Abstract
Hepatitis C virus core protein is the viral nucleocapsid of hepatitis C virus. Interaction of core with cellular membranes like endoplasmic reticulum (ER) and lipid droplets (LD) appears to be involved in viral assembly. However, how these interactions with different cellular membranes are regulated is not well understood. In this study, we investigated how palmitoylation, a post-translational protein modification, can modulate the targeting of core to cellular membranes. We show that core is palmitoylated at cysteine 172, which is adjacent to the transmembrane domain at the C-terminal end of core. Site-specific mutagenesis of residue Cys172 showed that palmitoylation is not involved in the maturation process carried out by the signal peptide peptidase or in the targeting of core to LD. However, palmitoylation was shown to be important for core association with smooth ER membranes and ER closely surrounding LDs. Finally, we demonstrate that mutation of residue Cys172 in the J6/JFH1 virus genome clearly impairs virion production.
Introduction
Hepatitis C virus (HCV)2 is a major causative agent of chronic hepatitis (1). HCV is an RNA virus of the Flaviviridae family and has a single-stranded, positive sense RNA genome of 9.6 kb (2). The HCV RNA genome encodes a polyprotein of ∼3000 amino acids (aa) that is processed by host and viral proteases into 10 different components (3). Core protein is the only virus-encoded nucleocapsid protein involved in assembly and packaging of the viral plus-strand RNA genome (3). The C-terminal signal sequence (aa 173–191) facilitates channeling of the nascent HCV polyprotein to the endoplasmic reticulum (ER) (4). After cleavage, core protein (191 aa) is released and further processed by an intramembrane protease, the signal peptide peptidase (spp), to yield a protein of 177 aa (5, 6). The fully processed core protein interacts mainly with lipid droplets (LD) and ER membranes and was also reported to be translocated into the nucleus (7–9). The C-terminal part of core (aa 120–191) includes a predicted amphipathic α-helix that is responsible for core association with LD and ER membranes (8, 10). Recent studies have indicated that assembly of HCV particles occurs on ER membranes that are associated closely with LD (11). Core protein on LD recruits the viral proteins of the replication complex and is translocated to ER-associated membranes where it interacts with HCV RNA to produce assembled viral particles (11). To facilitate HCV assembly, core protein also promotes LD accumulation when expressed in cells (12, 13). HCV core protein as well as the replication complex are also found in the detergent-resistant membrane (DRM) fraction, which is distinct from the classical lipid rafts (14–16).
Because HCV core is targeted to different organelle membranes during the viral life cycle, we investigated whether post-translational modification of core in the form of palmitoylation could be involved in this trafficking. Palmitoylation or S-acylation is the covalent attachment of a fatty acid group, usually the saturated 16 carbon palmitate to cysteine residues via a thioester bond (17). Protein palmitoylation enhances surface hydrophobicity and membrane affinity and plays an important role in modulating protein trafficking (18).
In this study, we identified a site for palmitoylation in HCV core protein. The cysteine modified by a palmitate moiety is adjacent to the ER-targeting domain located at the C terminus. We demonstrate that spp processing and LD targeting was unaffected by impairment of this palmitoylation generated by mutation. However, mutated HCV core protein accumulated differently in the ER membrane and was poorly associated with ER surrounding LD. It is of high importance that we observed that a mutation in the palmitoylation site of HCV core impaired viral infectivity.
EXPERIMENTAL PROCEDURES
Construction of pPIC3.5Kcore, pcDNA3.1core, and J6/JFH1 Mutants
Clones of Pichia pastoris expressing HCV core C (aa 1–191) of strain H77 (genotype 1a) and spp, a signal peptide peptidase of human cell origin, have been described previously (19). Mutations were introduced by PCR and conventional cloning methods. Cys172 and Cys91 were replaced by Ser and Leu, respectively. Yeast cells were transformed, and protein expression was induced with methanol as described previously (20). The coding sequences of the HCV core were inserted into the polylinker site of pcDNA3.1 (Invitrogen) with BamHI and EcoRI restriction sites (New England BioLabs). Vaccinia virus expressing HCV core-E1 protein (Sc59 6C/Ss) was kindly provided by Chiron (Emeryville, CA). Vaccinia Ankara strain expressing T7 polymerase was generously provided by Bernard Moss (NIAID, National Institutes of Health, Bethesda, MD). The plasmid FL-J6/JFH-5′C19Rluc2AUbi, which consists of the full-length HCV genome and expresses Renilla luciferase, was kindly provided by Charles M. Rice (The Rockefeller University, New York, NY). The substitution of Cys for Ser in J6 core protein was introduced by PCR and cloned into the J6/JFH1 clone as a BglII/BsiWI fragment. All of the plasmid HCV sequences were verified by sequencing.
Biotin Switch Assay to Detect Palmitoylation
Palmitoylation of core was examined by a recently developed biotin switch assay technique (21). In this protocol, acylation groups attached to cysteine residues via thioester bonds are replaced with biotin moieties. Yeast cell cultures (200 ml) expressing spp or co-expressing spp and core proteins were induced as previously described (20). The cells were collected and ground to a fine powder in liquid nitrogen. For analysis of human hepatoma cells, Huh7.5 cells were infected with vaccinia virus (C-E1). After 24 h, the cells were washed with phosphate-buffered saline (PBS) and harvested using a rubber policeman. The cells were recovered by centrifugation and frozen at −80 °C. The samples were resuspended in lysis buffer (150 mm NaCl, 5 mm EDTA, 1× complete protease inhibitors (Roche Applied Science), 50 mm Tris, pH 7.4) containing 10 mm N-ethylmaleimide (Pierce) and 1.7% Triton X-100. The samples were incubated for 1 h at 4 °C followed by centrifugation at 4 °C at 500 × g to remove insoluble material. The proteins were precipitated with methanol/chloroform, and the air-dried pellet was resuspended in 3.6 ml of SDS buffer (1% SDS, 100 mm NaCl, 0.2% Triton X-100, 50 mm Tris·HCl, pH 7.4) and incubated overnight at 4 °C with 5 mm N-ethylmaleimide. The proteins were precipitated three times with methanol/chloroform and resuspended in 1.5 ml of SDS buffer to be further divided into two equal aliquots. One aliquot was combined with 2.9 ml of 0.7 m fresh hydroxylamine, 1× complete protease inhibitors, 0.2% Triton X-100, and 1 mm biotin-N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (HPDP) (Thermo Scientific). As a control, the remaining aliquot was treated using the same procedure, but hydroxylamine was replaced with 50 mm Tris, pH 7.4. Both samples were incubated at room temperature for 1 h. The proteins were precipitated using methanol and chloroform and resuspended in 1.2 ml of biotin buffer (0.4% SDS, 120 mm NaCl, 0.16% Triton X-100, 5 mm EDTA, 0.16 mm biotin-HPDP 1× inhibitor mixture, 50 mm Tris·HCl, pH 7.4) and incubated for 1 h at room temperature. The proteins were precipitated three times using the same procedure and resuspended in 2.5 ml of lysis buffer containing 0.2% Triton X-100 and 0.1% SDS. The samples were centrifuged at 15,000 × g for 1 min, and aliquots were removed as controls. The remaining reactions were incubated with 15 μl of neutravidin-agarose beads (Thermo Scientific) at room temperature for 90 min. The beads were washed four times with lysis buffer containing 0.2% Triton X-100 and 0.1% SDS, and the proteins were eluted by adding 1% β-mercaptoethanol to the washing buffer and incubating for 15 min at 37 °C. The samples were centrifuged at 15,000 × g for 1 min, and aliquots were removed as controls (22).
