Abstract
The morphology and curvature of biological bilayers are determined by the packing shapes and interactions of their participant molecules. Bacteria, except photosynthetic groups, usually lack intracellular membrane organelles. Strong overexpression in Escherichia coli of a foreign monotopic glycosyltransferase (named monoglycosyldiacylglycerol synthase), synthesizing a nonbilayer-prone glucolipid, induced massive formation of membrane vesicles in the cytoplasm. Vesicle assemblies were visualized in cytoplasmic zones by fluorescence microscopy. These have a very low buoyant density, substantially different from inner membranes, with a lipid content of ≥60% (w/w). Cryo-transmission electron microscopy revealed cells to be filled with membrane vesicles of various sizes and shapes, which when released were mostly spherical (diameter ≈100 nm). The protein repertoire was similar in vesicle and inner membranes and dominated by the glycosyltransferase. Membrane polar lipid composition was similar too, including the foreign glucolipid. A related glycosyltransferase and an inactive monoglycosyldiacylglycerol synthase mutant also yielded membrane vesicles, but without glucolipid synthesis, strongly indicating that vesiculation is induced by the protein itself. The high capacity for membrane vesicle formation seems inherent in the glycosyltransferase structure, and it depends on the following: (i) lateral expansion of the inner monolayer by interface binding of many molecules; (ii) membrane expansion through stimulation of phospholipid synthesis, by electrostatic binding and sequestration of anionic lipids; (iii) bilayer bending by the packing shape of excess nonbilayer-prone phospholipid or glucolipid; and (iv) potentially also the shape or penetration profile of the glycosyltransferase binding surface. These features seem to apply to several other proteins able to achieve an analogous membrane expansion.
Introduction
Bacteria have well defined membrane and envelope ultrastructures, where changes during cell division, for example, are orchestrated by a range of proteins interacting sequentially in a coordinated manner. Gram-negative species, like Escherichia coli, have no internal membranes but only the inner and outer membranes on each side of the enclosing peptidoglycan layer. In contrast, photosynthetic species like Synechocystis sp. PCC 6803 (1, 2) and Rhodobacter sphaeroides (3) have elaborate and extensive internal membrane systems, of different organization, housing the photosystem and antenna assemblies. We report here that overexpression of a foreign lipid glycosyltransferase (GT)4 can yield massive formation of membrane vesicles in the cytoplasm of E. coli, which seems coupled to the sequence and structural features of this GT enzyme.
Induction of internal membrane systems in E. coli have occasionally been observed before for a handful of proteins in connection with routine overexpression, but not to the same extent as here, and in no instance were the molecular mechanisms for this analyzed or understood. First, overexpression of a few endogenous integral membrane proteins, i.e. the fumarate reductase (4), complete ATP synthase (5), or its b-subunit (6), the mannitol permease MtlA (7), sn-glycerol-3-phosphate acyltransferase PlsB (8), and chemotaxis receptor Tsr (9), induce the formation of membrane stacks or tubules in the cytoplasm, close to and continuous with the inner (cytoplasmic) membrane. Some of these proteins may also rely on the integral YidC chaperon for membrane integration (10). Second, overexpression of some “foreign” membrane proteins, such as certain viral ones (11, 12) or an alkane hydroxylase (13), can yield analogous features. Third, deletion mutants for a number of proteins in cell division (14), cell shape maintenance (15), protein secretion (9, 16), lipopolysaccharide inner membrane transporters (17), and the thermosensitive strain 0111a (18) can induce formation of a few membrane vesicles, membrane stacks, or whorls. In addition, E. coli like many other Gram-negative bacteria can form and shed to the surroundings vesicles from their outer membranes (19, 20).
In eukaryotic cells, a number of dedicated endogenous proteins seem to be able to increase membrane curvature and curl up intracellular membranes to tubules and vesicles (21–23). A foreign bacterial (Shiga) toxin can achieve similar features (24). The mechanisms seem to involve a combination of electrostatic and hydrophobic interactions by certain segments in the responsible proteins, sometimes in combination with an arc shape of the protein like in Bin-Amphiphysin-Rvs domain ones. Penetration of hydrophobic segments into the bilayer and lipid molecular packing shapes can also be involved (25). Finally, a large number of “bioactive” peptides may yield analogous morphological membrane changes in both cells and liposome systems, e.g. temporins, penetratin, and polyarginines (26, 27).
The morphology and macroscopic curvature of a biological membrane should be determined by several intrinsic and complex factors, like the dynamic packing geometries and charge of the various lipids constituting the liquid-crystalline bilayer, the geometries of all the integral, much stiffer proteins, the extent of long range protein-protein binding, the weight fractions of lipids and proteins in the membrane, and the transmembrane asymmetries of the latter components. Extrinsic factors like a cytoskeleton can also be involved, but this seems not the case in bacteria. However, the high osmotic concentrations in many bacteria yield a turgor pressure keeping the inner membrane tight against the surrounding peptidoglycan mesh. The spontaneous curvature of the lipid bilayer in E. coli, given by the lipid packing shapes and dominated by the nonbilayer-prone phosphatidylethanolamine (PE), should be negative according to the established phase equilibria (28) and may facilitate vesicle formation. Likewise, cardiolipin (CL), with a large intrinsic curvature, is enriched in the two curved cell pole cap regions (29, 30).
In the small bacterium Acholeplasma laidlawii, the bilayer spontaneous curvature is extensively regulated by two closely related lipid GTs, consecutively synthesizing one nonbilayer-prone and one bilayer-forming glucolipid (31). These two enzymes are both monotopic, i.e. anchored in the membrane cytoplasmic interface by hydrophobic and charge interactions (32). We recently discovered that the “first” GT enzyme monoglucosyldiacylglycerol synthase (alMGS), for the nonbilayer glucolipid α-glucosyl-diacylglycerol (GlcDAG) synthesis, can be overproduced in E. coli to an extent rarely seen for other membrane proteins (33). We report here that this is accompanied by a massive formation of small intracellular membrane vesicles in E. coli, yielding more cytoplasmic membranes than observed for any other protein or condition, cf. above. Similar features were recorded for the “second” lipid GT diglucosyldiacylglycerol synthase (alDGS) and an inactive alMGS mutant. Furthermore, on the basis of structural features for the alMGS and alDGS proteins, we suggest potential general mechanisms for this membrane reorganization phenomenon.
EXPERIMENTAL PROCEDURES
Cloning and Expression Hosts
The cloning of the monoglycosyldiacylglycerol synthase (ALmgs) and the diglycosyldiacylglycerol synthase (ALdgs) genes from A. laidlawii has been described previously (34, 35). Both the ALmgs and the ALdgs genes were ligated into a pET-15b vector (Novagen), containing an N-terminal His6 tag, followed by a thrombin cleavage site before the ALmgs or ALdgs genes. The calculated molecular mass of the expressed alMGS protein is 48,000 Da, and for alDGS it is 41,000 Da. The antibiotic selection marker for both alMGS and alDGS was 100 μg/ml carbenicillin. The E. coli strain BL21-AITM (Invitrogen) was used as expression host for both genes. For alDGS, the E. coli strain BL21-Star (Invitrogen) was also used. As a control experiment, BL21-AITM was used without any plasmid and with 15 μg/ml tetracycline for antibiotic selection.
Growth Conditions
For large scale expression with E. coli BL21-AITM and BL21-Star, batches were grown using either Terrific Broth (TB) or Luria Broth (LB) media. GT expression was started with a 1% (v/v) inoculum from an overnight culture to 250 ml of TB media (12 g/liter bacto-tryptone (Difco), 24 g/liter bacto-yeast extract (Difco), 4 ml/liter glycerol, 2.3 g/liter KH2PO4, and 12.5 g/liter K2HPO4), supplemented with appropriate antibiotics, in a 2-liter baffled flask at 37 °C with shaking at 200 rpm. At A600 ∼0.6, the cultures were transferred to 22 °C, and gene expression was induced after 30 min with 0.2% (w/v) l-arabinose and 1 mm isopropyl 1-thio-β-d-galactopyranoside for the BL21-AI cells and 1 mm isopropyl 1-thio-β-d-galactopyranoside only for BL21-Star cells. After 21–22 h at 22 °C, the cells were collected by centrifugation at 6000 rpm (∼4000 × g) at 4 °C for 30 min, washed with 50 mm HEPES (pH 8), centrifuged, and finally stored at −80 °C until use.
