed that Tud, Aub, and AGO3 co-sedimented even in relatively heavy fractions (Supplementary Figure 3B). These results suggest that Tud might, to some extent, simultaneously associate with Aub and AGO3 and form a heteromeric complex. However, even in tud mutant ovaries Aub was able to interact with AGO3 (Supplementary Figure 3C). Although we cannot exclude the possibility that loss of tud could somehow promote association of Aub with AGO3, we speculated that Aub and AGO3 might not constantly require Tud as their bridging molecule. Taken together, we concluded that Tud, Aub, and AGO3 are likely to form multiple, heterogeneous complexes in different combinations in vivo.
Figure 3.
The sDMA-dependent association of Tud with AGO3. (A) Two AGO3 peptides, AGO3-1 and AGO3-2, which correspond to methionine1 to lysine25 (M1–K25) and to threonine58 to histidine82 (T58–H82), respectively, were synthesized with and without sDMA modification and pull-down assays were performed from ovary lysates as in Figure 2A. Rs in red indicates those Rs that were sDMA modified. The protein pools obtained were probed with anti-Tud and anti-Spn-E antibodies. Signals for Tud, but not for Spn-E, were detected in pools pulled-down with Aub-sDMA and AGO3-2-sDMA, but not with AGO3-1-sDMA peptides, indicating that the AGO3 region, T58–H82, is the binding domain for Tud. (B) Proteins bound with Aub, Aub-sDMA, AGO3-2, and AGO3-2-sDMA peptides were probed with anti-Aub and anti-AGO3 antibodies. Both proteins were detected specifically with the sDMA peptides. These results suggest that Tud is able to simultaneously associate with Aub and AGO3 to form a heteromeric complex. (C) The anti-AGO3 immunopurified complexes from wt ovary lysates were probed with anti-Tud, anti-Aub, and anti-AGO3 antibodies.
Tud function in the piRNA processing pathway
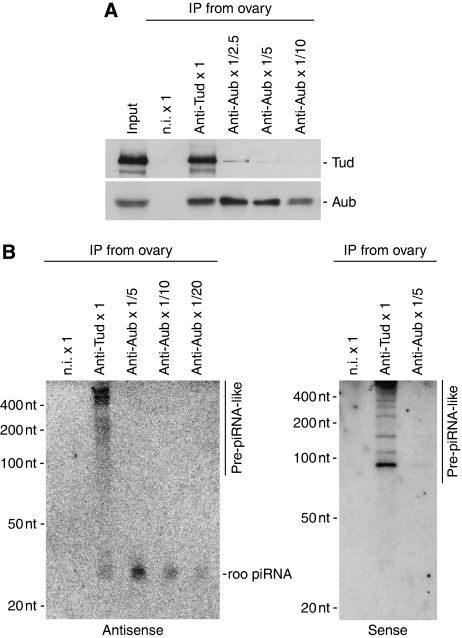
The observations that Tud associates with Aub and AGO3 in an sDMA-dependent manner (this study) and that the PRMT5-SMN-system serves as a chaperone system that promotes proper assembly of U snRNP (Pellizzoni et al, 2002; Yong et al, 2004; Chari et al, 2008; Neuenkirchen et al, 2008) raised the possibility that the sDMA-dependent association of Tud with Aub and AGO3 might promote association of piRNAs with the PIWI proteins. We first compared the amounts of piRNAs in anti-Aub and anti-Tud immunoprecipitates. When approximately equal amounts of antibodies and ovary lysates were used, the anti-Aub immunoprecipitates contained a greater abundance of Aub compared with the anti-Tud immunoprecipitates (data not shown); thus, to equalize Aub quantities, the anti-Aub immunoprecipitates were sequentially diluted before western blot analysis. Quantified results showed that a 1 in 5 dilution equalized the amount of Aub in the anti-Aub and anti-Tud immunoprecipitates (Figure 4A). We then performed northern blot analysis to visualize the roo piRNAs associated with both complexes. Far fewer roo piRNAs were found to be associated with Aub residing in the anti-Tud immunoprecipitates compared with Aub in the anti-Aub immunoprecipitates (which had been diluted 1:5 before analysis) (Figure 4B). This suggested that Aub, in a form associated with Tud, is not significantly loaded with mature piRNAs. We also noticed that the anti-Tud immunoprecipitates (but not Aub immunoprecipitates) contained a smeary band, spanning ∼80 to 1000 nt, which was positively recognized by roo piRNA probes (Figure 4B). The simplest explanation for these observations is that the smear corresponds to roo piRNA precursors. RT–PCR, using primers recognizing roo transcripts in total RNAs isolated from the anti-Tud immunoprecipitates, showed that part of the roo antisense transcript (∼500 nt), which supposedly gives rise to multiple roo piRNAs, exists in the complex (Supplementary Figure 4A). Therefore, antisense roo piRNA precursor-like molecules were confirmed in the Tud complex. Interestingly, probes in the opposite orientation, which should recognize sense roo piRNAs, also identified the smeary band and also some distinct bands within the smeary band (Figure 4B). Sense roo piRNAs were barely detected in the anti-Aub immunoprecipitates, as expected (Figure 4B). RT–PCR using primers that only detect the roo sense transcript showed that the anti-Tud immunoprecipitates contain sense roo piRNA precursor-like molecules (Supplementary Figure 4B). These results suggest that the Tud complex is able to hold piRNA precursor-like molecules in both sense and antisense orientations, but most likely as single-stranded forms. A mechanism analogous to U snRNP assembly may occur for piRNA precursor recruitment to the heteromeric Tud/Aub/AGO3 complexes, previously assembled through Tud recognition of sDMA-Aub and sDMA-AGO3. Tud may dissociate from the complex after piRNA processing and association with PIWI proteins because Aub, in a form associated with Tud, contains only a low level of mature piRNAs.