[3H]Palmitic Acid Labeling
Yeast cultures were labeled with [3H]palmitic acid as previously described (23). P. pastoris was cultured overnight in 200 ml of minimal glucose medium (Invitrogen) at 30 °C to an optical density of 2 and, after a short centrifugation, transferred to 20 ml of minimal glucose medium + 0.1% methanol (Invitrogen) for 6 h in the presence of the fatty acid synthesis inhibitor cerulenin (2 mg/ml). In some cell cultures, 50 μm 2-bromopalmitate (2-BP) or Me2SO were also added. The cell cultures were then incubated for 2 h with 1 mCi of [9,10-3H(N)]palmitic acid (Amersham Biosciences) (50 Ci/mmol). The cells were disrupted with sarkosyl (0.5%), and the proteins were denatured with 1% SDS. The protein samples were resolved on nonreducing 10% SDS-PAGE. The gel was blotted onto a nitrocellulose filter and exposed to a [3H] intensifying screen for 7 days. The signals were revealed using phosphorimaging. The blot was then analyzed further by Western blotting using anti-core antibodies 537 (19). The Huh7.5 cells labeling with [3H]palmitic acid was performed as previously described (24). The cells were infected with vaccinia virus and vaccinia virus expressing HCV core protein and incubated overnight in medium (Dulbecco's modified Eagle's medium, 2% fetal bovine serum, nonessential amino acids, 5 mm sodium pyruvate) supplemented with 37 μCi/ml of [9,10-3H(N)]palmitic acid (PerkinElmer). The cells were washed with PBS and lysed in radioimmune precipitation assay buffer (50 mm Tris·HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× complete protease inhibitors). The cell lysate was resolved in a nonreducing SDS-PAGE and analyzed with phosphorimaging.
Flotation Assay
The flotation assay was carried out as previously described (25). Two hours after infection of Huh7.5 cells with vaccinia virus carrying the T7 polymerase gene, the cells were transfected with pcDNA3.1 constructs using FuGENE 6 transfection reagent (Roche Applied Science). In some cases, 50 μm of 2-BP was added to the medium. Twenty-four hours after transfection, the cells were washed with ice-cold PBS and then harvested using a rubber policeman. The collected cells were suspended in 0.6 ml of TNEi buffer (150 mm NaCl, 2 mm EDTA, 1× complete protease inhibitors, 50 mm Tris, pH 7.4), homogenized with a Dounce homogenizer and then resuspended using a 25-gauge needle. The samples were split into two equal portions, and each was incubated for 30 min on ice with or without 1% Triton X-100. The lysates were mixed with 0.6 ml of Optiprep (Sigma) to a final concentration of 40% iodixanol. This mixture was overlaid with 2.9 ml of 30% iodixanol and 400 μl of TNEi and then centrifuged at 40,000 rpm, 4 °C for 4 h in a SW60ti rotor (Beckman Coulter, Fullerton, CA). The fractions (0.4 ml) were collected from the top of the centrifuging tube and then precipitated with 4 volumes of cold acetone. The pellets were resuspended in loading buffer, boiled, and subjected to SDS-PAGE and Western blotting. The proteins were revealed with anti-core, anti-caveolin-1 (Sigma), and anti-calnexin (Sigma) antibodies.
Immunofluorescence Microscopy
Huh7.5 cells were transfected with pcDNA3.1 plasmids using FuGENE 6 transfection reagent and grown on glass coverslips. Two days after transfection, the cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature followed by 15 min in 0.1% Triton X-100 in PBS. Primary antibodies (anti-core 537) were diluted in 5% bovine serum albumin and incubated with cells for 2 h at room temperature. After three washes in PBS, Alexa fluor 488 goat anti-rabbit IgG (Invitrogen) were added to cells at a 1:200 dilution for 1 h at room temperature. To stain lipids, the slides were rinsed in 60% isopropanol, incubated in 5 mg/ml oil red O (Fisher Scientific, Pittsburgh, PA), in 60% isopropanol for 2 min at room temperature, and rinsed in 60% isopropanol again. After staining, the slides were washed in PBS and mounted in ProLong Antifade (Molecular Probes). Microscopy was performed using a Zeiss confocal laser scanning microscope LSM 510, and the images were captured with a Nikon Eclipse TE 300. Image analysis of LD content was performed using Cell profiler software (26) on 35 micrographs for each sample. The area occupied by LD was evaluated for each positive cell.
ER Extraction and Fractionation
For extraction of ER membranes, Huh7.5 cells infected with vaccinia T7 polymerase and transfected for 24 h with pCDNA3.1 expressing core or core mutant C172S were washed with ice-cold phosphate-buffered saline (PBS), scraped into homogenization buffer (250 mm sucrose, 1 mm EDTA, 1× complete protease inhibitors, 10 mm Hepes-NaOH, pH 7.4) to be disrupted by Dounce homogenization and repeated passages through a fine syringe needle. The extract was spun down at 1,500 × g for 10 min, and the supernatant was further centrifuged at 150,000 × g for 1 h in a 70.1ti rotor (Beckman Coulter, Fullerton, CA). The resulting pellet, representing the membrane fraction, was resuspended in homogenization buffer and layered (0.5 ml) on top of an iodixanol step gradient composed of 0.5 ml of 10%, 0.5 ml of 15%, 1 ml of 20%, 0.5 ml of 25%, and 1 ml of 30% iodixanol and centrifuged at 200,000 × g at 4 °C for 2.5 h in a SW60Ti rotor (Beckman Coulter). Fractions (0.5 ml) were collected from top to bottom, and the fraction proteins were analyzed by SDS-PAGE and immunoblotting using anti-core 537 and anti-calnexin (Sigma) antibodies.
Immunoelectron Microscopy
Yeast cells were fixed as described in Ref. 27 with 3% paraformaldehyde containing 0.2% glutaraldehyde in 0.1 m phosphate buffer, pH 7.2, at room temperature for 2 h and then washed well with 0.1 m phosphate buffer, pH 7.2. Fixed cells were treated with 1% metaperiodate for 30 min followed by 30 min of incubation with 50 mm NH4Cl/PO4, pH 7.2. After washing, the cells were embedded in LR white. Ultrathin sections were incubated with anti-core 537 (1:2000) and then with gold-labeled anti-rabbit IgG (Amersham Biosciences). The ultrathin sections were stained with uranylacetate and lead citrate and analyzed with a JEOL 1010 80-kV transmission electron microscope. Image analysis of the LD content was performed with cell profiler software (26) on 20 micrographs taken at low magnification (10,000× and 12,000×), which included over 300 cells for each clone. The area occupied by LD on the micrograph was normalized to the area occupied by all the cells on the same micrograph.
Production of Infectious HCV and Infection of Huh7.5 Cells
The plasmid FL-J6/JFH-5′C19Rluc2AUbi wt and mutated (C172S) were linearized with XbaI and treated with mung bean nuclease (New England Biolabs) to yield the exact HCV 3′ end (28). In vitro transcription was performed with a T7 RiboMAX express large scale RNA production system (Promega) following the manufacturer's instructions. Subconfluent Huh7.5 cells were trypsinized, washed twice with ice-cold OptiMEM (Invitrogen), and resuspended in OptiMEM at 10 × 106/ml. Four hundred microliter of cells were mixed with 10 μg of in vitro transcribed RNA or not (mock) in a cuvette with a gap width of 0.4 cm (Bio-Rad) and immediately pulsed at 960 farads and 270 V using a Gene Pulser system (Bio-Rad). Electroporated cells were transferred in 4 ml of complete medium, and 300 μl were cultured for 4 or 72 h in a 24-well plate prior to be lysed in Renilla lysis buffer (Promega). For infection, 5 × 104 naïve Huh7.5 cells, cultured in 24-well plate, were inoculated overnight by 250 μl of filtered supernatant harvested 3 days post-electroporation. After being washed twice in PBS, infected cells were left in culture for 48 h additional and lysed in Renilla lysis buffer. Luciferase activity was measured for 10 s, using a luminometer.
