Abstract
Secretins are an unusual and important class of bacterial outer membrane (OM) proteins. They are involved in the transport of single proteins or macromolecular structures such as pili, needle complexes, and bacteriophages across the OM. Secretins are multimeric ring-shaped structures that form large pores in the OM. The targeting of such macromolecular structures to the OM often requires special assistance, conferred by specific pilotins or pilot proteins. Here, we investigated HxcQ, the OM component of the second Pseudomonas aeruginosa type II secretion system. We found that HxcQ forms high molecular mass structures resistant to heat and SDS, revealing its secretin nature. Interestingly, we showed that HxcQ is a lipoprotein. Construction of a recombinant nonlipidated HxcQ (HxcQnl) revealed that lipidation is essential for HxcQ function. Further phenotypic analysis indicated that HxcQnl accumulates as multimers in the inner membrane of P. aeruginosa, a typical phenotype observed for secretins in the absence of their cognate pilotin. Our observations led us to the conclusion that the lipid anchor of HxcQ plays a pilotin role. The self-piloting of HxcQ to the OM was further confirmed by its correct multimeric OM localization when expressed in the heterologous host Escherichia coli. Altogether, our results reveal an original and unprecedented pathway for secretin transport to the OM.
Introduction
The presence of an outer membrane (OM)4 in Gram-negative bacteria constitutes a second barrier for the secretion of exoproteins into the extracellular medium. At least six different secretion pathways have evolved in these bacteria for the secretion of a very diverse pool of extracellular proteins (1–2). Among them, the type II secretion pathway is a two-step process in which exoproteins with an N-terminal signal peptide (SP) are first exported through the cytoplasmic membrane by either the Sec or Tat translocons. Following removal of the SP, they are released into the periplasm (3–4). The periplasmic intermediates are specifically recognized by the type II secretion system (T2SS), also called secreton, for their transport across the OM. This pathway, therefore, promotes the specific transport of exoproteins requiring intracellular folding, like periplasmic disulfide bridge formation, and, in some cases, assembly into multimeric complexes prior to their secretion. Such a requirement implies that the secretion process uses a large and tightly controlled secretion channel in the OM. The T2SS is a highly complex nanomachine embedded in the bacterial envelope consisting of 12–16 different proteins, depending on the organism (1, 5). Interestingly, there is only one integral OM protein in this system, which therefore constitutes the only candidate for the OM translocation channel. This OM component belongs to a family of proteins generically designated as secretins (6). This family also includes members that are involved in type III protein secretion (T3SS), type IV pilus assembly, type IV bundle-forming pili, toxin co-regulated pili, and assembly and export of filamentous phage (7–12). Therefore, secretins constitute an important group of transporters specialized in the translocation of bulky macromolecules or macromolecular complexes across the OM.
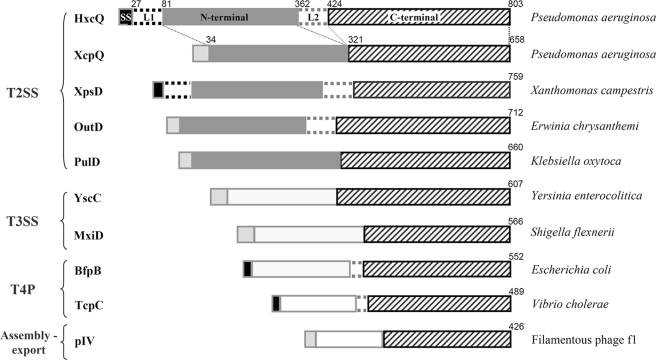
Several secretins have been purified and analyzed by electron microscopy, revealing that 12–14 identical secretin monomers form ring-like complexes with a central channel large enough to accommodate their substrates (7, 13–14). The homology between the members of the secretin family is contained within the C-terminal half of the protein (see Fig. 2) (15). Therefore, this domain has been proposed to form the secretion channel, whereas the much less conserved N-terminal domain that largely protrudes into the periplasm probably undertakes more specific functions, such as substrate recognition and/or interaction with the other components of the corresponding machineries (13, 16).
FIGURE 2.
Primary sequence comparison of HxcQ and other secretins listed in this study. Secretin primary sequence are presented as followed from their N terminus to their C terminus: the signal sequence (SS), an L1 domain (when present), followed by the variable N-terminal domain, the L2 domain (when present) and finally, the C-terminal domain. Linker amino acids are indicated on the top of each schematic representation. Type I signal sequences are indicated in light gray whereas type II lipoprotein signal sequences are indicated in black. The L1 domain, present only in HxcQ and XpsD primary sequences is represented with black dotted lines. The typical N-terminal domain found in T2SS secretins (43) is indicated in gray, whereas N-terminal domains from other types of secretins are indicated in white. The L2 domain, bridging the N and C terminus domains of OutD and the four lipidated secretins (HxcQ, XpsD, BfpB, and TcpC) is indicated with gray dotted lines. The highly conserved C-terminal domain among all secretins is striped. Homologous domains between HxcQ and XcpQ are connected with small dotted lines. The transport systems to which each secretin belongs are indicated on the left.
Among the large diversity of identified secretins, most of them depend on a small pilot protein for their correct final insertion into the outer membrane. In most cases, pilot proteins are outer membrane-linked lipoproteins called pilotins. To date, characterized secretin/pilotin couples are: PulD/PulS of Klebsiella (17–18), OutD/OutS of Erwinia (19) for T2SS; YscC/YscW of Yersinia (8), InvG/InvH of Salmonella (12), MxiD/MxiM of Shigella (20) for T3SS, and PilQ/Tgl of Myxococcus (21) for Type IV pilus systems. For T2SS secretin/pilotin couples, the specific pilotin binding domain is localized at the extreme C terminus of the secretin (19, 22). The majority of the genes encoding pilotins are found in the same cluster as the genes encoding the corresponding secretion systems. However, in several secretin-containing systems, a pilotin gene has yet to be identified, suggesting the existence of possible alternatives to the pilotin biogenesis pathway. Recently, a soluble nonlipidated periplasmic protein has been shown to be important for the OM localization of XcpQ secretin in P. aeruginosa (23). Interestingly, three secretins are themselves lipoproteins, but no function has so far been attributed to their atypical N-terminal lipid anchor. One, XpsD of Xanthomonas campestris pv. campestris, belongs to a T2SS (24), and two others, BfpB of enteropathogenic E. coli (25) and TcpC of Vibrio cholerae (11) are members of type IV pili systems.
