Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) and poly(ADP-ribose) glycohydrolase (PARG) are enzymes that modify target proteins by the addition and removal, respectively, of ADP-ribose polymers. Although a role for PARP-1 in gene regulation has been well established, the role of PARG is less clear. To investigate how PARP-1 and PARG coordinately regulate global patterns of gene expression, we used short hairpin RNAs to stably knock down PARP-1 or PARG in MCF-7 cells followed by expression microarray analyses. Correlation analyses showed that the majority of genes affected by the knockdown of one factor were similarly affected by the knockdown of the other factor. The most robustly regulated common genes were enriched for stress-response and metabolic functions. In chromatin immunoprecipitation assays, PARP-1 and PARG localized to the promoters of positively and negatively regulated target genes. The levels of chromatin-bound PARG at a given promoter generally correlated with the levels of PARP-1 across the subset of promoters tested. For about half of the genes tested, the binding of PARP-1 at the promoter was dependent on the binding of PARG. Experiments using stable re-expression of short hairpin RNA-resistant catalytic mutants showed that PARP-1 and PARG enzymatic activities are required for some, but not all, target genes. Collectively, our results indicate that PARP-1 and PARG, nuclear enzymes with opposing enzymatic activities, localize to target promoters and act in a similar, rather than antagonistic, manner to regulate gene expression.
Introduction
Poly(ADP-ribosyl)ation (PARylation)6 is a post-translational modification involving the polymerization of ADP-ribose (ADPR) units from donor NAD+ molecules on target proteins (1, 2). PARylation occurs on a variety of target proteins in all cellular compartments and plays roles in a wide array of cellular processes, such as stress responses, DNA repair, and transcriptional regulation (1, 2). Nuclear targets include core histones, the linker histone H1, and an assortment of transcription factors (3). The synthesis and degradation of poly(ADP-ribose) (PAR) is catalyzed by two types of enzymes: PAR polymerases (PARPs) and PAR glycohydrolases (PARGs), respectively (4, 5). Although recent studies have begun to explore the functional interplay between these two types of enzymes (1–3, 5), a clear picture of how they cooperate to regulate cellular processes remains unclear.
PARP-1, a ubiquitous 116-kDa nuclear enzyme, is the founding member of the PARP superfamily (2, 4). It is a highly abundant protein (1–2 million molecules per cell) that is likely responsible for the majority of PAR synthesis in cells (1). PARP-1 has three major structural and function domains: (i) an amino-terminal DNA binding domain, (ii) a central automodification domain, and (iii) a carboxyl-terminal catalytic domain with low basal activity (1, 6). The catalytic activity of PARP-1 is potently allosterically activated by the binding of PARP-1 to certain forms of DNA (7–10), nucleosomes (11–13), and protein binding partners (14–16). In vivo, PARP-1 is the major target for PARP-1-mediated PARylation through an automodification reaction involving the automodification domain (1, 17), although an array of other nuclear targets has been described (1–3). Its DNA binding, catalytic, and automodification functions allow PARP-1 to modulate a wide variety of processes involving genomic DNA.
Cellular PARG activities in mammals are mediated by multiple PARG isoforms encoded by a single gene (5, 18, 19). The predominant isoforms, all of which have catalytic activity, include: (i) a set of long (∼100–∼110-kDa) isoforms that may shuttle between the nucleus and cytoplasm and (ii) a short (∼65-kDa) isoform that resides in the cytoplasm (18, 19). These PARG isoforms catalyze the hydrolysis of PAR to produce ADPR monomers and short ADPR polymers (5). The longest PARG isoforms contain two major functional domains: (i) a regulatory domain and (ii) a catalytic domain (2, 3, 5). The regulatory domain contains both nuclear localization and nuclear export signals, which mediate shuttling between the nuclear and cytoplasmic compartments (20, 21). The catalytic domain contains the enzyme active site, which confers both endoglycosidic and exoglycosidic activities, allowing for the rapid hydrolysis of PAR (5).
PARP-1 and PARG play important roles in an overlapping set of biological processes. For example, gene-specific and genomic studies have revealed a clear role for PARP-1 in transcriptional regulation (2, 3, 22–24), and PARP-1 localizes to the promoters of more than 90% of expressed genes in MCF-7 human breast cancer cells (25). Likewise, although more limited, recent gene-specific studies have also implicated PARG in transcriptional regulation (11, 26, 27). Few studies, however, have directly examined the interplay between PARP-1 and PARG in the regulation of their biological endpoints in side-by-side experiments in the same cell type. As such, the means by which PARP-1 and PARG coordinate their enzymatic activities to regulate gene expression across the genome are unknown. Based on the opposing enzymatic activities of PARP-1 and PARG, one might expect PARG to counter the gene regulatory actions of PARP-1 by degrading the PAR chains synthesized by PARP-1. However, PARP-1 and PARG have similar effects on many biological endpoints (2, 23, 27–35), suggesting that this simple model is unlikely to be correct.
In the current studies, we used a series of genomic and gene-specific assays to explore the coordinated regulation of gene expression by PARP-1 and PARG, including the role of their respective enzymatic activities. Our results indicate that PARP-1 and PARG localize to target promoters and act in a similar, rather than antagonistic, manner to regulate global patterns of gene expression.
EXPERIMENTAL PROCEDURES
Antibodies
The custom rabbit polyclonal antibodies against PARP-1 and PARG used for Western blotting and chromatin immunoprecipitation (ChIP) assays were generated by using a purified fragment of human PARP-1 (amino-terminal, PARP-N (11)) and full-length rat PARG as antigens (Pocono Rabbit Farm and Laboratory, Inc.). The antibodies were screened for: (i) specificity by Western blotting MCF-7 cell extracts, (ii) the ability to immunoprecipitate their cognate antigens from formaldehyde cross-linked chromatin samples by a ChIP-Western protocol (11) (supplemental Fig. S1), and (iii) a reduction in Western blot signal upon knockdown of PARP-1 or PARG (see Fig. 1A). The custom rabbit polyclonal antibody against SIRT1 used for Western blotting was generated by using full-length mouse SIRT1 as an antigen (36). The mouse monoclonal PAR antibody was purchased from Trevigen.
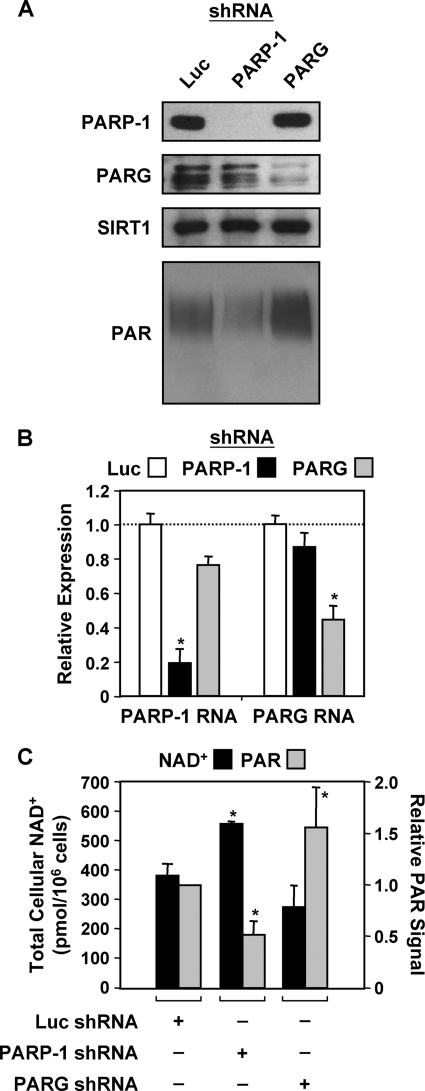
FIGURE 1.
shRNA-mediated knockdown of PARP-1 and PARG in MCF-7 cells. A, whole cell lysates collected from Luc, PARP-1, and PARG stable shRNA-mediated knockdown cell lines were subjected to Western blotting analyses for PARP-1 and PARG. SIRT1 was also analyzed as a loading control. PAR levels were analyzed by Western blotting using nuclear extracts from the same cell lines. B, RT-qPCR analysis confirms the knockdown PARP-1 and PARG mRNA in MCF-7 cells. Total RNA was isolated from Luc, PARP-1, and PARG knockdown cells, reverse-transcribed, and subjected to qPCR using gene-specific primers to PARP-1 and PARG. Each bar is the mean + S.E. (error bars) for three independent RNA isolations. Bars that are marked with an asterisk are statistically different from the Luc control, as determined by a Student's t test with a p value threshold of <0.05. C, total cellular NAD+ levels (black bars) in Luc, PARP-1, and PARG knockdown cells were measured using a quantitative HPLC/mass spectrometry method with 18O standards. PAR levels (gray bars) were quantified from Western blots (from A, above) by densitometry. The data are shown as the mean + range or S.E. (error bars) from two or more independ ent biological replicates. Bars that are marked with an asterisk are statistically different from the Luc control, as determined by a Student's t test with a p value threshold of <0.05.
Oligonucleotides
Information about the oligonucleotide sequences used for the short hairpin RNA (shRNA) constructs, site-directed mutagenesis, RT-qPCR, and ChIP-qPCR can be found in the supplemental data.
PARP-1 and PARG Knockdown and Expression Constructs
shRNA expression constructs for retroviral-mediated knockdown of PARP-1 and PARG were made using the pSUPER.retro vector (OligoEngine). Double-stranded oligonucleotides containing shRNA sequences targeting luciferase (Luc control), PARP-1, or PARG were cloned into the vector (puromycin- or neomycin-resistant) using BglII and XhoI restriction sites as described by the manufacturer. The shRNA sequences (one for Luc, two for PARP-1, and two for PARG) were based on sequences reported in the literature (15, 37) or designed using the Dharmacon siDESIGN® Center software (the specific sequences used can be found in the supplemental data). All constructs were confirmed by sequencing.
