Abstract
The contractile phenotype of smooth muscle (SM) cells is controlled by serum response factor (SRF), which drives the expression of SM-specific genes including SM α-actin, SM22, and others. Myocardin is a cardiac and SM-restricted coactivator of SRF that is necessary for SM gene transcription. Growth factors inducing proliferation of SM cells inhibit SM gene transcription, in a manner dependent on the activation of extracellular signal-regulated kinases ERK1/2. In this study, we found that ERK1/2 phosphorylates mouse myocardin (isoform B) at four sites (Ser812, Ser859, Ser866, and Thr893), all of which are located within the transactivation domain of myocardin. The single mutation of each site either to alanine or to aspartate has no effect on the ability of myocardin to activate SRF. However, the phosphomimetic mutation of all four sites to aspartate (4×D) significantly impairs activation of SRF by myocardin, whereas the phosphodeficient mutation of all four sites to alanine (4×A) has no effect. This translates to a reduced ability of the 4×D (but not of 4×A) mutant of myocardin to stimulate expression of SM α-actin and SM22, as assessed by corresponding promoter, mRNA, or protein assays. Furthermore, we found that phosphorylation of myocardin at these sites impairs its interaction with acetyltransferase, cAMP response element-binding protein-binding protein, which is known to promote the transcriptional activity of myocardin. In conclusion, we describe a novel mode of modulation of SM gene transcription by ERK1/2 through a direct phosphorylation of myocardin.
Introduction
Myocardin (MyoCD)2 and myocardin-related transcription factors (MRTF-A and MRTF-B) represent a family of coactivators of serum response factor (SRF) that drives the transcription of multiple genes containing CArG consensus sequences CC(AT)6GG (1, 2). Whereas MRTFs are ubiquitously expressed (3), MyoCD expression is restricted to cardiac and smooth muscle (SM) (4, 5). MyoCD is a powerful activator of SM gene transcription (5–8), and knock-out of the MyoCD gene leads to a defective differentiation of vascular SM cells (9).
Although the function of MyoCD and MRTFs is quite established, regulation of their activity is poorly understood. Activation of MRTFs occurs by a complex mechanism involving a small GTPase RhoA, actin polymerization, and translocation of MRTFs from the cytoplasm to the nucleus where they bind and activate nuclear-localized SRF (10–12). In contrast, MyoCD is localized constitutively to the nucleus (4), and regulation of its activity probably occurs by a mechanism distinct from that of MRTFs. One potential mechanism may involve AKT-dependent phosphorylation and cytoplasmic retention of the inhibitor of MyoCD, forkhead transcription factor Foxo4 (13). However, the activities of both MyoCD and MRTFs are negatively regulated by extracellular signal-regulated protein kinases (ERK1/2) through a similar mechanism involving phosphorylation of Elk1 transcription factor by ERK1/2 and competition of phosphorylated Elk1 with MyoCD or MRTFs for SRF binding (14, 15).
Emerging evidence suggests that MyoCD and MRTFs are themselves targets of protein phosphorylation. Mouse MyoCD was shown to be phosphorylated by glycogen synthase kinase-3 at serines 455, 459, 463, 467 and 624, 628, 632, 636; and this phosphorylation by glycogen synthase kinase-3 attenuates the transcriptional activity of MyoCD by an unknown mechanism (16). Human MRTF-A, also called megakaryoblastic leukemia 1, is phosphorylated by ERK1/2 at Ser454, which inhibits its activity through a nuclear extraction of phosphorylated MRTF-A (17). In the present study, we found that ERK1/2 phosphorylates MyoCD at four novel sites within its transactivation domain, and we examined the functional consequences of MyoCD phosphorylation by ERK1/2 at these sites.
EXPERIMENTAL PROCEDURES
Cell Culture and Transient Transfection of DNA
10T1/2 cells and HT1080 cells (American Type Culture Collection) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml streptomycin, 250 ng/ml amphotericin B, and 100 units/ml penicillin. Chinese hamster ovary (CHO) cells were grown in Ham's F-12 medium supplemented as above. In experiments described in Figs. 1 and 2, CHO cells were serum-deprived for 24 h in Ham's F-12 medium containing 0.1% bovine serum albumin and 2 mm l-glutamine. Transient transfection of DNA was performed using Lipofectamine-PLUS reagent (Invitrogen) following the manufacturer's standard protocol.
