Abstract
The regulated degradation of proteins within eukaryotes and bacterial cells is catalyzed primarily by large multimeric proteases in ATP-dependent manner. In eukaryotes, the 26 S proteasome is essential for the rapid destruction of key regulatory proteins, such as cell cycle regulators and transcription factors, whose fast and tuned elimination is necessary for the proper control of the fundamental cell processes they regulate. In addition, the 26 S proteasome is responsible for cell quality control by eliminating defective proteins from the cytosol and endoplasmic reticulum. These defective proteins can be misfolded proteins, nascent prematurely terminated polypeptides, or proteins that fail to assemble into complexes. These diverse activities and its central role in apoptosis have made the proteasome an important target for drug development, in particular to combat malignancies.
Marking Proteins for Degradation
Targeting of most substrates to the 26 S proteasome requires their prior marking by a covalently linked polyubiquitin chain(s). During association with the proteasome, the substrate is directed into the catalytic core, where it is digested, whereas most of the ubiquitin molecules are recycled.
Protein ubiquitination is a multistep process orchestrated by the concerted action of three enzymes. The reaction begins with E1,2 which initially adenylates the C-terminal glycine of ubiquitin and then forms a thioester bond between the activated glycine residue and a cysteine residue on the E1 catalytic site. Next, E2 acquires the activated ubiquitin through a transthioesterification reaction to form a similar thioester bond between the E2 active-site cysteine and the activated ubiquitin. Finally, E3 recruits the target protein and guides the transfer of the activated ubiquitin from the E2 enzyme to the substrate. In most cases, an ϵ-NH2 group of a lysine residue on the substrate attacks the thioester bond between the ubiquitin and E2, and an isopeptide bond is formed, linking the activated C-terminal glycine of ubiquitin to the amino group in the attacking lysine of the target substrate (1). Ubiquitin transfer from the E2 enzyme to the substrate is catalyzed directly by RING (really interesting new gene) finger-containing E3 enzymes and indirectly when a HECT (homologous to E6-AP carboxyl terminus) domain-containing E3 is mediating the transfer. The process is repeated in a cyclic manner where, in each step, a new moiety of ubiquitin is conjugated to an internal lysine residue (typically Lys48) of the previously conjugated molecule. This generated polyubiquitin chain is regarded as the targeting signal for the downstream 26 S proteasome. However, in view of recent findings, several alternative mechanisms have been proposed (for a recent review, see Ref. 2). Li et al. (3) demonstrated in a reconstituted cell-free system that a preformed polyubiquitin chain can be initially assembled on the active-site cysteine of E2 (UBE2G2), presumably by the action of an “exogenous” E2 acting in trans. Once assembled, an E3 enzyme (gp78) catalyzes the transfer of the polyubiquitin module to a lysine residue of the target substrate (the C terminus of HERP, a known substrate of these E2/E3 enzymes). In a related study, Ravid and Hochstrasser (4) proposed that the polyubiquitin chain generated on the E2 Ubc7 (the yeast ortholog of UBE2G2) is recognized by the proteasome and may serve as a degradation signal in an autoregulatory feedback mechanism.
Several forms of ubiquitination have been identified (5). Single or multiple monoubiquitinations have been described where single or multiple ubiquitin moieties are conjugated to distinct lysine residues on the substrates, but they do not polymerize. These forms of ubiquitination were implicated in various cell pathways, which include endocytosis and sorting of proteins to different cell compartments (6, 7), but also in several cases of proteasomal activity, such as the processing of the p105 precursor of the NF-κB transcription regulator (8). However, polyubiquitination is the most common form of post-translational modification of proteins destined for degradation (9). Because ubiquitin has seven lysine residues (positions 6, 11, 27, 29, 33, 48, and 63) (Fig. 1A), in principle, polyubiquitin chains can be formed based on any of these residues. Accordingly, seven different topologies of polyubiquitination can be generated (excluding mixed or more than singly branched topologies) (10). The current view holds that proteasomal degradation is mediated mainly by polyubiquitination based on Lys48 as the conjugated residue (11), although chains based on all other lysines have been implicated in targeting proteins to the proteasome (12, 13).
