Abstract
Serotonin transporter (SERT) is responsible for the re-uptake of 5-hydroxytryptamine (5-HT) from the synaptic cleft after release from serotonergic neurons. We show here that cysteine residues at positions in transmembranes 1 and 3 of SERT, like the corresponding positions in the γ-aminobutyric acid transporter, can be cross-linked using copper(II)(1,10-phenanthroline)3. The presence of a cross-link was detected by a novel methionine mutagenesis strategy. A change in mobility for an N-terminal cyanogen bromide fragment accompanied disulfide cross-linking of the two cysteine residues. Cross-linking also inhibited transport, and this process was blocked by cocaine, which is expected to stabilize SERT in conformations where the two positions are separated, but cocaine did not decrease accessibility of either of the two cysteines to modification by 2-aminoethyl methanethiosulfonate. Cysteine was required at both positions on the same molecule for efficient cross-linking, indicating that the reaction was intramolecular.
Introduction
The neurotransmitter:sodium symporter family (also known as SLC6) contains transporters for many neurotransmitters including serotonin (5-HT),2 γ-aminobutyric acid, dopamine, norepinephrine, and glycine (1). It also contains hundreds of bacterial and archaeal proteins including the amino acid transporters TnaT, Tyt1, and LeuT (2–4). Several crystal structures for LeuT in complex with amino acids and inhibitors have been published (3, 5–7). Crystal structures of transporters from several other families that were previously thought to be unrelated to the neurotransmitter:sodium symporter family were observed to adopt a very similar structure to that of LeuT (8–11).
In the LeuT structures, especially in complex with the competitive inhibitor tryptophan (7), a permeation pathway is visible leading from the extracellular medium toward the site of amino acid and Na+ binding. However, none of the LeuT structures indicates the location of the permeation pathway leading from the binding site to the cytoplasm.
Studies employing cysteine scanning mutagenesis of serotonin transporter (SERT) identified a potential permeation pathway leading from the binding site to the cytoplasm, composed of TMs 1, 5, 6, and 8 (12–13). This pathway is apparently accessible in a cytoplasm-facing conformation of SERT. Accessibility of reactive cysteine residues in this cytoplasmic pathway was modulated by 5-HT and SERT inhibitors (12, 14). Cocaine, a competitive inhibitor of SERT, norepinephrine, and dopamine transporters, decreased accessibility of the cytoplasmic pathway residues (12–13) but increased reactivity of some positions in the extracellular pathway (15–16). 5-HT and the non-competitive inhibitor ibogaine increased accessibility in the cytoplasmic pathway and decreased it in the extracellular pathway (12–14).
We generated a model of LeuT in a cytoplasm-facing conformation that was consistent with accessibility information from the cysteine scanning studies (13). LeuT could interconvert between conformations represented by the crystal structures and the cytoplasm-facing model by tilting or rocking of a 4-helix bundle composed of TMs 1, 2, 6, and 7 within a scaffold formed by the rest of the protein. We proposed that this “rocking bundle” mechanism could account for alternating access in the neurotransmitter: sodium symporter transporter family (13).
These accessibility changes are consistent with cocaine stabilizing conformations of SERT similar to those of the LeuT crystal structures (with the bundle tilted so as to open the extracellular pathway and close the cytoplasmic pathway). A model was recently proposed for cocaine binding to dopamine transporter in a conformation similar to that of the occluded LeuT crystal structure (17). Ibogaine is expected to stabilize a conformation more like the cytoplasm-facing model of LeuT (with the bundle tilted so as to close the extracellular pathway and open the cytoplasmic one).
Information about proximity between residues within a polypeptide can be obtained by site-directed cross-linking experiments (18–21). In the homologous γ-aminobutyric acid transporter GAT-1, an interaction between TM1 and TM3 was proposed based on paired cysteine mutagenesis and cross-linking (22). In a mutant containing cysteines at positions 68 and 143, transport was sensitive to inhibition by Cd2+ and copper(II)(1,10-phenanthroline)3 (CuPh3), which can chelate or cross-link, respectively, two cysteine thiol groups. Cross-linking by CuPh3 was not shown chemically but only by inhibition of transport activity (22).
Detection of cross-linking events in membrane proteins has been challenging. In other systems, where disulfide cross-links were formed between polypeptide chains, detection by a change in mobility on SDS-PAGE was sufficient (21). Alternatively, protease-sensitive sites have been inserted into proteins to allow cleavage into peptides that would separate on SDS-PAGE unless cross-linked (23). However, to be useful, these sites must be inserted in locations where they are accessible to the specific protease and also do not interfere with protein function, requirements that may be difficult to satisfy. In this study, we employed a novel strategy of methionine mutagenesis to generate specific cleavage sites for cyanogen bromide.
According to our analysis of LeuT (13), cysteine cross-linking between Cys-68 and Cys-143 in GAT-1 represented disulfide formation between the 4-helix bundle and the scaffold comprising the remainder of the protein. We reasoned that the distance between these positions might change with the bundle orientation, and might be affected by binding of specific ligands that affect transporter conformation. Accordingly, we tested the corresponding positions in SERT, where cocaine and ibogaine are known to have different effects on protein conformation.
EXPERIMENTAL PROCEDURES
Materials
HeLa cells (CCL-2) were obtained from American Type Culture Collection (Manassas, VA). Recombinant VTF7-3 vaccinia virus encoding T7 RNA polymerase was prepared as described previously (24). [3H]5-HT (27.1 Ci/mmol) was purchased from PerkinElmer Life Sciences. CNBr was from Sigma; formic acid was from J. T. Baker (Phillipsburg, NJ); ECL reagent was from Thermo Scientific (Rockford, IL); anti-myc antibody was from Upstate (Temecula, CA); 4–15% precast SDS-PAGE gels were from Bio-Rad; and all restriction endonucleases were from New England Biolabs Inc. (Ipswich, MA).
