Abstract
The Lung Cancer Survivability Prediction Tool (LCSPT) is a web-based system that predicts the survival possibility of a lung cancer patient based on the status of the patient and the treatments provided. In order to make the LCSPT more accessible and convenient to doctors working in a clinical setting, we have developed a new interface for mobile devices. The display size along with wireless data transfer speeds pose the most significant challenges to porting a software application to a mobile device. We have addressed these issues by redefining the interface and limiting the amount of data that is required. The resultant tool provides doctors with the flexibility of mobility while maintaining the effectiveness of the desktop version of the LCSPT.
I. INTRODUCTION
With advancements in genomics research over the past decade, personalized medicine is expected to play a significant role in providing quality patient care. With personalized medicine, information such as a patient's genotype, molecular profiles, or clinical phenotypes (e.g., imaging, laboratory, and clinical information) is used to make decisions on medication, provisions of therapies, or the initiation of preventative measures. To make such decisions, health care teams need to be able to access the most current treatment plans and adjust their strategies accordingly with confidence. A feedback loop exists between the patient and a health care team. An effective feedback loop will improve the quality of health care and lead to an improved outcome. To be effective, this loop should be direct, rapid and specific.
The next generation of medical systems will integrate multimodality data from a variety of sources to aid individual physicians in providing the best treatment and care for their patients and to help researchers understand patterns of disease in collections of large databases [2]. Recently, we developed the Lung Cancer Survivability Prediction Tool (LCSPT). The LCSPT is a web-based system that predicts the survival possibility of a lung cancer patient based on the status of the patient and the treatments provided [12]. In order to make the LCSPT more accessible and convenient to doctors working in a clinical setting, we have developed a new interface for mobile devices. The display size along with wireless data transfer speeds pose the most significant challenges to porting a software application to a mobile device. We have addressed these issues by redefining the interface and limiting the amount of data that is required. The resultant tool provides doctors with the flexibility of mobility while maintaining the effectiveness of the desktop version of the LCSPT.
II. LUNG CANCER SURVIVABILITY PREDICTION TOOL
Different treatment models have been investigated and published, but few of them have ever been tested in clinical settings and even fewer have been used eventually in clinical practice. Most of the models are based on statistics and involve many clinical and genomic factors. They are researched and developed in a statistical programming environment. As a statistical programming environment is intended to be used in biomedical research and not for clinical application, a final software translation is usually required to code the model into a computer system appropriate for a clinical setting. Due to costly software development processes, most of the models remain in the literature and their translation never actually happens. In order to bring these models to clinical practice, a tool is needed for automatically translating the statistical models. To ensure the system fits in a clinical setting, such a software translation tool must
work inside the clinical network with a centralized database management system on the server;
have a web-based data submission process;
enable easy and quick submission and testing processes for different statistics-based medical treatment models;
facilitate easy maintenance and upgrading of the system; and
work effectively on a mobile device.
Although the original LCSPT [11] was designed to work with a centralized database management system, enable web-based data submission, support multiple statistical models, and was designed to be easily upgraded, it was not designed for a mobile device. The LCSPT has recently been enhanced to run effectively on mobile devices like the iPhone.
A. Lung Cancer Prediction Model
We developed two models for predicting lung cancer patient survival. The first model predicts the patient’s survival probability using only his/her histological information including age, gender, stage, cell type, and tumor grade. The second model uses additional information, including the treatment options and the patient’s smoking status.
In building these models, we examined the association of age at diagnosis, gender, smoking history, stage, histological cell type, histological grade, and treatment with the patient’s survival using the Kaplan-Meier method. A multivariable Cox proportional hazards model was applied to evaluate all of the above-mentioned variables for their independent predictive value on the patient’s survival. Among the variables evaluated, only smoking status was not significantly associated with survival. Both models were evaluated for their prediction accuracies on a test set of 1,518 patients [9]. The evaluation was done by comparing the predicted and observed survival curves. The predicted survival curves from Model 1 and 2 are both virtually identical to the observed survival curve. No statistical difference between the predicted and observed survivals was observed, suggesting the adequateness of each model.
B. Survival Probability Prediction Architecture
Since the introduction of Internet-based solutions, several electronic data collection and visualization applications have been developed in medical applications [3, 5, 7, 11]. Web tools have also been developed to make personalized predictions of conditional survival for individual patients depending on the patient’s age, gender, race and tumor site and stage. One of these web tools focuses on the development of a regression model [10]. The distinguishing goal for our tool was to build an extensible software platform in which different prediction models could be easily added. Using such a system, it is possible to integrate research and clinical practice into one software platform [12].