Wild type and catalytically inactive point mutants of human PARP-1 and rat PARG were used in the studies described herein. The catalytically inactive human PARP-1 contained a change at Glu-988 to Lys (E988K) (38, 39), whereas the catalytically inactive rat PARG contained changes at Tyr-788 and -791 to Phe and Ala, respectively (Y788F/Y791A) (40). Cytomegalovirus-based mammalian expression constructs for full-length wild-type or catalytically inactive human PARP-1 (with a carboxyl-terminal His6/FLAG tag) or rat PARG (with a carboxyl-terminal FLAG tag) were generated by PCR-based cloning of the tagged cDNAs into pCMV5. The pCMV5-hPARP-1-His/FLAG and pCMV5-rPARG-FLAG vectors were then used as templates to generate cDNAs resistant to the shRNAs noted above by site-directed mutagenesis (QuikChange® site-directed mutagenesis kit from Stratagene). The following nucleotides relative to the first nucleotide of the first codon were changed in the human PARP-1 (hPARP-1) cDNA: g2835a, c2838t, t2841c, and c2844t (for shRNA#1) and c2946g, t2949g, a2952g, and a2955g (for shRNA#2). The following nucleotides were changed in the rat PARG (rPARG) cDNA: a1827g, g1833c, and a1836g (for shRNA#1). Note that differences between the human-based PARG shRNA#2 sequence and the rat PARG cDNA eliminated the need to alter the #2 recognition site). Positive clones were identified by sequencing and then cloned into the pQCXIH retroviral expression vector (BD Biosciences; hygromycin-resistant) using NotI and BamHI restriction sites.
Generation and Culture of MCF-7-derived Cell Lines
Parental MCF-7 human breast cancer cells, kindly provided by Dr. Benita Katzenellenbogen (University of Illinois at Urbana-Champaign), were maintained in MEM Eagle's medium containing Hanks' salts, l-glutamine, and non-essential amino acids (Sigma) supplemented with 5% bovine calf serum (Sigma), 20 mm HEPES (pH 7.6), 100 units/ml penicillin, 100 μg/ml streptomycin, 25 μg/ml gentamicin, and 0.22% sodium bicarbonate. The shRNA knockdown (“knockdown”) and shRNA knockdown + shRNA-resistant re-expression (“knockdown/add-back”) cell lines used in these studies were generated by sequential retroviral infections of parental MCF-7 cells with the appropriate shRNA and cDNA expression vectors (see below). In each case, the final cell line constructions were populations of individual transformants, not clonal lines. A minimum of two independently generated populations were made and tested for each cell line. The Luc, PARP-1, and PARG knockdown cells used in Figs. 1–7 express two distinct shRNA sequences targeting the intended factor. These same shRNA sequences were also tested individually in gene-specific expression studies (supplemental Fig. S4).
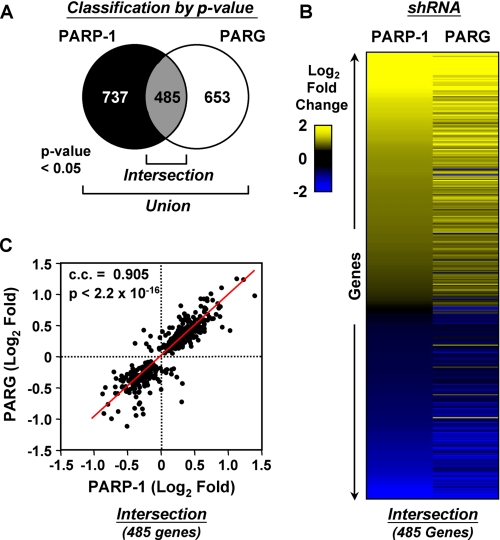
FIGURE 2.
PARP-1 and PARG coordinately regulate global patterns of gene expression in MCF-7 cells. A, Venn diagram of PARP-1- and PARG-regulated genes in MCF-7 cells as defined by shRNA-mediated knockdown and expression microarrays. Genes passing both present call and p value <0.05 criteria for at least one factor represent the union (see also supplemental Fig. 1), whereas genes regulated by both factors represent the commonly regulated genes or intersection. B, heat map showing the expression profiles of the commonly regulated genes (i.e. intersection; 485 genes) from A. The genes are ranked in the heat map by log2 -fold change in the PARP-1 knockdown cell line (see color scale). C, correlation analysis of the commonly regulated genes (i.e. intersection; 485 genes) from A. The Spearman correlation coefficient (c.c.) and p value are indicated.
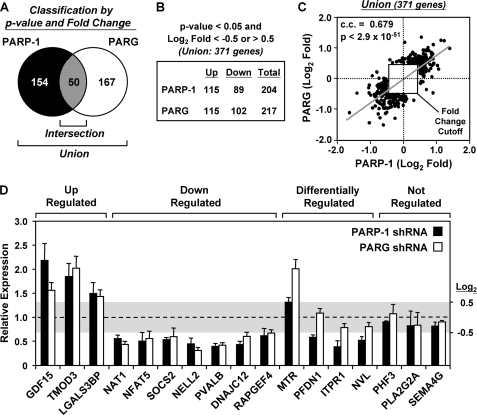
FIGURE 3.
Defining the genes most robustly regulated by PARP-1 and PARG. A, Venn diagram of PARP-1- and PARG-regulated genes after applying a -fold change cutoff of log2 <−0.5 or >0.5. Of the union of 371 genes most robustly regulated by either PARP-1 or PARG, 50 are commonly regulated. B, the distribution of the most robustly up-regulated or down-regulated genes upon knockdown of PARP-1 or PARG is indicated. C, correlation analysis comparing the magnitude and direction of regulation of the 371 most robustly regulated genes shown in A and B. The Spearman correlation coefficient (c.c.), p value, and -fold change cutoff are indicated. D, gene-specific confirmation of the expression microarray results by RT-qPCR. Luc, PARP-1, and PARG knockdown MCF-7 cells were seeded and grown under the same conditions used for the expression microarrays. Total RNA was isolated, reverse-transcribed, and subjected to qPCR using gene-specific primers. Genes were considered to be regulated if the log2 -fold change was <−0.5 or >0.5 (values falling outside of the shaded box). Each bar represents the mean + S.E. (error bars) from three or more independent determinations. All bars with a mean value falling outside of the shaded box are statistically different from the Luc control, as determined by a Student's t test with a p value threshold of <0.05.
FIGURE 4.
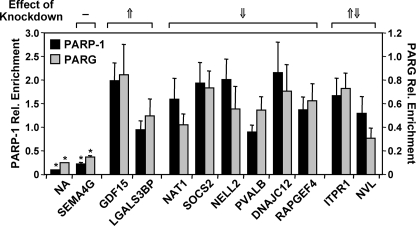
PARP-1 and PARG localize to the promoters of regulated target genes. PARP-1 and PARG show similar patterns of localization at the promoters of commonly regulated target genes. The occupancy of PARP-1 (black bars) and PARG (gray bars) at the promoters of coregulated genes was examined by ChIP analyses. Each bar represents the mean + S.E. (error bars) from three or more independent determinations. Bars that are not marked with an asterisk are statistically different from the no antibody (NA) control, as determined by a Student's t test with a p value threshold of <0.05. SEMA4G is a gene not regulated by knockdown of PARP-1 or PARG (Fig. 3D). The effects of PARP-1 and PARG knockdown (from Fig. 3D) are indicated for comparison: 1) −, no effect; 2) ↑, up-regulated by both PARP-1 and PARG knockdown; 3) ↓, down-regulated by both PARP-1 and PARG knockdown; and 4) ↑ ↓, differentially regulated by PARP-1 and PARG knockdown. Rel. Enrichment, relative enrichment.
FIGURE 5.
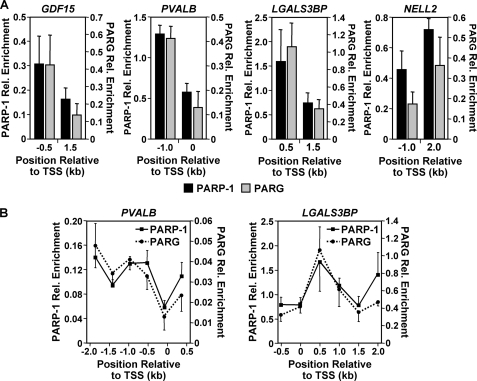
PARP-1 and PARG binding patterns are similar across individual target gene promoters. PARP-1 and PARG binding patterns are similar across individual promoters with both high and low occupancy. A, the occupancy of PARP-1 (black bars) and PARG (gray bars) at two regions (∼1 kb apart) of the GDF15, PVALB, LGALS3BP, and NELL2 promoters was determined by ChIP analyses. Each bar represents the mean + S.E. (error bars) from three or more independent determinations. Rel. Enrichment, relative enrichment. TSS, transcription start site. B, PARP-1 (solid line) and PARG (dotted line) occupancy was examined by ChIP-tiling analyses over a 2.5-kb region of the PVALB and LGALS3BP promoters. Each point represents the mean + S.E. (error bars) from three or more independent determinations; all values are statistically different from the no antibody control, as determined by a Student's t test with a p value threshold of <0.05.
FIGURE 7.
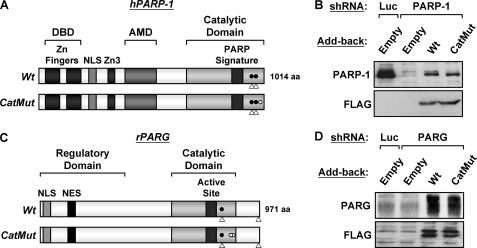
Stable re-expression of shRNA-resistant PARP-1 and PARG in their cognate knockdown cell lines. A, schematic diagram of the human PARP-1 structural and functional domains. The DNA binding domain (DBD), zinc fingers (Zn Fingers), nuclear localization signal (NLS), third zinc binding domain (Zn3), automodification domain (AMD), and PARP “signature” motif are shown. Open triangles indicate the location of the 21-nucleotide shRNA recognition sequences used for knockdown. Filled circles indicate the location of the silent point mutations engineered into the cDNAs to make them resistant to the shRNAs. The open circle indicates the location of the inactivating point mutation (Glu 988 to Lys) that inhibits PARP-1 enzymatic activity in the catalytically inactive mutant (CatMut). Wt, wild type. B, FLAG-tagged RNA interference-resistant wild-type or catalytically inactive PARP-1 was stably expressed in MCF-7 PARP-1 knockdown cell lines. Re-expression was confirmed by Western blotting for PARP-1 and FLAG. An empty vector was used as a control (Empty) in both Luc and PARP-1 knockdown cell lines. C, schematic diagram of the rat PARG structural and functional domains. The regulatory domain, catalytic domain, active site, nuclear localization signal, and nuclear export signal (NES) are shown. Open triangles indicate the location of the 21-nucleotide shRNA recognition sequences used for knockdown. Filled circles indicate the location of the silent point mutation engineered into the cDNAs to make them resistant to the shRNA. The open circles indicate the location of the inactivating point mutations (Tyr-788 to Phe and Tyr-791 to Ala) that inhibit PARG enzymatic activity in the catalytically inactive mutant. D, FLAG-tagged RNA interference-resistant wild-type or catalytically inactive PARG was stably expressed in MCF-7 PARG knockdown cell lines. Re-expression was confirmed by Western blotting for PARG and FLAG. An empty vector was used as a control (Empty) in both Luc and PARP-1 knockdown cell lines.