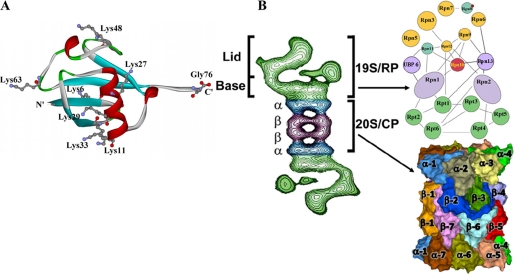
FIGURE 1.
A, ribbon representation of the three-dimensional structure of ubiquitin (Protein Data Bank code 1UBI). The seven lysine residues and the C-terminal glycine are depicted as balls and sticks. Note that six of the lysine residues (positions 6, 11, 27, 29, 33, and 48) are located around the midplane of the structure, whereas Lys63 and the C-terminal glycine define the long axis of the molecule. B, the 26 S proteasome. Left, two-dimensional model of the 26 S proteasome generated through averaging of electron micrographs. Right, schematic representation of the 26 S proteasome. The 26 S proteasome is composed of one 20 S complex (lower), which is made of four seven-membered rings. The active sites reside on the β-rings in the center of the structure (β1, β2, and β5). The 26 S proteasome contains one or two 19 S regulatory particles. The regulatory particle can be further subdivided into the lid and the base. The subunit organization of the 19 S particle was adapted from Ref. 68. RP, regulatory particle; CP, core particle.
The Proteasome
The 26 S proteasome is a large complex of ∼2.5 MDa (Fig. 1B). Based on distinct functions and biochemical analyses (mainly in the presence of salt), this complex can be divided (and dissociated) into two smaller subcomplexes, the 20 S core catalytic particle, which is the proteolytic component, and the 19 S regulatory particle, which appears to be responsible for recognizing, unfolding, and translocating the polyubiquitinated substrates into the 20 S proteasome, where they are degraded (Fig. 1B).
The core particle is a 670-kDa barrel-shaped protein complex made of four stacked seven-membered rings (four × seven subunits), two outer α-rings and two inner β-rings (α1–7β1–7β1–7α1–7). The proteolytic active sites are located on the two identical β-rings, which are positioned at the center of this complex (14, 15). The catalytic activities of these active β-type subunits are associated with their N-terminal threonine residue that acts as a nucleophile in hydrolysis. Assembly of the 26 S proteasome is thought to start with spontaneous chaperone-assisted formation of the seven-membered α-rings. Once mature, an α-ring serves as a template for assembly of the complementary β-rings from individual β-subunits. When all of the β-subunits associate to generate the ring, the resulting half-proteasomes dimerize. A latent 20 S core particle is formed, as the active β-subunits (β1, β2, and β5) still contain N-terminal propeptides. In addition, the narrow entry port in the α-ring (11–15 Å) is blocked by a mesh made of the N-terminal domains of the individual α-subunits (16). The propeptides are removed autocatalytically, leading to maturation of the 20 S proteasome subcomplex and degradation of the set of chaperones that directed the process and prevented non-regulated activity. Presumably, the mature 20 S proteasome conjugates with a pre-assembled 19 S regulatory complex (see below) to form the biologically active 26 S proteasome (for a review, see Ref. 17). Several studies suggested that following association with the 20 S particle, the C termini of the ATPases in the 19 S complex (see below), which are located on the perimeter of the 19 S ATPase ring (Fig. 1B), project out of the ring plane and protrude into the 20 S proteasome α-ring (18, 19). This interaction invokes a conformational transition in the proteasome α-subunits, leading to dissociation of the mesh created by the seven-α-subunit N-terminal domains that gate the proteasome. This transition appears to flip the N termini of the α-ring outwards into the cavity of the ATPase ring, stabilizing the complex and allowing substrate entry from the 19 S particle into the 20 S proteasome.