In Gram-negative bacteria, most lipoproteins are periplasmic proteins anchored to the inner or outer membrane through a lipid moiety attached to their invariable N-terminal cysteine residue (3). Lipidation and maturation of lipoproteins take place after their translocation through the inner membrane via Sec machinery (3). Lipoprotein-specific signal peptides (SPs) are characterized by a specific consensus motif (V/L)XXC called the Lipobox (26). The Lipobox is both the lipidation site and the maturation site recognized by the lipoprotein signal peptidase II, which cleaves the SP upstream of the cysteine (27–28).
P. aeruginosa strain PAO1 possesses two complete and nonredundant T2SS, referred to as the Xcp and Hxc systems (1). While more than a dozen exoproteins utilize the Xcp T2SS for their secretion, the Hxc T2SS, which is induced under phosphate starvation, is dedicated to the secretion of one single low molecular mass protein, the alkaline phosphatase LapA (29).
In the present work, we reveal that the atypical HxcQ secretin of the Hxc T2SS of P. aeruginosa is a lipoprotein. Moreover, we demonstrate that the HxcQ liposecretin is self-piloted to the OM via its N-terminal lipid anchor, therefore revealing a new pathway for secretin biogenesis.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains, vectors, and plasmids used in this study are listed in Table 1. Recombinant DNA methods were performed essentially as described previously (30). Oligonucleotides used for PCR are listed Table 2. P. aeruginosa and E. coli strains were grown at 37 °C in Luria-Bertani medium. To induce LapA production and secretion via the P. aeruginosa Hxc T2SS, cells were grown at 30 °C under phosphate-limiting conditions using proteose peptone medium (Difco Laboratories) containing 0.4% glucose, with horizontal shaking (29). When required, media were supplemented with the following antibiotics used at the indicated concentrations: 50 μg·ml−1 kanamycin, 20 μg·ml−1 gentamicin; and 50 μg·ml−1 ampicillin; 50 μg·ml−1 for E. coli and 250 μg·ml−1 carbenicillin; 50 μg·ml−1 gentamycin; and 2,000 μg·ml−1 streptomycin for P. aeruginosa. Bacterial growth was measured by optical density at 600 nm (A600). 1 A600 unit corresponds to 109 cells/ml. The E. coli CC118λpir strain was used to propagate pKNG101 and derivative plasmids, while TG1 and the DH5α strains were used for other plasmid manipulations. Plasmids were transferred to P. aeruginosa using the conjugative properties of the helper plasmid pRK2013 in triparental matings (31). Transconjugants were selected on Pseudomonas isolation agar (Difco) containing 2.5% glycerol (v/v) supplemented with corresponding antibiotic(s). For classical arabinose induction, bacterial cultures were induced with 0.2% filtered l-arabinose (Sigma) at A600 0.8 for 2.5 h.
TABLE 1.
Strains and plasmids used in this study
Ap, ampicillin; Gm, gentamicin; Km, kanamycin; Sm, streptomycin.
| Strains, vectors, and plasmids | Relevant characteristics | Source or Ref. |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | SupE Δ(lac-proAB) thi hsd RΔ5 (F':traD36 rpo A+B+ lacIq lacZΔM15) | Lab collection |
| DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab collection |
| CC118λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 RfR(λpir) | Ref. 46 |
| P. aeruginosa | ||
| PAO1 | Prototrophe, chl-2 | B. Holloway |
| PAO1ΔxcpQ | Non polar deletion of the xcpQ gene in PAO1 | Ref. 47 |
| PAO1ΔhxcQ | Non polar deletion of the hxcQ gene in PAO1 | This study |
| PAO1ΔhxcQΔxcpQ | Non polar deletion of the hxcQ gene in PAO1ΔxcpQ | This study |
| PAO1ΔhxcQΔxcpQΔpAΔqA | Non polar deletion of the xphA, and xqhA genes in PAO1ΔhxcQxcpQ | This study |
| Vectors and Plasmids | ||
| pKNG101 | SmR, mobRK2, sacBR+ (suicide vector) | Ref. 48 |
| pRK2013 | KmR, ColE1, Tra+ Mob+ (RK2) | Ref. 30 |
| pCR2.1 | ApRKmR, ColE1, f1 ori | Invitrogen |
| pJN105 | GmR, araC-pBAD, (broad-host-range vector) | Ref. 49 |
| pMMB67HE | ApR, IncQ tac promoter; lacIq, (broad-host-range vector) | Ref. 50 |
| pMMB67HEhxcQV5 | hxcQV5 cloned into pMMB67HE (XbaI/SmaI) | This study |
| pMB4 | xcpQ cloned in pMMB67HE (PstI-SmaI) | Ref. 47 |
| pET-DEST42 | ApR CmR, Gateway destination vector | Invitrogen |
| pET-DEST42xcpQV5 | xcpQV5 cloned in the Gateway vector pET-DEST42 | This study |
| pET-DEST42hxcQV5 | hxcQV5 cloned in the Gateway vector pET-DEST42 | This study |
| pKNGΔhxcQ | Mutator plasmid for hxcQ deletion | This study |
| pKNGΔpAqA | Mutator plasmid for xphA-xqhA deletion | Ref. 31 |
| pJNhxcQV5 | hxcQV5 cloned into pJN105, pBAD | This study |
| pJNhxcQnlV5 | hxcQnlV5 cloned into pJN105, pBAD | This study |
| pJNhxcQnl | hxcQnl cloned into pJN105, pBAD | This study |
TABLE 2.
Oligonucleotides used in this study
| Name | Nucleotide sequence (5′→ 3′) |
|---|---|
| hxcQ-500 | CAGGCCTACTGGCGGCAACTGGCGCCG |
| hxcQ for | CCCATGAGGCGTCGGCGACATGCAGGC |
| hxcQ rev | TCGCCGACGCCTCATGGGGAATCCTTG |
| hxcQ +500 | AATGGGTCTCGAAGGGCTCGATGTGGA |
| petDEST42 for | TCTAGAAATAATTTTGTTTAACTTTAA |
| xcpQ114 rev | GACCTTCGGCGTCTCGCTGCCGCTGTTTTCGGCGTGCG |
| hxcQ85 for | CACGCCGAAAACAGCGGCAGCGAGACGCCGAAGGTCCC |
| petDEST42 rev | TGTTAGCAGCCGGATCAAACTCAATGG |
| hQnl_1 for | GTCGACGAGCGCGCCAGCACCGCCGCCGGG |
| hQnl_4 rev | ATGGATATCTGCGCAGAATTCGCCTCATAGCGGCGCCGCCTCGCCC |
Construction of P. aeruginosa Mutants and Plasmids
Details are available under supplemental “Experimental Procedures.”