Retroviruses were generated by transfection of the pSUPER.retro or pQCXIH vectors described above with an expression vector for the vesticular stomatitis virus G envelope protein into Phoenix Ampho cells using GeneJuice transfection reagent (Novagen) according to the manufacturer's protocol. The resulting viruses were collected, filtered through a 0.45-μm syringe filter to remove any remaining cells, and used to infect the parental MCF-7 cells. Stably transduced cells were isolated under appropriate selection with puromycin (Sigma; 0.5 μg/ml), G418 sulfate (Invitrogen; 800 μg/ml), or hygromycin (Cellgro; 200 μg/ml), expanded, and frozen in aliquots for future use. The cells were grown under subconfluent conditions for routine maintenance and most experimental procedures.
For experiments, cells from the various lines were plated in MEM-modified Eagle's medium with Earle's salts and non-essential amino acids, without phenol red (Sigma), supplemented with 5% charcoal/dextran-treated bovine calf serum (CDCS; Sigma) and the other additives noted above. Subconfluent populations of cells were collected for analysis between 3 and 4 days after plating.
PAR and NAD+ Measurements
Stable Luc, PARP-1, and PARG knockdown cells were seeded at ∼6 × 105 cells/15-cm plate and grown for at least 3 days in MEM containing 5% CDCS and the additives noted above. After 3 days (at ∼60–80% confluence), the cells were collected by trypsinization, washed with ice-cold phosphate-buffered saline, and frozen in liquid nitrogen. The levels of PAR were determined by Western blotting of whole cell or nuclear extracts prepared in the presence of gallotannin to prevent degradation of the PAR polymers by PARG. The signals were quantified by densitometry using the ImageQuant software (Molecular Dynamics). The concentration of NAD+ in whole cell extracts for each cell line was determined by using a quantitative HPLC/mass spectrometry method (HPLC/matrix-assisted laser desorption ionization/mass spectrometry) with 18O Standards (41, 42).
Expression Microarrays
Stable Luc, PARP-1, and PARG knockdown cells were seeded at ∼8 × 105 cells/10-cm diameter dish and grown for at least 3 days in MEM containing 5% CDCS and the additives noted above. Three independently generated populations of cells were used for these experiments. The cells were collected, and total RNA was isolated using TRIzol reagent (Invitrogen) followed by an RNeasy column (Qiagen) according to the manufacturers' protocols. The RNA quality was assessed using an Agilent Bioanalyzer 2100, and suitable RNA was analyzed for global patterns of gene expression at the Cornell University Microarray Core Facility. Briefly, 7 μg of total RNA was labeled using the Affymetrix standard one-cycle amplification and labeling protocol. The labeled cRNA was then hybridized to Affymetrix human U133A 2.0 GeneChips, which were scanned using a GeneChip Scanner 3000. The raw array data were processed by the Affymetrix GeneChip Operating Software (GCOS) to obtain detection calls and signal values. The signals were normalized by scaling to a target value of 500 using GCOS. To adjust for batch effects due to day-to-day differences in RNA isolations, the empirical Bayes method was applied to the data set (43). After adjusting any values less than 0.01–0.01, the data were log2-transformed, median-centered for each array, and median-centered for each individual probe set. Filters were then applied to obtain final gene lists. Specifically, the criteria for a gene to be considered regulated by PARP-1 or PARG were: (i) detection call flagged as present or marginal in two out of three array replicates for both control and factor knockdown cell lines and (ii) significance of values between control and knockdown cell lines for any given gene had a two-tailed Student's t test with a p value <0.05 (see Fig. 2A, Union). To determine the genes most robustly regulated by PARP-1 or PARG, we added a -fold change criterion, namely a log2 -fold change cutoff between 0.5 and −0.5 when compared with the Luc control knockdown cells (the final gene lists can be found in supplemental Tables S1, S2, and S3). The heat maps used to visualize the microarray expression data (see Fig. 2B and supplemental Fig. S3B) were generated using Java Treeview (44).
Gene Ontology Analyses
Gene ontology (GO) analyses of the microarray expression data were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resource web site for gene ontology analysis (45). PARP-1- and PARG-regulated gene lists (consisting of 204 and 217 genes, respectively; see Fig. 3, A and B) were generated according to the present call, p value, and -fold change criteria as noted above. The commonly regulated gene list (50 genes) represents the overlap between the PARP-1- and PARG-regulated gene lists. Affymetrix gene ID numbers for each list were entered into the DAVID web site for gene ontology analysis. The resulting terms were grouped together under each category, and duplicate probe sets were removed for accurate percentage representation. Only those GO terms yielding a p value <0.05 using a Fisher's exact test were considered significantly enriched in each gene list.
mRNA Expression Analyses by RT-qPCR
For gene-specific mRNA expression analyses, knockdown or knockdown/add-back MCF-7 cells were grown under standard conditions (see above). For all expression experiments, the cells were seeded at ∼1.5 × 105 cells/well in 6-well plates and grown for 3 days in MEM containing 5% CDCS and the additives noted above. Total RNA was isolated using TRIzol reagent (Invitrogen), reverse-transcribed, and subjected to qPCR using a set gene-specific primers (see below; the primer sequences can be found in the supplemental data). All target gene transcripts were normalized to the β-actin transcript, which was unaffected by PARP-1 or PARG knockdown (data not shown). All experiments were conducted a minimum of three times with independent RNA isolations to ensure reproducibility.
Chromatin Immunoprecipitation Assays
Parental or knockdown MCF-7 cells were seeded at ∼6 × 105 cells/15-cm plate and grown for at least 3 days in MEM containing 5% CDCS and the additives noted above. The cells were cross-linked with 1% formaldehyde in phosphate-buffered saline at 37 °C for 10 min immediately prior to harvesting. ChIP assays were performed as described previously (25, 46) using polyclonal antibodies against PARP-1 and PARG (see above), as well as “no antibody” controls. The no antibody signals from the ChIP assays are comparable with preimmune sera in both ChIP-Western and ChIP-qPCR assays (data not shown). The resulting ChIP DNA material was used in gene-specific qPCR analyses (see below; the primer sequences are listed in the supplemental data). For ChIP-Western analysis (supplemental Fig. S1A), a small aliquot was removed from the input and ChIP samples prior to reversing the cross-links for conventional Western blotting.
Quantitative PCR Analyses (RT-qPCR and ChIP-qPCR)
Gene-specific mRNA expression and ChIP analyses were analyzed by quantitative PCR in a similar manner. Briefly, reactions containing DNA from either source, 1× SYBR Green PCR master mix, and forward and reverse primers (250 nm for ChIP, 500 nm for expression) were used in 40–45 cycles of amplification (95 °C for 15 s, 60 °C for 1 min) using an MJ Research DNA engine Opticon 2 (96-well) or an Applied Biosystems 7900 HT sequence detection system (384-well) following an initial 10-min incubation at 95 °C. Melting curve analysis was performed to ensure that only the targeted amplicon was amplified.
Statistical Analyses
For the NAD+ measurements in Fig. 1C, the ChIP-qPCR assays in Figs. 4, 5, and 6, and the RT-qPCR assays in Figs. 1B and 3D, a paired Student's t test with a p value threshold of <0.05 was used to determine the significance of differences between the control and experimental samples. For the gene-specific expression analyses in Fig. 8, analysis of variance with a p value threshold of <0.05 was used to determine the significance of differences between samples.
FIGURE 6.
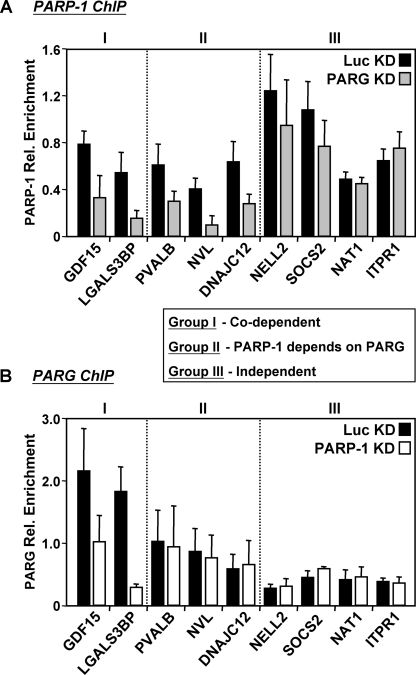
PARP-1 and PARG can affect each other's binding at target gene promoters. To determine whether PARP-1 and PARG can affect each other's binding, the occupancy of PARP-1 and PARG was examined by ChIP analyses in the control and knockdown (KD) cell line of the opposing factor. A, the occupancy of PARP-1 was examined by ChIP analyses in the Luc (black bars) and PARG (gray bars) knockdown cell lines. Each bar represents the mean + S.E. (error bars) from three or more independent determinations. B, the occupancy of PARG was examined by ChIP analyses in the Luc (black bars) and PARP-1 (white bars) knockdown cell lines. Each bar represents the mean + S.E. (error bars) from three or more independent determinations. The data revealed three groups of PARP-1 and PARG binding patterns, as indicated. All changes in occupancy for PARP-1 (Group I and Group II) and PARG (Group I) are statistically different between the control and knockdown cell line, as determined by a Student's t test with a p value threshold of <0.05.
FIGURE 8.
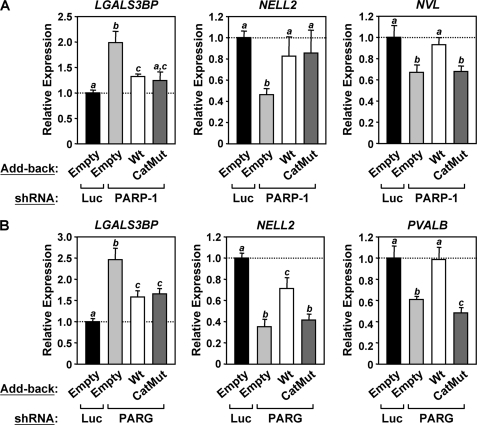
Catalytically inactive mutants of PARP-1 and PARG support the wild-type expression patterns of some, but not all, target genes. RNA interference-resistant wild-type (Wt) or catalytically inactive (CatMut) PARP-1 and PARG were stably expressed in their respective MCF-7 knockdown cells using retrovirus-mediated gene transfer, as shown in Fig. 7. Total RNA was isolated from the cells, reverse-transcribed, and subjected to qPCR using gene-specific primers. Each bar represents the mean + S.E. (error bars) from three or more independent determinations. Bars that do not share at least one lowercase letter marking (a, b, or c) within each graph are statistically different, as determined by analysis of variance with a p value threshold of <0.05. A, re-expression of PARP-1 after PARP-1 knockdown restores the wild-type expression pattern of some PARP-1 target genes. B, re-expression of PARG after PARG knockdown restores the wild-type expression pattern of some PARG target genes.