Under certain conditions, the 20 S proteasome forms complexes with non-ATPase activators, such as PA28 and Blm10/PA200. The precise configurations of these alternative regulators in the cell are not well understood. PA28-containing proteasomes (immune proteasomes) are induced by interferon γ and are rich in organs of the immune system. They have been shown to contain also alternative β-subunits in place of the three catalytic β-subunits, β1i, β2i, and β5i. These subunits provide the proteasome with stronger trypsin- and chymotrypsin-like activities, which augment the ability of the proteasome to produce degradation products suitable for major histocompatibility complex class I presentation (20).
Although proteasomes can cleave after most amino acid residues, proteolytic activity measured using fluorogenic substrates suggests three distinct (although not exclusive) cleavage preferences. The β2-subunit possesses a tryptic activity (i.e. cleaving after basic residues), the β5-subunit has a chymotryptic activity (i.e. cleaving after hydrophobic residues), and the β1-subunit has a caspase-like or post-acidic activity (supplemental Fig. S1). The proteolytic active sites of the proteasome are facing the lumen of the barrel and are sequestered from the bulk solution. In addition, the gated channel in the α-ring, through which substrates enter the 20 S particle, is narrow. Thus, only unfolded polypeptides can enter the 20 S proteasome. Consequently, a globular substrate must be unfolded (probably by the ATPases of the 19 S complex) to be translocated and digested within the 20 S particle.
The 19 S regulatory particle is a large complex of ∼1 MDa and consists of at least 19 different subunits (supplemental Table S1). Nine of these subunits form a “lid,” whereas the other 10 subunits, including the six ATPases, compose the “base” of the 19 S particle (Fig. 1B). Electron micrographs (21–23) and cross-linking experiments (24, 25) demonstrated that the six homologous ATPases are associated with the α-rings of the 20 S particle. In addition to these six ATPases, which are termed (in yeast) Rpt1–6, the base contains four non-ATPase subunits (Rpn1, Rpn2, Rpn10, and Rpn13). Despite the high sequence homology between the six ATPases, some of their amino acid sequences are significantly divergent (mainly at the N-terminal domains), and the different subunits may have distinct functions as indicated previously (26). One ATPase, Rpt2, was shown to play a role in opening the gated α-ring to facilitate substrate entry (27), whereas Rpt5 was implicated in the recognition of the substrate-linked polyubiquitin chain (28). Another base component, Rpn10, is also a ubiquitin receptor (29, 30) but was found to be nonessential in yeast. Rpn1 was found to interact with a series of ubiquitin chain receptors that shuttle ubiquitinated proteins to the proteasome. These proteins, Rad23, Dsk2, and Ddi1, are not integral components of the proteasome but associate with it substoichiometrically. They share a common ubiquitin-like domain at their N termini that probably mediates their recognition by Rpn1. In addition, Ubp6, which is a 19 S particle-associated deubiquitinating enzyme that is thought to have a central role in recycling ubiquitin by hydrolyzing polyubiquitin chains on the target substrate (31), is also recognized by Rpn1 through a similar domain. The function(s) of Rpn2 are less clear. The yeast Rpn2 was reported to bind certain ubiquitin ligases (E3) such as Ubr1 (32) and Hul5 (33). Eight subunits were originally assigned to the lid subcomplex of the regulatory particle (Fig. 1B). This complex is characterized by high sequence homology to two other cell complexes: the COP9 signalosome, a conserved protein complex that is made of eight subunits (CSN1–8) and that is involved in regulating the translation initiation factor eIF3 (34) and the activity of cullin-RING E3 complexes via its ability to remove the NEDD8 modifier from the cullin component of these ubiquitin ligases (35). Unlike the other resident deubiquitinating enzyme found in the 19 S particle (Ubp6), Rpn11 is essential for viability in yeast. This deubiquitinating activity is similar to the deNEDDylating activity of the COP9 signalosome.