SDS-PAGE and Immunoblotting
Protein samples were analyzed under denaturing or semi-native conditions as described previously (32). Protein samples were solubilized in SDS-PAGE sample buffer (33) containing 2% SDS and mercaptoethanol (denaturing) or 0.2% SDS without mercaptoethanol (semi-native). Samples were heated for 10 min at 95 °C (denaturing) or stored at 4 °C (semi-native). Electrophoresis was performed using 11% SDS-polyacrylamide gel at room temperature and 25 mA/gel (denaturing) or 3.5–9% gradient SDS-polyacrylamide-free gel at 4 °C and 100 V (semi-native conditions) or a different percentage of polyacrylamide when indicated. For Western blotting, proteins were transferred from gels onto nitrocellulose membranes. The membranes were blocked overnight in Tris-buffered saline (pH 7.6), 5% milk, and 0.05% Tween 20 and incubated with primary antibodies directed against the V5 epitope (Bethyl/Interchim), LapA (laboratory stock), TolR (laboratory stock), XcpY and XcpR (laboratory stock), HxcQ (EUROGENTEC peptides based polyclonal antibody protocol (AS-DOUB-LXP), peptide 28 (H2N-GGEGNEGDQQRARLSG-CONH2) for specific multimer detection, and peptide 29 (H2N-SSVDERASTAAGVC-CONH2) for specific monomer detection in blocking buffer, followed by a second incubation with horseradish peroxidase-conjugated goat anti-rabbit IgG antibodies (A6154, Sigma) in blocking buffer. Membranes were developed using the enhanced chemiluminescence protocol (Pierce).
Preparation of Culture Supernatants
P. aeruginosa strains were grown under phosphate-limiting conditions to an A600 of 1.5. Cells and extracellular medium were separated by centrifugation; proteins contained in the supernatants were precipitated by adding trichloroacetic acid (10% (w/v) final concentration) and incubated overnight at 4 °C. Samples were subsequently centrifuged (30 min at 15,000 × g), the pellets were washed with 90% (v/v) acetone, resuspended in SDS-PAGE sample buffer, and analyzed under denaturing conditions.
Inhibition of Lipoprotein Signal Peptidase with Globomycin
Bacteria were grown in Luria-Bertani medium to an A600 of 0.8, and arabinose at 0.2% final and globomycin at 100 μg·ml−1 final were added to the culture, and incubation was continued for 30 min at 37 °C. Bacteria were harvested by centrifugation and resuspended in SDS sample buffer, and solubilized proteins were examined by SDS-PAGE and immunoblotting.
[3H]-Palmitic Acid Labeling
Bacteria were grown at 37 °C in a liquid minimal medium, proteose peptone 1× (supplemented with 0.4% glucose as carbon source) to an A600 of 0.3. Gene expression was induced with 0.2% l-arabinose. At the same time, 50 μCi of [3H]-palmitic acid was added to 1 ml of the culture. Cells were grown for 150 min and collected, and total proteins were analyzed by SDS-PAGE. Gels were then either dried on filter paper and subjected to autoradiography for 5 months at −80 °C and revealed or blotted for HxcQ detection.
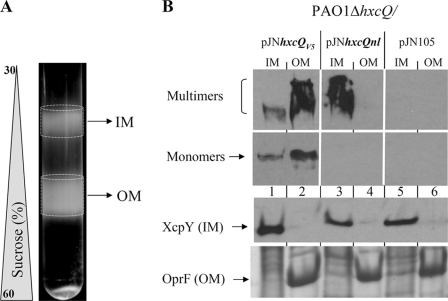
Isolation and Separation of P. aeruginosa Membranes by Density Sucrose Gradient Centrifugation
250 A600 units equivalent of bacterial cells were harvested by centrifugation at 2,000 × g. The pellet was resuspended in 1.5 ml buffer A (10 mm Tris, pH 7.4, 1 mm p-toluenesulfonyl fluoride (Sigma-Aldrich); 10 μg·ml−1 DNase and RNase (Roche); and sucrose 20% (w/w)). The cells were passed twice through a French press cell disrupter (Thermo) at 15,000 pressure units using a 3/8-inch diameter piston (20K French pressure cell, AMINCO). Unbroken cells were removed by centrifugation at 4 °C for 15 min at 1,600 × g. The supernatant was centrifuged at 4 °C for 30 min at 125,000 × g. The crude membrane pellet was resuspended in 0.5 ml buffer M (10 mm Tris, pH 7.4, “Complete EDTA-free” proteases inhibitor mixture (Roche) and 5 mm EDTA) containing 20% (w/w) sucrose and then loaded on top of a discontinuous sucrose gradient consisting of 1.5 ml layers of buffer M solution containing 60 (bottom), 55, 50, 45, 40, 35, and 30% (w/w) sucrose. The membrane separation was performed by centrifugation at 4 °C for 65 h at 39,000 rpm in a Beckman SW41 rotor. The gradients were visually checked, and predicted inner membrane (IM) (upper disc) and OM (lower disc) fractions were collected for experiments as presented (Fig. 5A). Fractions were electrophoresed in (i) 11% denaturing SDS-polyacrylamide gel followed by Coomassie Blue staining and visual identification of the OM protein OprF; (ii) 11% denaturing SDS-polyacrylamide gel followed by Western blotting for XcpY detection; (iii) 3.5–9% gradient seminative polyacrylamide gel followed by Western blotting with antibody against HxcQ peptide 28 for detection of multimers of secretins; and (iv) 9% denaturing SDS-polyacrylamide gel followed by Western blotting with antibody against HxcQ peptide 29 for detection of monomers of secretins.
FIGURE 5.
Membrane localization of HxcQV5 and HxcQnl recombinant secretin in P. aeruginosa. Inner and outer membrane fractions were visually detectable after sucrose gradient sedimentation (A). IM and OM fractions were collected, and their proteins content analyzed by semi-native PAGE for immunodetection of secretin monomers and multimers with the HxcQ antibodies or denaturing SDS-PAGE for OprF and XcpY detection (B).