RESULTS
Generation of PARP-1 and PARG Knockdown Cell Lines
To explore the role of PARP-1 and PARG in the regulation of gene expression, we used shRNA-mediated knockdown to deplete each of the proteins individually in MCF-7 human breast cancer cells. Two distinct shRNA sequences targeting luciferase (Luc, used as a control), PARP-1, or PARG were stably introduced into MCF-7 cells using sequential retrovirus-mediated gene transfer, creating cells doubly targeted for a given factor. Stably transduced cell populations, rather than clonal lines, for each double knockdown were isolated by appropriate drug selection (see “Experimental Procedures”) and tested for knockdown by Western blot analysis. The PARP-1 and PARG proteins were depleted by ∼90 and 70%, respectively, when compared with the Luc control (Fig. 1A). A Western blot for SIRT1 was included as a loading control. Similar effects were also observed on the levels of PARP-1 and PARG mRNA (Fig. 1B). Notably, knockdown of either PARP-1 or PARG in MCF-7 cells had no discernable effect on cell proliferation or cell cycle progression relative to the Luc control for cells maintained under subconfluent growth conditions (supplemental Fig. S2, A and B).
To further characterize these knockdown cell lines, we analyzed PAR (Fig. 1, A and C) and NAD+ (Fig. 1C) levels. Knockdown of PARP-1 in MCF-7 cells reduced, whereas knockdown of PARG enhanced cellular PAR levels, as determined by Western blot with a PAR-specific antibody. Using a quantitative HPLC/mass spectrometry method with 18O-labeled standards (41, 42), we observed an increase in total cellular NAD+ levels upon PARP-1 knockdown and a slight decrease upon PARG knockdown. Together, these results demonstrate that depletion of PARP-1 and PARG alters NAD+ and PAR levels in MCF-7 cells in a predictable manner. Thus, we have generated cell lines with stable shRNA-mediated knockdown of PARP-1 or PARG that can serve as useful models for studying gene regulation.
PARP-1 and PARG Regulate Global Patterns of Gene Expression in MCF-7 cells
To determine the effects of PARP-1 and PARG knockdown on global patterns of gene expression in MCF-7 cells, we isolated total RNA from three control-matched sets of independently generated Luc, PARP-1, and PARG knockdown cell populations. The use of three independently generated populations of cells represents a stringent approach to control for experimental variability and limits the complications that may be observed when using clonally selected lines. Total RNA was isolated, labeled, and hybridized to Affymetrix U133A 2.0 human expression microarrays, which contain more than 22,000 probe sets, including 14,500 well characterized human genes. The raw data were normalized using the Affymetrix GCOS software and adjusted for batch effects using an empirical Bayes method (43). Next, the data were log2-transformed, median-centered, and filtered as described under “Experimental Procedures.” In our initial analyses, we performed filtering based on (i) a detection call of present or marginal in two of the three replicates for both Luc and PARP-1/PARG and (ii) a Student's t test p value cutoff of <0.05 (Fig. 2 and supplemental Fig. S3). In subsequent analyses, we also applied a log2 -fold change cutoff of >0.5 or <−0.5 (Fig. 3). The fully processed and analyzed data yielded lists of genes regulated by PARP-1 or PARG (i.e. genes whose expression changed upon PARP-1 or PARG knockdown; see supplemental Tables S1, S2, and S3 for the regulated gene lists). These data sets contain genes both directly regulated (i.e. primary effects) and indirectly regulated (i.e. secondary effects) by PARP-1 or PARG. For the mechanistic studies described herein (see below), we chose a set of target genes based on the criteria discussed below.
The application of a p value cutoff without a -fold change cutoff is one useful approach to define sets of significantly regulated genes in expression microarray experiments (47–49). Analysis of our expression microarray data in this manner defined ∼1200 genes (∼8.7% of all genes tested) regulated by PARP-1 knockdown and ∼1100 genes (∼8.1% of all genes tested) regulated by PARG knockdown at 95% confidence, with a commonly regulated gene set (i.e. intersection) of ∼500 genes (∼3.5% of all genes tested) (Fig. 2A). A heat map representation (Fig. 2B) and a Spearman correlation analysis (Fig. 2C) of the PARP-1 and PARG commonly regulated gene set (i.e. intersection) show that the majority of genes affected by the knockdown of one factor were similarly affected by the knockdown of the other factor. That is, the changes (i.e. either up-regulated or down-regulated) generally occurred in the same direction and with the same magnitude for both PARP-1 and PARG knockdown (Fig. 2, B and C). This pattern was also evident in the union of regulated gene sets (supplemental Fig. S3), indicating that even for genes not passing the p value cutoff in one condition (i.e. PARP-1 or PARG knockdown), the pattern of regulation was similar. Together, these data indicate that PARP-1 and PARG regulate the expression of a common set of genes in a largely similar manner.
Next, to define the sets of genes most robustly regulated by PARP-1 and PARG knockdown (i.e. the top 15–20% of the regulated genes), we applied a -fold change cutoff (log2 -fold change of >0.5 or <−0.5) to data prefiltered for both detection call and p value, as in Fig. 2A. By these criteria, we identified 204 PARP-1-regulated genes and 217 PARG-regulated genes (Fig. 3A). About half of each group of the most robustly regulated genes was up-regulated, whereas the other half was down-regulated (Fig. 3B). Fifty of the most robustly regulated genes were in both the PARP-1-regulated and the PARG-regulated lists (i.e. the intersection). As expected, based on Fig. 2C and supplemental Fig. S3C, the most robustly regulated genes were regulated in the same direction and with the same magnitude, as illustrated by a Spearman correlation analysis (Fig. 3C). Lists of the most robustly regulated genes can be found in supplemental Tables S1, S2, and S3.
Gene-specific mRNA determinations by RT-qPCR were used to confirm the microarray expression data for a subset of genes falling into four classes: (i) up-regulated by both PARP-1 and PARG knockdown, (ii) down-regulated by both PARP-1 and PARG knockdown, (iii) differentially regulated by both PARP-1 and PARG knockdown, and (iv) not regulated by PARP-1 or PARG knockdown (Fig. 3D). This analysis confirmed a group of more than 20 genes with consistent and well characterized patterns of regulation in response to PARP-1 and PARG knockdown. We focused on these confirmed genes for further analysis in the experiments described below. Additional gene-specific RT-qPCR assays comparing gene expression profiles from single knockdown cell populations indicate that the gene regulatory effects of the shRNAs used in our assays are unlikely to be due to off-target effects (supplemental Fig. S4).
Genes Commonly Regulated by PARP-1 and PARG Are Enriched in Metabolism and Stress-response Functions
To explore the function of the genes most robustly regulated by PARP-1 and PARG knockdown, we performed GO analyses using the DAVID Bioinformatics Resource (Table 1). Both the PARP-1-regulated genes and the PARG-regulated genes from Fig. 3A (i.e. genes passing the present call, p value, and -fold change criteria noted above) are enriched in cell structure and metabolism functions (e.g. ITPR1 for PARP-1, SOCS2 and MTR for PARG). The 50 commonly regulated genes are enriched in stress-response and metabolism functions (e.g. LGALS3BP), consistent with the stress- and metabolic-related phenotypes of PARP-1 and PARG knock-out mice or cells from the knock-out animals grown in culture (28–31).
TABLE 1.
Gene ontology of PARP-1-, PARG-, and commonly-regulated genes in MCF-7 cells
The PARP-1- and PARG-regulated gene lists from Fig. 3B and supplemental Tables 1, 2, and 3, which include genes passing the present call, p value, and -fold change criteria noted under “Results,” were subjected to GO analyses using the DAVID Bioinformatics Resource. Resulting terms were grouped together under each category, and duplicate probe sets were removed for accurate percentage representation of individual genes. Only those GO terms yielding a p value < 0.05 by a Fisher's exact test were considered significantly enriched in each gene list.
| Category | Count | Percentage of total | p value | Examplea |
|---|---|---|---|---|
| PARP-1-regulated genes | ||||
| Cell structureb | 33 | 16% | <0.045 | ITPR1* |
| Metabolismc | 24 | 12% | <0.049 | ALDH5A1 |
| PARG-regulated genes | ||||
| Cell structured | 49 | 23% | <0.049 | SOCS2*, TMOD3* |
| Metabolisme | 24 | 11% | <0.028 | MTR*, NAT1* |
| GTPase regulationf | 9 | 4% | <0.046 | VAV3 |
| Commonly-regulated genes | ||||
| Stress responseg | 8 | 16% | <0.033 | LGALS3BP* |
| Metabolismh | 8 | 16% | <0.049 | PRODH |
a Examples of representative gene(s) for each ontological category. Those marked with an asterisk are genes whose expression patterns in the PARP-1 and PARG knockdown cell lines were confirmed by RT-qPCR, as shown in Fig. 3D.
b GO terms: Plasma membrane.
c GO terms: Organic acid metabolism, carboxylic acid metabolism, amine metabolism, nitrogen compound metabolism, aromatic compound metabolism, and regulation of transferase activity.
d GO terms: Actin binding, cytoplasmic membrane-bound vesicle, cytoplasmic vesicle, membrane-bound vesicle, vesicle, cytoskeletal protein binding, cytoskeleton, endomembrane system, cell organization and biogenesis, and nuclear envelope.
e GO terms: Amino sugar metabolism, amine metabolism, cofactor biosynthesis, coenzyme metabolism, nitrogen compound metabolism, cofactor metabolism, carboxylic acid metabolism, and organic acid metabolism.
fGO terms: GTPase regulator activity and GTPase activator activity.
g GO terms: Response to stress.
h GO terms: Amine metabolism, nitrogen compound metabolism, carboxylic acid metabolism, organic acid metabolism, amino acid metabolism, and amino acid and derivative metabolism.