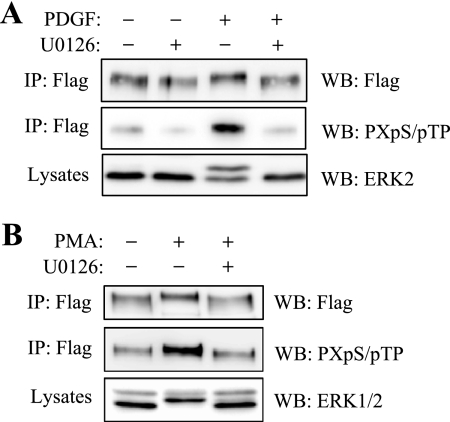
FIGURE 1.
Phosphorylation of MyoCD by ERK1/2. A, CHO cells were transfected with FLAG-tagged MyoCD and stimulated with 1 ng/ml platelet-derived growth factor (PDGF; A) or 1 μm PMA (B) for 10 min in the presence or absence of a MEK inhibitor U0126 (10 μm). Myocardin was immunoprecipitated (IP) from cell lysates with FLAG antibodies, and the immune complexes or total cell lysates were examined by Western blotting (WB) with antibodies against FLAG, phosphorylated MAP kinase substrates (PXpS/pTP), or ERK1/2 as indicated.
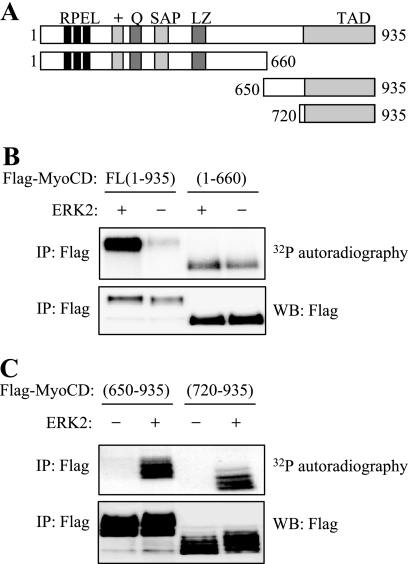
FIGURE 2.
In vitro phosphorylation of MyoCD by ERK2. A, diagram of MyoCD deletion mutants used in these experiments depicting the established functional domains of MyoCD. B and C, CHO cells were transfected with FLAG-tagged fragments of MyoCD as indicated. The fragments were immunoprecipitated (IP) with FLAG antibodies, and the immune complexes were subjected to a kinase reaction with [32P]ATP in the presence (+) or absence (−) of recombinant-activated ERK2. The samples were resolved by electrophoresis and analyzed by 32P autoradiography or by Western blotting (WB) with FLAG antibodies. PRFL, proline, arginine, phenylalanine, leucine motifs; Q, polyglutamine region; SAP, SAP (SAF-AB, Acinus, PIAS) domain; TAD, transactivation domain.
DNA Reagents and Antibodies
The cDNA for mouse MyoCD (isoform B, accession number AF384055) was provided by Dr. Eric Olson. All MyoCD deletion constructs were generated by PCR, cloned into FLAG-tagged CMV-Tag2B vector (Stratagene), and confirmed by sequencing. Point mutations were introduced by using the QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's instruction and confirmed by sequencing. The luciferase reporter plasmids for SRF (CArG-Luc), SM α-actin promoter (α-SMA-Luc), and SM22 promoter (SM22-Luc) were used previously (17–19). The antibodies were from the following sources: anti-SM α-actin (antibody A2547), anti-β-actin (A5441), and anti-FLAG (F1804) were from Sigma. Anti-SM22 (Ab14106) was from Abcam. Anti-ERK1/2 was a gift from Dr. Michael Dunn. Anti-MAP kinase substrate antibodies (2325) were from Cell Signaling. Anti-CREB-binding protein (CBP) (sc-369), anti-SRF(sc-335), and anti-Myc (sc-40), were from Santa Cruz Biotechnology. U0126 was from EMD Biosciences. Phorbol 12-myristate 13-acetate (PMA) was from Sigma.