Quantitative Detection of HCV RNA by Quantitative Reverse Transcription-PCR
Viral RNA was isolated from 50 μl of supernatants harvested 3 days post-electroporation using the MagMAX-96 viral RNA isolation kit as recommended by the manufacturer (Ambion, Austin, TX). 7.5 μl of sample were used for quantitative reverse transcription-PCR analysis employing an Applied Biosystems 7500 sequence detection system (Applied Biosystems, Foster City, CA). Amplifications were conducted in duplicate with the TaqMan RNA-to-Ct 1 step as described by the manufacturer (Applied Biosystems) using 0.5 μm of primers amplified a conserved segment of the 5′-untranslated region of HCV genotype 2a (reverse, 5′-GAGTGGGTTTATCCAAGAAAG-3′, and forward, 5′-TCTGCGGAACCGGTGAGT-3′) and 0.2 μm of the TaqMan probe HCV 2a (5′-FAM-CCGGAATTGCCGGGAAGACTG-BHQ-1-3′) (Biosearch Technologies, Novato, CA). The amounts of HCV RNA were calculated by comparison with serially diluted in vitro transcripts purified as described above. This assay was linear between 107 and 101 RNA copies/microliter.
Statistical Analysis
For the data depicted in Figs. 9 and 11, statistical significance between groups was determined by analysis of variance. Calculations were made with Prism version 3.03 software. p values < 0.05 were considered statistically significant. The statistical significance of the results was defined by performing a one-way analysis of variance combined with Turkey's post tests to compare all pairs of columns.
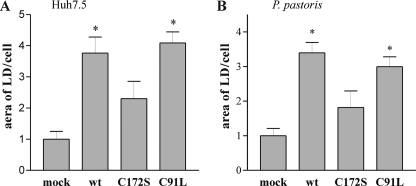
FIGURE 9.
Effect of HCV core expression on LD accumulation in Huh7.5 cells (A) and P. pastoris (B). A, after immunostaining of Huh7.5 cells with anti-core antibodies and LD coloration with Red Oil, the images (35 micrographs/sample) were analyzed for LD content using Cell profiler software (25). B, EM micrographs of P. pastoris cells expressing spp and co-expressing spp and HCV core, C172S and C91L were analyzed with Cell profiler software to determine the area occupied by LD/cell. 20 micrographs for each sample, representing ∼300 cells in total, were used for the analysis. The values were normalized, with mock cells assigned a value of 1. The asterisks denote significant differences versus mock cells (p < 0.05).
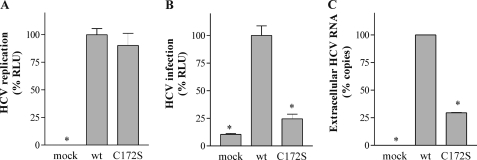
FIGURE 11.
Effect of Cys172 mutations on HCV infectivity. A, Huh7.5 cells were transfected in parallel with RNA transcripts from J6/JFH1 and J6/JFH1-C172S. Replication was assayed by luciferase activity at 72 h post-electroporation. B, infectious virus production at 72 h post-electroporation was also assayed by luciferase activity 3 days post-infection. C, detection by quantitative reverse transcription-PCR of the total viral RNA released 72 h after electroporation of the transcripts transfected in A. The means and S.E. of data from quadruplicate of three different electroporations are shown. The values were normalized on the J6/JFH1 virus data. The asterisks denote significant differences versus J6/JFH1 (wt) infected cells (p < 0.05).
RESULTS
HCV Core Is Palmitoylated
Upon cleavage of HCV polyprotein by signal peptidase, HCV core stays anchored to ER membranes through its C-terminal transmembrane domain. A subsequent maturation event of core meditated by spp cleaves this hydrophobic domain to produce the mature core, which is predicted by algorithm for amphiphilicity index (29) to be a soluble protein. However, most of the processed core remains tightly associated to intracellular membranes (30). This association depends on the integrity of an amphipathic α-helix spanning aa 116–134 (8). In this study, we investigated whether the affinity of core to membranes was enhanced by post-translational modification such as S-acylation. Although there is no unique canonical motif for acylation sites, the computer algorithm CSS-Palm 2.0 predicts potential palmitoylation sites by clustering and scoring known palmitoylated proteins (31). This algorithm was applied to the sequence of the mature HCV core protein genotype 1, which contains three cysteine residues (i.e. Cys91, Cys128, and Cys172). The program highlights a possible palmitoylation site at Cys172 using a threshold for prediction corresponding to 89% accuracy and 94% specificity (Table 1). The palmitoylation site is predicted only for the 177-aa core that has been processed by spp. Palmitoylation of Cys172 is not predicted in the context of the HCV polyprotein, when core protein is still fused to E1, or in its immature form comprising 191 aa that is generated upon signal peptidase cleavage.
TABLE 1.
Palmitoylation site prediction in HCV core protein
The amino sequence of the mature core protein was analyzed for palmitoylation site probability using the updated software CSS-Palm (28). Score 1 was the result obtained with core protein in the context of the HCV polyprotein (unprocessed core protein (191 aa) gave the same results; not shown). Score 2 reflects the result for the form of core protein processed by spp. The cut-off with low threshold is 0.6 (accuracy, 76%; specificity, 75%) and with high threshold is 1 (accuracy, 89%; specificity, 94%).
| Position | Peptide | Score 1 | Score 2 |
|---|---|---|---|
| 91 | NEGCGWA | 0.139 | 0.139 |
| 128 | TLTCGFA | 0.052 | 0.052 |
| 172 | LPGCSFS | 0.417 | 1.122a |
a Only Cys172 in the processed form of core has a score indicating a high likelihood of palmitoylation.
To determine whether HCV core protein is indeed modified by the addition of a lipid moiety (e.g. palmitoylation), we measured its endogenous acylation state using the biotin switch assay. This method relies on the exchange of thioester-linked protein acyl modifications for biotin moieties (21). HCV core (191 aa) was co-expressed with human spp and extracted from P. pastoris as described previously (20). As a first step, free sulfhydryl groups of proteins were blocked by incubation with N-ethylmaleimide. Then fatty acid groups were specifically removed using hydroxylamine to generate a free cysteine residue at the palmitoylation site. The newly exposed sulfhydryl groups were labeled with thiol-reactive biotin. The proteins were then precipitated with neutravidin and analyzed by Western blotting. Incomplete blockage of the cysteine residue prior to acyl group exchange could lead to false positive results. To exclude this possibility, hydroxylamine was omitted and replaced by Tris in half the samples. These control samples should give negative signals if the protein has been adequately blocked by N-ethylmaleimide. As shown in Fig. 1A, biotinylated core protein was present in the hydroxylamine-treated sample and absent from the nonhydroxylamine-treated sample. Prior to extraction, yeast cell samples were also incubated for 24 h with the palmitoylation inhibitor 2-BP. As shown in Fig. 1A, for the same amount of initial core protein, 2-BP reduced the amount of biotinylated protein recovered in the neutravidin-purified samples in a dose-dependent manner.
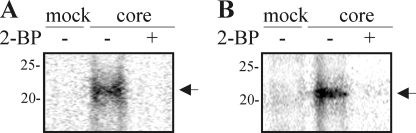
FIGURE 1.