Mutagenesis
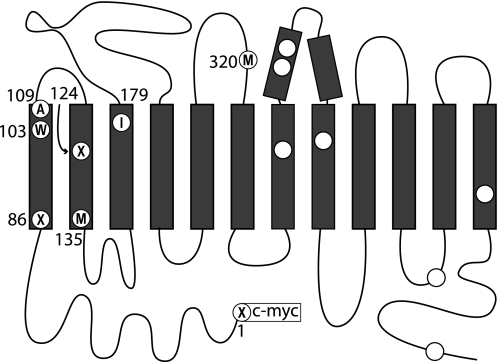
All rSERT mutants were generated by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). After introduction of the desired mutations into pBluescript II SK(−) containing rSERT, the mutated region was excised by double digestion with suitable restriction endonucleases and subcloned individually back into the original pBluescript-rSERT through the same restriction sites. All mutants contained the C109A mutation and a c-myc epitope tag at the N terminus of the protein (residue numbers are for un-tagged rSERT). The C109A mutation was used because this single cysteine is the only residue that reacted with MTS (methanethiosulfonate) reagents applied to the cell exterior (25). X3M denotes M1V/M86V/M124V/L320M/C109A, with the first three methionines in the rSERT sequence changed to valine to remove all sites for CNBr cleavage in the N-terminal half of the protein except for Met-135 (see Fig. 1). This background facilitated analysis of CNBr cleavage products. W103C, I179C, and X3M2C designate X3M/W103C, X3M/I179C, and X3M/W103C/I179C, respectively. Leu-320 was replaced with a methionine to introduce a CNBr cleavage site in the third extracellular loop (between TM5 and TM6). The transport activity and sensitivity of these mutants to inhibitors is shown in Table 1. All mutations were screened by restriction mapping and confirmed by DNA sequencing.
FIGURE 1.
Topological diagram of SERT showing locations of mutated residues. The three methionine residues mutated to valine in X3M are labeled as “X,” the alanine replacing cysteine at 109 as “A,” and Leu-320 converted to methionine as “M.” Met-135 near the cytoplasmic end of TM2 is also labeled M. All the remaining methionine residues are shown as open circles. Trp-103 and Ile-179 are labeled “W” and “I,” respectively. The top of the diagram represents the extracellular medium and the bottom represents the cytoplasm.
TABLE 1.
Relative uptake of 5-HT and IC50 of pseudo-wild type (C109A) and mutants for cocaine, ibogaine, and CuPh3
5-HT influx was measured as described under “Experimental Procedures” in the absence of inhibitors and after incubation with a range of concentrations of each agent for 10 min at 20 °C. Half-maximal inhibition was determined from fitting the resulting rate versus concentration curves. The rate for C109A was 0.53 ± 0.1 pmol min−1 mg−1.
| Mutants | Relative uptake of 5-HT | IC50 |
||
|---|---|---|---|---|
| Cocaine | Ibogaine | CuPh3 | ||
| % of C109A | μm | |||
| C109A | 100 | 1.7 ± 0.4 | 6.3 ± 1.3 | No effect |
| X3M | 93 | 0.68 ± 0.19 | 13.1 ± 0.6 | No effect |
| X3M2C | 72 | 1.37 ± 0.23 | 21.0 ± 5.0 | 0.24 ± 0.06 |
| W103C | 69 | 0.03 ± 0.01 | 14.7 ± 1.9 | 2.86 ± 0.01 |
| I179C | 67 | 0.48 ± 0.09 | 11.9 ± 1.8 | No effect |
Expression of rSERT
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a humidified 5% CO2 incubator. Cells plated in 96-well culture plates (∼25,000 cells per well) were infected with recombinant VTF7-3 virus and transfected with plasmid bearing rSERT C109A or other mutant cDNA under control of the T7 promoter (24). Transfected cells were incubated for ∼24 h at 37 °C with 5% CO2 before assaying transport.
5-HT Transport Assay
[3H]5-HT transport activity was assayed in monolayer cultures at 20 °C. Transfected HeLa cells in 96-well plates were washed once with 100 μl of PBS (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, and 1.4 mm KH2PO4, pH7.3) containing 0.1 mm CaCl2 and 1 mm MgCl2 (PBSCM) and incubated in PBSCM for the indicated time at 20 °C with or without the indicated reagents. 5-HT influx was initiated by addition of [3H]5-HT (20 nm final concentration), and the incubation was continued for 10 min. The assays were terminated by three rapid washes with ice-cold PBS. The cells were then solubilized in 30 μl of 0.1 m NaOH for 30 min. The extent of [3H]5-HT accumulated was determined by liquid scintillation spectrometry in a PerkinElmer Microbeta plate counter. All influx measurements were corrected by subtracting blank values measured in the presence of 1 mm cocaine.
Transport Inhibition by Copper(II)(1,10-Phenanthroline)3 and Effect of Inhibitors
To measure inhibition by CuPh3, HeLa cells transfected with the indicated constructs were washed once with 20 °C PBSCM and then incubated with CuPh3 for the indicated time. Then the cells were washed three times with PBSCM at 20 °C before measuring transport. To measure the effect of inhibitors on CuPh3 modification, cells were preincubated with the indicated concentration of cocaine or ibogaine for 10 min before incubating with CuPh3 for the indicated time. Cells were then washed three times with PBSCM containing the same inhibitor concentration to remove CuPh3 and then washed three times with PBSCM alone to remove the inhibitor before measuring transport. The CuPh3 stock solution was freshly prepared for each experiment by mixing 0.4 ml of 1.25 m 1,10-phenanthroline in H2O:ethanol (1:1) with 0.6 ml of 250 mm CuSO4. CuPh3 working solutions were prepared by diluting the stock solution with PBSCM.