Exploring the Mechanisms of PARP-1- and PARG-mediated Gene Expression at Target Gene Promoters
After examining the control of global patterns of gene expression in MCF-7 cells by PARP-1 and PARG, we sought to determine the underlying mechanisms of regulation at specific target gene promoters. We considered a variety of criteria that might help us identify target genes for mechanistic studies. Ultimately, we focused on a set of genes (i) showing the most robust alterations in gene expression upon knockdown of PARP-1 and PARG (i.e. from the list of the 50 most robustly regulated genes; Fig. 3A and supplemental Table S3), (ii) exhibiting binding of PARP-1 and PARG at their promoters by ChIP assays (see the next section), and (iii) whose expression can be complemented by re-expression of PARP-1 or PARG in the corresponding knockdown cell lines (see below). We also considered other existing evidence of regulation at the promoters of possible target genes, including changes in chromatin composition (e.g. linker histone binding) and structure (promoter chromatin architecture; see “Discussion”). Together, these criteria were used as a set of parameters to identify targets for further analysis.
PARP-1 and PARG Localize to the Promoters of Target Genes and Can Affect Each Other's Binding
Previous studies have shown that PARP-1 localizes (and in some cases, is recruited in a signal-dependent manner) to the promoters of target genes (14, 15, 25, 50). In fact, a recent genomic analysis from our laboratory has shown that PARP-1 localizes to the promoters of most expressed genes in the genome of MCF-7 cells (25). Whether PARG also localizes to target genes has not been determined.
To address this question, we first needed to validate the use of a new custom PARG antibody for ChIP assays. ChIP, coupled with Western blotting of the immunoprecipitated material (i.e. ChIP-Western), demonstrated that our previously validated PARP-1 antibody (25), as well as a new custom PARG antibody, specifically immunoprecipitate PARP-1 and PARG, respectively, under ChIP conditions (supplemental Fig. S1A). Notably, although our custom antibody recognizes many of the known PARG isoforms in a whole cell lysate (i.e. ChIP input), it specifically enriches for two isoforms in a ChIP assay (supplemental Fig. S1A). The longest of the PARG isoforms is 110 kDa and has been shown to be nuclear, whereas other slightly shorter isoforms are both nuclear and cytoplasmic (18, 19). Our ChIP-Western results demonstrate that the longest PARG isoforms are chromatin-bound. Thus, these antibodies are useful for exploring the localization of PARP-1 and PARG at native promoters in ChIP assays.
We then used the gene expression data from Figs. 1–3, as well as our existing PARP-1 genomic localization data set from MCF-7 cells (25), to identify genes for further examination in ChIP-qPCR assays for PARP-1 and PARG. Our analyses showed that both PARP-1 and PARG localize to the promoters of genes from Fig. 3D with signals of ∼5–∼20-fold over the no antibody (NA) control but not to the promoter of an unregulated gene (i.e. SEMA4G) (Fig. 4). Specificity was demonstrated by a reduction of the ChIP signals upon PARP-1 or PARG knockdown (supplemental Fig. S1, B and C). PARP-1 and PARG occupy the promoters of both up-regulated and down-regulated target genes, indicating gene-specific effects for transcriptional regulation by these factors. Interestingly, the levels of PARG were proportional to the levels of PARP-1 across the genes we examined (Fig. 4). That is, a higher PARG signal corresponded to a higher PARP-1 signal (Spearman correlation coefficient of 0.645, p value <0.05), even for genes that were regulated by knockdown of one factor but not the other (e.g. ITPR1 and NVL). This latter result fits well with the striking correlation between the patterns of gene regulation by PARP-1 and PARG in the microarray expression experiments (Figs. 2C and 3C; supplemental Fig. S3).
Our previous genomic study showed that PARP-1 is enriched at promoters, with regions containing high levels of binding and regions containing low levels of binding (25). ChIP-qPCR analyses in “off-peak” regions confirmed this pattern of binding for both PARP-1 and PARG at selected target gene promoters (e.g. the transcription start site for PVALB) (Fig. 5A). ChIP “tiling” through the LGALS3BP and PVALB promoter regions further confirmed this pattern of binding for both PARP-1 and PARG (Fig. 5B). Together, these results indicate that PARG, like PARP-1, can localize to the promoters of target genes. Furthermore, they show that the localization of PARG correlates with the localization of PARP-1.
The similar localization patterns of PARP-1 and PARG at promoters raised the question of whether they might affect each other's binding (e.g. one recruits the other or they bind cooperatively). To address this issue, we examined the effect of knockdown of one protein on the promoter binding of the other in ChIP assays. Our results indicate that PARP-1 and PARG can indeed affect each other's binding but that they do so in a gene-specific manner (Fig. 6). For example, we found genes, such as NAT1, where the binding of PARP-1 and PARG appear to occur independently; genes, such as NVL, where the binding of PARP-1 requires the binding of PARG; and genes, such as GDF15, where PARP-1 and PARG require each other for binding. These results support the hypothesis that there is a functional interplay between PARP-1 and PARG at target gene promoters, although, not surprisingly, the details of the mechanisms may differ between promoters (see “Discussion”). Collectively, our ChIP analyses indicate that PARP-1 and PARG localize to promoters of target genes to regulate their expression.
Catalytically Inactive Mutants of PARP-1 and PARG Support the Wild-type Expression Patterns of Some, but Not All, Target Genes
Previous studies have provided mixed results about the requirement for the enzymatic activity of PARP-1 during gene regulation. Some studies have indicated that the enzymatic activity of PARP-1 is required (11, 14, 15, 34, 51–53), whereas others have indicated that it is not (50, 54–57). The role of PARG enzymatic activity in the regulation of gene expression has not been examined extensively. To determine the specific requirement of PARP-1 and PARG enzymatic activities in the regulation of target gene expression, we devised a system to stably re-express FLAG-tagged versions of wild type or catalytically inactive mutants of PARP-1 and PARG in their respective knockdown cell lines using retrovirus-mediated gene transfer. To prevent knockdown of the re-expressed proteins in the PARP-1 and PARG MCF-7 knockdown cell lines, we used shRNA-resistant versions of the PARP-1 and PARG cDNAs (Fig. 7, A and C; see “Experimental Procedures”). Using this approach, we were able to restore PARP-1 expression levels to about 20% of the levels in parental cells and PARG expression levels to about 10–20 times the levels in parental cells (Fig. 7, B and D). These levels of re-expression were consistent across multiple independent gene transfer experiments. Re-expression of wild-type PARP-1 modestly but reproducibly increased total PAR levels above the levels seen in the PARP-1 knockdown cells, whereas re-expression of the catalytically inactive PARP-1 mutant did not (supplemental Fig. S5A). Similarly, re-expression of wild-type PARG reduced total PAR levels below the levels seen in the PARG knockdown cells, whereas re-expression of the catalytically inactive PARG mutant did not (supplemental Fig. S5B). These results illustrate that wild-type PARP-1 and PARG and their catalytically inactive mutants alter cellular PAR levels as expected in vivo.
We used these knockdown/add-back cell lines to analyze the roles of PARP-1 and PARG catalytic activity in target gene expression. The expression of selected target genes from the gene-specific expression (Fig. 3D) and ChIP (Fig. 4) experiments was tested in the knockdown/add-back cell lines. For example, the levels of LGALS3BP mRNA, which increased in response to both PARP-1 and PARG knockdown, was largely restored to control levels upon re-expression of either wild-type or catalytically inactive PARP-1 or PARG (Fig. 8, A and B, left panels). These results suggest that neither PARP-1 nor PARG enzymatic activity is required for the proper expression of LGALS3BP. In contrast, the levels of NELL2 mRNA, which decreased in response to both PARP-1 and PARG knockdown, were largely restored to control levels upon re-expression of either wild-type or catalytically inactive PARP-1, as well as wild-type PARG, but not catalytically inactive PARG (Fig. 8, A and B, middle panels). These results suggest that the catalytic activity of PARG, but not PARP-1, is required for the proper expression of NELL2. For this gene, PARG may be required to degrade an alternate (i.e. non-PARP-1) source of PAR, perhaps from PARP-2, another nuclear PARP enzyme (4). Other genes (e.g. NVL and PVALB) also showed a requirement for PARP-1 or PARG catalytic activity for proper expression (Fig. 8, A and B). For the genes not dependent on PARP-1 or PARG catalytic activity, dominant negative effects of the catalytically inactive mutants may contribute to the gene expression outcomes.
The results for LGALS3BP and NELL2 were explored further using chemical inhibitors of PARP-1 (i.e. PJ34) and PARG (i.e. gallotannin; a.k.a. common tannic acid). After verifying the efficacy of the inhibitors using autoPARylation of PARP-1 as an end point (supplemental Fig. S6A), we examined their effects on gene expression (supplemental Fig. 6, B and C). As expected based on the knockdown/add-back experiments, PJ34 had no effect on the expression of LGALS3BP and NELL2, verifying that PARP-1 enzymatic activity is not required for the proper expression of these genes. Likewise, gallotannin inhibited proper expression of NELL2, but not LGALS3BP, verifying the requirement (or lack of requirement) for PARG enzymatic activity (supplemental Fig. 6, B and C). Overall, of the nine PARP-1- or PARG-regulated genes that we tested in detail, about half required the enzymatic activity of the regulating protein (Fig. 8 and data not shown). These results suggest that alternate non-enzymatic functions for PARP-1 and PARG (e.g. protein-protein interactions or scaffolding) in some gene contexts may be important, as suggested by other studies (50, 54–57).
DISCUSSION
Both PARP-1 and PARG, two enzymes functionally linked in the nuclear PAR metabolic pathway, have been implicated in the regulation of gene expression. However, the means by which they coordinate their gene regulatory actions, both globally and at specific target genes, as well as the role of their enzymatic activities in the regulation of gene expression, have not been clearly established. In the current study, we used both genomic and gene-specific assays to address both of these questions in MCF-7 cells. Collectively, our results indicate that PARP-1 and PARG, two nuclear enzymes with opposing enzymatic activities, localize to target promoters and generally act in a similar, rather than antagonist, manner to regulate gene expression.