Immunoprecipitation and Western Blot Analysis
Cells were washed twice with ice-cold phosphate-buffered saline and lysed in extraction buffer containing 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm sodium orthovanadate, 2.5 mm β-glycerophosphate, 1 mm EGTA, and protease inhibitors (1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride). The lysates were passed through a 27.5-gauge needle five times and cleared by centrifugation at 20,000 × g for 10 min. The immunoprecipitation of FLAG-tagged MyoCD and its mutants was performed by incubation of the cleared lysates with agarose-conjugated mouse anti-FLAG antibodies followed by three washes in the same buffer. The immune complexes were boiled in Laemmli buffer and subjected to Western blotting with the desired antibodies using the standard technique. The digital chemiluminescence images were taken by a Luminescent Image Analyzer LAS-4000 (Fujifilm).
In Vitro ERK2 Kinase Assay of MyoCD
FLAG-tagged MyoCD or its mutants were expressed in CHO cells as described above. Cells were lysed in a buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 0.5% Triton X-100, 1 mm sodium orthovanadate, 2.5 mm β-glycerophosphate, 1 mm EGTA, 1 mm dithiothreitol, and protease inhibitors (1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mm phenylmethylsulfonyl fluoride). The lysates were cleared from insoluble material by centrifugation at 20,000 × g for 10 min at 4 °C, and MyoCD or its mutants were purified using anti-FLAG-agarose beads. The beads were washed once with a kinase buffer (25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, 0.1 mm ATP, 0.01% Brij 35, 5 mm β-glycerophosphate) and subjected to a kinase reaction (45 μl) in the same buffer supplemented with 50 ng of recombinant activated ERK2 (Upstate, 14-173) and 10 μCi of [γ-32P] ATP at 30 °C for 30 min. The reaction was stopped by the addition of 15 μl of 4× Laemmli buffer and heating for 5 min at 95 °C. Proteins were subjected to SDS-PAGE followed by autoradiography or Western blotting with the desired antibodies.
32P Labeling of MyoCD in Intact Cells
Phosphorylation of MyoCD in intact cells was performed as described previously (20–22). CHO cells were transfected with cDNAs for FLAG-tagged MyoCD mutants and serum-starved for 24 h. Cells were incubated with 0.25 mCi/ml 32Pi in a phosphate-free and serum-free medium followed by three washes and stimulation with 1 μm PMA for 15 min. Cells were then lysed, and the cell lysates were subjected to immunoprecipitation with FLAG antibodies, followed by electrophoresis, 32P autoradiography, or Western blotting with FLAG antibodies as described above.
Luciferase Reporter Assay
Cells grown in 24-well plates were cotransfected with 20 ng of the desired firefly luciferase reporter plasmid, 10 ng of thymidine kinase-driven Renilla luciferase plasmid (Promega), and 20 ng of empty vector or cDNA for WT MyoCD or its mutants per well. Cells were serum-starved overnight following transfection, washed with phosphate-buffered saline, and lysed in protein extraction reagent. The lysates were assayed for firefly and Renilla luciferase activity using the Promega Dual luciferase assay kit. To account for differences in transfection efficiency, the firefly luciferase activity of each sample was normalized to Renilla luciferase activity.
Reverse Transcription-Quantitative Real Time PCR
Total RNA was harvested using RNA STAT-60 (Tell-Test, Inc.) following the manufacturer's protocol. One microgram of total RNA was used as a template for random-primed reverse transcription using an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's protocols. Real time PCR analysis was performed using iTaq SYBR Green supermix with ROX (Bio-Rad) in a MyIQ single color real time PCR detection system (Bio-Rad). Real time PCR primers were of the following sequences: α-SMA, forward AAAGACAGCTACGTGGGTGACGAA and reverse TTCCATGTCGTCCCAGTTGGTGAT; SM22, forward AAGAATGATGGGCACTACCGTGGA and reverse TGAAGGCCAATGACATGCTTTCCC. The 18 S transcript was used to normalize the amount and quality of the extracted RNA, using the following primer set: 18 S forward, GATTAAGTCCCTGCCCTTTG; reverse, GTTCACCTACGGAAACCTTG.
Statistics
Student's t test was utilized to test for statistically significant differences (p values are indicated in the figure legends).
RESULTS
Phosphorylation of MyoCD by ERK1/2
Fig. 1A shows phosphorylation of MyoCD in intact cells at consensus sites of phosphorylation by MAP kinases, as assessed by immunoprecipitation of MyoCD followed by Western blotting with antibodies against phosphorylated MAP kinase substrate motif (PXpS/pTP). Activation of MAP kinase by platelet-derived growth factor increased phosphorylation of MyoCD, and this was abolished by the inhibitor of ERK1/2 upstream activator MEK1 (U0126), which supports the role of ERK1/2 in this effect. The efficiency of U0126 was confirmed by inhibition of ERK1/2 phosphorylation, as assessed by electrophoretic mobility shift assay for phosphorylated ERK2 (Fig. 1A, bottom panel). ERK-dependent phosphorylation of MyoCD was also induced by PMA (Fig. 1B).