An important problem related to the proteasome is the identity of the subunits that bind ubiquitin. S5a/Rpn10 was the first ubiquitin-binding proteasomal subunit to be discovered (36, 37) and was shown to bind ubiquitin chains through ubiquitin-interacting motifs. Because inactivation of this subunit had almost no phenotype (37), researchers looked for additional/other subunits. Indeed, later findings showed that the 19 S ATPase Rpt5 (S6′) can also bind polyubiquitin chains in a manner that requires ATP hydrolysis (28). Recently, Rpn13/ADRM1/ARM1, which docks to the 19 S ATPase (Rpn2) through its N-terminal domain, was also shown to bind Lys48-based polyubiquitin chains (38, 39). Other receptors that are not integral proteasomal subunits but rather deliver ubiquitinated targets to the proteasome (40, 41) were also identified. The ubiquitin-associated domains of these proteins bind ubiquitin (42–44), whereas their ubiquitin-like domains interact reversibly with the proteasome through Rpn1 and potentially also through Rpn10 (30, 45, 46). Interestingly, although both ubiquitin and proteasomes are essential, the inactivation of Rpn10 and Rpn13 in yeast does not appear to be lethal. Rather, a synthetic phenotype results (38). These findings suggest that an additional, yet to be discovered ubiquitin/polyubiquitin-binding protein(s) that circumvents the absence of the already known ubiquitin receptors probably exists, yet it is also possible that polyubiquitinated non-proteasomal shuttling proteins and ubiquitin-binding proteasomal subunits act in parallel and that some of their functions are redundant. Therefore, it is not clear whether the existence of additional ubiquitin-binding proteasomal subunits is essential.
The Proteasome as a Drug Target
The structure of the UPS is rhomboid-like. The E1 at the top is required for all types of ubiquitinations and has no specificity. Underneath, the E2 enzymes (∼40 in mammals) appear to play a role in determining the type of the polyubiquitin chain synthesized (lysine specificity). The E3 enzymes (∼1000) occupy the broadest plane and endow the system with its high specificity for its substrates. The proteasome resides at the bottom, where the rhomboid tapers again. It recognizes most substrates via a common motif, the polyubiquitin chain. Interference with the association between an E3 and its cognate substrate(s) appears therefore to be the most obvious site for drug targeting, yet targeting an E3-substrate interaction has encountered many difficulties, including lack of information on the partnering motifs and the weak protein-protein interactions that occur along large surfaces that characterize many of these complexes. On the other hand, extensive mechanistic and structural studies of proteases (their active sites and substrates) led to the development of highly efficient and specific cell-permeable inhibitors, which facilitated the conversion of the proteasome to the first UPS drug target.
Most synthetic inhibitors are short peptides that mimic the substrates. Typically, the pharmacophore is bound to the carboxyl residue of the peptide and inhibits the threonine residue in the 20 S active site (47). Thus, some of the typical synthetic inhibitors are peptide aldehydes, peptide vinyl sulfones, peptide boronates, and peptide epoxyketones (48–50). Most notable among the natural bacterially derived non-peptide inhibitors is claso-lactacystin β-lactone (omuralide) (51). The related drugs salinosporamide A (NPI-0052) and carfilzomib (PR-171) are currently in advanced clinical trials (52). Unlike the synthetic compounds, these drugs share a core skeleton of bicyclic ring but differ from one another in the transforming groups. The different transformations appear to determine the specificity and probably the distinct effects of the various drugs.
The idea that proteasome inhibitors can become drug candidates emerged from the observation that they can specifically induce apoptosis in different leukemia- and lymphoma-derived cells (53, 54). Further development and clinical trials resulted in approval by the Food and Drug Administration (in May 2003) of the modified boronic dipeptide Pyz-Phe-boroLeu (where Pyz is 2,5-pyrazinecarboxylic acid; C19H25BN4O4; Bortezomib, Velcade®; known previously as PS-341, LDP-341, and MLM341) as a drug for the treatment of MM (55).