Light Microscopy
Ordinary phase contrast microscopy of various cultures, and fluorescence microscopy of cells labeled with 1 μm membrane stain FM4-64 (Molecular Probes), was performed as described (36).
Radiolabeled Cells
Cultures were started with a 1% (v/v) inoculum to 30 ml of TB media, supplemented with appropriate antibiotics and 1 μCi of [14C]acetic acid (GE Healthcare) in a 250-ml E-flask at 37 °C, and shaking at 200 rpm. At A600 ∼0.6, the culture was transferred to 22 °C and after 30 min induced with 0.2% (w/v) l-arabinose and 1 mm isopropyl 1-thio-β-d-galactopyranoside, followed by growth for 22 h before harvest by centrifugation at ∼5000 rpm for 20 min at 4 °C.
Density Centrifugation of Membranes
A cell pellet corresponding to ∼600 absorbance units (e.g. 30 ml of culture at A600 ∼20) was thawed and resuspended in 6 ml of a buffer consisting of 50 mm triethanolamine (TEA), 250 mm sucrose, 1 mm EDTA, and 1 mm dithiothreitol (DTT) (pH 7.5), supplemented with 1 mg/ml Pefabloc and 0.1 mg/ml DNase. The cell suspension was passed through a French press at 1100 p.s.i. for three cycles before the cell debris was pelleted at 8000 rpm at 4 °C for 20 min. An alternative method for breaking the cells was by adding 0.5 mg/ml lysozyme to the buffer and incubating at 30 °C for 1 h, before the cell debris was pelleted as above. The supernatant was loaded on top of a two-step sucrose gradient composed of 1 ml of 55% (w/w) and 5.5 ml of 8.8% (w/w) sucrose in a buffer containing 50 mm TEA, 1 mm EDTA, and 1 mm DTT (pH 7.5) (TEA buffer). The tubes where centrifuged for 2 h at 210,000 × g and 4 °C using a Beckman-Coulter SW-40 rotor. Thereafter, the membrane fraction was collected (from top of the 55% sucrose layer) using a syringe. The membrane fraction was loaded on top of a six-step sucrose gradient, consisting of (bottom to top) 0.8 ml of 55%, 1.2 ml of 49%, 2.0 ml of 43%, 2.5 ml of 37%, 1.5 ml of 32%, and 1.5 ml of 27% sucrose (w/w) in TEA buffer, respectively. Buoyant densities were obtained from Sigma. The tubes were centrifuged at 4 °C and at 210,000 × g for at least 16 h in a Beckman-Coulter SW-40 rotor. Fractions were collected using a syringe and diluted to double volume with TEA buffer before transfer to 1.5-ml ultracentrifuge tubes. These tubes were centrifuged at 150,000 × g for 40 min at 4 °C to collect the membranes, which were dispersed in 50 mm HEPES (pH 8) and stored at −80 °C until further analyzed. The six-step sucrose gradient tubes (above) were photographed, and the localization levels of the various membrane fractions were recorded. The latter were analyzed by cryo-transmission electron microscopy, SDS-PAGE, and Western blotting.
Small Scale Centrifugation of Radiolabeled Membranes
Cells were grown in small batches (30 ml), with and without 14C radiolabel, as described above. A cell pellet corresponding to ∼400 absorbance units was thawed and resuspended in 1 ml of a lysis buffer consisting of 50 mm TEA (pH 7.5), 1 mm EDTA, 1 mg/ml lysozyme, 0.1 mg/ml DNase, 20% (w/w) sucrose, and 1 mm DTT. The suspension was incubated on ice for 15 min before the addition of 2 ml of buffer containing 50 mm TEA (pH 7.5), 1 mm EDTA, 0.1 mg/ml DNase, 1 mm DTT, and 1.5 mg/ml Pefabloc. This suspension was passed through a French press at 1000 p.s.i. for three cycles before the cell debris was pelleted at 11,000 rpm and at 4 °C for 20 min. The supernatant was transferred to ultracentrifugation tubes for a Beckman-Coulter SW-60 rotor, which was centrifuged for 1 h at 210,000 × g at 4 °C. The supernatant was removed, and the membrane fraction was dispersed in ∼500 μl of buffer consisting of 50 mm TEA, 8.6% (w/w) sucrose, 1 mm EDTA, and 1 mm DTT (pH 7.5). The membrane fraction was loaded on top of a six-step sucrose gradient, consisting of (bottom to top) 0.25 ml of 55%, 0.5 ml of 49%, 0.6 ml of 43%, 1.0 ml of 37%, 0.75 ml of 32%, and 0.5 ml of 27% sucrose (w/w) in TEA buffer. The tubes were centrifuged at 4 °C at 210,000 × g for at least 16 h in a Beckman-Coulter SW-60 rotor. The fractions were collected using a syringe and diluted to double volume with TEA buffer before transfer to 1.5-ml ultracentrifuge tubes. The membranes were pelleted by centrifugation at 150,000 × g for 40 min at 4 °C, using a Beckman-Coulter TLA-55 rotor, and frozen at −20 °C. The six-step sucrose gradient tubes were photographed, and the localization levels of the membrane fractions were recorded. The final nonlabeled fractions were analyzed by SDS-PAGE and Western blot, whereas the radiolabeled fractions were analyzed for lipid composition.
Lipid Extraction and Analysis
Frozen cell pellets from small radiolabeled batches (above) were resuspended in 1.5 ml of buffer (1 m KCl, 0.2 m H3PO4) plus 3 ml of chloroform/methanol, 2:1 (v/v), and the lipids were extracted by thorough vortexing. After centrifugation at 10,000 rpm, the lower solvent (lipid) phase was transferred to a new tube and reduced under N2 (g) to half the volume (∼600 μl). Approximately 10 μl of this solution, together with appropriate lipid standards, was applied on a silica gel 60 TLC plate (Merck), which had been previously dried for 2 h at 110 °C. The TLC plate was developed (one dimension) in chloroform/methanol/acetic acid, 85:25:10 (v/v), and analyzed using a fluorescent image analyzer (Fuji FLA-300). Different 14C-labeled membrane fractions, obtained from the small scale density centrifugation procedure (above), were extracted, separated, and analyzed in a similar manner but with reduced solvent volumes.
Site-directed Mutagenesis
One of positions 1–4 and 9 in the conserved EXSE active site sequence motif of alMGS (300ETQGLTYVE308) was changed to Ala in five individual clones. Mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene), following the manufacturer's protocol, and confirmed twice by DNA sequencing (MWG Biotec).
Enzymatic Assay of GlcDAG Formation
E. coli cells expressing alMGS were solubilized in assay buffer (100 mm HEPES (pH 8.0), 20 mm MgCl2, and 20 mm CHAPS), followed by extensive vortexing for 1 min and sonication twice during incubation on ice for 30 min. The protein concentrations of solubilized cell extracts were around 6 mg/ml (determined by QuickStartTM Bradford Protein Assay, Bio-Rad). The mixed micelle solutions were prepared by mixing lipids dissolved in chloroform/methanol (2:1, v/v) to the final concentrations (2.5 mm diacylglycerol acceptor substrate, 5 mm 1,2-dioleoyl-sn-glycero-3-phosphoglycerol as activator lipid, and 17.5 mm 1,2-dioleoyl-sn-glycero-3-phosphocholine as matrix lipid). The solvent was evaporated under a stream of N2, and the lipid mixture was then solubilized to homogeneity in assay buffer (cf. above) by extensive vortexing, bath sonication for 5 min, and stored at 4 °C until use. The standard enzymatic assay was described previously by Berg et al. (34). Briefly, 25 μl of protein solution was added to 20 μl of mixed lipid micelle solution and incubated on ice for 30 min. The reaction was started by the addition of 25 nCi (1 μl) of UDP-[14C]glucose (GE Healthcare) and 4 μl water, yielding 1.7 μm glucose. After 30 min of incubation at 28 °C, the reaction was stopped with 375 μl of methanol/chloroform, 2:1 (v/v). The lipids were then extracted (37) and separated on TLC plates as described above. All assays were done in duplicate.
Cryo-TEM
Samples of membrane fractions isolated by sucrose gradient centrifugation and whole E. coli were resuspended in 50 mm HEPES (pH 8) or 1× phosphate-buffered saline buffer and investigated by cryogenic transmission electron microscopy (cryo-TEM).