MyoCD was also readily phosphorylated by ERK2 in vitro as determined by a kinase assay of immunoprecipitated MyoCD with recombinant activated ERK2 in the presence of [32P]ATP (Fig. 2B). Some basal phosphorylation was also observed in the absence of ERK2, which was likely a result of phosphorylation by endogenous kinase(s) coimmunoprecipitated with MyoCD. Deletion analysis showed that the N-terminal fragment-(1–660) of MyoCD was poorly phosphorylated by recombinant ERK2 in vitro, whereas the basal phosphorylation remained (Fig. 2B). Focusing on the MyoCD C-terminal region, we generated additional deletion mutants (Fig. 2A), and we found that the region comprising the transactivation domain of MyoCD (fragment-(720–935)) is phosphorylated by ERK2 in vitro at multiple sites, as viewed by 32P autoradiography as well as by electrophoretic mobility shift ladder (Fig. 2C).
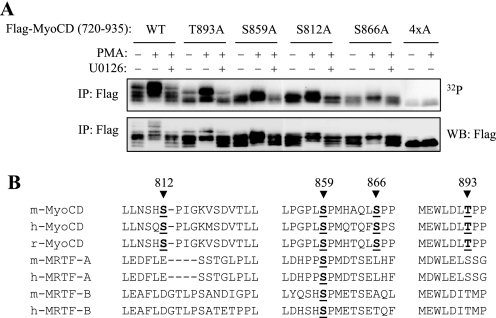
The fragment-(720–935) region of MyoCD contains a number of proline-directed putative ERK1/2 phosphorylation sites (S/TP), whose phosphorylation was examined by site-directed mutagenesis. To exclude the potential artifact of in vitro kinase assay, we examined the phosphorylation of the alanine mutants of fragment-(720–935) of MyoCD by 32P incorporation in intact cells. PMA was used as an ERK1/2 activator, whereas U0126 was used to dissect the role of ERK1/2-dependent phosphorylation. As shown in Fig. 3A, the single alanine mutation of Ser812, Ser859, Ser866, and Thr893 residues resulted in a clear reduction of PMA-induced and U0126-inhibited phosphorylation, whereas mutation of all four residues (4×A) nearly abolished phosphorylation, as evident by 32P autoradiography, as well as by a change in electrophoretic mobility shift ladder (indicative of phosphorylation). Fig. 3B shows the location of these sites in the sequences of MyoCD from different species aligned with MRTF-A and MRTF-B. All four phosphorylation sites of MyoCD are highly conserved among species. Interestingly, the homologous sequences of MRTF-A or MRTF-B lack putative ERK phosphorylation sites, with the exception of one site that is analogous to Ser859 of MyoCD. This may suggest that, within the family of these transcription factors, phosphorylation of transactivation domain by ERK1/2 is unique to MyoCD.
FIGURE 3.
Identification of ERK1/2 phosphorylation sites on MyoCD. A, CHO cells were transfected with cDNAs for FLAG-tagged MyoCD fragment-(720–935) or the corresponding alanine mutants as indicated. Cells were then labeled with 32Pi and stimulated with 1 μm PMA for 10 min in the presence or absence of a MEK inhibitor U0126 (10 μm). Myocardin fragments were immunoprecipitated with FLAG antibodies, followed by electrophoresis and 32P autoradiography or Western blotting with FLAG antibodies as indicated. B, alignment of MyoCD, MRTF-A, and MRTF-B at the sites of MyoCD phosphorylation by ERK1/2. The numbered sites correspond to the mouse MyoCD, isoform-B. m, mouse; h, human; r, rat.