Isolation and Separation of E. coli Membranes by Density Sucrose Gradient Centrifugation
500 A600 units equivalent of bacterial cells were harvested by centrifugation at 2,000 × g. The pellet was resuspended in 5 ml of buffer B (10 mm Tris, pH 7.4; 1 mm p-toluenesulfonyl fluoride; 10 μg·ml−1 DNase and RNase (Roche); sucrose 20% (w/w); and 400 μg·ml−1 lysozyme (Euromedex)). The cells were passed twice through a French press cell disrupter (Thermo) at 15,000 pressure units using a 3/8-inch diameter piston (20K French pressure cell, AMINCO). Unbroken cells were removed by centrifugation at 4 °C for 15 min at 1,600 × g. The supernatant was centrifuged at 4 °C for 30 min at 125,000 × g. The crude membrane pellet was resuspended in 0.5 ml buffer M (10 mm Tris, pH 7.4; “Complete EDTA-free” proteases inhibitor mixture (Roche), and 5 mm EDTA), containing 20% (w/w) sucrose and then loaded on top of a discontinuous sucrose gradient consisting of 1.5 ml layers of buffer M solution containing 60 (bottom), 55, 50, 45, 40, 35, and 30% (w/w) sucrose. The membrane separation was performed by centrifugation at 4 °C for 18 h at 39,000 rpm in a Beckman SW41 rotor. The gradient was further collected in 16 fractions of about 550 μl each. Fractions were electrophoresed in 3.5–11% denaturing SDS-polyacrylamide gel followed by (i) Coomassie Blue staining and visual identification of the OM porins; and (ii) Western blotting for TolR, HxcQ monomers, and multimers detection. NADH oxidase activity measurement was carried out essentially as described by Osborn et al. (51). Briefly, incubation mixtures containing 50 mm Tris HCl, pH 7.4, 0,12 mm β-nicotinamide adenine dinucleotide, reduced form (NADH) disodium salt hydrate (Sigma, N-8129), and 0.2 mm dithiothreitol (Sigma, D-0632), and the membrane fractions (20 μl) in a final volume of 200 μl were prepared. The rate of decrease in absorbance at 340 nm was measured in microplates at 23 °C in a Multiskan Ascent recording spectrophotometer (Thermo Labsystems).
Differential Detergent Solubilization of Membrane-associated Secretins
The equivalent of 10 A600 units of a crude membrane pellet (obtained following the density sucrose gradient protocol) was resuspended in 500 μl of either 2% Triton X-100 (w/v) (T-9284, Sigma) solution, 100 mm sodium carbonate pH 11 (S-6139 Sigma) solution, or 4 m urea (161-0731, Bio-Rad) in 20 mm MES, 99% (M-3023, Sigma) solution. Samples were incubated for 30 min at 4 °C with gentle shaking. Soluble and insoluble membrane proteins were separated by centrifugation at 4 °C for 30 min at 125,000 × g. Insoluble membrane proteins were recovered in the pellet fraction, whereas solubilized membrane proteins present in the soluble fraction were precipitated by adding tRNA (100 μg·ml−1 final) and trichloroacetic acid (10% (w/v) final) and incubated overnight at 4 °C. Samples were subsequently centrifuged (30 min at 15,000 × g), and the pellets washed with 90% acetone. Pellets containing insoluble or precipitated soluble membrane proteins were resuspended in denaturing or seminative SDS-PAGE sample buffer for SDS-PAGE and immunoblotting.
Transmembrane Potential Measurements
P. aeruginosa strains were grown exponentially for 2 h in the presence of 0.2% l-arabinose and harvested by centrifugation at room temperature. The transmembrane potential (Δψ) was measured essentially as described previously (34). Briefly, the cell pellet from two A600 units was resuspended in 100 μl of 100 mm Tris-HCl, pH 7.8, and 1 mm EDTA for outer membrane permeabilization and incubated for 3 min at 37 °C. The cell suspension was then diluted 20-fold in the transport buffer (100 mm phosphate buffer, pH 7.8, 1 mm KCl, and 0.4% glycerol). A tritiated triphenylphosphonium bromide (Br-TPP) solution ([3H]Br-TPP; Amersham Biosciences; diluted 40-fold in 2 mm cold Br-TPP) was added at the final concentration of 10 μm in 200 μl of the cell suspension and further incubated at 37 °C for 10 min. Cells were recovered by filtration (Whatman) and washed twice with transport buffer and once with transport buffer without glycerol. As a control for nonspecific TPP binding, cell aliquots were first incubated with 10 μm of carbonyl cyanide m-chlorophenylhydrazone for 15 min at room temperature before addition of the Br-TPP solution, incubation, and filtration.
RESULTS
HxcQ Is a Member of the Secretin Family
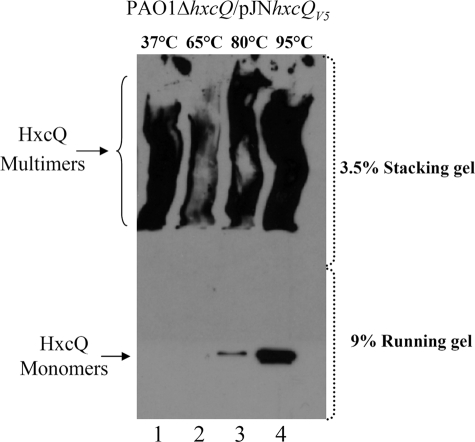
Among the different T2SS Hxc proteins encoded by the hxc cluster, HxcQ is predicted to be the secretin component of the system (29). As presented in Fig. 1, HxcQ forms SDS-resistant high molecular mass (HMM) complexes, which has been shown to be a general characteristic of secretins. We observed such HMM complexes for HxcQ secretin when total cell fractions of P. aeruginosa ΔhxcQ producing a C-terminal V5-hexahistidine tagged HxcQ (HxcQV5) were loaded on a standard SDS-polyacrylamide gel (Fig. 1). However, secretin complexes can show different behaviors in response to heat treatment. For example, HMM complexes formed by PulD or pIV secretins are fully resistant to heat (18, 35), whereas HMM complexes formed by XcpQ, BfpB, TcpC, or OutD secretins are totally dissociated after boiling (7, 10–11, 36). We found that HxcQ multimers are partially heat-resistant even when samples are incubated at up to 95 °C for 10 min (Fig. 1, lane 4).
FIGURE 1.
HxcQ forms SDS and heat-resistant HMM complexes. Immunodetection of HxcQ secretin with anti-HxcQ multimers and anti-HxcQ monomer antibodies. The PAO1 hxcQ mutant complemented with pJNhxcQV5 was grown under standard conditions to induce HxcQV5 production. Whole cell extracts were collected, resuspended in SDS-PAGE sample buffer containing 2% SDS and incubated at the indicated temperature for 10 min. The proteins were separated using 3.5% acrylamide stacking and 9% running gel. The positions of HxcQ multimers and monomers are indicated on the left.
HxcQ secretin encodes an 803-amino acid protein that is 30% identical and 49% similar to XcpQ, a well characterized T2SS secretin of P. aeruginosa. HxcQ primary sequence analysis revealed the typical two subdomains found in XcpQ, the highly conserved C-terminal domain (residues 424–803) involved in pore formation, and the dissimilar N-terminal domain (residues 81–362), predicted to be periplasmic (Fig. 2). Primary sequence comparison between HxcQ and XcpQ also revealed the presence of two supplemental linker regions on both sides of HxcQ N-terminal domain that are absent in XcpQ (Fig. 2). The region located between the signal peptide and the N-terminal domain is called L1. L1 is 71-amino acids long and is mostly composed of small amino acids such as alanine, serine, and glycine. A comparable linker region is also present in the Xanthomonas campestris XpsD T2SS secretin (Fig. 2). The second linker region, L2, located between the N- and the C-terminal domains, is 62- amino acids long and has a composition of 58% serine and glycine. A similar polyserine/glycine region has already been described for OutD and BfpB (16, 37) and is also present in XpsD and TcpC secretins (Fig. 2).