A Zeiss EM 902A transmission electron microscope (Carl Zeiss NTS, Oberkochen, Germany), operating at 80 kV and in zero loss bright field mode, was used. Digital images were recorded under low dose conditions with a BioVision Pro-SM Slow Scan CCD camera (Proscan Gmbh, Scheuring, Germany) and analysis® software (Soft Imaging System, Münster, Germany). To visualize as many details as possible, an underfocus of 1–2 μm was used to enhance the image contrast.
In short, the method for sample preparation was as follows, with a more comprehensive description available in Almgren et al. (38). Samples were equilibrated at 25 °C and ∼99% relative humidity within a climate chamber. A small drop of sample (∼1 μl) was deposited on a copper grid covered with a perforated polymer film and provided with thin evaporated carbon layers on both sides. Excess liquid was removed by means of blotting with a filter paper, leaving a thin film of the solution on the grid. Immediately after blotting, the sample was vitrified in liquid ethane and kept just above its freezing point, −183 °C. Samples were kept below −165 °C and protected against atmospheric conditions throughout the transfer to the TEM and examination.
Protein Electrophoresis and Western Blot
SDS-PAGE analysis of membrane proteins was performed with NuPAGE®, 4–12% BisTris, SDS-polyacrylamide gels (Invitrogen) and NuPAGE® MES SDS-running buffer. The SDS gels were stained using PageBlueTM protein staining solution (Fermentas), and the molecular mass marker was low molecular weight standard (GE Healthcare). Western blot analysis was done after transferring the proteins from the NuPAGE® gels to a nitrocellulose membrane. The membrane was blocked with 5% milk and 0.1% Tween 20 in phosphate-buffered saline. The primary antibody was a mouse anti-His monoclonal IgG (Novagen) diluted 1:5000, and the secondary antibody was a goat anti-mouse IgG horseradish peroxidase-conjugated antibody diluted 1:5000. The membrane was developed using ECL Plus Western blotting detection reagents (GE Healthcare), and spots were detected with an LAS-100 plus scanner (Fuji).
Fold Recognition and Prediction Methods
Structures of several proteins able to induce cytoplasmic membranes in E. coli were predicted using the Meta Server of the Polish Bioinformatic Institute, where the results from a number of good prediction servers are integrated and jointly evaluated. Membrane association of target proteins was predicted by the Membrane Protein Explorer. Data for the molecular composition of E. coli were taken from the CyberCell data base.
RESULTS
Overexpression of Foreign Lipid Glycosyltransferase
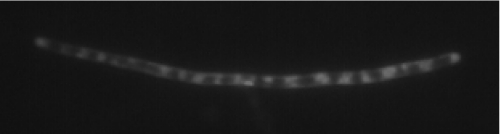
The lipid GT alMGS from A. laidlawii, synthesizing an α-glucosyl-diacylglycerol glucolipid (GlcDAG), is a membrane interface-associated (monotopic) protein of 45 kDa that can be overexpressed in E. coli to reach 170 mg of purified and concentrated enzyme per liter of growth medium (330 mg before concentration) (33). Previous fold recognition and modeling, including validation, have shown both the alMGS and the alDGS structures to be very similar to several established GT three-dimensional structures, like the peptidoglycan precursor GT MurG (Protein Data Bank code 1F0K) from E. coli, having a double Rossmann fold (35, 39). Both alMGS and alDGS enzymes have a strong affinity for a lipid bilayer interface, with the alMGS dissociation constant (KD) reaching 10−15 m for an E. coli bilayer mimetic system, and where binding is enhanced by anionic (PG and CL) and nonbilayer-prone lipids (PE and 1,2-dioleoyl-sn-glycerol) (32). The maximum lateral (cross-section) area occupied by one alMGS molecule in the bilayer interface is ≈2000 Å2, according to the structure model and MurG (data not shown). With an average area of ≈60 Å2/phospholipid, and ≈8 million phospholipids in the inner monolayer of the cytoplasmic membrane in one E. coli cell (CyberCell data base), there is simply not enough lipid interface to house all the alMGS molecules. 330 mg of alMGS enzyme/liter of culture corresponds to ∼220,000 molecules/cell, which should be compared with the totally ≈200,000 inner membrane protein molecules in a “standard” E. coli cell (CyberCell). FM4-64 is a vital fluorescent lipid membrane stain that can efficiently visualize internal membranes in bacteria (40). Fig. 1 shows an E. coli cell after overexpression of alMGS with a strong promoter; note the banded appearance of stained material throughout the elongated cell. FM4-64 is known to clearly visualize single bilayer membranes and also intracellular ones, like an in-growing septum or a spore membrane (41). An elongated, slightly thinner cell shape is common for strongly overexpressing cells. Hence, strong expression of alMGS yields large amounts of lipid-staining material in the E. coli cytoplasm.
FIGURE 1.
Membrane staining of E. coli strongly overexpressing the alMGS protein. E. coli cells were labeled with membrane stain FM4-64, which normally only visualizes the inner and outer membranes, and examined by fluorescence microscopy.
Separation of New Light Density Membranes
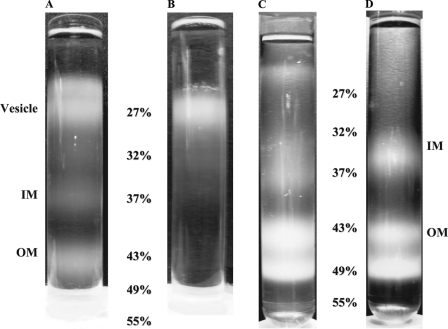
Bacterial inner (IM) and outer membranes (OM) can be separated by equilibrium centrifugation in sucrose density gradients (42), after breaking cells with an appropriate technique, like the French press. In most cases, two standard membrane fractions were obtained here, inner membranes in the 32–37% (w/w) sucrose layers and the outer membranes in the 43–49% sucrose layers (Fig. 2). For experiments with overexpressing alMGS cells, substantial amounts of lower density fractions were also obtained in the very top of the gradient above the IM fraction, either in the 27% sucrose layer or for the large scale preparations even on top of this layer. Moreover, for the large scale preparation, usually five fractions in total were obtained, two of these were in or above the 27% sucrose layer, one corresponding to the IM fraction and then two OM fractions in the 43–49% steps.
FIGURE 2.
Separation of membranes by density centrifugation. Sucrose density gradient tubes are as follows. A, fractions of alMGS-BL21-AI membranes from French press cell disruption. B, only one fraction was obtained from alMGS-BL21-AI when cells were disrupted with lysozyme only. C, fractions of alDGS-BL21-Star membranes. D, fractions from control BL21-AI cells. Vesicle fractions are present in alMGS and alDGS tubes in or above the 27% layer. Inner membrane fractions localize in 32–37% layer and outer membrane in 43–49% layers.
For the small scale preparation (see under “Experimental Procedures”), usually only one fraction was obtained in the 27% sucrose layer, one in the 32–37% layer (IM), and two in the 43–49% layers (OM). Similar features were observed for cells expressing the alDGS as for the alMGS enzyme, i.e. lighter density membrane fractions above the IM fraction were obtained for both but with smaller amounts for alDGS (Fig. 2). The top fraction from the large scale preparations of alMGS had a slightly lower density than in the small scale preparations, as seen from its distribution in the density gradient. The low buoyant density in the 27% layer (1.117 g/cm3) and above corresponds to a lipid fraction of ≈60% w/w or more in these membranes (cf. Ref. 43), compared with the ≈40% w/w in normal inner membranes, i.e. in the 32–37% sucrose interface layer. Pure (delipidated) membrane proteins have a density around 1.28 g/cm3, whereas membrane lipids are close to 1.0 g/cm3 (44). A pure light density preparation, with no IM or OM fractions, was obtained by omitting the French press breaking up step and releasing the membranes by a lysozyme treatment only (Fig. 2B). Cells without the expression plasmids or empty BL21-AI control cells confirmed that a normal gradient pattern was obtained with only two major membrane fractions at 32% (1.142 g/cm3) and 43% sucrose (1.198 g/cm3), corresponding to the IM and OM fractions, respectively (Fig. 2D). Hence, strong overexpression of alMGS and alDGS proteins yields large amounts of a new light density membrane fraction in E. coli, in addition to normal inner and outer membranes.