Phosphorylation of Myocardin Attenuates Its Transcriptional Activity
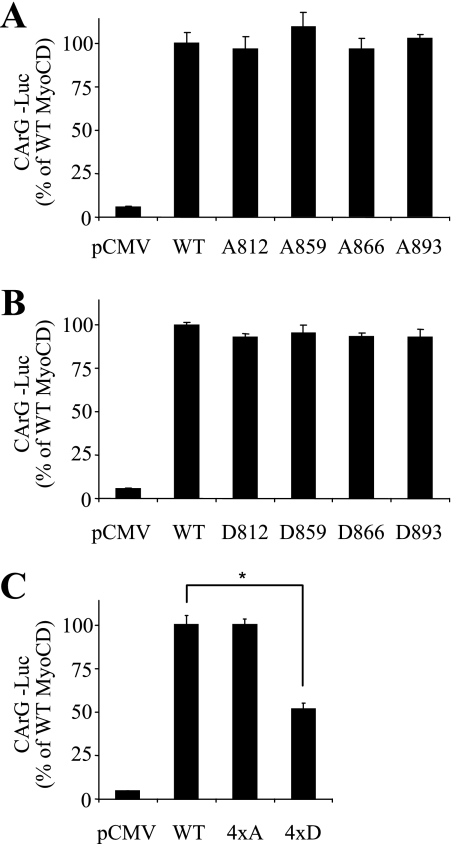
Having identified the four ERK phosphorylation sites, we then generated the corresponding phosphodeficient alanine (A) mutants or phosphomimetic aspartate (D) mutants of the full-length MyoCD, and we examined the ability of these mutants to activate SRF. As expected, overexpression of MyoCD resulted in a profound activation of SRF, as determined by CArG-luciferase reporter assay (Fig. 4). Apparently, a single mutation of each phosphorylation site of MyoCD (either to alanine or to aspartate) had no effect on its ability to activate SRF (Fig. 4, A and B). Mutation of all four phosphorylation sites to alanine (4×A) also had no effect on SRF reporter activity (Fig. 4C). In contrast, mutation of all four phosphorylation sites to aspartate (4×D) diminished the ability of MyoCD to activate SRF reporter (Fig. 4C), suggesting that phosphorylation of MyoCD attenuates its transcriptional activity.
FIGURE 4.
Activation of SRF-dependent gene transcription by ERK1/2 phosphorylation mutants of MyoCD. 10T1/2 cells were transfected with cDNAs for firefly luciferase driven by two copies of CArG elements (CArG-Luc), Renilla luciferase driven by thymidine kinase promoter, together with either empty vector (pCMV), or cDNAs for the full-length MyoCD or the corresponding single-site alanine mutants (A), single-site aspartate mutants (B), or quadruple mutants where all four indicated residues were mutated either to alanine or aspartate (C) for 24 h. Luciferase activity was measured as described under “Experimental Procedures.” Data represent mean ± S.D. from a representative (of three) experiment performed in triplicate (*, p < 0.05).
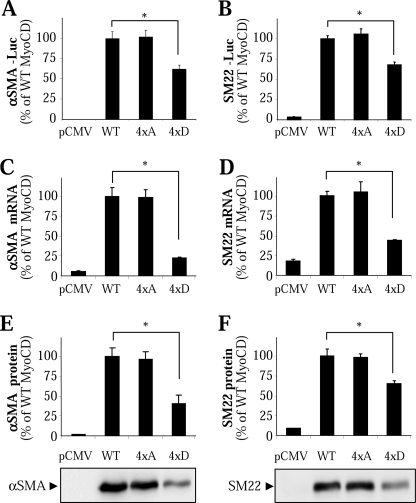
We then examined the ability of these MyoCD mutants to drive SM gene transcription, focusing on SM α-actin (α-SMA) and SM22. As shown in Fig. 5, A and B, activation of α-SMA and SM22 promoters by the 4×D mutant of MyoCD was significantly reduced as determined by the corresponding luciferase reporters. Furthermore, the 4×D mutant was significantly less active in induction of α-SMA and SM22 mRNA as assessed by real time PCR (Fig. 5, C and D). Finally, stimulation of α-SMA and SM22 protein expression by 4×D mutant was also significantly reduced compared with the WT (Fig. 5, E and F). In all of these assays, the activity of the phosphodeficient 4×A mutant was not significantly different from that of the WT MyoCD (Fig. 5). Together, these data suggest that phosphorylation of MyoCD at these four sites attenuates its ability to activate SRF and SM gene transcription.
FIGURE 5.