MM is a differentiated clonal B cell malignancy characterized by rapidly proliferating plasma cells in bone marrow. It is accompanied by osteoporosis, lytic skeletal lesions, pathological fractures, and hypercalcemia that result from erosion of the long bones and vertebrae. The hypercalcemia can cause severe renal damage. A hallmark of the disease is production of a monoclonal immunoglobulin that can be found in the circulation and that contributes to the renal damage. MM accounts for 1% of all malignancies and 10% of hematological malignancies. The annual incidence of the disease is four to five new cases/100,000 people, which mean that ∼14,000 new cases are diagnosed annually in the United States alone. The prevalence of the disease is ∼21/100,000 with a total number of ∼63,000 patients in the United States. Several clinical trials have shown that Bortezomib is efficient in patients with relapsed and refractory disease (50, 56). Interestingly, a constitutively increased proteasomal activity has been found in myeloma cells. This high level is reflected also in the appearance of the enzyme in the circulation of the patients, where it was identified as a prognostic factor for survival (57). It is highly probable that the treatment with proteasome inhibitors will not remain limited to MM. Trials using Bortezomib in related hematological disorders such as non-Hodgkin lymphoma have already shown promising results (56).
The inhibitors appear to exert their selective effect on malignant cells via multiple mechanisms (58). One is suppression of activation of the transcription regulator NF-κB. The factor is constitutively activated in MM, resulting in several tumor-promoting activities. It supports survival (by inducing IAP (inhibitor of apoptosis) proteins and Bcl-xL, for example), drug resistance (by inducing MDR1 and P-glycoprotein), growth and proliferation (by inducing Myc and cyclin D1), angiogenesis (by inducing vascular endothelial growth factor and COX2), motility and migration (by inducing matrix metalloproteinases), and inflammation (by inducing interleukins and tumor necrosis factor α). It also antagonizes p53 function, partially by cross-competition for transcriptional co-activators (59). Several steps along the NF-κB activation pathway are UPS-dependent, including upstream events that regulate signaling components such as the receptor-interacting protein but also downstream events such as the limited processing of the precursor protein p105 to yield the p50 active subunit and the signal-induced degradation of the inhibitor IκBα that is bound to the heterodimeric p50-p65 regulator. Its degradation unmasks a nuclear localization signal that allows translocation of active NF-κB to the nucleus (60, 61), yet it was clear that inhibition of NF-κB cannot explain all the antitumor effects of proteasome inhibitors, as down-regulation of the factor using more specific agents had a much smaller effect on myeloma cell proliferation than inhibition of the proteasome (62). It should be noted that by inhibiting NF-κB activation, proteasome inhibitors may chemosensitize malignant cells to the activity of other chemotherapeutic agents, such as daunorubicin and vinblastin, that activate NF-κB and by that attenuate their own activity (50, 60). Other mechanisms also contribute to the ability of proteasome inhibitors to overcome resistance developed to other drugs. For example, proteasomal activity is required for maturation of MDR1. The nonfunctional pumps that accumulate in the presence of the inhibitors cannot remove the chemotherapeutic agents from the malignant cells, and their toxicity increases (50). Thus, it appears that proteasome inhibitors can potentiate the activity of other drugs.
Proteasome inhibitors may act also by inducing JNK-mediated apoptosis. Inhibition of the proteasome can activate JNK, which leads to phosphorylation of 14-3-3 proteins, translocation of Bax into the mitochondria, and release of cytochrome c, which initiates the apoptotic cascade (63). The inhibitors can up-regulate p53 independently of inhibiting NF-κB by slowing down its degradation, which also sensitizes cells to apoptosis.
An organelle that can induce apoptosis when stressed is the ER. Because malignant plasma cells produce large amounts of immunoglobulins, they are dependent on intact ER-associated degradation and the UPR to degrade the increased load of misfolded proteins and to regulate the ER stress that these proteins induce (64, 65). Inhibition of the proteasome increases the mass of the misfolded proteins in the ER and up-regulates certain components of the UPR (66). Overall, however, the UPR is down-regulated: the drugs inhibit activation of the endoribonuclease/kinase IRE1α, which is responsible for generation of the spliced mRNA that codes for XBP-1, a critical transcription factor of many genes involved in the UPR. The combined effects of the increased load of misfolded proteins in the ER and suppression of the UPR result in efficient induction of apoptosis.