Preparation of Lyophilized Protein Sample
HeLa cells expressing rSERT mutants were treated, where indicated, with CuPh3 as described for transport inhibition experiments. Cells were washed once with ice-cold PBSCM, and then scraped into an appropriate volume of cell collecting buffer (20 mm Tris, 5 mm EDTA, pH 7.3) containing protease inhibitor mixture (Sigma) and transferred to a 1.5-ml tube. Cells were lysed by three cycles of freezing at −80 °C and thawing at 20 °C and then passed through a 26-gauge needle 10 times. Cell debris was removed by precipitation at 500 × g for 10 min at 4 °C and the membranes in the supernatant suspension were retained for analysis. Protein concentration was determined with the Micro BCA protein assay (Pierce). Protein samples containing ∼50 μg were placed in 1.5-ml tubes, evaporated to dryness in a SpeedVac evaporator, and stored at −20 °C until use.
Chemical Cleavage of rSERT by CNBr
For the cleavage reaction, 15 μl of water was added to each lyophilized rSERT protein sample followed by 85 μl of a saturated solution of CNBr in 88% formic acid (26). The sample was agitated in a Vortex mixer until the protein was completely solubilized in the CNBr solution. Samples were incubated 4 h in the dark at 20 °C. Addition of 500 μl of benzene to the sample tube allowed formic acid to evaporate as an azeotrope as the sample dried in a SpeedVac evaporator. For more complete removal of formic acid, an additional 500 μl of benzene was added and the sample dried again. Cleavage products were dissolved in 50 μl of Laemmli sample buffer (27) with or without dithiothreitol (DTT), and the samples were stored at −20 °C until use.
Electrophoresis and Immunoblotting
CNBr cleavage products (10–20 μg) were separated by 4–15% SDS-PAGE (Bio-Rad) and then transferred to a polyvinylidene difluoride membrane (Bio-Rad) (28) followed by Western Blot analysis (29). The N-terminal fragments were visualized by enhanced chemiluminescent reagent (ECL, Pierce) using the c-myc epitope tag.
Data Analysis
Nonlinear regression fits of experimental and calculated data were performed with Origin (OriginLab, Northampton, MA), which uses the Marquardt-Levenberg nonlinear least squares curve-fitting algorithm. The statistical analyses are from triplicate experiments. Data with error bars represent the mean ± S.E. for triplicate measurements. Statistical analysis was performed using Student's paired t tests. Rate constants (k) for 2-aminoethylmethane thiosulfonate (MTSEA) modification were calculated from the concentration of MTSEA (IC50) that led to half-maximal inactivation in 10 min (t½) using the equation: k = ln(2)/(IC50 × t½).
RESULTS
Effect of CuPh3 on SERT Mutants
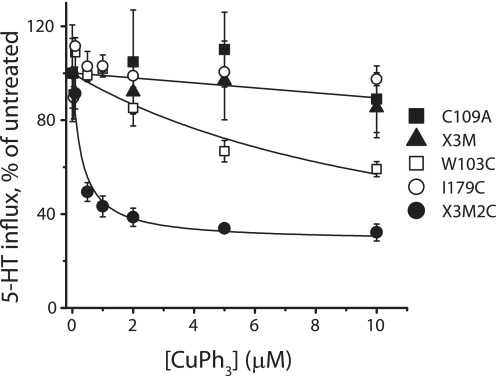
In previous work with GAT-1, Zomot et al. (22)showed that replacing Trp-68 in TM1 and Ile-143 in TM3 with cysteine caused the transporter to become sensitive to inactivation by CuPh3, which they interpreted as evidence for cross-linking the two cysteine sulfhydryl groups. We reasoned that if the structures of SERT and GAT-1 were sufficiently similar, introduction of cysteines at the corresponding positions of SERT would create a similar sensitivity to CuPh3. Accordingly, we mutated Trp-103 in TM1 and Ile-179 in TM3 in rSERT, to cysteine. Mutants with each of these mutations separately and with both together were created in the X3M/C109A background (see “Experimental Procedures” for details) to make single or double cysteine replacement mutants. Table 1 shows that these mutants all transported 5-HT at ∼70% the rate of SERT C109A (which itself was similar to wild-type SERT (25)). As shown in Fig. 2, CuPh3 strongly inhibited the 5-HT influx activity of HeLa cells transfected with the double cysteine replacement mutant X3M2C (filled circles). This inhibition was half-maximal at 0.24 ± 0.06 μm and saturated at ∼70% inhibition at the highest CuPh3 concentrations tested (5–10 μm). For rSERT X3M (triangles) or C109A (on which it is based, filled squares), or I179C (containing one of the two cysteines, open circles), CuPh3 had no effect. CuPh3 also weakly inhibited W103C (the mutant with the other inserted cysteine, open squares). The IC50 for CuPh3 inhibition of W103C was 2.9 ± 0.01 μm, over 10-fold higher than that of X3M2C. The activity was decreased 40% by incubation with 10 μm CuPh3. These data are similar to the corresponding results with GAT-1, indicating a similar structural relationship between the two positions in both transporters. The weak sensitivity of W103C also corresponds to the sensitivity of GAT-1 W68C to CuPh3.
FIGURE 2.