Phosphorylation of MyoCD by ERK1/2 reduces its induction of SM gene transcription. A and B, 10T1/2 cells were transfected with cDNAs for the full-length MyoCD or its quadruple phosphodeficient alanine (4×A) or phosphomimetic aspartate (4×D) mutant, along with firefly luciferase reporters for SM α-actin (A) or SM22 (B) promoters and Renilla luciferase driven by thymidine kinase promoter control reporter. Following 24 h of transfection, the luciferase activity was measured as described under “Experimental Procedures.” C and D, 10T1/2 cells were transfected with cDNAs for the full-length MyoCD or its quadruple 4×A or 4×D mutant for 24 h, and cells were analyzed for mRNA levels of SM α-actin (C) and SM22 (D) by quantitative real time PCR. E and F, HT-1080 cells were transfected with cDNAs for the full-length MyoCD or its quadruple 4×A or 4×D mutant for 24 h, and cells were analyzed for protein expression of SM α-actin (E) and SM22 (F) by Western blotting with corresponding antibodies. Data represent mean ± S.D. from a representative (of three) experiment performed in triplicate (*, p < 0.05).
Phosphorylation of MyoCD Inhibits Its Association with CBP
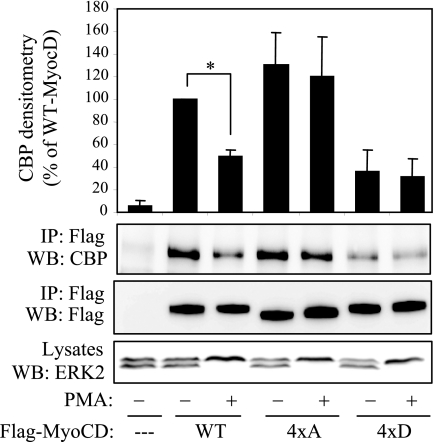
Multiple binding partners interact with various regions of MyoCD and modulate its activity (4, 13, 14, 23–26). Among those, the acetyltransferases CBP/p300 interact with transactivation domain of MyoCD (24). Given that all four phosphorylation sites that we have identified are located within the transactivation domain of MyoCD, we examined how their mutation affects the interaction of MyoCD with CBP by coimmunoprecipitation. As shown in Fig. 6, activation of ERK by PMA resulted in a significant reduction of CBP binding by WT MyoCD. The binding of 4×A mutant of MyoCD to CBP was higher than that of the WT, and it was not affected by PMA. In contrast, the binding of 4×D mutant was reduced significantly compared with that of WT MyoCD (Fig. 6). This suggests that phosphorylation of MyoCD at the sites that we have identified inhibits its interaction with CBP.
FIGURE 6.
Phosphorylation of MyoCD by ERK1/2 reduces its interaction with CBP. HT1080 cells were transfected with cDNA for CBP together with cDNAs for FLAG-tagged MyoCD or its quadruple 4×A or 4×D mutant for 24 h. Cells were stimulated with 1 μm PMA for 10 min and lysed, followed by immunoprecipitation (IP) of cell lysates with FLAG antibodies. The FLAG·immune complexes were immunoblotted (WB) with antibodies against CBP or against the FLAG epitope as indicated. The relative amounts of MyoCD-bound CBP were digitally quantified and expressed as percent of WT MyoCD binding (mean ± S.D. from three independent experiments; *, p < 0.05). Activation of ERK2 was confirmed by electrophoretic mobility shift assay of cell lysates.
DISCUSSION
It was reported previously that ERK1/2 may regulate SM gene transcription through phosphorylation of Elk1, which then competes with MyoCD for SRF binding (14). The present study demonstrates for the first time that MyoCD itself is phosphorylated by ERK1/2 at multiple sites (Ser812, Ser859, Ser866, and Thr893). Furthermore, our data suggest that phosphorylation of MyoCD by ERK1/2 attenuates its ability to activate SRF and SM gene transcription, at least in part, through inhibition of the interaction between MyoCD and CBP.
It was shown previously that mouse MyoCD (isoform B) is phosphorylated by glycogen synthase kinase-3 at serines 455, 459, 463, 467 and 624, 628, 632, 636, resulting in inhibition of MyoCD transcriptional activity by an unknown mechanism (16). In this study, we describe four novel sites that are simultaneously phosphorylated by ERK1/2, all of which are located within the C-terminal transactivation domain of MyoCD, downstream of those phosphorylated by glycogen synthase kinase-3 (Fig. 3). Glycogen synthase kinase-3-dependent phosphorylation at multiple sites often results in the proteasomal degradation of a target protein. However, we did not see any difference in the degradation rate of WT versus 4×A mutant of MyoCD (data not shown). Thus, we believe that phosphorylation of MyoCD by glycogen synthase kinase-3 or by ERK down-regulates its activity by distinct mechanisms.