The new generation of proteasome inhibitors appears to be more effective than Bortezomib, as their mechanisms of action are somewhat different. Unlike Bortezomib, salinosporamide and carfilzomib bind to the proteasome irreversibly. In addition, they appear to have different specificities for the different catalytic sites of the 20 S complex. For example, the β2-tryptic site that is not affected by Bortezomib appears to be inhibited by salinosporamide. It is possible that the repertoire of substrates affected by them is somewhat different from that affected by Bortezomib. Indeed, their effects on cells appear to be different. For example, salinosporamide is significantly less dependent on Bax and Bak for inducing mitochondrion-mediated cell death. On the other hand, Bortezomib relies less on the Fas-associated death domain-caspase 8 signaling axis than does salinosporamide (69). These differential mechanisms support a mode of action in which the two drugs will act synergistically.
The success of the proteasome inhibitors is probably due to the selective response of these malignancies to even partial inhibition of the enzyme. Because the malignant Ig-secreting cells are particularly sensitive to apoptosis, it is the resensitization of the cells to genotoxic stimuli that underlies the mechanism of action of the inhibitors.
Recently, inhibitors that affect other components of the system have started to emerge. One is the inhibitor of the NEDD8-activating enzyme MLN4924 (67). NEDD8 conjugation of the cullin component of many SCF (Skp2/cullin/F-box protein) RING finger-containing ubiquitin ligases that regulate processes such as the cell cycle and signal transduction is essential for their activity. Initial experiments demonstrated that MLN4924 induces apoptotic death in different human tumor models (67).
Supplementary Material
The work performed in the laboratory of A. N. was supported by the Israel Science Foundation, the Minerva Foundation (Germany), the German-Israeli Foundation for Scientific Research and Development, and a special gift from Rolando Uziel. The work performed in the laboratory of A. C. was supported by grants from the Dr. Miriam and Sheldon Adelson Foundation for Medical Research, the Israel Science Foundation, the German-Israeli Foundation for Scientific Research and Development, the European Union Networks of Excellence NeuroNE and Rubicon, a professorship from the Israel Cancer Research Fund USA, and a grant from the Foundation for Promotion of Research in the Technion. This is the eleventh article in the Thematic Minireview Series on Proteolytic Enzymes. The first article was published in the November 7, 2008 issue; the second and third articles in the May 22, 2009 issue; the fourth and fifth articles in the July 31, 2009 issue; the sixth and seventh articles in the August 14, 2009 issue; the eighth article in the August 28, 2009 issue; the ninth article in the November 6, 2009 issue; and the tenth article in the November 27, 2009 issue. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin-protein ligase
- UPS
- ubiquitin-proteasome system
- MM
- multiple myeloma
- JNK
- c-Jun N-terminal kinase
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response.
REFERENCES
- 1.Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. (2006) Cell 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 3.Li W., Tu D., Brunger A. T., Ye Y. (2007) Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 4.Ravid T., Hochstrasser M. (2007) Nat. Cell Biol. 9, 422–427 [DOI] [PubMed] [Google Scholar]
- 5.Hicke L. (2001) Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 6.Hicke L., Dunn R. (2003) Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 7.Haglund K., Di Fiore P. P., Dikic I. (2003) Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 8.Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. (2009) Mol. Cell 33, 496–504 [DOI] [PubMed] [Google Scholar]