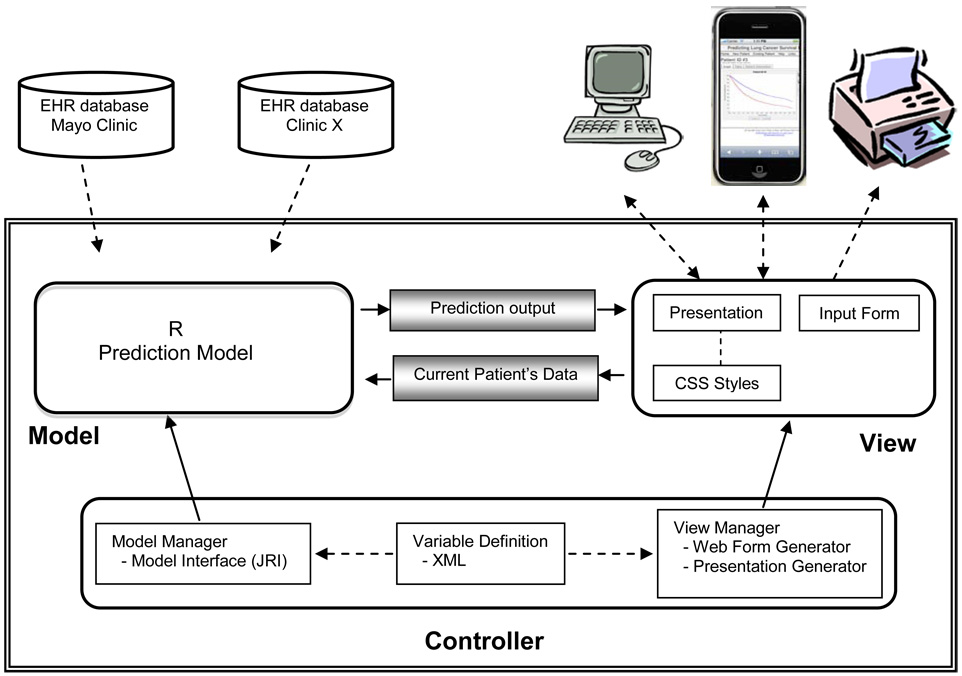
Our platform allows researchers to add and remove statistical models and to make changes to the input area of the user interface. TABLE I summarizes the functions that can be performed by researchers, clinicians, and data entry people. In designing the architecture of our framework, the guiding principle was to create an environment that researchers and diagnosticians could use to experiment with various diagnostic models and potential treatments without having to acquire expertise in a computer programming language and environment. The resulting Survival Probability Prediction Architecture (SPPA) was designed for experimentation with diagnostic models and survival prediction. SPPA is based on the Model-View-Controller architectural pattern, as shown in Fig. 1. It provides both a mechanism for defining models and a mechanism for testing the model in a clinical setting.
TABLE I.
Software Functions Provided to User Groups
| User Group | Functions Supported |
|---|---|
| Researchers | Add and remove prediction models in R; Change database for the model; Modify user interface |
| Clinicians | Add, view, and modify a patient record; Compare and select treatments |
| Data entry person | Add, view, and modify a patient record |
Fig. 1.
Survival Probability Prediction Architecture
The heart of the SPPA is the Controller. The Controller was designed to be sufficiently general enough to enable quick and seamless modification of the system by non-computer specialists. Although we expect researchers to be proficient in a statistical programming language like R, we do not expect them to be proficient in Java. Likewise, diagnosticians should be able to navigate a web page, but they should not have to generate one to be able to view the results of a model.
In order to insulate the researchers and clinicians from its internals, the Controller is subdivided into three components: the Model Manager, the View Manager, and the Variable Definition component. The Model Manager uses JRI to provide an interface between the Java methods of the Controller and the prediction model, which is currently written in R. However, the Model Manager enables researchers to build diagnostic models using any statistical programming language of their choice. At present, SPPA only supports statistical models written in R, but it can be easily modified to support any statistical modeling language by extending the Model Manager to provide an interface between Java and the new statistical modeling language.