Figure 4.
Mature piRNAs and piRNA precursor-like molecules contained in the Tud–Aub complex. (A) The immunoprecipitated complexes from ovary lysates using anti-Tud and anti-Aub antibodies were probed with anti-Tud and anti-Aub antibodies. Prior to the western blot analyses, the anti-Aub immunoprecipitate was diluted 1:2.5, 1:5, and 1:10. It should be noted that the ∼1:5 dilution of the Aub complex equalized the amounts of Aub in the anti-Tud and anti-Aub immunoprecipitated complexes. (B) RNA molecules isolated from the anti-Tud and anti-Aub immunoprecipitated complexes were probed with 32P-labelled DNA oligos that recognize roo piRNAs (antisense and sense on left and right panels, respectively). Although ∼1:5 dilution of the Aub complex equalized the amounts of Aub in both complexes, lower levels of roo piRNAs were detected in the anti-Tud immunoprecipitate (left panel), indicating that Aub, whereas bound to Tud, is associated with low levels of piRNAs. piRNA precursor-like signals (indicated as pre-piRNA-like) were observed in the anti-Tud immunoprecipitates with both sense and antisense probes, but sense roo piRNAs were not detected in Tud- or Aub-immunoprecipitated complexes (right). We speculate that the association of Tud with Aub and AGO3, through sDMA modification, recruits piRNA precursors to the complex, in a mechanism analogous to the recruitment of U snRNA into the SMN-Sm protein complex.
Tud provides quality-control of piRNA association with Aub and AGO3
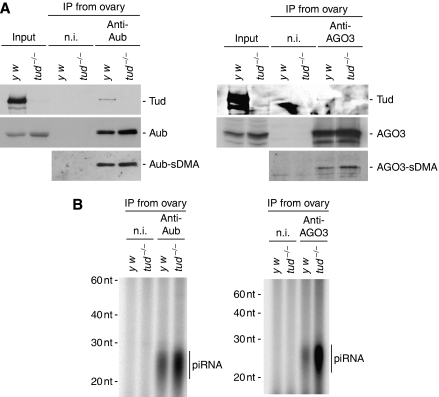
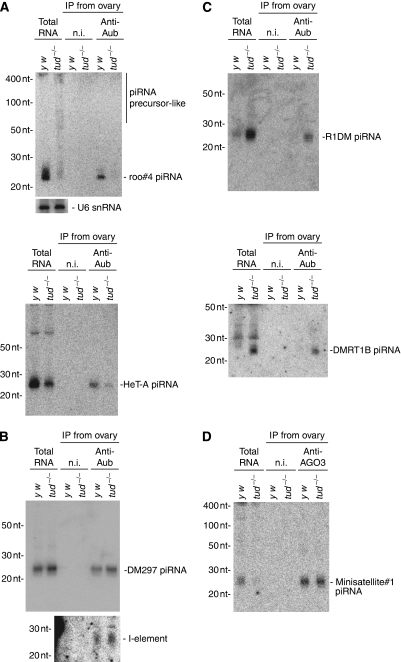
We then asked whether loss of tud affects piRNA loading onto Aub and AGO3. Western blot analysis of tud mutant and wt ovary lysates revealed that the expression levels of Aub and AGO3, along with their degree of sDMA modification, were not changed by mutations in tud (Figure 5A). 32P-labelled piRNAs in tud and wt ovaries showed that tud mutations approximately doubled and quadrupled the amounts of piRNAs loaded onto Aub and AGO3, respectively (Figure 5B). Northern blot analyses, however, in tud mutants, roo#4 piRNA, a piRNA derived from roo transcripts, was loaded onto Aub much less efficiently compared with that in wt ovaries (Figure 6A). A very similar result was obtained when probes recognizing piRNAs derived from Het-A transposon transcripts in the antisense orientation were used (Figure 6A). In contrast, an alkaline-treated, single-stranded RNA probe that detects I-element-derived piRNAs showed that a slightly larger, but still negligible amount of I-element piRNAs was loaded onto Aub in tud mutants (Figure 6B). A DM297-derived piRNA behaved in the same way (Figure 6B). Interestingly, some other piRNAs, derived from R1Dm and DMRT1B, were more abundantly associated with Aub in the tud mutants (Figure 6C), suggesting that loss of tud function might affect the population of piRNAs that associate with Aub. To comprehensively examine changes to the piRNA population due to mutations in the tud gene, deep sequencing was performed for small RNAs associated with Aub in tud ovaries and in wt controls. Strand bias was not significantly affected (Supplementary Figure 5A); however, as inferred from northern blotting results, we noticed that the population of transposon-derived piRNAs was altered by loss of tud (Supplementary Figure 5B). The degree of occupancy of small RNAs corresponding to other RNAs, such as rRNA and tRNA, were not changed by the loss of tud function (Supplementary Figure 5C). In this regard, Tud function does not simply mimic that of Tdrd1 in mice. In mice, loss of tdrd1 caused association of aberrant piRNAs with PIWI proteins, which originated largely from rRNA and/or tRNA (Reuter et al, 2009). A piRNA that was previously recognized to be loaded onto AGO3 (termed minisatellite#1) (Gunawardane et al, 2007) was detected at about equal levels in AGO3 complexes in tud and wt ovaries (Figure 6D), although far fewer piRNAs were detected to be associated with AGO3 in wt ovaries (Figure 5B). These results indicate that loss of tud function significantly affects the population of piRNAs loaded onto Aub and AGO3. Thus, Tud provides quality-control of PIWI-piRNAs by specifically associating with Aub and AGO3, through dPRMT5-mediated sDMA modification.