- 9.Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 10.Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. (2006) Mol. Cell 24, 701–711 [DOI] [PubMed] [Google Scholar]
- 11.Pickart C. M. (2000) Trends Biochem. Sci. 25, 544–548 [DOI] [PubMed] [Google Scholar]
- 12.Saeki Y., Kudo T., Sone T., Kikuchi Y., Yokosawa H., Toh-e A., Tanaka K. (2009) EMBO J. 28, 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Nature 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 15.Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. (1995) Science 268, 533–539 [DOI] [PubMed] [Google Scholar]
- 16.Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) Nat. Struct. Biol. 7, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 17.Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 18.Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medalia N., Beer A., Zwickl P., Mihalache O., Beck M., Medalia O., Navon A. (2009) J. Biol. Chem. 284, 22952–22960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aki M., Shimbara N., Takashina M., Akiyama K., Kagawa S., Tamura T., Tanahashi N., Yoshimura T., Tanaka K., Ichihara A. (1994) J. Biochem. 115, 257–269 [DOI] [PubMed] [Google Scholar]
- 21.Walz J., Erdmann A., Kania M., Typke D., Koster A. J., Baumeister W. (1998) J. Struct. Biol. 121, 19–29 [DOI] [PubMed] [Google Scholar]
- 22.Baumeister W., Walz J., Zühl F., Seemüller E. (1998) Cell 92, 367–380 [DOI] [PubMed] [Google Scholar]
- 23.Nickell S., Mihalache O., Beck F., Hegerl R., Korinek A., Baumeister W. (2007) Biochem. Biophys. Res. Commun. 353, 115–120 [DOI] [PubMed] [Google Scholar]
- 24.Hendil K. B., Hartmann-Petersen R., Tanaka K. (2002) J. Mol. Biol. 315, 627–636 [DOI] [PubMed] [Google Scholar]
- 25.Hartmann-Petersen R., Tanaka K., Hendil K. B. (2001) Arch. Biochem. Biophys. 386, 89–94 [DOI] [PubMed] [Google Scholar]
- 26.Braun B. C., Glickman M., Kraft R., Dahlmann B., Kloetzel P. M., Finley D., Schmidt M. (1999) Nat. Cell Biol. 1, 221–226 [DOI] [PubMed] [Google Scholar]
- 27.Köhler A., Cascio P., Leggett D. S., Woo K. M., Goldberg A. L., Finley D. (2001) Mol. Cell 7, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 28.Lam Y. A., Lawson T. G., Velayutham M., Zweier J. L., Pickart C. M. (2002) Nature 416, 763–767 [DOI] [PubMed] [Google Scholar]
- 29.Verma R., Oania R., Graumann J., Deshaies R. J. (2004) Cell 118, 99–110 [DOI] [PubMed] [Google Scholar]
- 30.Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., Finley D. (2002) Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 31.Hanna J., Leggett D. S., Finley D. (2003) Mol. Cell. Biol. 23, 9251–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Y., Varshavsky A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2497–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., Finley D. (2006) Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 34.Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) Cell 94, 615–623 [DOI] [PubMed] [Google Scholar]
- 35.Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. (2002) Science 298, 608–611 [DOI] [PubMed] [Google Scholar]
- 36.Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. (1994) J. Biol. Chem. 269, 7059–7061 [PubMed] [Google Scholar]
- 37.van Nocker S., Sadis S., Rubin D. M., Glickman M., Fu H., Coux O., Wefes I., Finley D., Vierstra R. D. (1996) Mol. Cell. Biol. 16, 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., Dikic I. (2008) Nature 453, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiner P., Chen X., Husnjak K., Randles L., Zhang N., Elsasser S., Finley D., Dikic I., Walters K. J., Groll M. (2008) Nature 453, 548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elsasser S., Finley D. (2005) Nat. Cell Biol. 7, 742–749 [DOI] [PubMed] [Google Scholar]