The View Manager is responsible for providing the researcher and/or the clinician with the results of the prediction model on a given patient. It consists of two components: the Web Form Generator and the Presentation Generator. Information (e.g., age, gender, etc.) associated with new patients is gathered through a web page form that is generated by the Web Form Generator. The Web Form Generator works with the XML definitions of the Variable Definition component to dynamically create the web form.
The glue that connects the Model with the View is the Variable Definition component. The Variable Definition component uses XML to define the type and structure of the inputs that describe the state of the patient, and defines which models use which inputs. The XML files are used by both the Model Manager and the View Manager. The View Manager uses the XML definitions within the Web Page Generator to create the Patient Information Entry Form like the one given in Fig. 1. Once the clinician has completed the form, the inputs that he/she entered about the patient are passed to the prediction model via the Model Manager.
III. PORTING LCSPT TO THE IPHONE
The use of an electronic health record (EHR) system may reduce medical errors by providing healthcare workers with decision support and providing fast access to current best practices in medicine. To make a decision on a patient’s treatment, healthcare teams need to be able to access the most current treatment plans and adjust their strategies accordingly and with confidence. Indeed, there are many published cancer treatment models. A quick search on pubMed (http://www.ncbi.nlm.nih.gov/pubmed/) turned out 84 publications on treatment models for lung diseases only in 2008 and 109 publications in 2007. However, there exist barriers in adopting these treatment models; these include software acquisition and implementation costs, slow and uncertain financial payoffs, and the lack of interoperability among different EHR systems. Often, a computer tool is developed for a particular disease treatment model and is very difficult to modify or adapt for another model [4]. This section of the paper describes the ease with which our software support architecture was adapted to a mobile device.
Facing several treatment choices for lung cancer patients, it is crucial for doctors to have a quantitative way of comparing the available treatments. The LCSPT gives doctors a clear idea of the possibility that a patient could survive under certain treatment and health conditions using a statistical prediction model that is based on data that has been collected from the Mayo Clinic over the past ten years. However, the current tool was developed for use on a desktop. In order to make the tool more useful in a clinical setting, it would be desirable if doctors could get access to the results from the statistical models in real-time on a mobile device while they are making their rounds. This would provide them with a more convenient way of comparing treatments and making decisions.
In practice, the doctors often need the data at times when a desktop computer is not available. For example, it is much more convenient to update the patients’ information while they gathering it rather than waiting until later to enter it. Furthermore, studies have shown that better results can be observed in both health status and clinical outcomes when patients are provided with proper tools that enable involvement and active management of their illness [8], so it is expected that the LCSPT will be released to patients. Therefore, accessing the system with a mobile device is a logical next step. The research suggests, however, that only about one-third of all mobile web users are satisfied with their experience. This dissatisfaction is due to a number of factors, including that it is a more time consuming process [1] and it is more difficult to locate the content in long narrow page [6]. Thus, we have focused on these shortcomings in the mobile version of LCSPT.
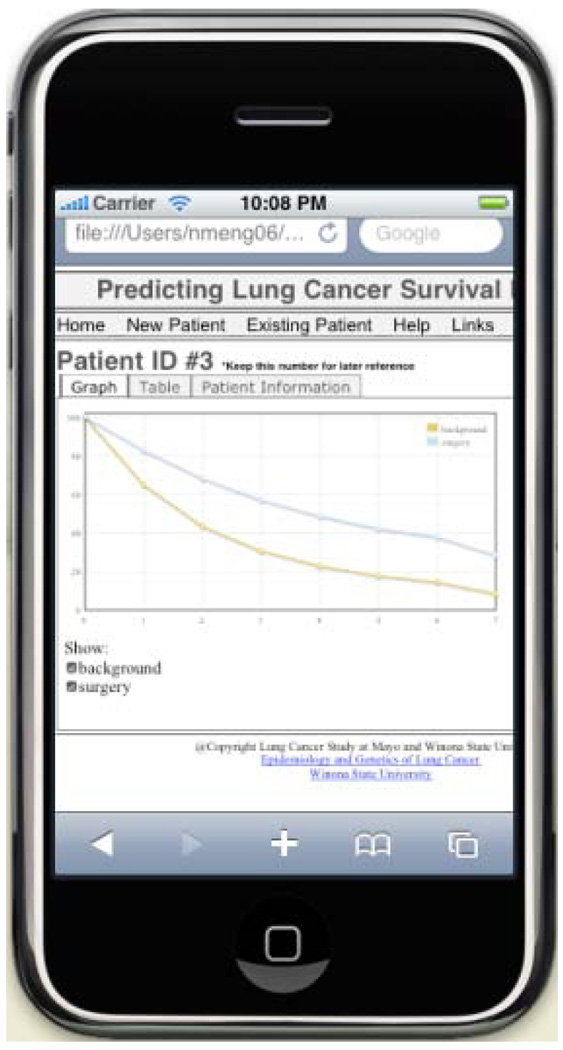