Formation of Intracellular Vesicles
Cryogenic transmission electron microscopy (cryo-TEM) was used to investigate both whole cells expressing alMGS and alDGS, control cells, and the different membrane fractions from the alMGS-expressing cells prepared by sucrose density centrifugation.
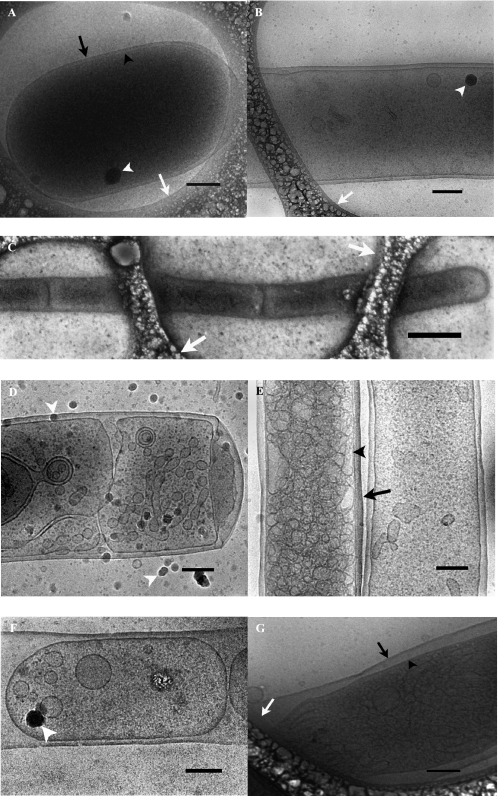
Fig. 3, A and B, shows whole BL21-AI E. coli without the expression plasmids. The normal size of an E. coli cell is at the limit of what can be investigated by cryo-TEM. Because of the scattering of the electron beam, samples thicker than ∼500 nm will appear dark. As can be seen from Fig. 3, A and B, whole cells can, however, be visualized by cryo-TEM, and the outer and inner membranes are clearly resolved.
FIGURE 3.
Cryo-TEM analyses of whole E. coli cells. A and B, control strain BL21-AI without expression plasmid. C and D, samples from cells expressing wild-type alMGS. E and F, cells expressing wild-type alDGS. G, cells expressing inactive alMGS mutant E308A. A, normal sized E. coli filling up a hole in the polymer film. B, part of an elongated cell of the control strain. C, elongated E. coli-expressing alMGS cell with continuous outer membrane and several inner membrane compartments, stretching over several polymer holes. D, E. coli cell expressing alMGS with intracellular membrane structures, e.g. vesicles. E, elongated cell expressing alDGS filled with intracellular membrane vesicles next to an almost empty E. coli. F, part of an elongated E. coli expressing alDGS, where several inner membrane compartments with intracellular vesicles share the same outer membrane. G, E. coli cell expressing the alMGS E308A mutant, unable to synthesize glucolipid, filled with intracellular membrane vesicles. The black arrowheads indicate inner membranes of whole cells. The black arrows indicate outer membranes of whole cells. The white arrowheads exemplify the appearance of ice crystals deposited on the sample surface after vitrification, and the white arrows point to the polymer film present on the grid. Scale bars, 200 nm except in C where the scale bar indicates 1 μm.
Fig. 3, C–F, shows cells expressing wild-type alMGS and alDGS, respectively. Most of the cells were elongated and often spanned several holes of the perforated polymer film on the TEM-grid (Fig. 3C). Therefore, Fig. 3, D–F, shows only parts of the complete E. coli cells. Elongated cells were also occasionally detected in the sample without the expression plasmids, Fig. 3B. Inspection of whole alMGS and alDGS cells confirmed the presence of numerous membrane vesicles formed inside the cells. In many cases the vesicles totally filled the cytoplasm (Fig. 3, D and E). As can be seen from the micrographs, the outer cell membrane was continuous, whereas the vesicles that filled up the cell interior space to a crowded state appeared to be pinched off from the inner membrane. In the elongated cells, the cytoplasmic compartment was divided into subcompartments, each surrounded by a joint inner membrane (Fig. 3, D and F). This fact most likely explains the striated feature of the intact cells stained with FM4-64 (cf. Fig. 1). The alDGS-expressing cells appeared more or less identical to the alMGS cells in the cryo-TEM images, and importantly, both cell types displayed inner membrane vesicles filling up the cytoplasmic space. The elongated vesicle-filled cells are in sharp contrast to the empty BL21-AI control cells where the cytoplasm only contained a few structures, probably granules and, in the case of elongated cells, an occasional vesicle (Fig. 3, A and B).
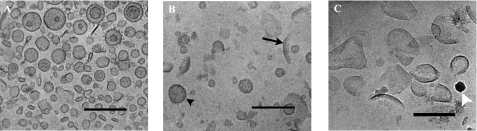
The micrographs representing the “new” 27% light density membrane fraction (Fig. 4A) show a selection of many closed vesicles with diameters of 50–100 nm. Some more or less irregularly shaped vesicles and open structures are also present. The normal inner membrane fraction, isolated from the 32 to 37% sucrose step, consisted of both vesicles formed by the E. coli inner membrane and a large amount of membrane fragments (Fig. 4B). The open structures seen in Fig. 4B most likely originate from the outer membrane, as they are very similar to the structures found in the 43–49% sucrose layers where the outer membrane fraction is normally found (Fig. 4C). Hence, the large amounts of new light density membranes formed by strong overexpression of the alMGS and alDGS proteins in E. coli consist of closed vesicles that have pinched off into the cytoplasm from the inner cell membrane.
FIGURE 4.
Cryo-TEM analyses of membrane fractions from cells expressing alMGS and isolated by sucrose gradient separations. A, vesicle fraction from the 27% sucrose step. B,“normal” inner membrane fraction from the 32 to 37% sucrose steps. C, normal outer membrane fraction from the 43 to 49% sucrose step. The black arrowheads indicate inner membrane vesicles, and the black arrows indicate outer membrane fragments in the isolated membrane fractions. The white arrowheads exemplify the appearance of ice crystals deposited on the sample surface after vitrification. Scale bars, 200 nm.
Protein Composition of Vesicle Membranes
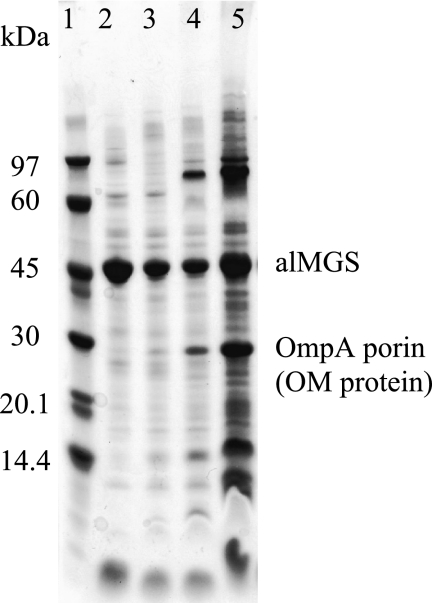
The various membrane fractions from both the small and large scale sucrose density gradient separations from overexpressing cells were analyzed with SDS-PAGE to study the protein composition and distribution among the types. The alMGS (48 kDa) protein constituted the dominating majority of the proteins in the vesicle and IM fractions but not as evident in the OM fraction, according to staining intensity (Fig. 5). Many proteins were observed in all fractions, and generally the light density membrane vesicles were very similar to the IM fraction except for a few minor differences. For the outer membrane fraction, from the 43 to 49% sucrose layers, the typical OmpA porin protein was used as an identity marker (Fig. 5) and a label for estimating the extent of separation and contamination of the various membranes after centrifugation. According to the cryo-TEM micrographs of the membrane fractions, OM fragments could also be seen in the IM fractions (cf. Fig. 4B) as well as some whole cells in the IM and OM fractions. This could explain the content of some OmpA porin protein in both IM and OM fractions. According to the analysis, the light density vesicle membrane fraction contained very little OM proteins.
FIGURE 5.