Detection of acylation of the HCV core protein by biotin switch assay. A, total cell lysates from yeast P. pastoris expressing spp (mock) or expressing spp and core were prepared and treated with N-ethylmaleimide to block free cysteines. The protein extracts were split in half and treated (+) or not (−) with hydroxylamine (NH2OH) to remove lipid moieties. The newly exposed cysteine residues were then biotinylated with biotin-HPDP. The biotinylated proteins (acylated proteins) were precipitated with neutravidin-agarose beads for subsequent immunoblotting analysis with anti-core antibodies. An inhibitor of palmitoylation, 2-BP (I), was added to the growth medium during induction at final concentrations of 100 (I100) or 400 (I400) μm. + and − above the lanes indicate hydroxylamine-treated (+) or untreated (−) protein, and neutravidin precipitation pellet (+) or total extract (−). B, Huh7.5 cells were infected with vaccinia virus alone (mock) or with vaccinia virus expressing HCV core protein and harvested after 24 h. The proteins were extracted and subjected to the biotin switch assay as described for A. Acylated proteins were analyzed by Western blot using anti-core antibodies.
To verify whether this modification occurs also in human cells, the biotin switch assay was performed on protein extracts of hepatocytes expressing HCV core (Fig. 1B). Huh7.5 cells were infected with vaccinia virus expressing HCV protein from 1 to 382 aa (C-E1). In this construct, the core protein is cleaved first with signal peptidase followed by spp maturation. Biotinylated core protein was present in the protein sample treated with hydroxylamine and extracted with neutravidin. These results indicate that HCV core protein is also an acylated protein in Huh7.5 cells.
To confirm the observations made with the biotin switch experiments, we performed a metabolic labeling assay with radioactive palmitate. P. pastoris expressing spp alone or co-expressing core and spp were grown in minimal medium supplemented with [3H]palmitate for 2 h. As well, Huh7.5 cells expressing core protein were also incubated with [3H]palmitate for 24 h. The proteins were extracted and resolved on SDS-PAGE under nonreducing conditions. A radioactive band of 21 kDa was present in cells expressing core but absent in mock transfected cells or cells incubated with 2-BP and that for yeast extracts (Fig. 2A) or the hepatocytes samples (Fig. 2B). Western blot analysis of these samples revealed that these protein bands corresponded to core protein (data not shown). These results confirmed that HCV core is indeed a palmitoylated protein.
FIGURE 2.
Palmitoylation of HCV core protein. A, P. pastoris cells expressing spp (mock) or co-expressing core and spp were induced in medium containing [3H]palmitic acid (50 Ci/mmol), supplemented in some cases with 2-BP (50 μm). B, Huh7.5 cells were infected with vaccinia virus alone (mock) or with vaccinia virus expressing HCV core protein in a medium supplemented with [3H]palmitic acid (37 μCi/ml) and, when mentioned, with 2-BP (25 μm). The cells were disrupted in nonreducing buffer. The samples were resolved on nonreducing SDS-polyacrylamide gels. The gel was blotted onto a nitrocellulose filter and exposed to a [3H] intensifying screen for 7 days. The bands were revealed using phosphorimaging. The arrow shows the position of the core protein revealed by Western blotting with anti-core antibodies. Molecular markers (Bio-Rad) are indicated on the left in kDa.
Identification of Cysteine Residue Involved in HCV Core Palmitoylation
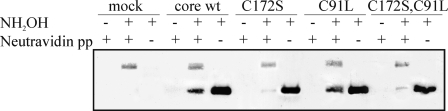
Cysteines Cys128 and Cys172 in core protein are well conserved among the sequences of different HCV genotypes. Only a JFH1 strain of genotype 2 shows a phenylalanine at position 172. Cys91 is not conserved in the core sequences of the different genotypes and is often replaced by Leu or Met (supplemental Table S1). To identify the cysteine involved in palmitoylation of core, we mutated Cys91 or Cys172 to Leu or Ser, respectively, to abolish putative acylation reaction at these sites. Residue Cys128 was mutated to Ala or Ser. Mutations of Cys128 generated a protein that was unstable in both P. pastoris and human cells and barely detectable by Western blot. Therefore, we excluded the Cys128 mutated clones from our investigation. The biotin switch test was performed on P. pastoris cells expressing core mutants. As seen in Fig. 3, C172S mutations reduced the amount of biotinylated core present in the neutravidin-purified samples, although the amount of core protein was essentially the same in all of the samples before affinity extraction. Mutated protein C91L reacted differently from the C172S mutation; the level of protein purified from the biotinylated C91L sample was similar to that of the wt protein sample. These results suggest that Cys172 of core is the major site of palmitoylation.
FIGURE 3.
Identification of the palmitoylated cysteine of HCV core by biotin switch assay. Total cell lysates from yeast P. pastoris expressing spp (mock) or expressing spp and mutant core protein were subjected to the biotin switch assay as described for Fig. 2. + and − above the lanes indicate hydroxylamine-treated (+) or untreated (−) protein and neutravidin precipitation pellet (+) or total extract (−). The samples assayed included core wt, core with C172S or C91L mutations and the double mutant C172S,C91L. Palmitoylated proteins were analyzed by Western blot using anti-core antibodies.
Effect of Cys172 Mutation on spp Cleavage
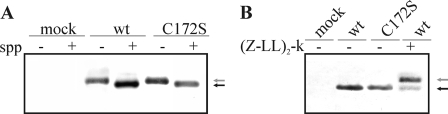
Processing of HCV core protein by spp is regulated by the C terminus of the core as well as by other internal sequences known to stabilize the protein at ER membranes (32, 33). To determine whether mutation of Cys172 could affect processing of core by spp, we expressed these proteins in P. pastoris in the presence or absence of human spp expression. In these cells, maturation of core by the resident yeast spp has previously been shown to be inefficient, but maturation of core can be brought to completion when the human spp is co-expressed with HCV core (20). As shown in Fig. 4A, wt core protein extracted from yeast co-expressing human spp migrated faster than the immature form. When residue Cys172 was mutated to Ser, similar results were obtained, suggesting that maturation of core was not influenced by palmitoylation of the protein. To confirm this result in human hepatocytes where resident spp can cleave efficiently core protein, we analyzed maturation of our constructs in the presence or absence of (Z-LL)2-ketone, an spp inhibitor. We observed that C172S mutant proteins migrated similarly to wt protein; unprocessed protein was not detected with the core protein sample in the absence of the inhibitor (Fig. 4B). From these results we concluded that palmitoylation of core at Cys172 is not required for maturation by spp.
FIGURE 4.
Effect of the Cys172 mutation on processing of core by spp. A, twenty-four hours after methanol induction, yeast cells expressing core and the C172S mutant protein with or without (mock) co-expressing spp were lysed. Protein extracts were separated on SDS-PAGE and revealed by Western blotting using anti-core antibodies. B, Huh7.5 cells were transfected with pcDNA3.1 plasmid DNA encoding core protein or the C172S mutant 24 h before protein extraction. Where indicated, hepatocytes were supplemented with 10 μm of (Z-LL)2-Ketone. Core proteins were analyzed by Western blot using anti-core antibodies. The arrows on the right of each panel indicate the unprocessed (gray) and mature (black) forms of the core protein.