Figure 5.
Effects of tud mutations on piRNA loading onto Aub and AGO3 in ovaries. (A) The anti-Aub (left) and anti-AGO3 (right) immunoprecipitates from wt (y w) and tud mutant ovaries were probed with anti-Tud, anti-Aub, andi-AGO3, and SYM11 (bottom panels). tud mutations did not change the stability or the sDMA modification of Aub or AGO3. (B) Small RNAs associated with Aub (left) and AGO3 (right) in wt and tud mutants were visualized by 32P-labelling. Immunoprecipitation of approximately equal amounts of Aub (left) and AGO3 (right) were checked by western blot analysis, as in (A). The total amounts of small RNAs associated with Aub and AGO3 in tud mutant ovaries were about two-fold and four-fold greater, respectively, compared with that in wt ovaries (this calculation was done with three individual sets of experimental data).
Figure 6.
Northern blot analyses for piRNAs associated with Aub and AGO3. (A) Northern blot analyses of RNAs illustrated in Figure 5B. DNA oligo probes for a roo piRNA, roo#4 piRNA, and HeT-A piRNAs were used. Although the total amount of small RNAs associated with Aub in tud mutant ovaries was two-fold greater compared with that in wt ovaries (Figure 5B), roo#4 and HeT-A-derived piRNAs in the Aub complex in tud mutant ovaries were at much lower levels compared with those in wt ovaries. Interestingly, roo piRNA precursor-like signals were observed to accumulate in tud mutant ovaries. (B) An alkaline-treated RNA probe (∼200 nt) for I-element-derived piRNAs and a DNA oligo for DM297-derived piRNA were used. (C) DNA oligos for R1DM- and DMRT1B-derived piRNAs were used. (D) Northern blot analyses of RNAs illustrated in Figure 5B (right). A DNA oligo that recognized a piRNA, minisatellite#1, was used. Similar signals were detected in the AGO3 lanes of both wt and tud mutant.
sDMA modification of Aub affects PIWI-piRNA loading
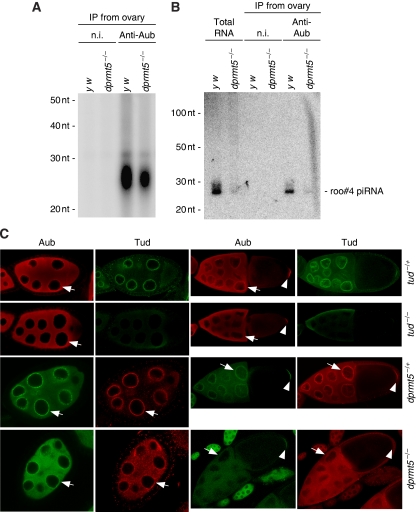
We then asked whether piRNA loading onto Aub is also affected by sDMA modification. We found that loss of dprmt5 does not affect the stability of Aub (Figure 2C), although this is in contrast to earlier findings (Kirino et al, 2009). We could, however, confirm that Aub is no longer dimethylated in dprmt5 mutants and thus did not co-immunoprecipitate with Tud (Figure 2C). 32P-labelling of small RNAs associated with Aub in dprmt5 and wt ovaries showed that loss of sDMA modification led to lower levels of piRNAs associated with Aub (Figure 7A), although the amounts of Aub in both immunoprecipitates were equal to each other (Figure 2C). Northern blot analysis for roo#4 piRNA confirmed that piRNA loading onto Aub was severely lowered by dprmt5 mutations (Figure 7B). These results imply that the sDMA modification itself has an impact on the association of piRNAs with Aub.
Figure 7.