Dr. Prywes and co-workers (17) reported recently that ERK1/2 phosphorylates human MRTF-A at Ser449, Thr450, and Ser454 sites and promotes nuclear extraction of MRTF-A, resulting in inhibition of its transcriptional activity. However, although MyoCD contains analogous conserved sites (Ser458 and Ser462), it is unlikely to be phosphorylated at these sites by ERK1/2 because ERK1/2 failed to phosphorylate fragment-(1–660) of MyoCD containing these sites (Fig. 2B). Furthermore, unlike MRTF-A, MyoCD localized to the nucleus of the cell under the basal conditions, which is consistent with the findings of others (4); and the 4×A or 4×D mutation did not affect its nuclear localization (data not shown). Finally, sequence alignment of MyoCD, MRTF-A, and MRTF-B suggests that MRTFs lack at least three of four sites within the transactivation domain that are phosphorylated by ERK1/2 in MyoCD (Fig. 3B). This suggests that, although both MyoCD and MRTF-A are directly phosphorylated by ERK1/2, the sites of phosphorylation and the mechanism of regulation are likely distinct.
Apparently, the single mutation of each ERK phosphorylation site that we have identified either to alanine or to aspartate has no effect on the activity of MyoCD (Fig. 4, A and B). However, the phosphomimetic mutation of all four of these sites to aspartate resulted in a diminished ability of MyoCD to activate SRF (Fig. 4C) and SM gene expression (Fig. 5). Even though we did not assess the effect of dual or triple mutation of MyoCD on its activity, one might think of a “titrated” regulation of MyoCD, dependent on the combination of phosphorylated sites and the duration of phosphorylation of each site. This will be addressed in the future once the phosphospecific antibodies against each phosphorylation site are generated.
Many transcription factors, including MyoCD, recruit acetyltransferases, such as CBP/p300, for an efficient transcriptional activity (24). Our data indicate that phosphorylation of MyoCD by ERK1/2 at the sites that we identified attenuates its interaction with CBP (Fig. 6), suggesting the mechanism of regulation. In contrast, we did not observe reproducible changes in coimmunoprecipitation of FLAG-MyoCD with Myc-SRF upon phosphorylation of MyoCD by ERK or upon 4×A mutation of MyoCD. This is consistent with the previous report showing that SRF interacts with the N terminus of MyoCD (4) which is distinct from transactivation domain phosphorylated by ERK1/2 (Fig. 2B). However, in the functional studies, the 4×A mutation of MyoCD did not translate to a stronger induction of SM gene transcription (Fig. 5). This may suggest that the basal level of CBP binding is sufficient for a full activation of SM gene transcription by MyoCD. Furthermore, although activation of ERK resulted in a reduced ability of WT MyoCD to stimulate SM gene transcription as expected, the 4×A mutation of MyoCD did not rescue this effect of ERK activation (supplemental Fig. S1). This suggests that other mechanisms of regulation of SM gene transcription by ERK must exist. One reported example is the regulation through Elk1 phosphorylation by ERK (14). However, it is possible that phosphorylation of MyoCD by ERK can also affect its interaction with partners other than CBP and hence, regulate a different function of MyoCD. These possibilities will be addressed in the future.
Supplementary Material
Acknowledgment
We thank Dr. Eric Olson for providing the cDNA for MyoCD.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL071755 (to N. O. D.), GM085058 (to N. O. D.), T32 HL07605 (to D. M. Y.), T32 HD07009 (to N. S.), and T32 HL07237 (to J. K.) and by American Heart Association Postdoctoral Fellowship Awards 0720151Z (to S. T.), 0520109Z (to N. S.), and 0825868G (to D. M. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- MyoCD
- myocardin
- CREB
- cyclic AMP response element-binding protein
- CBP
- CREB-binding protein
- CHO
- Chinese hamster ovary
- ERK
- extracellular signal-regulated kinase
- MAP
- mitogen-activated protein
- MEK
- MAP kinase/ERK kinase
- MRTF
- MyoCD-related transcription factor
- PMA
- phorbol 12-myristate 13-acetate
- SM
- smooth muscle
- SRF
- serum response factor
- WT
- wild type.