Effect of Cys172 Mutation on Targeting to DRMs
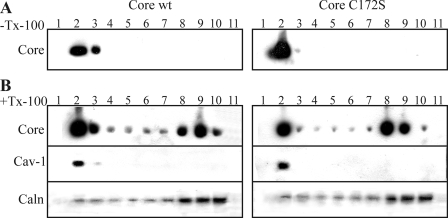
Recent studies have shown that HCV core is associated with DRMs or lipid rafts (6, 14, 30). DRM targeting has been recognized as one of the main functions of palmitoylation of proteins (18). DRMs are specialized membrane subdomains that are resistant to solubilization by cold nonionic detergents such as Triton X-100 (25). Therefore, we examined whether palmitoylation is responsible for localization of the HCV core protein to DRM. Wild type or mutated forms of C172S core proteins were expressed in Huh7.5 cells, solubilized at 4 °C in the presence or absence of 1% Triton X-100, and subjected to a flotation centrifugation assay. The fractions were collected from the top of the tube and analyzed by Western blotting. In the absence of detergent, the majority of the wild type and the mutated core were found in the membrane-containing fractions (fractions 2 and 3) (Fig. 5A). When treated with Triton X-100, a considerable amount of wt core protein remained associated with the DRMs (fraction 2), whereas the rest migrated to the bottom of the gradient, which corresponded to the detergent-soluble fraction (Fig. 5B). As markers for lipid rafts and ER, we identified the location in the gradient of caveolin-1 and calnexin by Western blot using specific antibodies on sample taken from the flotation assay (Fig. 5B). The pattern of the mutated protein C172S in the flotation assay was similar to that of wt core. C172S mutant core protein was also detected with DRMs and in the detergent-soluble fraction after Triton extraction. We concluded that palmitoylation of the Cys172 residue is not essential for association of the core to DRMs.
FIGURE 5.
Association of HCV core and C172S mutant proteins with DRMs. Huh7.5 cells expressing core or C172S were lysed, and the aliquots were incubated at 4 °C in the absence (A) or presence (B) of 1% Triton X-100 (Tx-100). The lysates were mixed with Optiprep to a final concentration of 40% iodixanol and then overlaid with 2.9 ml of 30% iodixanol and 400 μl of TNEi buffer. Following ultracentrifugation, the fractions were collected from the top of the tube, and the proteins were analyzed by Western blotting using anti-core, anti-caveolin-1 (cav-1), or anti-calnexin (caln) antibodies.
Effect of Cys172 Mutation on HCV Core Cellular Localization
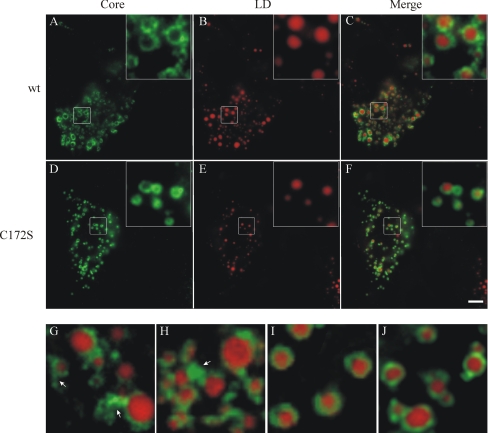
In hepatocytes, core is localized mainly on the surface of LD and on the ER surrounding LD (11). To examine the effect of palmitoylation of HCV core on LD association, Huh7.5 cells were transfected with plasmid pcDNA3.1 expressing wt core or the mutated form C172S. The hepatocytes were immunolabeled with anti-core antibodies, and LDs were revealed by Red Oil O staining. As shown in Fig. 6, C172S mutant core proteins were organized in ring-like structures around the LDs. At higher magnification, we noted that C172S protein was distributed evenly around the LDs with a smooth ring surface (Fig. 6, I and J), in 1contrast to the wt proteins, which showed irregular circle structures with dots and extra material on the surface of the ring (Fig. 6, G and H). It has been described before that core protein triggers ER membrane association to LDs with irregular distribution around the surface of the organelles (11, 34). Core protein was also detected with these ER membranes (11, 34). The difference in pattern distribution between the C172S mutant and the wt protein may suggest that C172S protein is less present on ER membranes that are closely associated with LDs.
FIGURE 6.
Immunofluorescence analysis of the intracellular distribution of HCV core. Huh7.5 cells were grown on coverslips and transfected with pcDNA3.1/core (A–C, G, and H) or pcDNA3.1/C172S (D–F, I, and J) using FuGENE 6 transfection reagent. Two days after transfection, the cells were stained with anti-core antibodies. Lipid droplets were stained with Red Oil O after immunostaining. Bar, 5 μm. Insets and G–J show magnified images. The arrows indicate ER membranes associated to LD.
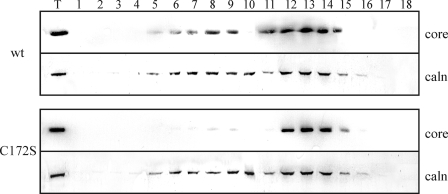
To analyze the distribution of C172S mutant protein to ER membranes, microsomal membranes were purified from extracts of Huh7.5 cells transfected with pcDNA3.1 plasmid expressing wt core or the mutated C172S protein and analyzed by Western blotting using anti-core antibodies. We did not detect any difference between the amount of wt and C172S protein present in fractions after ER extraction (Fig. 7, lane T). However, during ER purification, LD fractions are discarded by ultracentrifugation, possibly with the ER membranes closely associated with LDs. The ER fractions were further analyzed by density gradient fractionation. Interestingly, we noted a difference in core association with ER between the C172S and the wt protein. The wt protein was associated with the ER dense fraction (rough ER) as well as with the lighter ER membrane fraction (smooth ER). Core C172S protein was present only with dense ER membranes. It has been shown that ER membranes closely associated with LDs are free of ribosomes (35). So, according to the results of our density fractionation experiment, which showed that the C172S protein has less affinity for smooth ER, we expected that C172S protein to be absent from the smooth ER associated with LD.
FIGURE 7.
Isolation and fractionation of ER from Huh7.5 cells expressing HCV core and C172S mutant proteins. ER membranes from Huh7.5 cells were isolated by ultracentrifugation and then fractionated by 10–30% discontinuous iodixanol gradients. Lanes 1–18, fractions (0.5 ml) were collected from the top of the tube and analyzed by Western blotting using an antibody against HCV core protein and calnexin (caln). Lane T represents the total protein fraction before the Optiprep gradient.
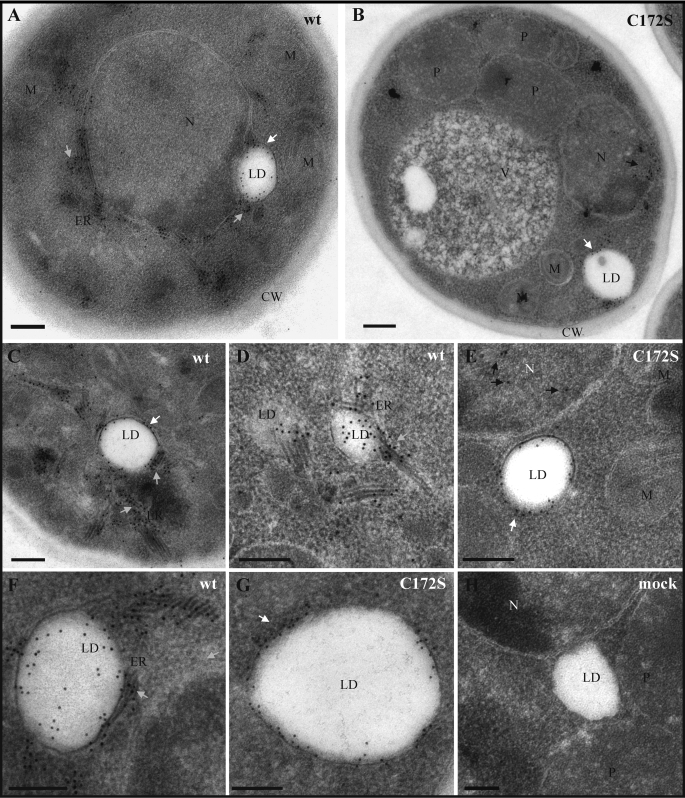
To investigate whether palmitoylation of Cys172 may have an impact on the retention of core in ER/LD-associated membranes, we examined EM micrographs of P. pastoris cells expressing core. P. pastoris cells have only a few LD/cell, but these organelles are larger (0.3–0.4, up to 1.6 μm in diameter) than what is seen in human cells (10–100 nm in diameter) (36). Therefore, it is easier to observe these organelles and their association to ER by immunogold labeling in yeast. In P. pastoris, wt core was localized around LD and also on ER membranes, which appear thicker than normal (Fig. 8, A, C, D, and F). However, C172S showed localization only around LD and not with ER membranes (Fig. 8, B, E, and G). In contrast to wt protein, C172S proteins were also detected in the nucleus. Stacks of ER membranes were easily observed around the LD of wt core (Fig. 8F). These patterns were not observed for the C172S mutant or for cells that were not expressing core (Fig. 8G); LDs appeared free of ER membranes. These results suggest that palmitoylation of core protein at residue Cys172 helps to recruit ER to the LDs and/or increases the affinity of core for ER-associated LDs.