Porting a software application to a mobile device introduces a number of challenges. In the mobile version of LCSPT, we focused on two of them: speed of the data transfer and the display size of the LCSPT on a small screen. Due to the transmission of the data over wireless connections, the amount of data that is transmitted needs to be minimized in order to ensure real-time response. Furthermore, the small screen size on mobile devices poses a problem with displaying the information in a useful and effective manner. In order to accelerate the loading process of the pages, we changed the way of generating the survival chart from server side to client side. The data calculated by the bioinformatic model is sent directly to the client side and the chart is generated by a JavaScript program locally, which saves the time in sending a large image file of the chart. At the same time, we have developed an algorithm to select the most meaningful and useful data from the massive results data to help the doctors make effective treatment decisions. Furthermore, the user interface was modified for mobile device users who would have a relatively smaller screen. The information layout was changed for displaying in a user-friendly way for smaller screen as shown in Fig. 2.
Fig. 2.
LCSPT on iPhone
The algorithm is based on the current study of lung cancer that only less than 30% of patients survived beyond 3 years. In non-small cell lung cancer, the average 5-year survival rate for a stage I patient is 65%. The rate declines to 41% for stage II, 5–55% for stage III, and 3–21% for stage IV patients. The one-year survival rate is only 25% for extensive small cell lung cancer. In clinical practice, a prolonged survival in terms of months would be significant for a patient in his/her first two years, and more detailed information about the treatment are expected. The amount of data to be transmitted is thus proportional to the survival rate and also dependent on the cell type of lung cancer.
A usability study on iPhone platform was performed by using a simulator of iPhone, which is an application developed for developing and testing new iPhone applications. Twenty students from Winona State University took part in the study. Only one of the participants had used an iPhone regularly and 4 of the participants have minimal experience with an iPhone, and the others have never used an iPhone before. Of the 20 participants, 11 participants had mobile device web browsing experience.
Participants were asked to accomplish two tasks first then evaluate their user experience on both versions of the LCSPT system. In both tasks, participants were asked to find the survival possibility of a patient in 3.5 years from the charts shown on the page. In task one, participants were asked to create a new patient with specified information. In task two, participants were asked to modify patient information for an existing patient and then get the result. Participants were divided into two groups, one group used iPhone version first and then PC version; the other one used the PC version first then the iPhone version.
All the participants successfully finished the tasks and wrote down the survival possibility data they found from the charts on PC and iPhone version. From TABLE II, the mean of the estimation from both versions were very close but with a big difference between the standard deviations. This data shows that PC version users may interpret data, which is much higher or lower than the average value, while the iPhone users are more concentrate on a smaller estimation range. The results from iPhone version are in a smaller range and have little volatility about the survivability than those from PC version, which means in the real medical situations, doctors would tend to agree with each other when using the iPhone version more than the PC version.
TABLE II.
Task Results
| Task I | Task II | |||
|---|---|---|---|---|
| PC | iPhone | PC | iPhone | |
| Mean | 58.025 | 57.269 | 66.585 | 64.963 |
| Standard Deviation | 4.948 | 1.158 | 2.712 | 1.266 |
| Range | 22 | 5 | 12 | 5 |
One of the advantages of the iPhone version is the chart on iPhone could interact with user, which provide users the data used to plot the line when they hover over the points on the curve. The iPhone users have more data to refer to when deciding the possibility of surviving. Furthermore, most of the participants were satisfied with the layout of the iPhone version. When asked about the ease with which to finish the tasks, 8 participants thought it is easy to accomplish, 11 said it took some time to figure it out but eventually they found everything in a reasonable time. Only 1 of them claimed that it was hard to finish the tasks on iPhone.