REFERENCES
- 1.Pipes G. C., Creemers E. E., Olson E. N. (2006) Genes Dev. 20, 1545–1556 [DOI] [PubMed] [Google Scholar]
- 2.Parmacek M. S. (2007) Circ. Res. 100, 633–644 [DOI] [PubMed] [Google Scholar]
- 3.Sasazuki T., Sawada T., Sakon S., Kitamura T., Kishi T., Okazaki T., Katano M., Tanaka M., Watanabe M., Yagita H., Okumura K., Nakano H. (2002) J. Biol. Chem. 277, 28853–28860 [DOI] [PubMed] [Google Scholar]
- 4.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 5.Chen J., Kitchen C. M., Streb J. W., Miano J. M. (2002) J. Mol. Cell. Cardiol. 34, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 6.Du K. L., Ip H. S., Li J., Chen M., Dandre F., Yu W., Lu M. M., Owens G. K., Parmacek M. S. (2003) Mol. Cell. Biol. 23, 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida T., Sinha S., Dandré F., Wamhoff B. R., Hoofnagle M. H., Kremer B. E., Wang D. Z., Olson E. N., Owens G. K. (2003) Circ. Res. 92, 856–864 [DOI] [PubMed] [Google Scholar]
- 8.Wang Z., Wang D. Z., Pipes G. C., Olson E. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7129–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Wang D. Z., Wang Z., Richardson J. A., Olson E. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9366–9370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miralles F., Posern G., Zaromytidou A. I., Treisman R. (2003) Cell 113, 329–342 [DOI] [PubMed] [Google Scholar]
- 11.Hinson J. S., Medlin M. D., Lockman K., Taylor J. M., Mack C. P. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H1170–H1180 [DOI] [PubMed] [Google Scholar]
- 12.Lockman K., Hinson J. S., Medlin M. D., Morris D., Taylor J. M., Mack C. P. (2004) J. Biol. Chem. 279, 42422–42430 [DOI] [PubMed] [Google Scholar]
- 13.Liu Z. P., Wang Z., Yanagisawa H., Olson E. N. (2005) Dev. Cell 9, 261–270 [DOI] [PubMed] [Google Scholar]
- 14.Wang Z., Wang D. Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N. (2004) Nature 428, 185–189 [DOI] [PubMed] [Google Scholar]
- 15.Yoshida T., Gan Q., Shang Y., Owens G. K. (2007) Am. J. Physiol. Cell Physiol. 292, C886–C895 [DOI] [PubMed] [Google Scholar]
- 16.Badorff C., Seeger F. H., Zeiher A. M., Dimmeler S. (2005) Circ. Res. 97, 645–654 [DOI] [PubMed] [Google Scholar]
- 17.Muehlich S., Wang R., Lee S. M., Lewis T. C., Dai C., Prywes R. (2008) Mol. Cell. Biol. 28, 6302–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis A., Hogarth K., Fernandes D., Solway J., Niu J., Kolenko V., Browning D., Miano J. M., Orlov S. N., Dulin N. O. (2003) Cell. Signal. 15, 597–604 [DOI] [PubMed] [Google Scholar]
- 19.Hogarth D. K., Sandbo N., Taurin S., Kolenko V., Miano J. M., Dulin N. O. (2004) Am. J. Physiol. Cell Physiol. 287, C449–C456 [DOI] [PubMed] [Google Scholar]
- 20.Dulin N. O., Sorokin A., Reed E., Elliott S., Kehrl J. H., Dunn M. J. (1999) Mol. Cell. Biol. 19, 714–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu J., Scheschonka A., Druey K. M., Davis A., Reed E., Kolenko V., Bodnar R., Voyno-Yasenetskaya T., Du X., Kehrl J., Dulin N. O. (2002) Biochem. J. 365, 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taurin S., Sandbo N., Qin Y., Browning D., Dulin N. O. (2006) J. Biol. Chem. 281, 9971–9976 [DOI] [PubMed] [Google Scholar]
- 23.Oh J., Wang Z., Wang D. Z., Lien C. L., Xing W., Olson E. N. (2004) Mol. Cell. Biol. 24, 8519–8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao D., Wang Z., Zhang C. L., Oh J., Xing W., Li S., Richardson J. A., Wang D. Z., Olson E. N. (2005) Mol. Cell. Biol. 25, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creemers E. E., Sutherland L. B., Oh J., Barbosa A. C., Olson E. N. (2006) Mol. Cell 23, 83–96 [DOI] [PubMed] [Google Scholar]
- 26.Long X., Creemers E. E., Wang D. Z., Olson E. N., Miano J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16570–16575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.