FIGURE 8.
Electron micrograph sections of P. pastoris expressing HCV core. Immunogold labeling with anti-core antibodies on thin sections of P. pastoris cells producing HCV core wt (A, C, D, and F) or C172S (B, E, and G). H, immunostaining of mock transfected cells. N, nucleus; P, peroxisome; M, mitochondria; V, vacuole: CW, cell wall. Gray, white, and black arrows indicate the core proteins in ER, LD, and nucleus, respectively. Bars, 0.2 μm.
Effect of Cys172 Mutation on LD Accumulation
Earlier studies have shown that lipid accumulation is significantly greater in cells expressing core protein (12, 37). The LD induced by core protein genotype 1a tend to be larger than those present in naive cells (37). Lipid droplets observed in sections of Huh7.5 cells expressing C172S (Fig. 6) appeared smaller than LD of cells expressing wt core. To evaluate whether the C172S mutation affects LD accumulation, the cumulative area of LD in Huh7.5 cell sections was evaluated using Cell profiler software (26). As expected, the cumulative area of LD was significantly greater for cells expressing wt core protein than for cells expressing the plasmid alone (p < 0.01) (Fig. 9A). In contrast, sections of cells expressing the mutant C172S did not reveal any significant increase in LD area as compared with mock transfected cells. Mutant C91L showed an accumulation of LD that was similar to that of wt core.
The analysis of LD accumulation was also performed on yeast cells. EM micrographs were analyzed for LD area variations using Cell profiler software. Similar results were obtained in the P. pastoris system; cumulative areas of LD were significantly higher in cells expressing wt core and C91L than in naive cells or those transfected with the mutant C172S (Fig. 9B). Thus, the C172S mutation clearly affected the ability of core to induce the accumulation of LD in both yeast and human cells.
Effect of Cys172 Mutation on Particle Formation
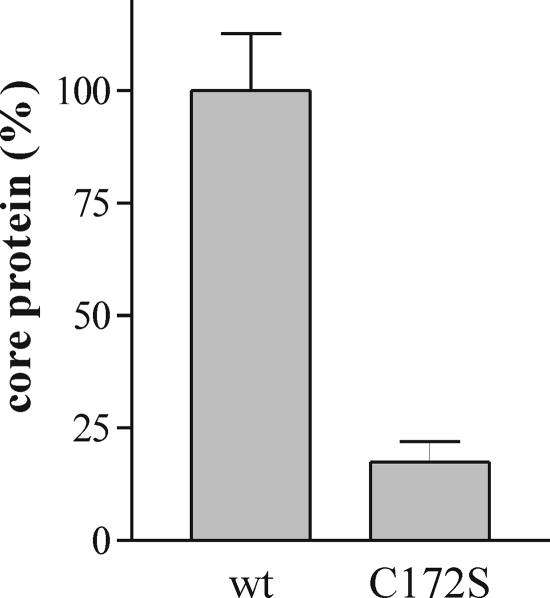
To determine the contribution of palmitoylation of core protein to particle formation, we analyzed the production of nucleocapsid-like particles (NLPs) (density, 1.11 g/ml) in yeast expressing core or its mutated form. P. pastoris expressing wt core was previously showed to produce NLPs enclosed in ER membranes that are similar in size and density to HCV virus particles (20, 38). Protein extracts from P. pastoris co-expressing spp with core or core mutant were isolated on sucrose gradients and analyzed by enzyme-linked immunosorbent assay with anti-core antibodies. As observed in Fig. 10, proteins present in the fraction at a density of 1.11 g/ml (20) were drastically reduced by the C172S mutation as compared with wt protein. From this results we concluded that palmitoylation of the core is necessary for the formation of NLPs in yeast cells.
FIGURE 10.
Effect of Cys172 mutations in HCV core on particle formation in yeast. Yeast extracts with similar core protein concentration were loaded on a 10–60% (w/w) sucrose gradient. After sedimentation, fractions containing NLPs at 1.11 g/ml were analyzed by enzyme-linked immunosorbent assay with anti-core antibodies. The results were normalized on the value of wt protein content.
Effect of Cys172 Mutation on HCV Infectivity
To determine the contribution of palmitoylation of core protein to virus infectivity, we mutated residue Cys172 in the context of the J6/JFH1 genome. The J6/JFH1 construct used (i.e. FL-J6/JFH1 C19Rluc2AUbi clone) expresses a luciferase protein that is cleaved from the polyprotein to generate a full-length HCV protein (28). Huh7.5 cells were electroporated with in vitro transcribed RNA generated from each construct. Luciferase activity in the cell lysates was measured 4 h post-electroporation as control of transfection efficiency and 72 h post-electroporation as RNA replication efficiency. We observed a similar level of transfection for both constructs (data not shown). Furthermore, the mutation C172S was not significantly affected by RNA replication compared with the wild type J6/JFH1 construct (Fig. 11A). The production of infectious virions was tested at 72 h post-electroporation by inoculation of naive Huh7.5 cells with filtered cell culture supernatants. Cell-associated luciferase activity was measured 72 h post-inoculation to quantify infectivity. In contrast to the robust infectious virus production of wild type J6/JFH1, the genome containing the C172S mutation in core failed to produce detectable levels of infectious virus (Fig. 11B). To determine whether the C172S mutation affected the release of the virions in the supernatant, we quantified by quantitative reverse transcription-PCR the amount of viral RNA released from the transfected cells 72 h after electroporation. As showed in Fig. 11C, the level of viral RNA detected in J6/JFH1-C172S construct was significantly reduced as compared with the wt. This result indicated that palmitoylation of core is important for virion assembly or/and for efficient release of the virion outside the infected cells.
DISCUSSION
For many proteins, the primary function of palmitoylation is to modify surface hydrophobicity and enhance membrane affinity, allowing the modified protein to interact with membranes (39). This post-translational modification plays an important role in protein stability (40), intracellular protein trafficking, and targeting to membrane microdomains (39). Palmitoylation is also crucial for assembly and budding of many viruses (41–44). Unlike other lipid modifications, palmitoylation is a reversible covalent modification, allowing dynamic regulation of multiple complex cellular systems. Palmitoylation occurs at multiple subcellular sites, from the point of synthesis in the ER, along the secretory pathway, and at the plasma membrane (45). Lipid modifications of a given protein, as well as palmitoylating enzymes, are conserved from yeast to humans (17, 46).