Protein composition in vesicle membranes. SDS-PAGE analysis of fractions from sucrose density gradient separation of membranes prepared from alMGS-expressing cells (small scale). Gel was stained with PageBlueTM protein staining solution. Lane 1, low molecular mass marker; lane 2, vesicle, 27% layer; lane 3, IM, 32% layer; lane 4, IM/OM, 37% layer; and lane 5, OM, 43% layer. Less amounts of proteins were applied from the vesicle and IM fractions to highlight the dominating presence of the alMGS protein (close to the 45-kDa reference), which was identified by Western blot (data not shown). The OM porin OmpA (31 kDa) was identified due to its change in migration ± boiling the sample (data not shown).
Hence, for cells overexpressing the alMGS, this protein strongly dominates the composition in vesicle and inner membrane fractions. These membranes also have a similar repertoire of other membrane proteins.
Polar Membrane Lipid Composition
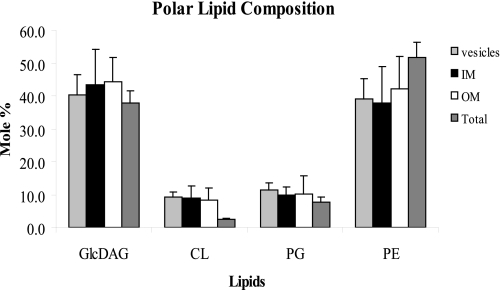
The membrane lipid compositions of the alMGS- and alDGS-overexpressing cells, and the empty BL21-AI (control) strain, were investigated by incorporation of radiolabeled acetic acid into the lipid acyl chains during growth. The lipids were extracted from the various density-fractioned membranes, separated by TLC, and quantitated as described under “Experimental Procedures.” Intact whole cells were also analyzed. The results showed that the overexpressed enzyme alMGS can produce a fair amount of the foreign glucolipid GlcDAG (≈40 mol %; Fig. 6), which does not exist naturally in E. coli. Previously, maximum amounts of only 5–10 mol % GlcDAG have been obtained with the alMGS enzyme in a lipid wild-type background but with much lesser extents of overexpression (34). The increase of GlcDAG lipid amounts here were similar in all the density-separated membrane fractions, with a concomitant decrease of PE to ≈40–50 mol % (Fig. 6) compared with the normal 75 mol % in wild-type E. coli. Similar features were valid for PG and CL. The latter seems lower in intact whole cells, potentially because of binding or less efficient extraction here. Note that GlcDAG is synthesized from diacylglycerol released from the normal turnover of PG (36). Hence, all GlcDAG here initially comes from PG; this indicates that the large amounts of alMGS enzyme may up-regulate PG synthesis substantially. For the alDGS cells, the lipid composition was very similar to BL21-AI control cells, i.e. 75–80 mol % PE, 15 mol % PG, and 5–10 mol % CL (data not shown), because the foreign substrate lipid GlcDAG needed is not present here, and diglucosyl-diacylglycerol can consequently not be synthesized.
FIGURE 6.
Polar lipid composition in vesicle membranes. Membrane lipid acyl chains were labeled with radioactive acetic acid during alMGS overexpression. After cell harvest and membrane separation, lipids were extracted from vesicles, inner and outer membranes, and whole cells controls, separated by TLC, and quantitated by electronic autoradiography. Wild-type cells contain ∼75 mol % PE (zwitterionic) and 25 mol % PG plus CL (both anionic); the latter is increased in the stationary growth phase. The foreign GlcDAG is synthesized by glucosylation of diacylglycerol, released from the turnover of PG (36). alDGS-expressing cells have wild-type lipid composition, because its substrate GlcDAG is not synthesized here (data not shown). S.D. ± σ.
Hence, cells strongly overexpressing the alMGS protein contain substantial amounts of the foreign nonbilayer-prone lipid GlcDAG in all membrane and vesicle fractions. However, this lipid seems not essential for vesicle formation per se, because alDGS-expressing cells also yield vesicles without the diglucosyl-diacylglycerol (or GlcDAG) lipid present.
Potential Effects of alMGS Membrane-binding Segment
Bilayer binding of purified alMGS is strongly stimulated by anionic lipids like PG and CL, in addition to nonbilayer-prone ones (32). An identified amphipathic helix with multiple plus charges (65SLKGFRLVLFVKRYVRKMRKLKL87) was experimentally proven to exhibit strong bilayer binding, with similar lipid preferences as the full-size protein (45). Because fairly short peptides, usually with a cationic character, can also induce bilayer reorganization (27), the potential influence of the alMGS-binding peptide was analyzed here. To efficiently express the peptide in E. coli, and also monitor membrane binding in situ, it was fused at the gene level C-terminally behind the green fluorescent protein and a proper linker (46). As controls, plain green fluorescent protein and a highly plus-charged nonhydrophobic segment from the yeast protein Myr1 with proven bilayer binding properties (46) were used. However, according to cryo-TEM analyses, these short and plus-charged peptides could not elicit formation of membrane vesicles in the E. coli cytoplasm in situ during growth, as could alMGS and alDGS proteins (data not shown). Hence, this ability seemed coupled to sequence and structural features from a larger part of alMGS.
Inhibition of Glycolipid Synthesis
Potentially, synthesis of GlcDAG by the alMGS enzyme may influence or enhance formation of the intracellular vesicles. Several GT sequence families with a retaining enzyme mechanism, of the total 91 in the CAZy GT data base, have a conserved EX7E sequence motif in their catalytic C-domains, including the large GT-4 family to which alMGS and alDGS belong (47). This sequence region is involved in binding the donor sugar substrate, as visualized, for example, in the three-dimensional structures of the GT-4 enzymes PimA (48) and WaaG (49). On the basis of the latter, and the effects of amino acid point changes in the related alDGS5 and Arabidopsis thaliana DGD25 lipid GTs (all from the GT-4 family), positions 1–4 and 9 (underlined) in the alMGS motif 300ETQGLTYVE308 were individually changed to Ala by site-specific mutagenesis. For all five mutations, enzyme activity (i.e. GlcDAG formation) was completely abolished in a standard in vitro-mixed micelle assay (Ref. 34 and data not shown). Analysis of membrane distribution from E300A and E308A mutants by sucrose density centrifugation revealed large amounts of membranes in the same low density fraction, above the inner membranes, as for the wild-type alMGS (data not shown). Furthermore, these two clones produced no GlcDAG in vivo even after overproducing alMGS overnight, according to a TLC analysis. Cryo-TEM (as above) of the E308A mutant cells overexpressing the modified alMGS protein showed that it contained intracellular membranes and vesicles, similar to the wild-type alMGS (see Fig. 3G). Hence, the notion that the vesicle-forming capacity is inherent in the alMGS or alDGS protein structure, without any essential contribution by the GlcDAG lipid, seems valid.
DISCUSSION
Formation of Intracellular Membranes
The large amounts of the alMGS protein synthesized, in combination with its sequence and structural properties, must be the reason for the extensive membrane vesicle formation, reaching higher levels than observed for other proteins in E. coli causing occasional formation of extra membranes. A key factor for incorporation of overexpressed integral membrane proteins in the membrane is sufficient amounts of the SecYEG translocon and associated chaperons. However, the monotopic alMGS and alDGS proteins are anchored in the IM cytoplasmic interface, without passing the translocon. Another factor is spatial requirements; more lipid bilayer is needed to accommodate large amounts of new proteins. The type and properties of the lipids may also be important, especially for heterologous membrane proteins. Four partially overlapping mechanisms for how heterologous or endogenous proteins can expand and/or “bend” membranes in E. coli cells, eventually leading to formation of closed intracellular membrane compartments, e.g. membrane vesicles (“vesiculation”) on a large scale, can be suggested as follows.
Packing Shapes of Transmembrane Proteins
Certain protein complexes have extending cytoplasmic domains that are substantially larger (space-demanding) laterally than their smaller transmembrane parts. This is very evident for a handful of E. coli proteins causing formation of extra cytoplasmic membranes upon overexpression, i.e. the FtsY-receptor-ribosome complex (9), the complete ATP synthase (5), the fumarate-reductase complex (4), the chemotaxis Tsr receptor, the MtlA permease (7), and the ATP synthase b-subunit (6), listed here in order of decreasing size/volume for the extruding cytoplasmic domain(s). All of these do use the SecYEG translocon for integration, and some also use the chaperon YidC (50). For these proteins, the larger space demands of the extruding part, in relation to the transmembrane part (cf. Fig. 7, schematic), should be a major cause for the membrane to bend, invaginate, and potentially also to pinch off from the E. coli inner membrane.
FIGURE 7.