HxcQ Secretin Is a Lipoprotein
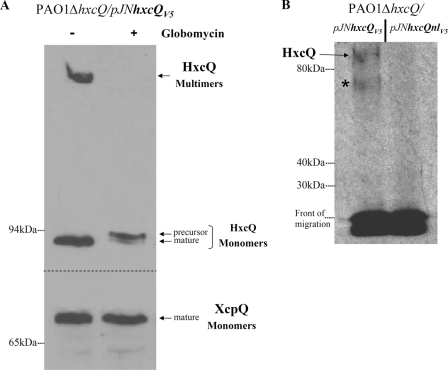
The comparison of Xcp and Hxc SPs revealed that, in contrast to XcpQ, which has a classical type I SP, HxcQ presents a characteristic type II or lipoprotein SP ending with a typical lipobox (supplemental Fig. S1). This observation was also supported by the lipoprotein prediction program DOLOP (38). To experimentally demonstrate the lipoprotein nature of HxcQ, we treated P. aeruginosa cells with globomycin, a specific lipoprotein signal peptidase II inhibitor (39). As shown in Fig. 3A, the maturation of HxcQV5 was significantly affected by the globomycin treatment, leading to the accumulation of the precursor form and loss of HxcQ multimers. As a control, we found that XcpQ remained unaffected in agreement with the resistance of signal peptidase I to globomycin. The lipidation of HxcQ was furthermore confirmed by the recovery of radiolabeled HxcQV5 when the bacteria were grown in the presence of [3H]palmitic acid (Fig. 3B). As a negative control, no radiolabeling was observed for a nonlipidated form of HxcQV5 called HxcQnlV5 (see below, and for description, see supplemental Fig. S1). A control experiment where proteins from palmitic acid-treated cells were blotted following SDS-PAGE and probed with antibody against the V5 epitope indicated that both HxcQV5 and HxcQnlV5 were equally produced (data not shown) and that HxcQV5 did migrate at the position corresponding to the band designated as HxcQV5 in Fig. 3B. In conclusion, both globomycin treatment and [3H]palmitic acid-labeling assays clearly demonstrated that, in contrast to XcpQ, HxcQ is a lipoprotein. From now on, we will refer to this variant of secretin as liposecretin.
FIGURE 3.
HxcQV5 globomycin sensitivity and [3H]-palmitic acid labeling. Globomycin inhibition of HxcQV5 maturation. Immunoblotting of total cell proteins from strain PAO1ΔhxcQ/pJN105hxcQV5 probed with either V5 antibody (top panel) for HxcQV5 detection or XcpQ antibody (bottom panel) for XcpQ detection. In the absence of globomycin (−), mature HxcQV5 monomers as well as HxcQV5 multimers are observed, whereas in the presence of globomycin (+), the maturation of HxcQV5 is inhibited leading to the loss of multimers and accumulation of the precursor form of HxcQV5 monomers. In contrast, mature XcpQ is detected with or without globomycin treatment. A, PAO1ΔhxcQ cells producing HxcQV5 or HxcQnlV5 from plasmids were labeled with [3H]-palmitic acid. Cell samples were electrophoresed on an 8% stacking/9% running SDS-polyacrylamide gel and radiolabeled lipoproteins were detected by autoradiography. Similar amounts of low molecular mass radiolabeled proteins are detected at the migration front in both samples. Full-length HxcQV5 protein and molecular mass markers (in kDa) are indicated on the left. The asterisk indicates a specific radiolabeled protein that might correspond to an HxcQV5 degradation product (B).
Lipidation of HxcQ Is Essential for Its Function
Given that most secretins, including the P. aeruginosa type II secretin XcpQ, are not lipoproteins (6), we wanted to determine if the N-terminal lipid anchor of HxcQ is required for its function. For this purpose, we constructed a nonlipidated HxcQV5 variant (HxcQnlV5). This construction was made by substituting the type II SP of the HxcQ wild type for the type I SP of XcpQ. To maintain a compatible environment for type I signal peptidase recognition, we also included the four amino acids downstream of the XcpQ SP cleavage site (supplemental Fig. S1).
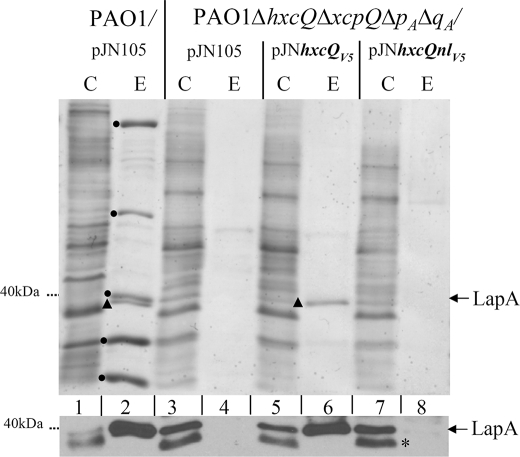
As both the wild type and non-lipidated HxcQV5 possess a C-terminal V5-hexahistidine tag, we first tested the influence of the tag on HxcQV5 function in the quadruple PAO1ΔhxcQΔxcpQΔpAΔqA mutant that is deficient in Hxc, Xcp, and hybrid Xcp T2SSs (40, 5) (Fig. 4, lane 4 versus lane 2). The expression of hxcQV5 from pJNhxcQV5 in this mutant specifically restored secretion of the unique Hxc substrate LapA in the extracellular medium (Fig. 4, lane 6), indicating a functional complementation and no influence of the V5 tag on HxcQ function. We then tested the functionality of the nonlipidated recombinant HxcQ. Although the amount of HxcQnlV5 produced by P. aeruginosa was similar to that of the lipidated form (data not shown), HxcQnlV5 was unable to restore secretion of LapA (Fig. 4, lane 8). Instead, LapA accumulated within the cells (Fig. 4, lower panel, lane 7), which indicates that the HxcQ N-terminal lipid anchor fulfills an essential secretion function. We constructed a tag-free HxcQnl to definitely exclude a possible effect of the V5 tag in HxcQnl nonfunctionality. We did not observe any phenotypic differences between tagged and untagged HxcQnl variants (data not shown).
FIGURE 4.