In this paper, we present evidence that the HCV core is modified by palmitoylation in yeast. Importantly, we demonstrate that this post-translational modification is also occurring in human cells, which provides additional credence to our findings. Consistent with our results, numerous studies have proven the reliability of yeast cells for palmitoylation studies (46–48). The prediction of core palmitoylation by the algorithm CSS-Palm 2.0 highlighted the fact that Cys172 is modified only in the context of the mature core protein after cleavage by spp and not in the context of the polyprotein before sp or spp cleavage. Accordingly, we observed that the C172S mutant could be processed by spp cleavage as efficiently as the wt core, suggesting that Cys172 palmitoylation does not influence spp cleavage.
We identified core protein residue Cys172 as the major palmitoylation site. Cys172 is present at the C terminus of the protein, in close proximity to the hydrophobic amino acid sequence that anchors the protein to the ER membrane (aa 174–191). Interestingly, palmitoylation sites are often found in close proximity to hydrophobic amino acid stretches of a transmembrane domain (TMD) (18, 48). Palmitoylation at these sites is predicted to increase the effective hydrophobic strength of the TMD and/or modify orientation of the TMD with respect to the plane of the lipid bilayer (18). Because the TMD of core is cleaved by spp to generate a protein of 177 aa (6), the length of the hydrophobic tail is reduced by 4 amino acids and is likely to be less efficient at stabilizing attachment of the protein to ER membranes. Palmitoylation of residue Cys172 probably helps maintain the core in close association with ER membranes. Other structures within core are involved in core-ER stability; the protein domain from aa 116 to 167 with predicted amphipathic α-helix structures is known to contribute to interaction of core with the ER after maturation of the protein by spp (8, 49). It is likely that this domain and palmitoylation of Cys172 ensure a stable interaction between core and the ER.
The Cys172 residue of core protein is well conserved among different HCV genotypes. Only strain JFH1 has an Phe at residue 172. For certain proteins, the substitution of the palmitoylated Cys by a hydrophobic residue could compensate for the absence of the lipid moiety (18, 50). We have some evidence that it is also the case for the core of JFH1, and this is under investigation.3
Palmitoylation has been proposed to enhance the affinity of proteins, both soluble and transmembrane, to cholesterol-rich raft-like domains (51). Recently core protein was shown to be associated with DRMs or rafts (14, 30). The DRMs where core accumulates have properties that distinguish them from classical plasmalemmal lipid rafts (14). However, our results show that palmitoylation of core was not involved in DRM targeting because the C172S mutant was also present in the DRM fraction after Triton solubilization.
We found that, as for the wt core protein, the mutant core protein C172S co-localized to the surface of LD. The C172S protein produced in Huh7.5 showed a uniform pattern of distribution around the LDs as compared with the wt protein, which exhibited irregular ring structures. It was suggested that core protein was present on the surface of LDs as well as in the ER in close proximity to the LD (11, 34); the outside surface of the large ring structures corresponds to stacks of ER membranes (11). Because the C172S mutant showed sharper and uniform rings around the LD, we deduced that C172S was absent from the ER and/or was deficient in recruiting ER membranes around LD structures. This result was confirmed by EM analysis of yeast expressing core; in yeast, mutation C172S affected the accumulation of core at ER membranes. Also, in these cells, we did not observe the stacking of ER membranes around LD that was easily identifiable in cells expressing wt core protein. We also found accumulation of C172S core, but not wt, in the nucleus. Consistently, accumulation of core in the nucleus was previously described in cells expressing mutant core proteins that have low affinity for ER membranes (9, 33, 52). In density gradient centrifugation of the ER-purified fraction from hepatocytes, we observed that affinity for smooth ER was affected by the C172S mutation. ER membranes associated with LD are smooth ER (35); therefore, C172S mutant has a reduced capacity to associate with ER membranes surrounding LD.
Core protein has been shown to trigger lipid accumulation in cell culture as well as in live animals (53). HCV core protein may interfere with lipid metabolism on three levels: impaired secretion, increased neosynthesis, and impaired degradation (53). Triglyceride accumulation is critical for HCV infection (54). The accumulation of lipid induced by core is certainly helpful in the establishment of viral infection. We found that the C172S mutant protein was less efficient in inducing LD accumulation in Huh7.5 than the wt protein; this impairment of lipid accumulation could have a direct impact on HCV infectivity.
We showed that palmitoylation of Cys172 was important for self-assembly of core in yeast cells. In vitro models of particle assembly do not require the C-terminal part of the protein (19, 55); however, assembly in yeast cells required the hydrophobic domain including Cys172 (19). Recently it was found that lipid modification, in the form of myristoylation, was essential for multimerization of human immunodeficiency virus Gag in mammalian cells (56), despite an in vitro model for human immunodeficiency virus assembly that does not require myristolated Gag protein. Membrane interactions could enhance protein-protein interactions by nucleation of the protein to a specific location that triggers the self-assembly process. In our yeast system, it is likely that this multimerization occurs at the ER membrane rather than at the LD because C172S core protein present on LD but absent from the ER cannot trigger particle formation.
Oligomerization of HCV core and virion assembly has been suggested to take place at the ER membranes that are closely associated with LD (11, 34). The association of core protein to the LD is an essential step in the production of infectious viral particles (57). According to our results, the association of core to LD is not sufficient per se for encapsidation. The movement of core toward ER associated with LD is also a critical parameter for the efficiency of virus production. One possible function of the palmitoylation of core is the retention of core in smooth ER membranes or the redistribution of core proteins from LD to ER membranes. Because we observed a decrease in ER membrane stacking around LD in C172S-expressing cells, the retention of ER in LD is possibly improved upon palmitoylation of core. Impairment of core palmitoylation also affects the amount of LD accumulation. All of these factors can contribute to reducing the formation and secretion of HCV particles in cells infected with the mutated virus C172S.
In conclusion, we showed for the first time that HCV core protein is modified by palmitoylation at residue Cys172, which is in proximity to ER membranes. Palmitoylation of Cys172 controls the association of core to ER membranes following spp processing, and this association is essential for virion production.
Supplementary Material
Acknowledgment
We thank Helen Rothnie for editing of the manuscript.
This work was supported by grants from the Canadian Institutes of Health Research of Canada and the Réseau Sida et Maladies Infectueuses of the Fonds de la Recherche en Santé du Québec.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
N. Majeau, R. Fromentin, M. J. Tremblay, and D. Leclerc, manuscript in preparation.
- HCV
- hepatitis C virus
- ER
- endoplasmic reticulum
- LD
- lipid droplets
- spp
- signal peptide peptidase
- aa
- amino acid(s)
- DRM
- detergent-resistant membrane
- PBS
- phosphate-buffered saline
- 2-BP
- 2-bromopalmitate
- wt
- wild type
- NLP
- nucleocapsid-like particle
- TMD
- transmembrane domain.