“Packing shapes” for vesicle-inducing membrane proteins. Contour models of the complete E. coli ATP synthase (A), chemotaxis receptor Tsr (B), a hydrophobic wedge (or cone)-shaped hydrophobic protein (C), and a monotopic glycosyltransferase (with double Rossmann folds, like alMGS) (D) in a schematic membrane. All these (except C) have been shown to be able to cause formation of extra intracellular membranes in E. coli (see text).
The eukaryotic proteins cytochrome b5 and PMA2/PMA1 ATPases also seem to conform to this principle of bulkiness and formation of extra membranes upon overexpression in Saccharomyces cerevisiae (51, 52). In mitochondria, dimer formation of the many bulky ATP synthases (cf. above) may be involved in the bending of cristae membranes (53). Likewise, controlled dimerization of artificial membrane proteins in mammalian tissue culture cells caused the generation of cubic, highly curved membranes in the endoplasmic reticulum (54).
In an analogous manner, transmembrane proteins where the integral part (in the membrane) is much more bulky inside one of the monolayers, compared with the other monolayer, and with no or very small protruding parts, and hence effectively hydrophobic “wedges” (or “cones”), should also cause membrane bending and potential vesiculation. To the best of our knowledge, the proposed effects of such “‘wedge” proteins have not been reported, but it may be valid for certain membrane channels and transporters having this shape.
Interface-interacting Proteins
Several monotopic membrane proteins also induce membrane formation, like the E. coli glycerol-3-phosphate acyltransferase PlsB (8), the peptidoglycan precursor glycosyltransferase MurG with an established structure (39, 55), and the two heterologous (A. laidlawii) lipid glycosyltransferases alMGS and alDGS in this work (cf. results above). For PlsB, fold recognition (at the Meta Server), as described before (34), yielded a partial similarity to a squash glycerol-3-phosphate acyltransferase structure (data not shown). These are all localized on the cytoplasmic monolayer of the inner membrane in E. coli, and they do not use the Sec translocon. Furthermore, they are all devoid of transmembrane segments but have several segments intercalating into the bilayer interface according to MPEx analyses (data not shown), where hydrophobicity is predicted based upon experimental thermodynamic principles.
Furthermore, the anchor segments of all these monotopic proteins, and in the archetypal monotopic membrane protein prostaglandin H2 synthase-1 (56), span a similar range of ΔG values for interface association (data not shown). These monotopic proteins will expand the inner monolayer upon incorporation at sufficient amounts, prevent transmembrane protein localization in the corresponding (juxta-positioned) outer monolayer area, and eventually cause convex bending and vesicle pinching.
A natural system with highly curved membranes of small dimensions is the prolamellar body in dark-grown plants and normal seed shoots, eventually transformed to the chloroplast thylakoids upon metabolic greening by light. Here, large amounts of a monotopic dimer for the enzyme protochlorophyllide oxidoreductase (with a Rossmann fold, according to our Meta Server prediction) seem to be involved in formation/maintenance of the cubic three-dimensional organization in these strongly lipid-enriched membranes (57, 58). Hence, Rossmann fold proteins only penetrating the membrane interfaces, like alMGS, alDGS, and MurG, may achieve an analogous curvature modification of membranes when present in large amounts.
Positive Charges at the Interface
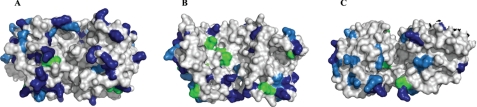
Some of the proteins are also characterized by having a substantial number of positively charged amino acids (Lys and Arg) close to the membrane interface (e.g. PlsB, ATP-b, MurG, and especially alMGS). It is well established that in peripheral and integral proteins, these residues can bind to anionic phospholipids like PG and CL, especially when occurring as pairs in the sequence (59, 60). Lys and Arg are also common in the interface regions of typical monotopic membrane proteins, e.g. prostaglandin H2 synthase-1 (56), signal peptide peptidase (61), and glycerol-3-phosphate dehydrogenase (62). Of the three monotopic proteins here with very similar three-dimensional structures (i.e. alMGS, alDGS, and MurG), alMGS is by far the most efficient vesiculation protein. alMGS has a substantially larger number of Lys and Arg residues than the others (61 residues), and only one Trp, whereas MurG has 36 Lys/Arg and 8 Trp. The bulkier Arg is also more frequent in MurG compared with alMGS (see Fig. 8 schematic for a comparison of surface charges). Intriguingly, it was recently reported from a molecular dynamics comparison of a dozen monotopic membrane proteins that the more Lys and Arg residues and to some extent His, in addition to hydrophobic ones in the binding surfaces, the deeper into the bilayer the proteins seem able to penetrate, potentially even reaching anionic phospholipids in the opposite monolayer (63). His is frequently found interacting with phospholipid headgroups in membrane protein three-dimensional structures, especially in sites for cardiolipin (64). In addition, His often contributes a pH dependence for binding (65, 66), becoming cationic at a slightly reduced pH. The more deeply penetrating monotopic proteins also caused a local convex change in bilayer curvature, which was absent for less penetrating species (63). Likewise, positively supercharged (soluble) green fluorescent protein can penetrate through mammalian membranes but where correspondingly charged peptides failed (67). Hence, numbers of cationic amino acids and protein penetration profiles of full-sized proteins seem connected. The penetration of alMGS into the bilayer is supported by a recorded decrease in acyl chain order (32).
FIGURE 8.
Positive charges on alMGS, alDGS, and MurG binding surfaces. The amounts of cationic charge on interface-penetrating surfaces in monotopic proteins seem to correlate with extent (depth) of intercalation according to a recent analysis (63). Validated three-dimensional models of alMGS and alDGS (from fold recognition and modeling (35)) and x-ray structure of MurG (Protein Data Bank code 1F0K) show the distribution of the positively charged residues arginine, lysine, and histidine. Analogous bottom (binding) surface areas are visualized. A, alMGS; B, alDGS; and C, MurG. The lysines are in dark blue, arginines in light blue, and histidines in green.
Membrane Expansion by Lipid Synthesis
Membrane lipid biosynthesis is frequently taking place in one monolayer in a membrane, and lipids are then distributed to the other monolayer by special transporters or by spontaneous but slower “flip-flop.” A strong stimulation of lipid synthesis may either bend the bilayer due to monolayer expansion (cf. “Interface-interacting Proteins” above) or, if transmembrane equilibrium is fast, bend the entire membrane inward into the cytoplasm if the lateral expansion is prevented by a surrounding barrier, like the peptidoglycan cell wall in bacteria. For erythrocytes, recorded shape changes by introduction of certain drugs and polar lipids with various packing geometries in one of the monolayers is explained by the “bilayer-couple hypothesis” (68, 69). Analogous features have been analyzed in liposomes and can be interpreted in terms of differences in lateral areas of the two monolayers, the enclosed volume and bilayer bending energies (70).
In E. coli the membrane lipid PG (anionic) seems to be a pace-keeper for polar lipid synthesis, the pathway branch of which can dramatically increase in rate to keep up with demands (71). The synthesis of the other major lipid PE (zwitterionic) is governed by the interface surface charge given by PG and CL (72, 73); hence, more PG yields synthesis of more PE. Withdrawal laterally of PG from the membrane pool by binding to plus-charged clusters in overexpressed membrane proteins may be a signal for increased membrane lipid biosynthesis, where a shortage of PG is sensed and counteracted by increased synthesis. Interestingly, PlsB (cf. above) is considered to function as a sensor for coordination of membrane protein synthesis and phospholipid formation in E. coli (74). It has been established recently that plus-charged proteins and peptides can bind and thereby withdraw the minus-charged phospholipids PG and CL from the free pool in a bilayer (59, 75). By binding to PG (and CL), the many alMGS molecules may withdraw these lipids laterally from the bilayer; alMGS and its binding helix have an established binding capacity for and potential sequestration of PG and CL (45). If the remaining unbound lipid has a molecular packing shape that promotes bending, like PE, then as a consequence the bilayer may also start to bend in addition to the metabolic lateral expansion.