Loss of dprmt5 function reduces the loading of piRNAs onto Aub in ovaries. (A) Small RNAs associated with Aub in wt and dprmt5 mutants were visualized by 32P-labelling. Immunoprecipitation of approximately equal amounts of Aub were checked by western blot analysis (as in Figure 2C). The total amount of small RNAs associated with Aub in dprmt5 mutant ovaries was approximately three-fold less compared with that in wt ovaries (the calculation was done with three individual sets of experimental data). (B) Northern blot analyses of RNAs illustrated in (A). A DNA oligo that recognized roo#4 piRNA was used. The signal was hardly detected in the dprmt5 mutant lanes. Like tud, loss of dprmt5 function seems to cause aberrant piRNA loading onto Aub, but the extent was lower than that in tud mutants. (C) Immunohistochemical analyses of tud and dprmt5 mutant egg chambers using anti-Aub and anti-Tud antibodies. In tud mutants, accumulation of Aub and Tud in the nurse cell nuage (indicated with an arrow) was severely impaired and the two proteins were distributed more evenly in the cytoplasm. In stage 10 egg chambers of dprmt5 and tud mutants, Aub accumulates to similar extents at the posterior pole; thus, Aub accumulation at the posterior pole (indicated with an arrow head) is not entirely dependent on both sDMA modification and association with Tud. Tud accumulation at the posterior pole in dprmt5 mutants was totally abolished as is seen in vls mutants (Anne and Mechler, 2005).
Aub localization in tud and dprmt5 mutants
How does the impairment in binding Tud and Aub in dpmrt5 and tud mutants affect the localization of Aub in egg chambers? To investigate this question, we immunostained ovaries of dprmt5 and tud mutants using anti-Aub and anti-Tud antibodies. Earlier studies have shown that both Aub and Tud are highly enriched both in the nuage, a germline-specific electron-dense material associated with nurse cell nuclei, and the posterior pole (Bardsley et al, 1993; Snee and Macdonald, 2004; Thomson and Lasko, 2004). In the germarium, wild-type staining patterns of Aub were seen in dpmrt5 and tud mutants (data not shown). However, in stage 8 nurse cells, the accumulation of Aub in the nuage was greatly impaired in both mutants and Aub was evenly dispersed in the cytoplasm of mutant nurse cells (Figure 7C). These data suggest that both the sDMA modification and the association with Tud are required for Aub to accumulate in the nuage. Furthermore, Tud accumulated at very low levels in the dprmt5 nuage, as is also seen in valois (vls) mutants (Anne and Mechler, 2005). Valois belongs to the superfamily of WD-repeat proteins and forms a complex with dPRMT5. Valois also accumulates in the nuage and the posterior pole in egg chambers and associates with Tud in vitro (Anne and Mechler, 2005). Although it is not known if Aub is unmethylated in vls mutants, as it is in dprmt5 mutants (this study and Kirino et al, 2009), it is speculated that Tud accumulation in the nuage might be reciprocally dependent on the sDMA modification of Aub and/or AGO3. However, in stage 10 egg chambers of dprmt5 and tud mutants, we observed that Aub accumulates at the posterior pole to a similar extent to that in wt; thus, Aub accumulation at the posterior pole only partially depends on both sDMA modification and association with Tud (Figure 7C). Tud accumulation to posterior pole in dprmt5 mutants was totally abolished (Figure 7C), as is seen in vls mutants (Anne and Mechler, 2005), meaning that Tud accumulation at the posterior pole depends on the sDMA modification of Aub, or of other, still unknown but dPRMT5-dependently sDMA-modified proteins.
Discussion
Argonaute proteins are non-sequence-specific RNA-binding proteins. In an in vitro environment, recombinant Argonaute produced from Escherichia coli can be artificially loaded with small RNAs of a variety of sequences by simple incubation in solution (Miyoshi et al, 2005; Rivas et al, 2005). This incubation results in the formation of an ‘active enzyme' that can cleave any given RNA molecule harboring a sequence completely complementary to the small RNA. If this disorderly process occurred in vivo, it would instantly threaten the life of a cell by possibly targeting important cellular RNAs. Therefore, to avoid such a dangerous situation, the RNA silencing system mediated by Argonaute proteins and small RNAs requires ‘gatekeepers' to ensure that Argonaute proteins are loaded with functional small RNAs. An important step towards a detailed understanding of piRNA biogenesis in vivo is to elucidate how Argonaute proteins specifically distinguish their guide small RNAs among the myriad of RNA sequences in cells. In this study, we biochemically characterized germline-specific Argonaute proteins, Aub and AGO3, and demonstrated that, at least in ovaries, a regulatory system to control piRNA association, or loading, is available and that the system is elaborate. It consists of two consecutive steps: the post-translational modification of Argonaute proteins, namely dPRMT5-mediated sDMA modification of Aub and AGO3, and the selective association of Aub and AGO3 with a specific protein, namely sDMA-dependent association of Aub and AGO3 with Tud.