Inhibition by CuPh3 of 5-HT influx by rSERT mutants. HeLa cells expressing the indicated transporter constructs were washed once with 20 °C PBSCM and then incubated with the indicated concentration of CuPh3 for 10 min at 20 °C. 5-HT influx was measured after three additional washes with PBSCM. Values at each CuPh3 concentration are reported as the percentage of activity in the absence of CuPh3. Data are averages of at least three experiments. Untreated influx was 0.53 ± 0.1 pmol min−1 mg−1 for C109A (filled squares), 0.48 ± 0.03 pmol min−1 mg−1 for X3M (triangles), 0.36 ± 0.1 pmol min−1 mg−1 for I179C (open circles), 0.37 ± 0.07 pmol min−1 mg−1 for W103C (open squares), and 0.38 ± 0.1 pmol min−1 mg−1 for X3M2C (filled circles). Error bars represent S.E. for multiple experiments.
CNBr Cleavage of rSERT Mutants
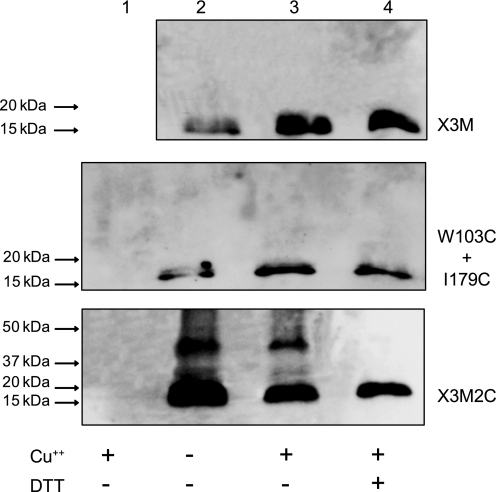
It has been assumed that inhibition by CuPh3 represents oxidation of the two cysteine residues to a cystine disulfide (18). To verify that this was indeed the case, we took advantage of the X3M background to measure disulfide linking of TM1 with other parts of SERT using CNBr cleavage. In X3M, and mutants generated in that background, the first methionine after the N-terminal c-myc tag is Met-135, near the cytoplasmic end of TM2, and the next endogenous methionine is in TM7 (see “Experimental Procedures” for mutant descriptions). CNBr is expected to generate a 14.9-kDa fragment containing the c-myc tag by cleaving these mutants at Met-135 (Fig. 1).
Fig. 3 shows that a myc-positive fragment of approximately this mass was generated from CNBr cleavage of X3M. The pattern was not significantly affected by treatment with CuPh3 or DTT. CNBr treatment of I179C and W103C, expressed together (Fig. 3) or separately (supplemental Fig. S1), also yielded a single N-terminal product at 15–20 kDa that was not affected by CuPh3 or DTT. A faint band at ∼30 kDa was occasionally observed with W103C, especially if the cells expressing this mutant were treated with CuPh3 before digestion (supplemental Fig. S1). DTT added after CNBr cleavage eliminated this band, suggesting that it resulted from disulfide formation (supplemental Fig. S1). In cells expressing the X3M2C mutant (with cysteines at both 103 and 179), a prominent band at ∼40 kDa was found in CNBr digests. Treatment with CuPh3 did not increase the intensity of this band, but it was absent in DTT-treated samples, indicating that it resulted from disulfide formation. If the N-terminal fragment was bonded through a 103–179 disulfide to the 185-amino acid peptide expected from cleavage at Met-135 and Met-320 (see Fig. 1), we would expect a product of 36.1 kDa (see supplemental Table S1). The absence of this 40-kDa band when the two single cysteine replacement mutants W103C and I179C were co-expressed, indicates that the disulfide bonded peptide did not result from an intermolecular cross-linking event.
FIGURE 3.
CNBr cleavage analysis of cross-linking. HeLa cells expressing the indicated mutants were treated with the following concentrations of CuPh3: 10 μm for X3M and X3M2C; 30 μm for W103C+I179C. The cells were treated as described under “Experimental Procedures” to prepare lyophilized membranes, which were cleaved with CNBr. Cleavage products were dissolved in Laemmli sample buffer with or without DTT as indicated, separated on 4–15% SDS-PAGE, and detected by immunobloting. Lane 1 was a control with untransfected HeLa cells; samples in lane 2 were not treated; samples in lane 3 were treated with CuPh3; samples in lane 4 were treated with CuPh3 and then reduced with DTT prior to SDS-PAGE. Incubation with 10 mm N-ethylmaleimide from 5 min prior to cell lysis through lyophilization did not affect the band patterns.
Conformation Dependence of Cross-linking Reaction
Previous studies suggest that SERT and related transporters exist in conformations that expose the binding site either to the extracellular or the cytoplasmic side of the membrane (12–15). The inhibitors cocaine and ibogaine apparently stabilize extracellular- and cytoplasm-facing SERT conformations, respectively, as demonstrated by their ability to increase or decrease accessibility of positions in the extracellular or cytoplasmic permeation pathways (12–14, 16). In our proposed mechanism for transport (13), the distance between TMs 1 and 3 should increase and decrease during this process. Therefore, we examined the effect of cocaine and ibogaine on cross-linking between the cysteine residues inserted at positions 103 and 179. Cocaine, by stabilizing SERT in a conformation similar to that of the LeuT crystal structure, was expected to hold the two cysteine residues apart, thereby decreasing cross-linking. Ibogine was expected to stabilize SERT conformations similar to a cytoplasm-facing conformation, and therefore was not expected to inhibit cross-linking, possibly even facilitating the process.