A calculation based on 220,000 alMGS molecules per E. coli cells indicates that, in addition to the shortage of lipid lateral areas for insertion, there is not enough PG plus CL molecules in the inner monolayer of the cytoplasmic membrane to match all the plus charges of the alMGS binding surface (data not shown). The surface of a 100-nm vesicle corresponds to maximally 1500 surface-bound alMGS molecules, but given the large fractions of lipids and alMGS constituting a maximum of 50% of the proteins (Fig. 5), a few hundred alMGS per vesicle seem plausible. This would in turn correspond to a similar number of vesicles per cell. Vesicles of similar but more fixed size are formed by the photosynthetic assemblies in R. sphaeroides membranes (3). Packing, geometry, and interactions of the core complexes are the driving forces for these stalked vesicles (76), with a long range organization that seems more complex than the varying alMGS vesicles here.
Mechanism(s) of Vesicle Formation
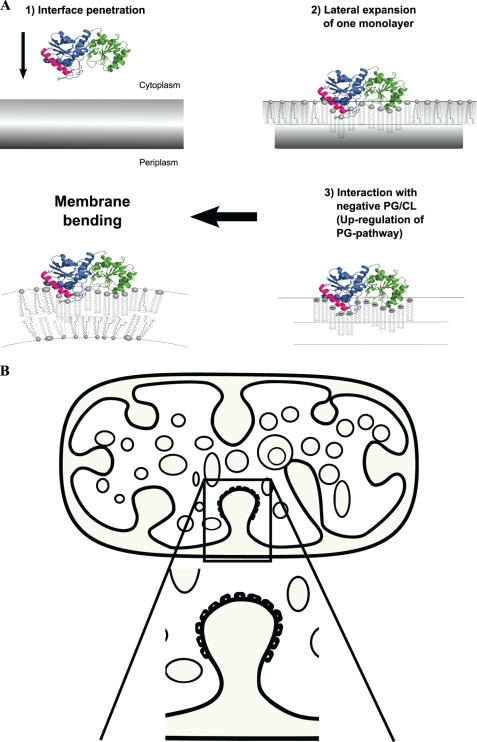
Hence, the increase in cytoplasmic membrane amounts, and eventually membrane binding and vesicle formation in E. coli caused by strong overexpression of alMGS, may be a combination of the following: (a) lateral expansion of one (the inner) monolayer by all alMGS binding, where bending may be enhanced by intercalation into the interface of one or several helices or segments in alMGS, changing the molecular packing of the bilayer (23, 25, 77); (b) stimulation of phospholipid synthesis by PG (and CL) binding to alMGS, causing membrane expansion by more polar lipids being made; (c) bilayer bending caused by the packing shape of the PE (or GlcDAG) lipid; and (d) the shape and penetration profile of the alMGS interacting surface may also be involved in bending, cf. the curvature change that the monotopic prostaglandin synthase COX-2 seems to cause in a bilayer (63, 78). In Fig. 9, the schematics indicate the individual steps (and mechanisms) for how many alMGS proteins may eventually achieve “bending” of the membrane, leading to pinching-off of vesicles.
FIGURE 9.
Vesicle formation by alMGS protein. Schematic illustrates the individual steps for how membrane interface intercalation of large amounts of alMGS or alDGS, and binding to anionic lipids, expand the inner membrane and cause bending (A), eventually leading to vesiculation (B). The nonbilayer-prone PE or GlcDAG lipids (cone-shaped) are synthesized on the IM inner monolayer but must be distributed to both monolayers. GlcDAG is not needed for vesiculation (see “Results”).
The mechanisms for alMGS-induced vesiculation are analogous to a number of shape changes taking place during membrane traffic in the eukaryotic cell. These can be induced by insertion of amphipathic helices, interface binding of proteins, binding of long, curved helical bundle proteins (e.g. Bin-Amphiphysin-Rvs-like (79)), and remodeling of lipid packing shapes. Most of these mechanisms expand laterally the area of one monolayer and thereby force the bilayer to bend (23, 25, 77). These E. coli membrane vesicles should be advantageous for a number of practical processes, like overexpression of transmembrane proteins and examination of mechanisms for membrane transporters and protein translocation (secretion), respectively.
Acknowledgments
We thank Dr. Malin Wikström and Prof. Lars Wieslander for fluorescence microscopy assistance and access, Filippa Stenberg for assistance with sucrose gradients, and Hanna Wieslander for assistance with figures.
This work was supported in part by grants from the Swedish Research Council (to Å. W. and K. E.) and by the European Commission 6th Framework Programme, Marie-Curie Action BIOCONTROL (to Å. W.).
A. Kelly, C. Ge, and Å. Wieslander, unpublished experiments.
- GT
- glycosyltransferase
- alMGS
- monoglucosyldiacylglycerol synthase from A. laidlawii
- alDGS
- diglucosyldiacylglycerol synthase from A. laidlawii
- GlcDAG
- monoglucosyl-diacylglycerol
- TEM
- transmission electron microscopy
- IM
- inner membrane
- OM
- outer membrane
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- CL
- cardiolipin (diphosphatidylglycerol)
- DTT
- dithiothreitol
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- MES
- 4-morpholineethanesulfonic acid
- TEA
- triethanolamine.
REFERENCES
- 1.Liberton M., Howard Berg R., Heuser J., Roth R., Pakrasi H. B. (2006) Protoplasma 227, 129–138 [DOI] [PubMed] [Google Scholar]
- 2.Nevo R., Charuvi D., Shimoni E., Schwarz R., Kaplan A., Ohad I., Reich Z. (2007) EMBO J. 26, 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sener M. K., Olsen J. D., Hunter C. N., Schulten K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15723–15728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner J. H., Lemire B. D., Elmes M. L., Bradley R. D., Scraba D. G. (1984) J. Bacteriol. 158, 590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Meyenburg K., Jørgensen B. B., van Deurs B. (1984) EMBO J. 3, 1791–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arechaga I., Miroux B., Karrasch S., Huijbregts R., de Kruijff B., Runswick M. J., Walker J. E. (2000) FEBS Lett. 482, 215–219 [DOI] [PubMed] [Google Scholar]
- 7.van Weeghel R. P., Keck W., Robillard G. T. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2613–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkison W. O., Walsh J. P., Corless J. M., Bell R. M. (1986) J. Biol. Chem. 261, 9951–9958 [PubMed] [Google Scholar]
- 9.Herskovits A. A., Shimoni E., Minsky A., Bibi E. (2002) J. Cell Biol. 159, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price C. E., Driessen A. J. (2008) J. Biol. Chem. 283, 26921–26927 [DOI] [PubMed] [Google Scholar]
- 11.Armour G. A., Brewer G. J. (1990) FASEB J. 4, 1488–1493 [DOI] [PubMed] [Google Scholar]
- 12.Weber S., Granzow H., Weiland F., Marquardt O. (1996) Virus Genes 12, 5–14 [DOI] [PubMed] [Google Scholar]
- 13.Nieboer M., Vis A. J., Witholt B. (1996) Eur. J. Biochem. 241, 691–696 [DOI] [PubMed] [Google Scholar]
- 14.Allison D. P. (1971) J. Bacteriol. 108, 1390–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bendezú F. O., de Boer P. A. (2008) J. Bacteriol. 190, 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Cock H., Meeldijk J., Overduin P., Verkleij A., Tommassen J. (1989) Biochim. Biophys. Acta 985, 313–319 [DOI] [PubMed] [Google Scholar]
- 17.Ruiz N., Gronenberg L. S., Kahne D., Silhavy T. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigand R. A., Shively J. M., Greenawalt J. W. (1970) J. Bacteriol. 102, 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J. Y., Doody A. M., Chen D. J., Cremona G. H., Shuler M. L., Putnam D., DeLisa M. P. (2008) J. Mol. Biol. 380, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mashburn-Warren L., McLean R. J., Whiteley M. (2008) Geobiology 6, 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drin G., Morello V., Casella J. F., Gounon P., Antonny B. (2008) Science 320, 670–673 [DOI] [PubMed] [Google Scholar]
- 22.Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M. M., Rapoport T. A., Prinz W. A. (2008) Science 319, 1247–1250 [DOI] [PubMed] [Google Scholar]
- 23.McMahon H. T., Gallop J. L. (2005) Nature 438, 590–596 [DOI] [PubMed] [Google Scholar]
- 24.Römer W., Berland L., Chambon V., Gaus K., Windschiegl B., Tenza D., Aly M. R., Fraisier V., Florent J. C., Perrais D., Lamaze C., Raposo G., Steinem C., Sens P., Bassereau P., Johannes L. (2007) Nature 450, 670–675 [DOI] [PubMed] [Google Scholar]