PARP-1 and PARG Act in Concert to Regulate a Largely Overlapping Gene Set in a Similar Manner
Our microarray experiments have revealed a number of interesting and unexpected facets of global gene regulation by PARP-1 and PARG that were not revealed in previous studies focusing on one factor or the other. First, PARP-1 and PARG regulate the expression of a common gene set generally in the same direction and with the same magnitude (Figs. 2 and 3; supplemental Fig. S3). Based on the seemingly opposing enzymatic activities of PARP-1 and PARG, this result was unexpected. Second, the most robustly regulated common genes are enriched for stress-response and metabolic functions (Table 1), which fits well with the known biological roles of PARP-1 and PARG from animal studies. Third, PARG localizes to the promoters of its target genes (Figs. 4 and 5), as has been demonstrated previously for PARP-1 (14, 15, 25, 50). Furthermore, the levels of chromatin-bound PARG at a given promoter generally correlate with the levels of PARP-1, at least across the subset of promoters tested herein. Finally, PARP-1 and PARG enzymatic activities are required for the regulation of some, but not all, target genes (Fig. 8).
In a simple model of gene regulation, PARG opposes the actions of PARP-1 by degrading the PAR chains synthesized by PARP-1. With respect to the expression analyses presented herein, this model fails in at least two ways. First, PARP-1 and PARG enzymatic activities are required for the regulation of a subset of target genes (Fig. 8). Second, as noted above, gene regulation by PARP-1 and PARG generally occurs in the same direction and with the same magnitude. Thus, our results point to concerted, rather than opposing, actions of PARP-1 and PARG in gene regulation. Interestingly, PARP-1 and PARG animal models demonstrate similarities in the biological endpoints analyzed (2, 23, 27–35). Although they have not yet been explored with respect to gene expression, perhaps the concerted actions of PARP-1 and PARG revealed by our gene expression analyses can provide an explanation to these biological phenomena.
Gene Ontology Analyses Reveal Common Functions for PARP-1- and PARG-regulated Genes
Both PARP-1-regulated and PARG-regulated genes are enriched in cell structure and metabolism functions (e.g. ITPR1 for PARP-1, SOCS2 and MTR for PARG), with the common genes enriched in stress-response and metabolism functions (e.g. LGALS3BP) (Table 1). These results are consistent with (i) the stress- and metabolic-related phenotypes of PARP-1 and PARG knock-out animals (mice, flies, and worms) or cells from knock-out mice grown in culture (28–31, 33–35, 58–66) and (ii) previous gene ontology analyses from microarray expression experiments examining PARP-1-dependent gene regulation. With respect to the latter, PARP-1-regulated genes in mouse embryonic fibroblasts were found to be enriched in functions related to apoptosis, cell cycle control, DNA synthesis/repair, stress and immune responses, chromosomal integrity, and protein processing (67–69). Likewise, PARP-1-regulated genes from mouse embryonic stem cells and livers were found to be enriched in functions related to metabolism, signal transduction, cell cycle control, and transcription (70).
The known functions of the ITPR1, SOCS2, MTR, and LGALS3BP gene products fit with the roles of PARP-1 and PARG in cellular physiology. ITPR1 encodes the type 1 inositol 1,4,5-trisphosphate receptor (71), which plays a critical role in Ca2+ signaling in neuronal, immune, and other cell types (72, 73). SOCS2 encodes a cytokine-inducible SH2 protein that functions as a suppressor of cytokine signaling 2 and plays a role in insulin-signaling pathways (74, 75). MTR encodes 5-methyltetrahydrofolate-homocysteine methyltransferase (a.k.a. methionine synthase), an enzyme that catalyzes the remethylation of homocysteine to methionine (76, 77). Impaired methionine synthase activity leads to elevated levels of plasma homocysteine, which is a risk factor in both birth defects and vascular disease (78). LGALS3BP encodes lectin galactoside-binding soluble 3-binding protein (a.k.a. Mac-2-binding protein and tumor-associated antigen 90K) (79)), a protein that promotes integrin-mediated cell adhesion and is found in increased levels under pathophysiological conditions (e.g. cancer and viral infections) (80). Collectively, our studies and others indicate a clear role for PARP-1- and PARG-regulated genes in metabolism, stress, DNA repair, and signaling functions.
How Might PARP-1 and PARG Act in Concert to Regulate Gene Expression?
The specific mechanisms of PARP-1- and PARG-dependent gene regulation are likely to differ depending on the requirement for PARP-1 and PARG catalytic activity at a particular gene. Previous studies have used chemical inhibitors and mutants to explore the roles of PARP-1 and PARG catalytic activities in the regulation of gene expression (2, 3). In some gene-specific studies, PARP-1 enzymatic activity was required for its gene regulatory functions (11, 14, 15, 34, 51–53), whereas in others, it was not (50, 54–57). The results for PARG have also been variable (11, 26, 27). These results suggest gene-specific (and perhaps cell type-specific) requirements for the PARP-1 and PARG catalytic activities, a result supported by our observations described herein.
For genes requiring PAR metabolism, the goal of the combined PARP-1 and PARG enzymatic activities may be the production of ADPR, rather than the addition and removal of PAR per se. ADPR may function as a signaling molecule in the nucleus by binding as a small molecule ligand to the macro domain of the histone variant macroH2A1.1 (81, 82), which may act to regulate gene expression in a chromatin-dependent manner. For genes that do not require PAR metabolism, PARP-1 and PARG may function as classical coregulators, as has been described previously for PARP-1 (14, 15, 22, 50, 83). Whether they act within common coregulatory complexes is unknown, although a physical interaction between PARP-1 and PARG has been reported in vitro and in vivo, under certain cellular conditions (84). Interestingly, PARP-1 and PARG localize to a similar set of target gene promoters at proportional levels (i.e. higher levels of PARG correspond to higher levels of PARP-1; Figs. 4 and 5). In addition, PARP-1 and PARG facilitate each other's binding to certain promoters (Fig. 6), which is consistent with the presence of both factors in the same complex. However, we also observe independent binding of PARP-1 and PARG to some promoters, suggesting that these factors are also able to function distinctly in some cases. Unlike PARP-1, PARG does not have a DNA binding domain. Thus, when it binds to chromatin independently of PARP-1, it must do so by binding to histones or through interactions with other DNA- or chromatin-binding proteins.
Our results demonstrating non-enzymatic functions for PARP-1 and PARG suggest that they may also function as scaffolding proteins. Indeed, PARP-1 has been shown to participate as a component of promoter-bound coregulatory complexes, perhaps serving as a protein scaffold or “exchange factor” within those complexes (22, 24, 50). Such a scaffolding role has not yet been described for PARG. Furthermore, PARP-1 and PARG may be subject to post-translational modifications that modulate their gene regulatory actions. For example, PARP-1 is known to be phosphorylated by various cellular kinases that can, in some cases, enhance its DNA binding or catalytic activity (85–87).
The regulation of chromatin structure may also be a common component of PARP-1- and PARG-dependent gene expression outcomes (24). We have shown previously that PARP-1 can bind specifically to nucleosomes and modulate chromatin structure in the absence of NAD+ (11, 13), although the release of PARP-1 from the nucleosomes requires its enzymatic activity (11, 13). The binding of PARP-1 and PARG at promoters can also affect the binding of other factors. For example, PARP-1 can regulate the binding of the linker histone H1 at target gene promoters. Specifically, we showed that RNA interference-mediated knockdown of PARP-1 increases the levels of H1 at the ITPR1, NAT1, NELL2, PVALB, and SOCS2 promoters concomitant with reduced expression of the genes (25).7 Given the inhibitory effect of H1 on transcription, the increase in H1 upon the knockdown of PARP-1 is likely to be accompanied by the formation of less accessible and more repressive chromatin structures. These actions of PARP-1 are consistent with previous biochemical assays, suggesting a role for both PARP-1 and PARG in the regulation of chromatin structure and transcription (11).
PARP-1 and PARG Act in Concert to Regulate a Largely Overlapping Gene Set in a Similar Manner
Our microarray experiments have revealed a number of interesting and unexpected facets of global gene regulation by PARP-1 and PARG that were not revealed in previous studies focusing on one factor or the other. First, PARP-1 and PARG regulate the expression of a common gene set generally in the same direction and with the same magnitude (Figs. 2 and 3; supplemental Fig. S3). Based on the seemingly opposing enzymatic activities of PARP-1 and PARG, this result was unexpected. Second, the most robustly regulated common genes are enriched for stress-response and metabolic functions (Table 1), which fits well with the known biological roles of PARP-1 and PARG from animal studies. Third, PARG localizes to the promoters of its target genes (Figs. 4 and 5), as has been demonstrated previously for PARP-1 (14, 15, 25, 50). Furthermore, the levels of chromatin-bound PARG at a given promoter generally correlate with the levels of PARP-1, at least across the subset of promoters tested herein. Finally, PARP-1 and PARG enzymatic activities are required for the regulation of some, but not all, target genes (Fig. 8).
In a simple model of gene regulation, PARG opposes the actions of PARP-1 by degrading the PAR chains synthesized by PARP-1. With respect to the expression analyses presented herein, this model fails in at least two ways. First, PARP-1 and PARG enzymatic activities are required for the regulation of a subset of target genes (Fig. 8). Second, as noted above, gene regulation by PARP-1 and PARG generally occurs in the same direction and with the same magnitude. Thus, our results point to concerted, rather than opposing, actions of PARP-1 and PARG in gene regulation. Interestingly, PARP-1 and PARG animal models demonstrate similarities in the biological endpoints analyzed (2, 23, 27–35). Although they have not yet been explored with respect to gene expression, perhaps the concerted actions of PARP-1 and PARG revealed by our gene expression analyses can provide an explanation to these biological phenomena.
Gene Ontology Analyses Reveal Common Functions for PARP-1- and PARG-regulated Genes
Both PARP-1-regulated and PARG-regulated genes are enriched in cell structure and metabolism functions (e.g. ITPR1 for PARP-1, SOCS2 and MTR for PARG), with the common genes enriched in stress-response and metabolism functions (e.g. LGALS3BP) (Table 1). These results are consistent with (i) the stress- and metabolic-related phenotypes of PARP-1 and PARG knock-out animals (mice, flies, and worms) or cells from knock-out mice grown in culture (28–31, 33–35, 58–66) and (ii) previous gene ontology analyses from microarray expression experiments examining PARP-1-dependent gene regulation. With respect to the latter, PARP-1-regulated genes in mouse embryonic fibroblasts were found to be enriched in functions related to apoptosis, cell cycle control, DNA synthesis/repair, stress and immune responses, chromosomal integrity, and protein processing (67–69). Likewise, PARP-1-regulated genes from mouse embryonic stem cells and livers were found to be enriched in functions related to metabolism, signal transduction, cell cycle control, and transcription (70).