Without dPRMT5, Aub is not modified to contain sDMAs. Loss of sDMA modification does not change the expression levels of Aub in ovaries, suggesting that sDMA itself does not function as an index for Aub stabilization in the cellular environment. However, in dprmt5 mutants, although Tud is still present, Aub is not recognized by Tud as a binding substrate and instead of accumulating in the nuage of nurse cells, Aub disperses in the cytosol, where it would be free to bind, in a rather non-specific manner, with single-stranded RNA molecules. Interestingly, deep-sequencing results show that the majority of the single-stranded RNA molecules selected by Aub are fragments of transposon transcripts. These RNAs captured by Aub might then be trimmed by unknown ribonucleases to the size of piRNAs in the same way that piRNA precursors are processed by ribonucleases in the processing pathway. Even in dprmt5 mutants, Aub shows substantial association with small RNAs the size of piRNAs. This small RNA population, however, seems to be different from that seen in the wt as northern blot analysis revealed that Aub in dprmt5 mutants is loaded with fewer transposon-originating piRNAs. Once Aub is loaded with small RNAs, regardless of whether they are ‘genuine' or ‘aberrant', it accumulates at the posterior pole of oocytes. This suggests that the transport machinery for accumulating Aub at the posterior pole cannot distinguish between Aub associated with genuine piRNA and Aub associated with other non-genuine piRNA molecules. Like Aub, Tud also lost the ability to accumulate in the nuage of dprmt5 mutants, indicating that the ability of Tud to accumulate in the nuage depends on its association with Aub (and AGO3) through sDMA modification.
How does Aub behave in tud mutants? Our data showed that in tud mutant ovaries, Aub is as stable as it is in wt ovaries. Also, Aub in tud mutants was sDMA modified to the same extent as Aub in wt; thus, in tud mutants, Aub, and most likely AGO3 as well, maintain the ability to be recognized as binding substrates by Tud. However, Tud itself is not available. This is why the tud mutant is largely a phenocopy of the dprmt5 mutant, although the severity of the phenotype was higher in the dprmt5 mutant (MCS and KMN, unpublished data). The only clear phenotypic difference found between tud and dprmt5 mutants in our study was that the total amount of small RNAs loaded onto Aub in tud mutants was much greater compared with that in dprmt5 mutants. One clear difference at the molecular level between tud and dprmt5 mutants was the sDMA modification; in tud mutants, Aub is sDMA modified, as in wt, whereas Aub is not sDMA modified in dprmt5 mutants. These data suggest that the RNA-binding ability of Aub is significantly affected by the presence or absence of sDMA modification. More precisely, Aub sDMA modification elevates the affinity of Aub for piRNAs.
As described earlier, in the piRNA biogenesis system, and especially in the amplification loop system, Aub and AGO3 should be located in intimate proximity to each other to enable the reciprocal passing of piRNA intermediates that have been cleaved from their primary transposon transcripts by the Slicer (endonuclease) activity of Aub and AGO3. This Aub and AGO3-mediated RNA cleavage is thought to be necessary for determining and forming the 5′ ends of piRNAs. However, the physical interaction between Aub and AGO3 in Drosophila has not been demonstrated. In this study, we show for the first time that in ovaries, Aub and AGO3 are able to be simultaneously associated with Tud. Even in tud mutants, Aub and AGO3 interacted with each other, indicating that Tud is not constantly required for the association between Aub and AGO3. However, in tud mutants, Aub does not accumulate in the nuage of nurse cells and Aub-associated piRNAs showed a clear difference in population from those of the wt control. Thus, Tud is clearly necessary for maintaining the proper association of piRNAs with PIWI proteins. It should be emphasized that Tud function does not equate to that of Tdrd1 in mice, because Mili piRNAs devoid of Tdrd1 accept the entry of abundant cellular transcripts, such as rRNA and tRNA, into the piRNA pathway (Reuter et al, 2009).
The heteromeric complexes consisting of Tud, Aub, and/or AGO3 held piRNA precursor-like molecules a few hundred nt long. This size range suggests that they do not correspond to the primary precursors of piRNAs or to the primary transcripts of transposons. However, the specificity shown by the northern blot analysis indicates that they might correspond to piRNA precursors that are ready to be processed by Aub and AGO3. This was further confirmed by RT–PCR experiments using primers for certain transposon transcripts. These findings suggest that Tud stringently scrutinizes RNAs for their specific features and passes them to PIWI proteins that are physically associated with Tud. Thus, the cooperation of dPRMT5 with Tud can be envisaged as a two-step regulatory unit for PIWI function in germline cells. The requirement of Tud in the primary piRNA processing pathway remains unclear, and is under investigation in our laboratory.
Materials and methods
Trypsin digestion and LC-MS/MS analysis
The method for trypsin digestion of protein has been described earlier (Fujinoki et al, 2003). LC-MS/MS analysis was performed using a LTQ orbitrap XL electron transfer dissociation (ETD) mass spectrometer (Thermo Fisher Scientific). The methods used for LC-MS/MS were slightly modified from those described earlier (Fujii et al, 2004). The mass spectrometer was operated in a data-dependent acquisition mode in which the MS acquisition with a mass range of m/z 420–1600 was automatically switched to MS/MS acquisition under the automated control of Xcalibur software. The top four precursor ions in an MS scan were selected by Orbitrap, with resolution R=60 000 and in subsequent MS/MS scans by ion trap in the automated gain control (AGC) mode where the AGC values were 5.00 × 105 and 1.00 × 104 for full MS and MS/MS, respectively. To analyse dimethylation sites, ETD was used.