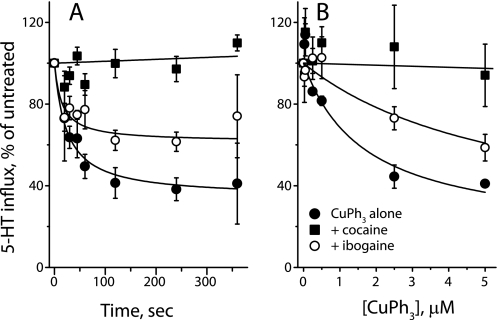
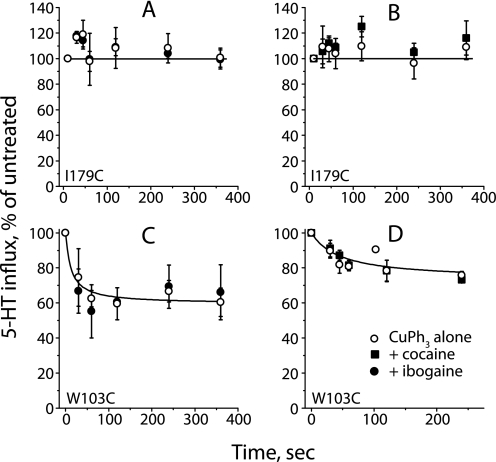
Fig. 4 shows that CuPh3 inactivation of X3M2C was dramatically inhibited by the presence of cocaine, and much less so by ibogaine. At 1 μm, CuPh3 inactivated 60% of transport activity within 5 min (Fig. 4A). The presence of 100 μm ibogaine did not noticeably change the time course of inactivation, but decreased the extent to less than 40%. Cocaine (7 μm) completely blocked inactivation by this concentration of CuPh3. In a 2-min incubation, the extent of inactivation by CuPh3 increased up to 5 μm CuPh3 (Fig. 4B). Again, inactivation was completely blocked by 5 μm cocaine and to a much lesser extent by 100 μm ibogaine, which decreased the extent of inactivation by roughly half. The concentrations of cocaine and ibogaine used in this experiment were equivalent to 5 times IC50 in terms of their inhibitory potency for each mutant (see Table 1). In contrast to the effect of these agents on X3M2C, CuPh3 did not inactivate I179C (see also Fig. 2) and there was no additional effect of either ibogaine (Fig. 5A) or cocaine (Fig. 5B). Although W103C was mildly sensitive to CuPh3 (Fig. 2), neither ibogaine (Fig. 5C) nor cocaine (Fig. 5D) influenced this sensitivity.
FIGURE 4.
Effect of SERT inhibitors on CuPh3 cross-linking. A, time course of CuPh3-dependent inactivation of X3M2C and the effect of cocaine and ibogaine. HeLa cells expressing X3M2C were incubated with or without inhibitor (7 μm for cocaine and 100 μm for ibogaine) for 10 min at 20 °C, 1 μm CuPh was added for the indicated time, and cells were washed six times to remove inhibitor and CuPh and assayed for 5-HT influx. B, effect of cocaine and ibogaine on the dependence of X3M2C inactivation on CuPh3 concentration. Inactivation of X3M2C was measured as in A over a range of CuPh3 concentrations using an incubation time of 2 min. Filled circles, CuPh3 alone (K0.5 ≈ 1.5 μm); filled squares, CuPh3 plus 5 μm cocaine (K0.5 > 80 μm); open circles, CuPh3 plus 100 μm ibogaine (K0.5 ≈ 7.8 μm). Error bars represent S.E. for multiple experiments.
FIGURE 5.
Effect of CuPh3 on W103C and I179C in the presence of SERT inhibitors. HeLa cells expressing the single cysteine replacement mutants I179C and W103C were preincubated, as in Fig. 4, with ibogaine or cocaine before adding 5 μm CuPh3. SERT activity was measured at the indicated times following CuPh3 addition for I179C in the presence of 70 μm ibogaine (A) and 2.5 μm cocaine (B) and for W103C with 75 μm ibogaine (C) and 0.15 μm cocaine (D). Error bars represent S.E. for multiple experiments.
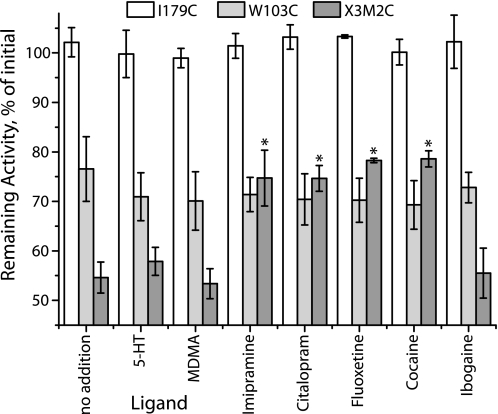
Several additional inhibitors and substrates were tested for their ability to modify the reactivity of I179C, W103C, and X3M2C. As shown in Fig. 6, I179C was not sensitive to inactivation by CuPh3 in the presence or absence of any of the tested ligands, including 5-HT, MDMA, imipramine, citalopram, fluoxetine, cocaine, or ibogaine. W103C was moderately sensitive to CuPh3, but this sensitivity did not change in the presence of any of the added ligands. X3M2C was even more sensitive to CuPh3, as shown also in Fig. 2. Although the transport substrates 5-HT and MDMA had no significant effect on this inactivation, imipramine, citalopram, and fluoxetine, like cocaine, all partially protected this mutant from inactivation. Of the inhibitors, only ibogaine did not protect against CuPh3.
FIGURE 6.
Effects of substrates and other inhibitors on CuPh3-mediated inactivation of X3M2C. HeLa cells expressing I179C (open bars), W103C (light gray), and X3M2C (dark gray) were preincubated for 10 min with 5-HT (1 μm), MDMA (1 μm), imipramine (0.5 μm), citalopram (0.1 μm), fluoxetine (0.1 μm), cocaine (5 μm), or ibogaine (100 μm) in PBSCM, followed by addition of CuPh3 to a final concentration of 5 μm and incubation for another 5 min. The cells were then washed 6 times with PBSCM and 5-HT transport was measured. Values represent the rate of transport as a percentage of the control rate with no CuPh3. Asterisks indicate values significantly different from no addition (p < 0.05). Error bars represent S.E. for multiple experiments.