N-terminal lipid anchor of HxcQ is required for function. Analysis of LapA secretion in supernatant fractions from P. aeruginosa PAO1 wild type and ΔhxcQΔxcpQΔxphAΔxqhA mutant strains producing HxcQV5 and HxcQnlV5. Cellular (C) and extracellular (E) proteins separated on a 12% (w/v) SDS-polyacrylamide gels were stained with Coomassie Blue (upper panel) or immunoblotted for LapA detection (lower panel). The dots indicate Xcp-dependent exoproteins, whereas LapA is indicated by a triangle. The asterisk indicates a nonspecific band reacting with the LapA antiserum. This band is also present in a lapA mutant (not shown).
The HxcQ Lipid Anchor Has a Pilotin Function
In order to understand why HxcQnlV5 was not functional, we examined its cellular localization. These studies were carried out in P. aeruginosa PAO1ΔhxcQ producing wild type or nonlipidated HxcQ from plasmids and under arabinose-inducing conditions. Bacterial cells were disrupted and both HxcQ secretins localized in the total membrane fraction (data not shown). In order to investigate the presence of the secretin multimers in the inner membrane (IM) or the OM, total membrane fractions were then separated by centrifugation on a sucrose density gradient. Regions corresponding to the IM and OM (Fig. 5A) were directly sampled from the tube and analyzed. The quality of the fractionation procedure was verified by the presence in the corresponding fractions of the integral IM protein XcpY and the major P. aeruginosa outer membrane protein OprF (Fig. 5B). Interestingly, we clearly detected both wild type and nonlipidated HxcQ multimers. However, whereas HxcQV5 multimers were correctly localized in the OM (Fig. 5B, lane 2), HxcQnl multimers were mislocalized and accumulated in the IM fraction (Fig. 5B, lane 3). In contrast to wild type HxcQ multimers, multimers of HxcQnl could only be detected under seminative conditions (see experimental procedure). Multimers of HxcQnl indeed appeared to be more sensitive to heat than wild type HxcQ multimers since they could not be detected under classical denaturing conditions (supplemental Fig. S2). We therefore used semi-native conditions for all HxcQnl multimers detection described in this study.
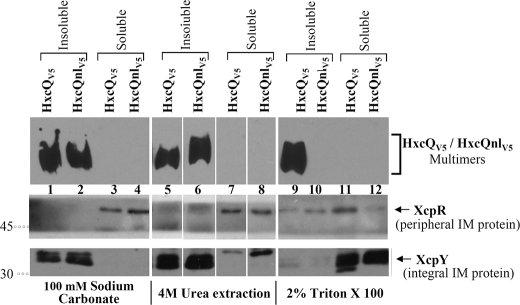
To determine whether IM-recovered HxcQnl multimers were integrated or peripherically associated with the IM, total membrane fractions containing HxcQV5 or HxcQnlV5 were treated with various solubilizing agents. As shown in figure 6, HxcQnlV5 behaves as an integral IM protein since it remained insoluble upon treatment of the membranes with 100 mm sodium carbonate or 4 m urea (Fig. 6, lane 2 and 6), both known to solubilize only peripheral membrane proteins, such as XcpR. In contrast, treatment with the non-ionic detergent Triton X-100, which typically solubilizes proteins inserted into the IM (XcpY), specifically affected the non-lipidated secretin (Fig. 6, lane 10), indicating its IM insertion. Since no HxcQnlV5 was recovered in the soluble fraction, its solubilisation probably led to its degradation or at least the degradation of the V5 epitope used for HxcQ detection. As a control, the wild type HxcQV5 was not found to be solubilized by Triton X-100 (Fig. 6, lane 9), which is congruent with its OM localization.
FIGURE 6.
Differential solubilization of membrane-associated HxcQV5 and HxcQnlV5. Membrane fractions containing HxcQV5 or HxcQnlV5 were treated with 2% (v/v) Triton X-100, 100 mm sodium carbonate, pH 11, or 4 m urea at pH 6.5 for differential solubilization. Soluble and insoluble fractions were analyzed under semi-native conditions for HxcQV5 and HxcQnlV5 multimer detection and under denaturing conditions on a 12% (w/v) SDS-polyacrylamide gel for XcpR and XcpY. HxcQV5 and HxcQnlV5 were probed with anti-V5 antibody. Molecular masses (in kDa) are indicated on the left.
The IM localization of secretin multimers in the absence of a functional pilotin has already been reported for the PulD and YscC secretins, respectively involved in type II and type III secretion (41, 8). As shown for PulD, the absence of the pilotin led to partial dissipation of the proton-motive force (pmf) indicative for IM perturbation. This increase in IM permeability was attributed to IM insertion of the mislocalized secretin multimers. Interestingly, we found similar and significant pmf dissipation when HxcQnlV5 was produced in P. aeruginosa ΔhxcQ (Table 3) and we attribute this effect to the integral IM insertion of HxcQnlV5 multimers. Together, our results show that lipidation of HxcQV5 is essential for correct localization of the protein in the OM. Moreover, the recovery of HxcQnlV5 multimers inserted in the IM suggests that the N-terminal lipid anchor of HxcQ plays a pilotin role, since such behavior was earlier reported for secretins produced in the absence of their cognate pilotin.
TABLE 3.
Measurement of Δψ in cells producing HxcQnlV5 or HxcQV5
Accumulation of [3H]-TPP+ was measured by the ratio of radioactivity inside and outside the cells (second column) and used to calculate the Δψ (third column), as described under “Experimental Procedures.”
| Strains | [3H]-TPP+ in/out | Δψ |
|---|---|---|
| mV | ||
| Wild type/pJN105 | 1,000 | 180 ± 4 |
| ΔhxcQ/pJN105 | 1,113.4 | 183 ± 2 |
| ΔhxcQ/pJN105hxcQV5 | 857.7 | 176 ± 8 |
| ΔhxcQ/pJN105hxcQnlV5 | 256 | 145 ± 11 |
HxcQ Correctly Localized in the OM of the Heterologous Host E. coli
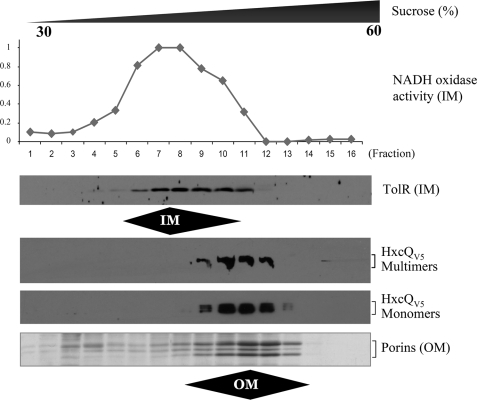
The biogenesis of secretins is impaired in the absence of their cognate pilotins or pilot proteins, and they either remain monomeric or mislocalize to the IM. We tested whether HxcQ liposecretin also requires the assistance of another P. aeruginosa protein for its biogenesis i.e. correct insertion of multimers into the OM. To this end, HxcQV5 was expressed in the heterologous host E. coli. Membrane samples were fractionated by sucrose density gradient and analyzed by SDS-PAGE. Data presented in Fig. 7 clearly indicates that in such a heterologous environment, HxcQ remains correctly localized in the OM as a multimer. This result contrasts with those observed for other secretins such as PulD which requires co-expression of a pilotin for proper localization in E. coli. This important finding demonstrates for the first time the pilotin-independent biogenesis of a member of the secretin family.