Database searching and protein identification
Database searches were performed using the MASCOT search engine (Matrix Science) against the NCBInr_20090513 database (selected for Drosophila), assuming trypsin as the digestion enzyme and allowing for trypsin specificity of up to four missed cleavages. The database was searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 3.0 p.p.m. The iodoacetamide derivative of cysteine was specified as a fixed modification and methylation of arginine, oxidation of methionine, dimethylation of arginine and acetylation of N-termini were specified as variable modifications. Scaffold (version Scaffold_2_02_03; Proteome Software) was used to validate MS/MS-based peptide and protein identifications. We accepted the peptide identifications when the Peptide Prophet algorithm (Keller et al, 2002) specified probabilities at >95.0%. Sequence coverage was defined as percentage of the protein in the identified peptide sequence.
Drosophila strains
The yellow white (y w) strain was used as a wild-type strain. The dprmt5 allele used was csule00797 (a kind gift from AG Matera). The PBac{RB}csule00797 insertion is located within exon 2, which generates a protein without the methyltransferase domain (Gonsalvez et al, 2006). The tud allele used was tud1 bw sp/CyO l(2)DTS5131 (Drosophila Genetic Resource Center stock number; 106505). tud1 has a stop codon (Lys1036UAG) mutation, which effectively abolishes protein expression, presumably because of the instability of the Tud fragment or because of degradation of tud mRNA through nonsense-mediated RNA decay (Arkov et al, 2006). All stocks were maintained at 25°C.
Pull-down assay
Aub, AGO3-1, and AGO3-2 peptides, with and without sDMA modification, were synthesized by Operon. All peptides were biotinylated at their N-terminal ends. The synthesized peptides (2 μg) were bound with 30 μl of streptavidin Dynabeads (Invitrogen) and incubated for 2 h with ovary lysates at 4°C. Ovaries lysates (from about 200 ovaries) were prepared by crushing in 200 μl of Binding buffer [30 mM HEPES-KOH (pH 7.3), 150 mM KOAc, 5 mM MgOAc, 5 mM DTT, 0.1% NP-40, 2 μg/ml Pepstatin, 2 μg/ml Leupeptin, and 0.5% Aprotinin]. After centrifugation at 14 000 g for 1 min at 4°C, the supernatant was transferred to a new microcentrifuge tube and kept on ice. The pellet was then crushed again in 200 μl of Binding buffer. After centrifugation as above, the supernatant was combined with the first supernatant and kept on ice. These steps were repeated several times. The protein concentration of the lysates was adjusted each time to 4 mg/ml with Binding buffer. After incubation with 1 ml of ovary lysate, the bead fractions were extensively washed with Binding buffer and the protein pools bound to the beads were eluted with SDS–PAGE sample buffer. After heating to 70°C for 10 min, protein samples were run on SDS–PAGE gels and either silver-stained or processed for western blot analysis. In all, 2% of the ovary lysate used for each pull-down assay was run as an input sample.
Western blot analysis
Western blot analysis was performed as described earlier(Miyoshi et al, 2005). Anti-Aub monoclonal antibody (Nishida et al, 2007) was used at 1 μg/ml. Culture supernatant of hybridomas producing anti-AGO3 monoclonal antibody (Gunawardane et al, 2007) was used without dilution as a primary antibody. Anti-tubulin antibody was obtained from the Developmental Studies Hybridoma Bank and used at 1:1000 dilution. Anti-Tud polyclonal antibodies were kind gifts from K Hanyu-Nakamura (Kobe-RIKEN CDB) and P Lasko (McGill University) and were used at 1:2000 dilution. To produce anti-Spn-E monoclonal antibodies, the N-terminal region of Spn-E (150 amino acids), fused with a GST peptide, was produced in E. coli and used to immunize mice. To produce anti-Tud monoclonal antibodies (Figures 3C and 5A), the C-terminal region of Tud (amino acids 2189-stop codon), fused with a His peptide, was produced in E. coli and used to immunize mice. The plasmid for His-tagged recombinant Tud was a kind gift from K Hanyu-Nakamura (Kobe-RIKEN CDB). Hybridomas producing anti-Spn-E and anti-Tud monoclonal antibodies were prepared essentially as described earlier (Ishizuka et al, 2002). Anti-sDMA antibody (SYM11) was purchased from Upstate and used according to the manufacturer's instructions.