- 41.Elsasser S., Chandler-Militello D., Müller B., Hanna J., Finley D. (2004) J. Biol. Chem. 279, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 42.Bertolaet B. L., Clarke D. J., Wolff M., Watson M. H., Henze M., Divita G., Reed S. I. (2001) Nat. Struct. Biol. 8, 417–422 [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. (2001) Nat. Cell Biol. 3, 939–943 [DOI] [PubMed] [Google Scholar]
- 44.Wang Q., Goh A. M., Howley P. M., Walters K. J. (2003) Biochemistry 42, 13529–13535 [DOI] [PubMed] [Google Scholar]
- 45.Hiyama H., Yokoi M., Masutani C., Sugasawa K., Maekawa T., Tanaka K., Hoeijmakers J. H., Hanaoka F. (1999) J. Biol. Chem. 274, 28019–28025 [DOI] [PubMed] [Google Scholar]
- 46.Walters K. J., Kleijnen M. F., Goh A. M., Wagner G., Howley P. M. (2002) Biochemistry 41, 1767–1777 [DOI] [PubMed] [Google Scholar]
- 47.Kisselev A. F., Goldberg A. L. (2001) Chem. Biol. 8, 739–758 [DOI] [PubMed] [Google Scholar]
- 48.Adams J. (2004) Cancer Cell 5, 417–421 [DOI] [PubMed] [Google Scholar]
- 49.Orlowski R. Z., Kuhn D. J. (2008) Clin. Cancer Res. 14, 1649–1657 [DOI] [PubMed] [Google Scholar]
- 50.Richardson P. G., Mitsiades C., Hideshima T., Anderson K. C. (2006) Annu. Rev. Med. 57, 33–47 [DOI] [PubMed] [Google Scholar]
- 51.Corey E. J., Li W. D. (1999) Chem. Pharm. Bull. 47, 1–10 [DOI] [PubMed] [Google Scholar]
- 52.Chauhan D., Catley L., Li G., Podar K., Hideshima T., Velankar M., Mitsiades C., Mitsiades N., Yasui H., Letai A., Ovaa H., Berkers C., Nicholson B., Chao T. H., Neuteboom S. T., Richardson P., Palladino M. A., Anderson K. C. (2005) Cancer Cell 8, 407–419 [DOI] [PubMed] [Google Scholar]
- 53.Imajoh-Ohmi S., Kawaguchi T., Sugiyama S., Tanaka K., Omura S., Kikuchi H. (1995) Biochem. Biophys. Res. Commun. 217, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 54.Orlowski R. Z., Eswara J. R., Lafond-Walker A., Grever M. R., Orlowski M., Dang C. V. (1998) Cancer Res. 58, 4342–4348 [PubMed] [Google Scholar]
- 55.Adams J., Kauffman M. (2004) Cancer Investig. 22, 304–311 [DOI] [PubMed] [Google Scholar]
- 56.Lee S. J., Richardson P. G., Sonneveld P., Schuster M. W., Irwin D., San Miguel J. F., Crawford B., Massaro J., Dhawan R., Gupta S., Anderson K. C. (2008) Br. J. Haematol. 143, 511–519 [DOI] [PubMed] [Google Scholar]
- 57.Jakob C., Egerer K., Liebisch P., Türkmen S., Zavrski I., Kuckelkorn U., Heider U., Kaiser M., Fleissner C., Sterz J., Kleeberg L., Feist E., Burmester G. R., Kloetzel P. M., Sezer O. (2007) Blood 109, 2100–2105 [DOI] [PubMed] [Google Scholar]
- 58.Voorhees P. M., Orlowski R. Z. (2006) Annu. Rev. Pharmacol. Toxicol. 46, 189–213 [DOI] [PubMed] [Google Scholar]
- 59.Nakanishi C., Toi M. (2005) Nat. Rev. Cancer 5, 297–309 [DOI] [PubMed] [Google Scholar]
- 60.Palombella V. J., Rando O. J., Goldberg A. L., Maniatis T. (1994) Cell 78, 773–785 [DOI] [PubMed] [Google Scholar]
- 61.Karin M., Ben-Neriah Y. (2000) Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 62.Hideshima T., Chauhan D., Richardson P., Mitsiades C., Mitsiades N., Hayashi T., Munshi N., Dang L., Castro A., Palombella V., Adams J., Anderson K. C. (2002) J. Biol. Chem. 277, 16639–16647 [DOI] [PubMed] [Google Scholar]
- 63.Lopes U. G., Erhardt P., Yao R., Cooper G. M. (1997) J. Biol. Chem. 272, 12893–12896 [DOI] [PubMed] [Google Scholar]
- 64.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 65.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee A. H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9946–9951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., et al. (2009) Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 68.Ferrell K., Wilkinson C. R., Dubiel W., Gordon C. (2000) Trends Biochem. Sci. 25, 83–88 [DOI] [PubMed] [Google Scholar]
- 69.Groll M., Berkers C. R., Ploegh H. L., Ovaa H. (2006) Structure 14, 451–456 [DOI] [PubMed] [Google Scholar]
- 70.Brannigan J. A., Dodson G., Duggleby H. J., Moody P. C., Smith J. L., Tomchick D. R., Murzin A. G. (1995) Nature 378, 416–419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.