Transporter ligands might influence cross-linking by altering the conformation of SERT or by sterically blocking one or both of the cross-linked residues. To assess the extent to which cocaine and ibogaine decrease access to Trp-103 or Ile-179, we measured inactivation by the cysteine reagent MTSEA. As previously described (16), W103C was accessible and sensitive to inactivation by MTSEA. This inactivation was unchanged in the presence of either cocaine or ibogaine (supplemental Fig. S2). MTSEA also inactivated I179C (Fig. 7A), as previously shown for MTSET (30). Although ibogaine had no significant effect on this inactivation, cocaine significantly potentiated the effect of MTSEA, increasing the rate constant for inactivation by 3-fold (Fig. 7B). Thus, rather than blocking access to Cys-179, cocaine actually increases the reactivity of this residue.
FIGURE 7.
Sensitivity of I179C to MTSEA inactivation in the presence of cocaine and ibogaine. A, concentration dependence. HeLa cells expressing I179C were treated for 10 min with the indicated concentrations of MTSEA in the presence of 2.5 μm cocaine (triangles) or 35 μm ibogaine (open circles) or neither (filled circles). 5-HT influx was measured at the indicated times after washing the cells free of MTSEA. Influx inactivation rate constants (B) were calculated from the MTSEA concentration giving half-maximal inactivation of 5-HT. Half-maximal MTSEA concentrations and corresponding rate constants were: MTSEA alone, 0.41 ± 0.06 μm and 2.9 ± 0.4 m−1 s−1; cocaine, 0.14 ± 0.05 μm and 10.0 ± 2.7 m−1 s−1; ibogaine, 0.35 ± 0.08 μm and 3.6 ± 0.6 m−1 s−1. Error bars represent S.E. for multiple experiments.
DISCUSSION
Protein conformational changes are thought to underlie the alternate exposure of binding sites to opposite sides of the membrane during transport (31, 32). We recently proposed a conformational mechanism for alternating access in the neurotransmitter transporter family (13). This mechanism proposed that tilting or rocking of a 4-helix bundle composed of TM helices 1, 2, 6, and 7 within a scaffold formed by the rest of the protein accomplished these accessibility changes. In the work described here, we provide additional experimental evidence in favor of that mechanism.
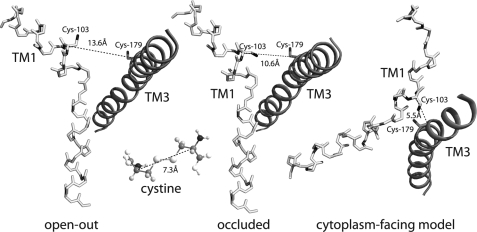
In the original LeuT crystal structure, proposed as a model for neurotransmitter transporter structure (3), the α-carbons of Leu-29 in TM1 and Ile-111 in TM3 are situated 10.6 Å from each other (Fig. 8) and ∼10 Å closer to the extracellular medium than the bound leucine molecule. In this structure, the bound leucine at the substrate site was occluded from both sides of the membrane. A more recent LeuT structure with tryptophan bound was presented as a possible model for the conformation of the transporter with the substrate site exposed to the extracellular medium (7). In this “open-out” structure, the C-α positions of the same two residues were 13.6 Å apart (Fig. 8). In the γ-aminobutyric acid transporter GAT-1, Kanner and co-workers (22) found that when both of the corresponding residues were replaced with cysteine, transport activity became sensitive to the sulfhydryl oxidizing agent CuPh3, and suggested that this inhibition resulted from disulfide formation between the two transmembrane helices. The C-α positions in the extended conformation of cystine are only 7.3 Å apart, significantly closer than the corresponding residues in either LeuT structure. Thus, CuPh3 inactivation of GAT-1 either occurred by another mechanism, or a conformational change was required to bring the two cysteine residues together.
FIGURE 8.
Transmembrane helices 1 and 3 in the open-out and “occluded” LeuT structures and in a model of the cytoplasm-facing conformation. The positions of Leu-29 and Ile-11 (labeled for the corresponding SERT residues Cys-103 and Cys-179, respectively) are shown as cysteine. Coordinates for these positions were taken from Protein Data Bank entries 3F3A (open-out) and 2A65 (occluded), and Forrest et al. (13). TM1 (light) is shown in stick representation and TM3 (dark) as a schematic helix. Distances between α-carbon atoms of Leu-29 and Ile-111 are shown along with, for comparison, the corresponding distance in the extended form of cystine, which is the product of disulfide formation between two cysteine molecules.
In the work presented here, we show that cysteine residues at the two corresponding positions in SERT led to chemical cross-linking between TM1 and TM3 (Fig. 3). This observation was facilitated by judicious mutation of methionine residues in SERT to create a background in which a single methionine in TM2 provided a CNBr cleavage site between the two reactive cysteine residues. We propose this method as a new application of CNBr cleavage to identify cross-linking reactions in proteins, especially in cases where the cross-linking event does not lead to a measurable change in activity.
The presence of cysteine at positions 103 and 179 was required for efficient CuPh3 inactivation (Figs. 2 and 4) and cross-linking (Fig. 3). However, the effect of CuPh3 on cross-linking was not visible in Fig. 3, and the extent of cross-linking was not measurably sensitive to ligand binding, although inactivation was dependent on CuPh3 (Figs. 2 and 4) and blocked by inhibitors (Figs. 4 and 7). The inability to detect a CuPh3 effect by CNBr cleavage may reflect the high extent of spontaneous cross-linking. Proteins destined for surface expression are subject to spontaneous disulfide oxidation in the endoplasmic reticulum (33). We surmise that only a minor fraction of SERT expressed in these cells catalyzed 5-HT influx, and was sensitive to oxidation by CuPh3, whereas the majority of cell surface SERT was cross-linked during biosynthesis.