Immunoprecipitation
Immunoprecipitation was performed using anti-Aub, anti-AGO3 (polyclonal), and anti-Tud antibodies immobilized on Dynabeads ProteinG (Invitrogen). The buffer used was Binding buffer. In Figures 2D and 4A, 1 ml ovary lysate (4 mg/ml) was used per immunoprecipitation. In Figures 2C and 5A and B, 1 ml ovary lysate (2 mg/ml) was used per immunoprecipitation. After incubation with ovary lysate, the bead fractions were extensively washed with Binding buffer and the protein pools bound to the beads were eluted with SDS–PAGE sample buffer. After heating to 70°C for 10 min, protein samples were run on SDS–PAGE gels and processed for western blot analysis. In all, 2% of the ovary lysate used for each immunoprecipitation was run as an input sample. Anti-AGO3 polyclonal antibodies were generated by immunizing mice with a His-tagged recombinant AGO3 peptide (amino acids 1–289) that had been produced in E. coli.
Northern blot analysis
Immunoprecipitation was performed using anti-Aub and anti-AGO3 (polyclonal) antibodies immobilized on Dynabeads ProteinG (Invitrogen). The buffer used was Binding buffer. In Figure 4B, 1 ml ovary lysate (4 mg/ml) was used per immunoprecipitation. In Figures 5B and C and 6A and B, 1 ml ovary lysate (2 mg/ml) was used per immunoprecipitation. After immunoprecipitation, total RNAs were isolated from the immunopurified complexes by phenol: chloroform extraction and precipitated with ethanol. RNAs were dephosphorylated with CIP (NEB) and labelled for visualization with [g-32P]ATP by T4 polynucleotide kinase (TaKaRa). Total RNAs from either y w, tud, or dprmt5 ovaries were isolated using ISOGEN (Nippon Gene). Total RNAs (10 μg) were used in Figures 6 and 7B. Northern blot analysis was performed as reported earlier (Saito et al, 2006). The probe used to recognize roo#4 piRNA was as described earlier (Nishida et al, 2007). Probes used in Figure 4B to detect antisense roo piRNAs were as follows:
- roo#1 piRNA-as:
5′-TGGGCTCCGTTCATATCTTATG-3′
- roo#3 piRNA-as:
5′-TGAGAGTTCGCTATTCGAAGAA-3′
- roo#7 piRNA-as:
5′-TCTGAGGCATCCGTTTGGTAAA-3′
- roo-primer-1701:
5′-TATCTAGAAGATACGTCTAAACTAATAGAC-3′
- roo-primer-1731:
5′-AGTAGTCCAGATTTCCTTAAAATAAGGAAA-3′
- roo-primer-1761:
5′-AATAAAATTGAATTTTTATGGCATAAAATA-3′
- roo-primer-1791:
5′-GATAACCTGATTGAACAGGTGAATAGTCGT-3′.
The probe used to detect minisatellite#1 piRNA (Figure 6D) was 5′-TCGTGTATTGTCTTTTTGGGTTTGCG-3′.
The DNA fragments used to detect piRNAs originating from the I-element retrotransposon (accession number: M14954.2) were generated by PCR of Drosophila melanogaster genomic DNA with the primers:
I-element-for: 5′-CAAGCAGAATACGATCGCTA-3′
I-element-rev: 5′-TGGTCCAATTTGGGTGGGAT-3′. Amplified fragments were then cloned into pBS SK+.
PCR was again performed using T7 and T3 promoter sequence primers, and the PCR products were used as templates for in vitro transcription using a MAXIscript T7 kit (Ambion) in the presence of 32P-UTP. Transcribed RNAs were extracted with phenol:chloroform, precipitated with ethanol, and partially hydrolyzed as described earlier (Saito et al, 2006).
Northern blots for HeT-A, DM297, DMRT1B, and R1DM piRNAs (in Figure 6) were performed according to Pall and Hamilton (2008). Probes used were as follows:
- HeT-A-2701:
5′-CTGCAGCCAAGCGGGATTTA-3′
- HeT-A-2801:
5′-TGCGGCACCCTGTGTCCCGG-3′
- HeT-A-2902:
5′-GCGCGACTTCCAACTTTGTAACTC-3′
- DM297:
5′-TAGTCTTAAGCTGAGATCCAAAGAA-3′
- DMRT1B:
5′-GGGCAGAGTGCCAACACAAATGCT-3′
- R1DM:
5′-CACGGGTTGAGCAGCTATCCAAGA-3′.
The DNA fragments used to detect pre-piRNAs originating from the roo retrotransposon (Figure 4B, right) were generated by PCR of D. melanogaster genomic DNA with the primers:
- T7-roo-for:
5′-TAATACGACTCACTATAGGGATATCTAGAAGATACGTCTA-3′
- T3-roo-rev:
5′-AATTAACCCTCACTAAAGGGCCGCCTTAACAACCGTCGAC-3′.
The PCR products were used as template for in vitro transcription using a MAXIscript T3 kit (Ambion) in the presence of 32P-UTP. Hybridizations were performed at 65°C in 0.2 M sodium phosphate (pH 7.2), 7% SDS and 1 mM EDTA and washed at 65°C in 2 × saline sodium citrate and 0.1% SDS.