FIGURE 7.
Membrane localization of HxcQV5 secretin in E. coli. Membrane fractions were collected after centrifugation on a density sucrose gradient and their protein content analyzed under denaturing SDS-PAGE conditions. HxcQV5 multimers and monomers were detected with HxcQ antibodies. IM fractions were defined by NADH activity and TolR detection. OM fractions were defined by the visual identification of E. coli OM porins on Coomassie Blue-stained gel.
DISCUSSION
Secretins are an unusual and important class of bacterial OM protein involved in various membrane transport pathways such as T2SS and T3SS, type IV pili assembly, and export and assembly of filamentous phage. They form, in the OM, about 1 MDa multimeric pore-forming structures that display relatively low β-strand content (13) and high resistance to dissociation in SDS (17). Such specialized and complex OM proteins require custom-made biogenesis pathways involving additional partners. Depending on the secretin, different routes and partners have been described (42), but so far no secretin has been shown to be self-transported to its final destination. In the present work we report on HxcQ liposecretin, the first example of a self-piloted secretin. Interestingly, we showed that the N-terminal lipid anchor of HxcQ which plays a critical role in its biogenesis might compensate for the lack of specific partner and directly participate in the proper targeting of HxcQ to the OM. Altogether our data reveal a new pathway for secretin transport.
As proposed earlier, the biogenesis of secretins sometimes requires special assistance conferred by pilotin lipoproteins (17–18, 19). In the case of the T2SS PulD/PulS secretin/pilotin pair, the pilotin binds to the secretin emerging from the IM translocon and either keeps it in a competent state, or prevents its non-productive aggregation, before its insertion into the OM. The pilotin may first maintain the secretin in its monomeric form and, second, assist its transport through the periplasm (41). Here, we demonstrate that the HxcQ N-terminal lipid moiety functions as a pilotin since a nonlipidated version of HxcQ behaves like a secretin in the absence of its cognate pilotin i.e. multimers accumulation in the IM. Given that HxcQ does not need any additional partner for its biogenesis, we propose that HxcQ liposecretin carries an intramolecular pilotin.
In type II secretion, the fatty-acylated pilotin binds the C-terminal domain of the secretin (22) whereas in HxcQ liposecretin, the secretin is fatty-acylated at its extreme N terminus. The C terminus of a secretin is embedded in the OM and is therefore well situated for interacting with a pilotin which is also anchored in the OM. The situation seems more conflicting for the N-terminal domain which needs some flexibility to interact with other periplasmic or inner membrane components (43). The extra glycine/alanine/serine rich domain between the lipid anchor and the N-terminal domain that we identified in HxcQ (Fig. 2) could give to the N-terminal extremity the flexibility necessary for its function. It is interesting to note that this domain, absent in nonlipidated PulD and XcpQ T2SS secretins, is also present in XpsD (Fig. 2), another T2SS-lipidated secretin.
HxcQ is the fourth secretin described to be a lipoprotein. Previously, BfpB, TcpC and XpsD were experimentally demonstrated to be lipoproteins (10–11, 24). The involvement of lipidation in secretin biogenesis was only tested for XpsD where this post translational fatty acylation turned out to be dispensable for secretin function (24). For BfpB and TcpC two small nonlipidated periplasmic proteins have been shown to be required for their stabilization and multimerization respectively (25, 11). N-terminal lipidation plays a key role for HxcQ transport and no additional specific partner is required. We therefore suggest that among the liposecretins, HxcQ defines a distinctive subclass whose biogenesis is guided by a new and unprecedented transport pathway.
Although the presence of a lipoprotein is often associated with secretin transport, the involvement of the Lol lipoprotein sorting pathway (44) in this process is still an open question. The discovery here that HxcQ is itself a lipoprotein might suggest that the Lol pathway is directly involved; however the Lol-dependent transport of HxcQ remains to be demonstrated. On the other hand and based on the broad diversity of secretin transport pathways it is also possible that certain secretins might follow an alternative Lol-independent pathway. This is particularly true when looking at XcpQ, another P. aeruginosa secretin. XcpQ is not a lipoprotein and so far, no cognate lipidated pilotin has yet been identified. The situation is also puzzling regarding the implication of the Bam general OM protein assembly machinery in secretin transport (32). Bam dependence was demonstrated for PilQ secretin (32) but invalidated for PulD (45). It would therefore be interesting to experimentally test the Bam- and Lol-dependence of HxcQ in order to reveal the contribution of these systems to the biogenesis of this liposecretin. Lol dependence should also be tested for other secretins, although it will be difficult to discriminate between the requirement of secretin and pilotin for Lol.
Supplementary Material
Acknowledgments
We thank Berengère Ize and Ben Field for careful reading of the manuscript; Jan Tommassen, Margot Koster, Steve Garvis, and Marc Despoints for helpful discussions; Elise Termine for Gateway cloning; Margot Koster for anti-XcpR antibody; Vincent Oréal for assistance with sucrose density gradients and discussions; and Shunichi Miyakoshi (Sankyo, Japan) for the generous gift of globomycin.
Research in the Romé Voulhoux laboratory was supported by Agence Nationale de la Recherche programs, “Young researcher” (Grant ANR-JC07-183230) and “ERA-NET Pathogenomics” (Grant ANR-08-PATH-004-01).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” additional references, and Figs. S1 and S2.
- OM
- outer membrane
- T2SS
- type II secretion system
- IM
- inner membrane
- MES
- 2-(N-morpholino)ethanesulfonic acid
- HMM
- high molecular mass
- SP
- signal peptide.