From our results with CNBr cleavage, we conclude that, at least in this system, the technique is useful to identify a pair of cysteine residues that are close enough to form a disulfide. However, we found that for determining the time course of cross-linking or measuring the effect of ligands on the process, it was more useful to follow changes in activity that accompanied the reaction.
The W103C mutant was weakly inactivated by CuPh3 (Figs. 2 and 5) and showed some evidence of cross-linking as indicated by the occasional formation of a minor DTT-sensitive band (supplemental Fig. S1). The corresponding mutant in GAT-1 also showed some sensitivity to CuPh3 (22). It is not clear what this cross-linking and CuPh3 sensitivity represent, although it is likely to be a different process from that observed with X3M2C. The mobility of the occasionally observed W103C cross-linked product was higher than with X3M2C, and cocaine, which blocked CuPh3 inactivation of X3M2C (Fig. 4), had no effect on W103C (Fig. 5). Whatever process was responsible for the reactivity of W103C, it is likely to represent a common feature of SERT and GAT-1. It is not clear why only W103C and not I179C was sensitive to inactivation and cross-linking by CuPh3. However, as part of the mobile 4-helix bundle, position 103 might have more access to other cysteine residues in SERT or adjacent proteins.
In our proposed mechanism for alternating access in this family, the outer half of the bundle, including TM1, tilts toward the scaffold, including TM3, as the extracellular permeation pathway closes (13). At the same time, the cytoplasmic half of the bundle tilts away from the scaffold, exposing the cytoplasmic permeation pathway. In our model, the distance between the α-carbons of Leu-29 and Ile-111 in the cytoplasm-facing conformation is only 5.5 Å, well within the distance required to form a disulfide between cysteines at the corresponding positions in GAT-1 and SERT (Fig. 8). Disulfide formation, therefore, would be facilitated by the conformational change that we proposed to interconvert these transporters from their open-out to cytoplasm-facing conformations. Moreover, covalent linkage of the corresponding positions in TMs 1 and 3 of GAT-1 and SERT would be expected to block transport by preventing the conformational change leading to the occluded or open-out conformations.
To provide further support for this interpretation, we utilized cocaine and ibogaine, two inhibitors that apparently stabilize different conformations of SERT. Using the reactivity of cysteine residues placed in the cytoplasmic permeation pathway, we previously showed that cocaine closed that pathway and ibogaine opened it (12, 14). At the same time, cocaine increased exposure, and ibogaine decreased exposure, of cysteine residues placed in the extracellular permeation pathway, consistent with a concerted conformational change resulting from tilting of the 4-helix bundle (14–15, 34). Fig. 4 shows that cocaine blocked the ability of CuPh3 to inactivate the SERT double cysteine mutant X3M2C, as expected if cocaine binding restricted SERT to conformations similar to the occluded or open-out structures of LeuT (Fig. 8). The effect of cocaine was not simply due to steric occlusion of the cysteine residues at positions 103 and 179, because the single cysteine replacement mutants were either equally (W103C, supplemental Fig. S2) or more sensitive (I179C, Fig. 7) to inactivation by MTSEA in the presence of cocaine. The increased accessibility of I179C is likely due to cocaine stabilizing the extracellular-facing conformation of SERT. We previously showed that cocaine also increased accessibility of the corresponding residue in norepinephrine transporter (15).
Other inhibitors, imipramine, fluoxetine, and citalopram (all antidepressant drugs that inhibit SERT by binding from the cell exterior), also protected SERT X3M2C from inactivation by CuPh3 (Fig. 6). In contrast, the transported substrates 5-HT and MDMA had little effect on inactivation. Ibogaine was the only inhibitor that did not significantly protect against CuPh3.
We interpret these results in the context of a conformational change associated with transport that brings positions 103 and 179 into close proximity. In the absence of ligands, the conformational flexibility of the transporter is high enough that the rate-limiting factor in cross-link formation is the rate of oxidation, itself dependent on CuPh3 concentration (Figs. 2 and 4B), rather than the proportion of SERT in a cross-linkable conformation. The presence of substrates (5-HT or MDMA) initiates catalytic cycling of the transporter, but does not inhibit cross-linking because intermediate conformations during transport also favor cross-linking. However, inhibitors such as cocaine and antidepressants bind to and stabilize SERT conformations in which positions 103 and 179 are separated enough not to cross-link (Figs. 4 and 6) and are accessible to MTSEA (Figs. 7 and supplemental S2).
Ibogaine, despite being a SERT inhibitor, did not inhibit CuPh3-dependent inactivation to the extent observed with other inhibitors. Compared with cocaine, ibogaine was less than 10% as effective at decreasing the sensitivity to CuPh3 (Fig. 4B) although both inhibitors were present at concentrations 5-fold higher than their IC50. Ibogaine apparently stabilizes a conformation of SERT different from that favored by other inhibitors. Thus, we conclude that when ibogaine is bound, SERT is in a conformation closer to that of the cytoplasm-facing intermediate, in which positions 103 and 179 are in close proximity. The observation that ibogaine slightly inhibited the rate of CuPh3-mediated inactivation relative to no addition suggests that the two cysteine residues at 103 and 179 may not be as close in the ibogaine-bound state as in the cytoplasm-facing conformation, or that the orientation of their sulfhydryl groups are constrained by ibogaine in a way that impedes their oxidation to cystine. Our model of the cytoplasm-facing conformation of LeuT (13) is not sufficiently accurate to predict the orientation of these residues in SERT, and we also do not know how closely the cytoplasm-facing conformation of SERT resembles the conformation with ibogaine bound.