The known functions of the ITPR1, SOCS2, MTR, and LGALS3BP gene products fit with the roles of PARP-1 and PARG in cellular physiology. ITPR1 encodes the type 1 inositol 1,4,5-trisphosphate receptor (71), which plays a critical role in Ca2+ signaling in neuronal, immune, and other cell types (72, 73). SOCS2 encodes a cytokine-inducible SH2 protein that functions as a suppressor of cytokine signaling 2 and plays a role in insulin-signaling pathways (74, 75). MTR encodes 5-methyltetrahydrofolate-homocysteine methyltransferase (a.k.a. methionine synthase), an enzyme that catalyzes the remethylation of homocysteine to methionine (76, 77). Impaired methionine synthase activity leads to elevated levels of plasma homocysteine, which is a risk factor in both birth defects and vascular disease (78). LGALS3BP encodes lectin galactoside-binding soluble 3-binding protein (a.k.a. Mac-2-binding protein and tumor-associated antigen 90K) (79)), a protein that promotes integrin-mediated cell adhesion and is found in increased levels under pathophysiological conditions (e.g. cancer and viral infections) (80). Collectively, our studies and others indicate a clear role for PARP-1- and PARG-regulated genes in metabolism, stress, DNA repair, and signaling functions.
How Might PARP-1 and PARG Act in Concert to Regulate Gene Expression?
The specific mechanisms of PARP-1- and PARG-dependent gene regulation are likely to differ depending on the requirement for PARP-1 and PARG catalytic activity at a particular gene. Previous studies have used chemical inhibitors and mutants to explore the roles of PARP-1 and PARG catalytic activities in the regulation of gene expression (2, 3). In some gene-specific studies, PARP-1 enzymatic activity was required for its gene regulatory functions (11, 14, 15, 34, 51–53), whereas in others, it was not (50, 54–57). The results for PARG have also been variable (11, 26, 27). These results suggest gene-specific (and perhaps cell type-specific) requirements for the PARP-1 and PARG catalytic activities, a result supported by our observations described herein.
For genes requiring PAR metabolism, the goal of the combined PARP-1 and PARG enzymatic activities may be the production of ADPR, rather than the addition and removal of PAR per se. ADPR may function as a signaling molecule in the nucleus by binding as a small molecule ligand to the macro domain of the histone variant macroH2A1.1 (81, 82), which may act to regulate gene expression in a chromatin-dependent manner. For genes that do not require PAR metabolism, PARP-1 and PARG may function as classical coregulators, as has been described previously for PARP-1 (14, 15, 22, 50, 83). Whether they act within common coregulatory complexes is unknown, although a physical interaction between PARP-1 and PARG has been reported in vitro and in vivo, under certain cellular conditions (84). Interestingly, PARP-1 and PARG localize to a similar set of target gene promoters at proportional levels (i.e. higher levels of PARG correspond to higher levels of PARP-1; Figs. 4 and 5). In addition, PARP-1 and PARG facilitate each other's binding to certain promoters (Fig. 6), which is consistent with the presence of both factors in the same complex. However, we also observe independent binding of PARP-1 and PARG to some promoters, suggesting that these factors are also able to function distinctly in some cases. Unlike PARP-1, PARG does not have a DNA binding domain. Thus, when it binds to chromatin independently of PARP-1, it must do so by binding to histones or through interactions with other DNA- or chromatin-binding proteins.
Our results demonstrating non-enzymatic functions for PARP-1 and PARG suggest that they may also function as scaffolding proteins. Indeed, PARP-1 has been shown to participate as a component of promoter-bound coregulatory complexes, perhaps serving as a protein scaffold or “exchange factor” within those complexes (22, 24, 50). Such a scaffolding role has not yet been described for PARG. Furthermore, PARP-1 and PARG may be subject to post-translational modifications that modulate their gene regulatory actions. For example, PARP-1 is known to be phosphorylated by various cellular kinases that can, in some cases, enhance its DNA binding or catalytic activity (85–87).
The regulation of chromatin structure may also be a common component of PARP-1- and PARG-dependent gene expression outcomes (24). We have shown previously that PARP-1 can bind specifically to nucleosomes and modulate chromatin structure in the absence of NAD+ (11, 13), although the release of PARP-1 from the nucleosomes requires its enzymatic activity (11, 13). The binding of PARP-1 and PARG at promoters can also affect the binding of other factors. For example, PARP-1 can regulate the binding of the linker histone H1 at target gene promoters. Specifically, we showed that RNA interference-mediated knockdown of PARP-1 increases the levels of H1 at the ITPR1, NAT1, NELL2, PVALB, and SOCS2 promoters concomitant with reduced expression of the genes (25).7 Given the inhibitory effect of H1 on transcription, the increase in H1 upon the knockdown of PARP-1 is likely to be accompanied by the formation of less accessible and more repressive chromatin structures. These actions of PARP-1 are consistent with previous biochemical assays, suggesting a role for both PARP-1 and PARG in the regulation of chromatin structure and transcription (11).
Acknowledgments
We thank members of the Kraus laboratory for critical reading of this manuscript; Michael Hottiger (University of Zurich) for PARP-1 cDNA; Mitsuko Masutani (National Cancer Center Research Institute, Japan) for the PARG cDNA; Mi Young Kim and Esther Chong for assistance in making some of the PARG expression constructs; Wei Wang and the Cornell University Microarray Core Facility for assistance with the microarray expression experiments; and James Smith and the Cornell University Biomedical Sciences Flow Cytometry Laboratory for assistance with the fluorescence-activated cell sorting analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant DK069710 throughout the NIDDK (to W. L. K.). This work was also supported by a grant from the Endocrine Society (to W. L. K.) and the Cornell University Nanobiotechnology Center, a Science and Technology Centers Program of the National Science Foundation (Agreement Number ECS-9876771).
This article was selected as a Paper of the Week.
The microarray expression data reported in this paper have been submitted to the National Institutes of Health Gene Expression Omnibus (GEO) under GEO accession number GSE12952.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables 1–3, and Figs. S1–S6.
R. Krishnakumar and W. L. Kraus, unpublished results.
- PARylation
- poly(ADP-ribosyl)ation
- PAR
- poly(ADP-ribose)
- PARP-1
- poly(ADP-ribose) polymerase-1
- PARG
- poly(ADP-ribose) glycohydrolase
- ADPR
- ADP-ribose
- shRNA
- short hairpin RNA
- ChIP
- chromatin immunoprecipitation
- qPCR
- quantitative real-time PCR
- RT-qPCR
- reverse transcription-quantitative real-time PCR
- Luc
- luciferase
- MEM
- minimal essential medium
- CDCS
- charcoal/dextran-treated bovine calf serum
- HPLC
- high pressure liquid chromatography
- GO
- gene ontology.
REFERENCES
- 1.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 2.Kim M. Y., Zhang T., Kraus W. L. (2005) Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 3.Kraus W. L., Lis J. T. (2003) Cell 113, 677–683 [DOI] [PubMed] [Google Scholar]
- 4.Amé J. C., Spenlehauer C., de Murcia G. (2004) BioEssays 26, 882–893 [DOI] [PubMed] [Google Scholar]
- 5.Davidovic L., Vodenicharov M., Affar E. B., Poirier G. G. (2001) Exp. Cell Res. 268, 7–13 [DOI] [PubMed] [Google Scholar]
- 6.Rolli V., Ruf A., Augustin A., Schulz G. E., Ménissier-de Murcia J., de Murcia G. (2000) in From DNA Damage and Stress Signalling to Cell Death: Poly ADP-Ribosylation Reactions (de Murcia G., Shall S. eds) pp. 35–79, Oxford Ubiversity Press, New York: [Google Scholar]
- 7.Lonskaya I., Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) J. Biol. Chem. 280, 17076–17083 [DOI] [PubMed] [Google Scholar]
- 8.Potaman V. N., Shlyakhtenko L. S., Oussatcheva E. A., Lyubchenko Y. L., Soldatenkov V. A. (2005) J. Mol. Biol. 348, 609–615 [DOI] [PubMed] [Google Scholar]
- 9.Kun E., Kirsten E., Mendeleyev J., Ordahl C. P. (2004) Biochemistry 43, 210–216 [DOI] [PubMed] [Google Scholar]
- 10.Kun E., Kirsten E., Ordahl C. P. (2002) J. Biol. Chem. 277, 39066–39069 [DOI] [PubMed] [Google Scholar]
- 11.Kim M. Y., Mauro S., Gévry N., Lis J. T., Kraus W. L. (2004) Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 12.Wacker D. A., Frizzell K. M., Zhang T., Kraus W. L. (2007) Subcell. Biochem. 41, 45–69 [DOI] [PubMed] [Google Scholar]
- 13.Wacker D. A., Ruhl D. D., Balagamwala E. H., Hope K. M., Zhang T., Kraus W. L. (2007) Mol. Cell. Biol. 27, 7475–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G. (2006) Science 312, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 15.Ju B. G., Solum D., Song E. J., Lee K. J., Rose D. W., Glass C. K., Rosenfeld M. G. (2004) Cell 119, 815–829 [DOI] [PubMed] [Google Scholar]
- 16.Oei S. L., Shi Y. (2001) Biochem. Biophys. Res. Commun. 284, 450–454 [DOI] [PubMed] [Google Scholar]
- 17.Ogata N., Ueda K., Kawaichi M., Hayaishi O. (1981) J. Biol. Chem. 256, 4135–4137 [PubMed] [Google Scholar]
- 18.Meyer-Ficca M. L., Meyer R. G., Coyle D. L., Jacobson E. L., Jacobson M. K. (2004) Exp. Cell Res. 297, 521–532 [DOI] [PubMed] [Google Scholar]
- 19.Meyer-Ficca M. L., Meyer R. G., Jacobson E. L., Jacobson M. K. (2005) Int. J. Biochem. Cell Biol. 37, 920–926 [DOI] [PubMed] [Google Scholar]