REFERENCES
- 1.Filloux A. (2004) Biochim. Biophys. Acta. 1694, 163–179 [DOI] [PubMed] [Google Scholar]
- 2.Remaut H., Waksman G. (2004) Curr. Opin. Struct. Biol. 14, 161–170 [DOI] [PubMed] [Google Scholar]
- 3.Pugsley A. P. (1993) Microbiol. Rev. 57, 50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voulhoux R., Ball G., Ize B., Vasil M. L., Lazdunski A., Wu L. F., Filloux A. (2001) EMBO J. 20, 6735–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michel G. P., Voulhoux R. (2009) Bacterial Secreted Proteins in Secretory Mechanisms and Role in Pathogenesis (Wooldridge K. ed). pp. 67–92, Caister Academic Press, Nottingham [Google Scholar]
- 6.Bitter W. (2003) Arch. Microbiol. 179, 307–314 [DOI] [PubMed] [Google Scholar]
- 7.Bitter W., Koster M., Latijnhouwers M., de Cock H., Tommassen J. (1998) Mol. Microbiol. 27, 209–219 [DOI] [PubMed] [Google Scholar]
- 8.Burghout P., Beckers F., de Wit E., van Boxtel R., Cornelis G. R., Tommassen J., Koster M. (2004) J. Bacteriol. 186, 5366–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins R. F., Frye S. A., Kitmitto A., Ford R. C., Tønjum T., Derrick J. P. (2004) J. Biol. Chem. 279, 39750–39756 [DOI] [PubMed] [Google Scholar]
- 10.Daniel A., Singh A., Crowther L. J., Fernandes P. J., Schreiber W., Donnenberg M. S. (2006) Microbiology 152, 2405–2420 [DOI] [PubMed] [Google Scholar]
- 11.Bose N., Taylor R. K. (2005) J. Bacteriol. 187, 2225–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crago A. M., Koronakis V. (1998) Mol. Microbiol. 30, 47–56 [DOI] [PubMed] [Google Scholar]
- 13.Chami M., Guilvout I., Gregorini M., Rémigy H. W., Müller S. A., Valerio M., Engel A., Pugsley A. P., Bayan N. (2005) J. Biol. Chem. 280, 37732–37741 [DOI] [PubMed] [Google Scholar]
- 14.Koster M., Bitter W., de Cock H., Allaoui A., Cornelis G. R., Tommassen J. (1997) Mol. Microbiol. 26, 789–797 [DOI] [PubMed] [Google Scholar]
- 15.Genin S., Boucher C. A. (1994) Mol. Gen. Genet. 243, 112–118 [DOI] [PubMed] [Google Scholar]
- 16.Bouley J., Condemine G., Shevchik V. E. (2001) J. Mol. Biol. 308, 205–219 [DOI] [PubMed] [Google Scholar]
- 17.Hardie K. R., Seydel A., Guilvout I., Pugsley A. P. (1996) Mol. Microbiol. 22, 967–976 [DOI] [PubMed] [Google Scholar]
- 18.Hardie K. R., Lory S., Pugsley A. P. (1996) EMBO J. 15, 978–988 [PMC free article] [PubMed] [Google Scholar]
- 19.Shevchik V. E., Condemine G. (1998) Microbiology 144, 3219–3228 [DOI] [PubMed] [Google Scholar]
- 20.Schuch R., Maurelli A. T. (2001) J. Bacteriol. 183, 6991–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nudleman E., Wall D., Kaiser D. (2006) Mol. Microbiol. 60, 16–29 [DOI] [PubMed] [Google Scholar]
- 22.Daefler S., Guilvout I., Hardie K. R., Pugsley A. P., Russel M. (1997) Mol. Microbiol. 24, 465–475 [DOI] [PubMed] [Google Scholar]
- 23.Seo J., Brencic A., Darwin A. J. (2009) J. Bacteriol. 191, 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu N. T., Hung M. N., Liao C. T., Lin M. H. (1995) Microbiology 141, 1395–1406 [DOI] [PubMed] [Google Scholar]
- 25.Schmidt S. A., Bieber D., Ramer S. W., Hwang J., Wu C. Y., Schoolnik G. (2001) J. Bacteriol. 183, 4848–4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sankaran K., Wu H. C. (1994) J. Biol. Chem. 269, 19701–19706 [PubMed] [Google Scholar]
- 27.Yamaguchi K., Yu F., Inouye M. (1988) Cell 53, 423–432 [DOI] [PubMed] [Google Scholar]
- 28.Seydel A., Gounon P., Pugsley A. P. (1999) Mol. Microbiol. 34, 810–821 [DOI] [PubMed] [Google Scholar]
- 29.Ball G., Durand E., Lazdunski A., Filloux A. (2002) Mol. Microbiol. 43, 475–485 [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Fritsch E. F., Maniatis T. (1989) in Molecular Cloning: A Laboratory Manual (Nolan C. ed) 2nd Ed., pp. 1.2–1.110, C. S. H. L. Press, New York [Google Scholar]
- 31.Figurski D. H., Helinski D. R. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. (2003) Science 299, 262–265 [DOI] [PubMed] [Google Scholar]
- 33.Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. (1975) FEBS Lett. 58, 254–258 [DOI] [PubMed] [Google Scholar]
- 34.Cascales E., Christie P. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17228–17233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linderoth N. A., Model P., Russel M. (1996) J. Bacteriol. 178, 1962–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shevchik V. E., Robert-Baudouy J., Condemine G. (1997) EMBO J. 16, 3007–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sohel I., Puente J. L., Ramer S. W., Bieber D., Wu C. Y., Schoolnik G. K. (1996) J. Bacteriol. 178, 2613–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madan Babu M., Sankaran K. (2002) Bioinformatics 18, 641–643 [DOI] [PubMed] [Google Scholar]
- 39.Hussain M., Ichihara S., Mizushima S. (1980) J. Biol. Chem. 255, 3707–3712 [PubMed] [Google Scholar]
- 40.Michel G. P., Durand E., Filloux A. (2007) J. Bacteriol. 189, 3776–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guilvout I., Chami M., Engel A., Pugsley A. P., Bayan N. (2006) EMBO J. 25, 5241–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayan N., Guilvout I., Pugsley A. P. (2006) Mol. Microbiol. 60, 1–4 [DOI] [PubMed] [Google Scholar]
- 43.Korotkov K. V., Pardon E., Steyaert J., Hol W. G. (2009) Structure 17, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokuda H., Matsuyama S. (2004) Biochim. Biophys. Acta. 1693, 5–13 [DOI] [PubMed] [Google Scholar]
- 45.Collin S., Guilvout I., Chami M., Pugsley A. P. (2007) Mol. Microbiol. 64, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 46.Herrero M., de Lorenzo V., Timmis K. N. (1990) J. Bacteriol. 172, 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Groot A., Koster M., Gérard-Vincent M., Gerritse G., Lazdunski A., Tommassen J., Filloux A. (2001) J. Bacteriol. 183, 959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaniga K., Delor I., Cornelis G. R. (1991) Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 49.Newman J. R., Fuqua C. (1999) Gene 227, 197–203 [DOI] [PubMed] [Google Scholar]
- 50.Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. (1986) Gene 48, 119–131 [DOI] [PubMed] [Google Scholar]
- 51.Osborn H. J., Gander J. E., Parisi E., Carson J. (1972) J. Biol. Chem. 247, 3962–3972 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.