The results presented here are consistent with a mechanism in which the extracellular half of the 4-helix bundle containing TM1 moves toward TM3 in the scaffold when the transporter is in the cytoplasm-facing conformation. Cocaine binding is proposed to block this movement and in our current results also prevented inactivation due to cross-linking. We anticipate that future structural and biochemical studies will provide further support for the rocking bundle mechanism.
Supplementary Material
Acknowledgments
We thank Drs. Sotiria Tavoulari and Lucy R. Forrest for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant DA007259 through the National Institute on Drug Abuse.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- 5-HT
- 5-hydroxytryptamine
- SERT
- serotonin transporter
- CuPh3
- copper(II)(1,10-phenanthroline)3
- TM
- transmembrane helix
- MTSEA
- 2-aminoethylmethane thiosulfonate
- GAT-1
- γ-aminobutyric acid transporter
- X3M
- SERT M1V/M86V/C109A/M124V/L320M
- X3M2C
- X3M/W103C/I179C
- PBS
- phosphate-buffered saline
- DTT
- dithiothreitol
- MDMA
- 3,4-methylenedioxymethamphetamine.
REFERENCES
- 1.Kanner B. I., Zomot E. (2008) Chem. Rev. 108, 1654–1668 [DOI] [PubMed] [Google Scholar]
- 2.Androutsellis-Theotokis A., Goldberg N. R., Ueda K., Beppu T., Beckman M. L., Das S., Javitch J. A., Rudnick G. (2003) J. Biol. Chem. 278, 12703–12709 [DOI] [PubMed] [Google Scholar]
- 3.Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 4.Quick M., Yano H., Goldberg N. R., Duan L., Beuming T., Shi L., Weinstein H., Javitch J. A. (2006) J. Biol. Chem. 281, 26444–26454 [DOI] [PubMed] [Google Scholar]
- 5.Singh S. K., Yamashita A., Gouaux E. (2007) Nature 448, 952–956 [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z., Zhen J., Karpowich N. K., Goetz R. M., Law C. J., Reith M. E., Wang D. N. (2007) Science 317, 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S. K., Piscitelli C. L., Yamashita A., Gouaux E. (2008) Science 322, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., Abramson J. (2008) Science 321, 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. (2008) Science 322, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ressl S., Terwisscha van Scheltinga A. C., Vonrhein C., Ott V., Ziegler C. (2009) Nature 458, 47–52 [DOI] [PubMed] [Google Scholar]
- 11.Fang Y., Jayaram H., Shane T., Kolmakova-Partensky L., Wu F., Williams C., Xiong Y., Miller C. (2009) Nature 460, 1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y. W., Rudnick G. (2006) J. Biol. Chem. 281, 36213–36220 [DOI] [PubMed] [Google Scholar]
- 13.Forrest L. R., Zhang Y. W., Jacobs M. T., Gesmonde J., Xie L., Honig B. H., Rudnick G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10338–10343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs M. T., Zhang Y. W., Campbell S. D., Rudnick G. (2007) J. Biol. Chem. 282, 29441–29447 [DOI] [PubMed] [Google Scholar]
- 15.Chen J. G., Rudnick G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1044–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry L. K., Adkins E. M., Han Q., Blakely R. D. (2003) J. Biol. Chem. 278, 37052–37063 [DOI] [PubMed] [Google Scholar]
- 17.Beuming T., Kniazeff J., Bergmann M. L., Shi L., Gracia L., Raniszewska K., Newman A. H., Javitch J. A., Weinstein H., Gether U., Loland C. J. (2008) Nat. Neurosci. 11, 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falke J. J., Koshland D. E., Jr. (1987) Science 237, 1596–1600 [DOI] [PubMed] [Google Scholar]
- 19.Pakula A. A., Simon M. I. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4144–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. (1996) Science 274, 768–770 [DOI] [PubMed] [Google Scholar]
- 21.Wu J., Kaback H. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14498–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zomot E., Zhou Y., Kanner B. I. (2005) J. Biol. Chem. 280, 25512–25516 [DOI] [PubMed] [Google Scholar]
- 23.Sun J., Kaback H. R. (1997) Biochemistry 36, 11959–11965 [DOI] [PubMed] [Google Scholar]
- 24.Blakely R. D., Clark J. A., Rudnick G., Amara S. G. (1991) Anal. Biochem. 194, 302–308 [DOI] [PubMed] [Google Scholar]
- 25.Chen J. G., Liu-Chen S., Rudnick G. (1997) Biochemistry 36, 1479–1486 [DOI] [PubMed] [Google Scholar]
- 26.Kaiser R., Metzka L. (1999) Anal. Biochem. 266, 1–8 [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28.Ursitti J. A., Mozdzanowski J., Speicher D. W. (2001) in Current Protocols in Protein Science (Coligan J. E., Dunn B. M., Speicher D. W., Wingfield P. T. eds) pp. 1–14, John Wiley & Sons, Inc., New York [Google Scholar]
- 29.Gallagher S. (2003) in Current Protocols in Protein Science (Coligan J. E., Dunn B. M., Speicher D. W., Wingfield P. T. eds) pp. 1–12, John Wiley & Sons, Inc., New York [Google Scholar]
- 30.Chen J. G., Sachpatzidis A., Rudnick G. (1997) J. Biol. Chem. 272, 28321–28327 [DOI] [PubMed] [Google Scholar]
- 31.Jardetzky O. (1966) Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 32.Mitchell P. (1990) Res. Microbiol. 141, 286–289 [DOI] [PubMed] [Google Scholar]
- 33.Frand A. R., Cuozzo J. W., Kaiser C. A. (2000) Trends Cell Biol. 10, 203–210 [DOI] [PubMed] [Google Scholar]
- 34.Barker E. L., Moore K. R., Rakhshan F., Blakely R. D. (1999) J. Neurosci. 19, 4705–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.