REFERENCES
- 1.Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. (1989) Science 244, 359–362 [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager R., Lohmann V. (2000) J. Gen. Virol. 81, 1631–1648 [DOI] [PubMed] [Google Scholar]
- 3.Irshad M., Khushboo I., Singh S., Singh S. (2008) Int. Rev. Immunol. 27, 497–517 [DOI] [PubMed] [Google Scholar]
- 4.Grakoui A., Wychowski C., Lin C., Feinstone S. M., Rice C. M. (1993) J. Virol. 67, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLauchlan J., Lemberg M. K., Hope G., Martoglio B. (2002) EMBO J. 21, 3980–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto K., Mori Y., Komoda Y., Okamoto T., Okochi M., Takeda M., Suzuki T., Moriishi K., Matsuura Y. (2008) J. Virol. 82, 8349–8361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hope R. G., McLauchlan J. (2000) J. Gen. Virol. 81, 1913–1925 [DOI] [PubMed] [Google Scholar]
- 8.Suzuki R., Sakamoto S., Tsutsumi T., Rikimaru A., Tanaka K., Shimoike T., Moriishi K., Iwasaki T., Mizumoto K., Matsuura Y., Miyamura T., Suzuki T. (2005) J. Virol. 79, 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki R., Matsuura Y., Suzuki T., Ando A., Chiba J., Harada S., Saito I., Miyamura T. (1995) J. Gen. Virol. 76, 53–61 [DOI] [PubMed] [Google Scholar]
- 10.Boulant S., Montserret R., Hope R. G., Ratinier M., Targett-Adams P., Lavergne J.-P., Penin F., McLauchlan J. (2006) J. Biol. Chem. 281, 22236–22247 [DOI] [PubMed] [Google Scholar]
- 11.Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 12.Abid K., Pazienza V., de Gottardi A., Rubbia-Brandt L., Conne B., Pugnale P., Rossi C., Mangia A., Negro F. (2005) J. Hepatol. 42, 744–751 [DOI] [PubMed] [Google Scholar]
- 13.Boulant S., Douglas M. W., Moody L., Budkowska A., Targett-Adams P., McLauchlan J. (2008) Traffic 9, 1268–1282 [DOI] [PubMed] [Google Scholar]
- 14.Matto M., Rice C. M., Aroeti B., Glenn J. S. (2004) J. Virol. 78, 12047–12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi S. T., Lee K. J., Aizaki H., Hwang S. B., Lai M. M. C. (2003) J. Virol. 77, 4160–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aizaki H., Lee K. J., Sung V. M., Ishiko H., Lai M. M. C. (2004) Virology 324, 450–461 [DOI] [PubMed] [Google Scholar]
- 17.Tsutsumi R., Fukata Y., Fukata M. (2008) Pflügers Arch. 456, 1199–1206 [DOI] [PubMed] [Google Scholar]
- 18.Charollais J., Van Der Goot F. G. (2009) Mol. Membr. Biol. 26, 55–66 [DOI] [PubMed] [Google Scholar]
- 19.Majeau N., Gagné V., Boivin A., Bolduc M., Majeau J. A., Ouellet D., Leclerc D. (2004) J. Gen. Virol. 85, 971–981 [DOI] [PubMed] [Google Scholar]
- 20.Majeau N., Gagné V., Bolduc M., Leclerc D. (2005) J. Gen. Virol. 86, 3055–3064 [DOI] [PubMed] [Google Scholar]
- 21.Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 22.Majeau N., Bolduc M., Duvignaud J. B., Fromentin R., Leclerc D. (2007) Biochem. Cell Biol. 85, 78–87 [DOI] [PubMed] [Google Scholar]
- 23.Hirschman J. E., Jenness D. D. (1999) Mol. Cell. Biol. 19, 7705–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G. Y., Lee K. J., Gao L., Lai M. M. C. (2006) J. Virol. 80, 6013–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingwood D., Simons K. (2007) Nat. Protoc. 2, 2159–2165 [DOI] [PubMed] [Google Scholar]
- 26.Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., Sabatini D. M. (2006) Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt M., Bowers B., Varma A., Roh D. H., Cabib E. (2002) J. Cell Sci. 115, 293–302 [DOI] [PubMed] [Google Scholar]
- 28.Tscherne D. M., Jones C. T., Evans M. J., Lindenbach B. D., McKeating J. A., Rice C. M. (2006) J. Virol. 80, 1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitaku S., Hirokawa T., Tsuji T. (2002) Bioinformatics 18, 608–616 [DOI] [PubMed] [Google Scholar]
- 30.Ogino T., Fukuda H., Imajoh-Ohmi S., Kohara M., Nomoto A. (2004) J. Virol. 78, 11766–11777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato T., Miyamoto M., Furusaka A., Date T., Yasui K., Kato J., Matsushima S., Komatsu T., Wakita T. (2003) J. Med. Virol. 69, 357–366 [DOI] [PubMed] [Google Scholar]
- 33.Okamoto K., Moriishi K., Miyamura T., Matsuura Y. (2004) J. Virol. 78, 6370–6380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roingeard P., Hourioux C., Blanchard E., Prensier G. (2008) Histochem. Cell Biol. 130, 561–566 [DOI] [PubMed] [Google Scholar]
- 35.Jack E. M., Stäubli W., Waechter F., Bentley P., Suter J., Bieri F., Muakkassah-Kelly S. F., Cruz-Orive L. M. (1990) Carcinogenesis 11, 1531–1538 [DOI] [PubMed] [Google Scholar]
- 36.Zweytick D., Athenstaedt K., Daum G. (2000) Biochim. Biophys. Acta 1469, 101–120 [DOI] [PubMed] [Google Scholar]
- 37.Hourioux C., Patient R., Morin A., Blanchard E., Moreau A., Trassard S., Giraudeau B., Roingeard P. (2007) Gut 56, 1302–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falcon V., Garcia C., Rosa M. C. d. l., Menendez I., Seoane J., Grillo J. M. (1999) Tissue Cell 31, 117–125 [DOI] [PubMed] [Google Scholar]
- 39.Greaves J., Prescott G. R., Gorleku O. A., Chamberlain L. H. (2009) Mol. Membr. Biol. 26, 67–79 [DOI] [PubMed] [Google Scholar]
- 40.Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 41.Boscarino J. A., Logan H. L., Lacny J. J., Gallagher T. M. (2008) J. Virol. 82, 2989–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen B. J., Takeda M., Lamb R. A. (2005) J. Virol. 79, 13673–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rousso I., Mixon M. B., Chen B. K., Kim P. S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13523–13525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vzorov A. N., Weidmann A., Kozyr N. L., Khaoustov V., Yoffe B., Compans R. W. (2007) Retrovirology 4, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baekkeskov S., Kanaani J. (2009) Mol. Membr. Biol. 26, 42–54 [DOI] [PubMed] [Google Scholar]
- 46.Nadolski M. J., Linder M. E. (2007) FEBS J. 274, 5202–5210 [DOI] [PubMed] [Google Scholar]
- 47.Linder M. E., Deschenes R. J. (2004) J. Cell Sci. 117, 521–526 [DOI] [PubMed] [Google Scholar]
- 48.Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hourioux C., Ait-Goughoulte M., Patient R., Fouquenet D., Arcanger-Doudet F., Brand D., Martin A., Roingeard P. (2007) Cell. Microbiol. 9, 1014–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greaves J., Prescott G. R., Fukata Y., Fukata M., Salaun C., Chamberlain L. H. (2009) Mol. Biol. Cell 20, 1845–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown D. A. (2006) Physiology 21, 430–439 [DOI] [PubMed] [Google Scholar]
- 52.Lo S. Y., Masiarz F., Hwang S. B., Lai M. M., Ou J. H. (1995) Virology 213, 455–461 [DOI] [PubMed] [Google Scholar]
- 53.Negro F. (2006) World J. Gastroenterol. 12, 6756–6765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aizaki H., Morikawa K., Fukasawa M., Hara H., Inoue Y., Tani H., Saito K., Nishijima M., Hanada K., Matsuura Y., Lai M. M., Miyamura T., Wakita T., Suzuki T. (2008) J. Virol. 82, 5715–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunkel M., Lorinczi M., Rijnbrand R., Lemon S. M., Watowich S. J. (2001) J. Virol. 75, 2119–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H., Dou J., Ding L., Spearman P. (2007) J. Virol. 81, 12899–12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. (2007) J. Biol. Chem. 282, 37158–37169 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.