- 25.Campelo F., McMahon H. T., Kozlov M. M. (2008) Biophys. J. 95, 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domanov Y. A., Kinnunen P. K. (2006) Biophys. J. 91, 4427–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamazière A., Burlina F., Wolf C., Chassaing G., Trugnan G., Ayala-Sanmartin J. (2007) PLoS One 2, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morein S., Andersson A., Rilfors L., Lindblom G. (1996) J. Biol. Chem. 271, 6801–6809 [DOI] [PubMed] [Google Scholar]
- 29.Huang K. C., Mukhopadhyay R., Wingreen N. S. (2006) PLoS Comput. Biol. 2, 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay R., Huang K. C., Wingreen N. S. (2008) Biophys. J. 95, 1034–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieslander A., Christiansson A., Rilfors L., Lindblom G. (1980) Biochemistry 19, 3650–3655 [DOI] [PubMed] [Google Scholar]
- 32.Li L., Storm P., Karlsson O. P., Berg S., Wieslander A. (2003) Biochemistry 42, 9677–9686 [DOI] [PubMed] [Google Scholar]
- 33.Eriksson H. M., Persson K., Zhang S., Wieslander K. (2009) Protein Expr. Purif. 66, 143–148 [DOI] [PubMed] [Google Scholar]
- 34.Berg S., Edman M., Li L., Wikström M., Wieslander A. (2001) J. Biol. Chem. 276, 22056–22063 [DOI] [PubMed] [Google Scholar]
- 35.Edman M., Berg S., Storm P., Wikström M., Vikström S., Ohman A., Wieslander A. (2003) J. Biol. Chem. 278, 8420–8428 [DOI] [PubMed] [Google Scholar]
- 36.Wikström M., Kelly A. A., Georgiev A., Eriksson H. M., Klement M. R., Bogdanov M., Dowhan W., Wieslander A. (2009) J. Biol. Chem. 284, 954–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsson O. P., Dahlqvist A., Wieslander A. (1994) J. Biol. Chem. 269, 23484–23490 [PubMed] [Google Scholar]
- 38.Almgren M., Edwards K., Karlsson G. (2000) Colloids Surf. A 174, 3–21 [Google Scholar]
- 39.Ha S., Walker D., Shi Y., Walker S. (2000) Protein Sci. 9, 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pogliano J., Osborne N., Sharp M. D., Abanes-De Mello A., Perez A., Sun Y. L., Pogliano K. (1999) Mol. Microbiol. 31, 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharp M. D., Pogliano K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborn M. J., Gander J. E., Parisi E., Carson J. (1972) J. Biol. Chem. 247, 3962–3972 [PubMed] [Google Scholar]
- 43.Letellier L., Moudden H., Shechter E. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupuy A. D., Engelman D. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2848–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lind J., Rämö T., Klement M. L., Bárány-Wallje E., Epand R. M., Epand R. F., Mäler L., Wieslander A. (2007) Biochemistry 46, 5664–5677 [DOI] [PubMed] [Google Scholar]
- 46.Georgiev A. (2008) Membrane Stress and the Role of GYF Domain Proteins. Ph.D. thesis, Stockholm University, Stockholm [Google Scholar]
- 47.Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 48.Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M., Alzari P. M. (2007) J. Biol. Chem. 282, 20705–20714 [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Fleites C., Proctor M., Roberts S., Bolam D. N., Gilbert H. J., Davies G. J. (2006) Chem. Biol. 13, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 50.Kol S., Nouwen N., Driessen A. J. (2008) J. Biol. Chem. 283, 31269–31273 [DOI] [PubMed] [Google Scholar]
- 51.Vergères G., Yen T. S., Aggeler J., Lausier J., Waskell L. (1993) J. Cell Sci. 106, 249–259 [DOI] [PubMed] [Google Scholar]
- 52.Supply P., Wach A., Thinès-Sempoux D., Goffeau A. (1993) J. Biol. Chem. 268, 19744–19752 [PubMed] [Google Scholar]
- 53.Vonck J., Schäfer E. (2009) Biochim. Biophys. Acta 1793, 117–124 [DOI] [PubMed] [Google Scholar]
- 54.Lingwood D., Schuck S., Ferguson C., Gerl M. J., Simons K. (2009) J. Biol. Chem. 284, 12041–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van den Brink-van der Laan E., Boots J. W., Spelbrink R. E., Kool G. M., Breukink E., Killian J. A., de Kruijff B. (2003) J. Bacteriol. 185, 3773–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Picot D., Loll P. J., Garavito R. M. (1994) Nature 367, 243–249 [DOI] [PubMed] [Google Scholar]
- 57.Selstam E. (1998) Lipids in Photosynthesis: Structure, Function and Genetics (Siegenthaler P. A., Murata N. eds) pp. 209–224, Kluwer Academic Publishers Group, Dordrecht, The Netherlands [Google Scholar]
- 58.Reinbothe C., Lebedev N., Reinbothe S. (1999) Nature 397, 80–84 [Google Scholar]
- 59.Epand R. F., Tokarska-Schlattner M., Schlattner U., Wallimann T., Epand R. M. (2007) J. Mol. Biol. 365, 968–980 [DOI] [PubMed] [Google Scholar]
- 60.Marius P., Zagnoni M., Sandison M. E., East J. M., Morgan H., Lee A. G. (2008) Biophys. J. 94, 1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim A. C., Oliver D. C., Paetzel M. (2008) J. Mol. Biol. 376, 352–366 [DOI] [PubMed] [Google Scholar]
- 62.Yeh J. I., Chinte U., Du S. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3280–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balali-Mood K., Bond P. J., Sansom M. S. (2009) Biochemistry 48, 2135–2145 [DOI] [PubMed] [Google Scholar]
- 64.Palsdottir H., Hunte C. (2004) Biochim. Biophys. Acta 1666, 2–18 [DOI] [PubMed] [Google Scholar]
- 65.He J., Vora M., Haney R. M., Filonov G. S., Musselman C. A., Burd C. G., Kutateladze A. G., Verkhusha V. V., Stahelin R. V., Kutateladze T. G. (2009) Proteins Struct. Funct. Bioinformat. 76, 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawai C., Pessoto F. S., Rodrigues T., Mugnol K. C., Tórtora V., Castro L., Milícchio V. A., Tersariol I. L., Di Mascio P., Radi R., Carmona-Ribeiro A. M., Nantes I. L. (2009) Biochemistry 48, 8335–8342 [DOI] [PubMed] [Google Scholar]
- 67.McNaughton B. R., Cronican J. J., Thompson D. B., Liu D. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6111–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christiansson A., Kuypers F. A., Roelofsen B., Op den Kamp J. A., van Deenen L. L. M. (1985) J. Cell Biol. 101, 1455–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheetz M. P., Singer S. J. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4457–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Svetina S., Zeks B. (2001) Cell. Mol. Biol. Lett. 6, 305–311 [PubMed] [Google Scholar]
- 71.Jackson B. J., Gennity J. M., Kennedy E. P. (1986) J. Biol. Chem. 261, 13464–13468 [PubMed] [Google Scholar]
- 72.Linde K., Gröbner G., Rilfors L. (2004) FEBS Lett. 575, 77–80 [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y. M., Rock C. O. (2008) Nat. Rev. Microbiol. 6, 222–233 [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y. M., Rock C. O. (2008) J. Lipid Res. 49, 1867–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Epand R. M., Rotem S., Mor A., Berno B., Epand R. F. (2008) J. Am. Chem. Soc. 130, 14346–14352 [DOI] [PubMed] [Google Scholar]
- 76.Sturgis J. N., Tucker J. D., Olsen J. D., Hunter C. N., Niederman R. A. (2009) Biochemistry 48, 3679–3698 [DOI] [PubMed] [Google Scholar]
- 77.Zimmerberg J., Kozlov M. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 9–19 [DOI] [PubMed] [Google Scholar]
- 78.Wan S., Coveney P. V. (2009) J. Comput. Chem. 30, 1038–1050 [DOI] [PubMed] [Google Scholar]
- 79.Ren G., Vajjhala P., Lee J. S., Winsor B., Munn A. L. (2006) Microbiol. Mol. Biol. Rev. 70, 37–120 [DOI] [PMC free article] [PubMed] [Google Scholar]