Immunohistochemistry
Ovaries were dissected manually from adult flies in 1 × PBS. Immunostaining was performed following standard procedures. For tud1, anti-Aub (1:1000 dilution) and anti-Tud (1:2000 dilution) antibodies were used as primary antibodies. Alexa 488-conjugated anti-rabbit IgG (Molecular Probes) (1:1000 dilution) and Cy3-conjugated anti-mouse IgG (Sigma) (1:100 dilution) were used as the secondary antibodies. For dprmt5 staining, anti-Aub antibody was directly labelled using a Fluorescein Labeling kit-NH2 (Dojindo Molecular Technologies). Anti-Tud antibody was used at a dilution of 1:500. Cy3-conjugated anti-rabbit IgG (Sigma) (1:100 dilution) was used as the secondary antibody. DNA was stained with DAPI. All images were collected using a confocal microscope (Zeiss LSM5 EXCITER).
Supplementary Material
Since the publication of this paper, the authors have noticed an omission in the Supplementary Information. They now provide the accession numbers for Aubergine-associated small RNAs in wild-type and tudor mutant ovaries in the replacement Supplementary Information. The file was corrected on 17 February 2010
Acknowledgments
We thank AG Matera, K Hanyu-Nakamura, A Nakamura, and P Lasko for providing reagents. We also thank the Drosophila Genetic Resource Center for kindly supplying Drosophila strains. For assistance with MS analysis, we acknowledge I Sagawa at the Institute of Health Biosciences University of Tokushima. We thank K Saito and K Miyoshi for technical advice and assistance, and other members of the Siomi laboratory for discussions and comments on the paper. This work was supported by MEXT (Ministry of education, Culture, Sports, Science, and Technology) grants to HS, and NEDO (New Energy and Industrial Technology Development Organization) grants to MCS. MCS is supported by CREST (Core Research for Evolutional Science and Technology) from JST (Japan Science and Technology Agency). MKN is supported by JSPS (Japan Society of the Promotion of Science). MCS is an Associate Professor of the Global Center of Excellence for Human Metabolomics Systems Biology, funded by MEXT.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anne J, Mechler BM (2005) Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuléen. Development 132: 2167–2177 [DOI] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM (2007) Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134: 137–146 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Arkov AL, Wang JY, Ramos A, Lehmann R (2006) The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 [DOI] [PubMed] [Google Scholar]
- Bardsley A, McDonald K, Boswell RE (1993) Distribution of tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development 119: 207–219 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG (2009) Protein arginine methylation in mammals: who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhäger M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U (2008) An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 135: 497–509 [DOI] [PubMed] [Google Scholar]
- Côté J, Richard S (2005) Tudor domains bind symmetrical dimethylated arginines. J Biol Chem 280: 28476–28483 [DOI] [PubMed] [Google Scholar]
- Fujii K, Nakano T, Hike H, Usui F, Bando Y, Tojo H, Nishimura T (2004) Fully automated online multi-dimensional protein profiling system for complex mixtures. J Chromatogr A 1057: 107–113 [DOI] [PubMed] [Google Scholar]
- Fujinoki M, Kawamura T, Toda T, Ohtake H, Ishimoda-Takagi T, Shimizu N, Yamaoka S, Okuno M (2003) Identification of 36-kDa flagellar phosphoproteins associated with hamster sperm motility. J Biochem 133: 361–369 [DOI] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA (1995) Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev 9: 2495–2508 [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG (2006) The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol 16: 1077–1089 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N (2007) Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol 301: 38–52 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H (2002) A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16: 2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Doudna JA (2009) A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412 [DOI] [PubMed] [Google Scholar]
- Keller BO, Wang Z, Li L (2002) Low-mass proteome analysis based on liquid chromatography fractionation, nanoliter protein concentration/digestion, and microspot matrix-assisted laser desorption ionization mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 782: 317–329 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z (2009) Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U (2008) Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett 582: 1997–2003 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC (2007) Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ (2008) Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G (2002) Essential role for the SMN complex in the specificity of snRNP assembly. Science 298: 1775–1779 [DOI] [PubMed] [Google Scholar]
- Ponting CP (1997) Tudor domains in proteins that interact with RNA. Trends Biochem Sci 22: 51–52 [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS (2009) Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC (2009) A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC (2008) Interactions between transposable elements and Argonautes have (probably) been shaping the Drosophila genome throughout evolution. Curr Opin Genet Dev 18: 181–187 [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC (2009) On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Kuramochi-Miyagawa S (2009) RNA silencing in germlines—exquisite collaboration of Argonaute proteins with small RNAs for germline survival. Curr Opin Cell Biol 21: 426–434 [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM (2004) Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci 117: 2109–2120 [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P (2004) Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 40: 164–170 [DOI] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P (2008) Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev 125: 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA (2009) Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Müller-Reichert T, Jessberger R (2009) Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 19: 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G (2004) Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol 14: 226–232 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Since the publication of this paper, the authors have noticed an omission in the Supplementary Information. They now provide the accession numbers for Aubergine-associated small RNAs in wild-type and tudor mutant ovaries in the replacement Supplementary Information. The file was corrected on 17 February 2010