- 20.Bonicalzi M. E., Vodenicharov M., Coulombe M., Gagné J. P., Poirier G. G. (2003) Biol. Cell 95, 635–644 [DOI] [PubMed] [Google Scholar]
- 21.Ohashi S., Kanai M., Hanai S., Uchiumi F., Maruta H., Tanuma S., Miwa M. (2003) Biochem. Biophys. Res. Commun. 307, 915–921 [DOI] [PubMed] [Google Scholar]
- 22.Hassa P. O., Hottiger M. O. (2002) Cell Mol. Life Sci. 59, 1534–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tulin A., Chinenov Y., Spradling A. (2003) Curr. Top. Dev. Biol. 56, 55–83 [DOI] [PubMed] [Google Scholar]
- 24.Kraus W. L. (2008) Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnakumar R., Gamble M. J., Frizzell K. M., Berrocal J. G., Kininis M., Kraus W. L. (2008) Science 319, 819–821 [DOI] [PubMed] [Google Scholar]
- 26.Rapizzi E., Fossati S., Moroni F., Chiarugi A. (2004) Mol. Pharmacol. 66, 890–898 [DOI] [PubMed] [Google Scholar]
- 27.Tulin A., Naumova N. M., Menon A. K., Spradling A. C. (2006) Genetics 172, 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z. Q., Auer B., Stingl L., Berghammer H., Haidacher D., Schweiger M., Wagner E. F. (1995) Genes Dev. 9, 509–520 [DOI] [PubMed] [Google Scholar]
- 29.Wang Z. Q., Stingl L., Morrison C., Jantsch M., Los M., Schulze-Osthoff K., Wagner E. F. (1997) Genes Dev. 11, 2347–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortes U., Tong W. M., Coyle D. L., Meyer-Ficca M. L., Meyer R. G., Petrilli V., Herceg Z., Jacobson E. L., Jacobson M. K., Wang Z. Q. (2004) Mol. Cell. Biol. 24, 7163–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh D. W., Lawler A. M., Poitras M. F., Sasaki M., Wattler S., Nehls M. C., Stöger T., Poirier G. G., Dawson V. L., Dawson T. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17699–17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher A. E., Hochegger H., Takeda S., Caldecott K. W. (2007) Mol. Cell. Biol. 27, 5597–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St-Laurent J. F., Gagnon S. N., Dequen F., Hardy I., Desnoyers S. (2007) DNA Repair 6, 329–343 [DOI] [PubMed] [Google Scholar]
- 34.Tulin A., Spradling A. (2003) Science 299, 560–562 [DOI] [PubMed] [Google Scholar]
- 35.Tulin A., Stewart D., Spradling A. C. (2002) Genes Dev. 16, 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T., Berrocal J. G., Frizzell K. M., Gamble M. J., DuMond M. E., Krishnakumar R., Yang T., Sauve A. A., Kraus W. L. (2009) J. Biol. Chem. 284, 20408–20417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah R. G., Ghodgaonkar M. M., Affar el B., Shah G. M. (2005) Biochem. Biophys. Res. Commun. 331, 167–174 [DOI] [PubMed] [Google Scholar]
- 38.Marsischky G. T., Wilson B. A., Collier R. J. (1995) J. Biol. Chem. 270, 3247–3254 [DOI] [PubMed] [Google Scholar]
- 39.Rolli V., O'Farrell M., Ménissier-de Murcia J., de Murcia G. (1997) Biochemistry 36, 12147–12154 [DOI] [PubMed] [Google Scholar]
- 40.Shimokawa T., Masutani M., Nagasawa S., Nozaki T., Ikota N., Aoki Y., Nakagama H., Sugimura T. (1999) J Biochem. 126, 748–755 [DOI] [PubMed] [Google Scholar]
- 41.Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., Sinclair D. A. (2007) Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang T., Sauve A. A. (2006) AAPS J. 8, E632–E643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson W. E., Li C., Rabinovic A. (2007) Biostatistics 8, 118–127 [DOI] [PubMed] [Google Scholar]
- 44.Saldanha A. J. (2004) Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 45.Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 46.Kininis M., Chen B. S., Diehl A. G., Isaacs G. D., Zhang T., Siepel A. C., Clark A. G., Kraus W. L. (2007) Mol. Cell. Biol. 27, 5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dow G. S. (2003) Malar. J. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hess A., Iyer H. (2007) BMC Genomics 8, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy D. (2002) Adv. Physiol. Educ. 26, 256–270 [DOI] [PubMed] [Google Scholar]
- 50.Pavri R., Lewis B., Kim T. K., Dilworth F. J., Erdjument-Bromage H., Tempst P., de Murcia G., Evans R., Chambon P., Reinberg D. (2005) Mol. Cell 18, 83–96 [DOI] [PubMed] [Google Scholar]
- 51.Butler A. J., Ordahl C. P. (1999) Mol. Cell. Biol. 19, 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nirodi C., NagDas S., Gygi S. P., Olson G., Aebersold R., Richmond A. (2001) J. Biol. Chem. 276, 9366–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyamoto T., Kakizawa T., Hashizume K. (1999) Mol. Cell. Biol. 19, 2644–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson M. G., Scoggin K. E., Simbulan-Rosenthal C. M., Steadman J. A. (2000) J. Virol. 74, 2169–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cervellera M. N., Sala A. (2000) J. Biol. Chem. 275, 10692–10696 [DOI] [PubMed] [Google Scholar]
- 56.Hassa P. O., Covic M., Hasan S., Imhof R., Hottiger M. O. (2001) J. Biol. Chem. 276, 45588–45597 [DOI] [PubMed] [Google Scholar]
- 57.Meisterernst M., Stelzer G., Roeder R. G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2261–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conde C., Mark M., Oliver F. J., Huber A., de Murcia G., Ménissier-de Murcia J. (2001) EMBO J. 20, 3535–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrison C., Smith G. C., Stingl L., Jackson S. P., Wagner E. F., Wang Z. Q. (1997) Nat. Genet. 17, 479–482 [DOI] [PubMed] [Google Scholar]
- 60.Tong W. M., Hande M. P., Lansdorp P. M., Wang Z. Q. (2001) Mol. Cell. Biol. 21, 4046–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mabley J. G., Suarez-Pinzon W. L., Haskó G., Salzman A. L., Rabinovitch A., Kun E., Szabó C. (2001) Br. J. Pharmacol. 133, 909–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oliver F. J., Ménissier-de Murcia J., Nacci C., Decker P., Andriantsitohaina R., Muller S., de la Rubia G., Stoclet J. C., de Murcia G. (1999) EMBO J. 18, 4446–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. (2002) Science 297, 259–263 [DOI] [PubMed] [Google Scholar]
- 64.Zong W. X., Ditsworth D., Bauer D. E., Wang Z. Q., Thompson C. B. (2004) Genes Dev. 18, 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao H., Coyle D. L., Meyer-Ficca M. L., Meyer R. G., Jacobson E. L., Wang Z. Q., Jacobson M. K. (2007) Exp. Cell Res. 313, 984–996 [DOI] [PubMed] [Google Scholar]
- 66.Hanai S., Kanai M., Ohashi S., Okamoto K., Yamada M., Takahashi H., Miwa M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zingarelli B., Hake P. W., O'Connor M., Denenberg A., Kong S., Aronow B. J. (2003) Mol. Med. 9, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simbulan-Rosenthal C. M., Ly D. H., Rosenthal D. S., Konopka G., Luo R., Wang Z. Q., Schultz P. G., Smulson M. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11274–11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simbulan-Rosenthal C. M., Rosenthal D. S., Iyer S., Boulares H., Smulson M. E. (1999) Mol. Cell Biochem. 193, 137–148 [PubMed] [Google Scholar]
- 70.Ogino H., Nozaki T., Gunji A., Maeda M., Suzuki H., Ohta T., Murakami Y., Nakagama H., Sugimura T., Masutani M. (2007) BMC Genomics 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamada N., Makino Y., Clark R. A., Pearson D. W., Mattei M. G., Guénet J. L., Ohama E., Fujino I., Miyawaki A., Furuichi T., Mikoshiba K. (1994) Biochem. J. 302, 781–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.deSouza N., Cui J., Dura M., McDonald T. V., Marks A. R. (2007) J. Cell Biol. 179, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue T., Kato K., Kohda K., Mikoshiba K. (1998) J. Neurosci. 18, 5366–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minamoto S., Ikegame K., Ueno K., Narazaki M., Naka T., Yamamoto H., Matsumoto T., Saito H., Hosoe S., Kishimoto T. (1997) Biochem. Biophys. Res. Commun. 237, 79–83 [DOI] [PubMed] [Google Scholar]
- 75.Dey B. R., Spence S. L., Nissley P., Furlanetto R. W. (1998) J. Biol. Chem. 273, 24095–24101 [DOI] [PubMed] [Google Scholar]
- 76.Li Y. N., Gulati S., Baker P. J., Brody L. C., Banerjee R., Kruger W. D. (1996) Hum. Mol. Genet. 5, 1851–1858 [DOI] [PubMed] [Google Scholar]
- 77.Leclerc D., Campeau E., Goyette P., Adjalla C. E., Christensen B., Ross M., Eydoux P., Rosenblatt D. S., Rozen R., Gravel R. A. (1996) Hum. Mol. Genet. 5, 1867–1874 [DOI] [PubMed] [Google Scholar]
- 78.Watkins D., Rosenblatt D. S. (1989) Am. J. Med. Genet. 34, 427–434 [DOI] [PubMed] [Google Scholar]
- 79.Koths K., Taylor E., Halenbeck R., Casipit C., Wang A. (1993) J. Biol. Chem. 268, 14245–14249 [PubMed] [Google Scholar]
- 80.Tinari N., Kuwabara I., Huflejt M. E., Shen P. F., Iacobelli S., Liu F. T. (2001) Int. J. Cancer 91, 167–172 [DOI] [PubMed] [Google Scholar]
- 81.Karras G. I., Kustatscher G., Buhecha H. R., Allen M. D., Pugieux C., Sait F., Bycroft M., Ladurner A. G. (2005) EMBO J. 24, 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A. G. (2005) Nat. Struct. Mol. Biol. 12, 624–625 [DOI] [PubMed] [Google Scholar]
- 83.Hassa P. O., Hottiger M. O. (1999) Biol. Chem. 380, 953–959 [DOI] [PubMed] [Google Scholar]
- 84.Keil C., Gröbe T., Oei S. L. (2006) J. Biol. Chem. 281, 34394–34405 [DOI] [PubMed] [Google Scholar]
- 85.Gagné J. P., Moreel X., Gagné P., Labelle Y., Droit A., Chevalier-Paré M., Bourassa S., McDonald D., Hendzel M. J., Prigent C., Poirier G. G. (2009) J. Proteome Res. 8, 1014–1029 [DOI] [PubMed] [Google Scholar]
- 86.Kauppinen T. M., Chan W. Y., Suh S. W., Wiggins A. K., Huang E. J., Swanson R. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7136–7141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mathieu J., Flexor M., Lanotte M., Besancon F. (2008) Oncogene 27, 3361–3370 [DOI] [PubMed] [Google Scholar]