IV. CONCLUSION
We developed a new interface for the LCSPT in order to make it more accessible and convenient to doctors working in a clinical setting. We have addressed the issues display size along with wireless data transfer speeds by redefining the interface and limiting the amount of data that is required. The resultant tool provides doctors with the flexibility of mobility while maintaining the effectiveness of the desktop version of the LCSPT.
Acknowledgments
This work was supported in part by the HealthForce Minnesota.
Footnotes
31st Annual International Conference of the IEEE EMBS Minneapolis, Minnesota, USA, September 2–6, 2009
Contributor Information
Tim Gegg-Harrison, Computer Science Department, Winona State University, Winona, MN 55987 USA (phone: 507-457-5381; tgeggharrison@winona.edu)..
Mingrui Zhang, Computer Science Department, Winona State University, Winona, MN 55987 USA (mzhang@winona.edu)..
Nan Meng, Computer Science Department, Winona State University, Winona, MN 55987 USA (nmeng06@winona.edu)..
Zhifu Sun, Department of Health Science Research, Mayo Clinic, Rochester, MN 55905 USA (Zhifu.Sun@mayo.edu)..
Ping Yang, Department of Health Science Research, Mayo Clinic, Rochester, MN 55905 USA (Yang.Ping@mayo.edu)..
REFERENCES
- 1.Gupta A, Kumar A, Mayank, Tripathi VN, Tapaswi S. Mobility'07. Singapore: 2007. Mobile Web: Web Manipulation for Small Displays using Multilevel Hierarchy Page Segmentation. [Google Scholar]
- 2.Halle MW, Kikinis R. Flexible framework for medical multimedia; Proceedings of the 12th annual ACM international conference on Multimedia; New York, USA: 2004. pp. 768–775. [Google Scholar]
- 3.Marks RG. Validating electronic source data in clinical trials. Controlled Clin. Trials. 2004;vol. 25:437–446. doi: 10.1016/j.cct.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Miller R, Sim I. Physicians’ Use of Electronic Medical Records: Barriers and Solutions. HEALTH AFFAIRS. 2004;vol. 23:116–126. doi: 10.1377/hlthaff.23.2.116. [DOI] [PubMed] [Google Scholar]
- 5.Pavlovic I, Miklavcic D. Web-based electronic data collection system to support electrochemotherapy clinical trial. IEEE Trans. Information Technology in Biomedicine. 2007;vol. 11:222–230. doi: 10.1109/titb.2006.879581. [DOI] [PubMed] [Google Scholar]
- 6.Shrestha S. Mibility'07. Singapore: 2007. Mobile Web Browsing: Usability Study. [Google Scholar]
- 7.Sippel H, Ohmann C. A web-based collection system for clinical studies using Java. Med. Inf. 1998;vol. 23:223–229. doi: 10.3109/14639239809001402. [DOI] [PubMed] [Google Scholar]
- 8.Stroetmann KA, Pieper M, Stroetmann VN. Understanding Patients: Participatory Approaches for the User Evaluation of Vital Data Presentation; ACM Conference on Universal Usability; British Columbia, Canada: Vancouver; 2003. pp. 93–97. [Google Scholar]
- 9.Sun Z, Aubry MC, Deschamps C, Marks RS, Okuno SH, Williams BA, Sugimura H, Pankratz VS, Yang P. Histologic grade is an independent prognostic factor for survival in non-small cell lung cancer: an analysis of 5018 hospital- and 712 population-based cases. J Thorac Cardiovasc Surg. 2006;vol. 131:1014–1020. doi: 10.1016/j.jtcvs.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 10.Wang SJ, Fuller CD, Kim J-S, Sittig DF, Thomas CRJ, Ravdin PM. Prediction Model for Estimating the Survival Benefit of Adjuvant Radiotherapy for Gallbladder Cancer. J Clin Oncol. 2008;vol. 26:2112–2117. doi: 10.1200/JCO.2007.14.7934. [DOI] [PubMed] [Google Scholar]
- 11.Wubbelt P, Femandez G, Heymer J. Clinical trial management and remote data entry on the Internet based on XML case report forms. Stud. Health Technol. Inf. 2000;vol. 77:333–337. [PubMed] [Google Scholar]
- 12.Zhang M, Olson S, Francioni J, Gegg-Harrison T, Meng N, Sun Z, Yang P. Integrating R Models with Web Technologies; International Conference on Health Informatics; Porto, Portugal: 2009. [Google Scholar]