Abstract
Insulin-degrading enzyme (IDE), a 110-kDa metalloendopeptidase, hydrolyzes several physiologically relevant peptides, including insulin and amyloid-β (Aβ). Human IDE has 13 cysteines and is inhibited by hydrogen peroxide and S-nitrosoglutathione (GSNO), donors of reactive oxygen and nitrogen species, respectively. Here, we report that the oxidative burst of BV-2 microglial cells leads to oxidation or nitrosylation of secreted IDE, leading to the reduced activity. Hydrogen peroxide and GSNO treatment of IDE reduces the Vmax for Aβ degradation, increases IDE oligomerization, and decreases IDE thermostability. Additionally, this inhibitory response of IDE is substrate-dependent, biphasic for Aβ degradation but monophasic for a shorter bradykinin-mimetic substrate. Our mutational analysis of IDE and peptide mass fingerprinting of GSNO-treated IDE using Fourier transform-ion cyclotron resonance mass spectrometer reveal a surprising interplay of Cys-178 with Cys-110 and Cys-819 for catalytic activity and with Cys-789 and Cys-966 for oligomerization. Cys-110 is near the zinc-binding catalytic center and is normally buried. The oxidation and nitrosylation of Cys-819 allow Cys-110 to be oxidized or nitrosylated, leading to complete inactivation of IDE. Cys-789 is spatially adjacent to Cys-966, and their nitrosylation and oxidation together trigger the oligomerization and inhibition of IDE. Interestingly, the Cys-178 modification buffers the inhibition caused by Cys-819 modification and prevents the oxidation or nitrosylation of Cys-110. The Cys-178 modification can also prevent the oligomerization-mediated inhibition. Thus, IDE can be intricately regulated by reactive oxygen or nitrogen species. The structure of IDE reveals the molecular basis for the long distance interactions of these cysteines and how they regulate IDE function.
Introduction
Reactive oxygen species (ROS)2 and reactive nitrogen species (RNS) are two major types of free radicals that have been identified in physiology. Superoxide anion radical (O2˙̄), hydrogen peroxide (H2O2), and the highly reactive hydroxyl radical (OH•), generated by cellular oxidases, are members of the ROS (1, 2). Nitric oxide (NO), generated by NO synthases, and its derivatives, peroxynitrite (ONOO−), dinitrogen trioxide (N2O3), and S-nitrosothiols, make up the RNS (3, 4). ROS and RNS play a critical role in regulating many key physiological responses such as signaling processes, immune responses, and aging (1, 5–8). Many of these effects are exerted via the reversible modification of thiol-containing molecules, such as cysteines, resulting in a change of structure, activity, or both. Prominent post-translational modifications of cysteines induced by ROS and RNS include sulfenic acid formation and S-nitrosylation, respectively (3, 4). Excessive ROS and RNS can also lead to nonspecific modifications of proteins that can damage their native functions and may in part be responsible for various human diseases (6, 9). Although this is apparently a widespread phenomenon, little is known about the molecular basis for the alteration of protein function by oxidative/nitrosative stress. Thus, the need to study ROS/RNS-mediated modifications in proteins is evident.
Insulin-degrading enzyme (IDE, EC 3.4.24.56), crystallized as a homodimer, consisting of 110-kDa subunits (10–13), is a zinc metalloprotease known to cleave multiple short polypeptides that vary considerably in sequence (10, 14–16). Considerable interest in IDE has been stimulated due to the discovery that IDE can degrade insulin (17), amyloid-β (Aβ) (14, 18, 19), and atrial natriuretic peptide (20), peptides implicated in the pathogenesis of diabetes, Alzheimer disease, and cardiovascular disease, respectively. An interesting feature of the human form of this enzyme is the presence of 13 cysteines that do not directly contribute to catalysis, as demonstrated by the production of fully active forms of IDE that were engineered free of all 13 cysteines (13, 21).
IDE was originally mis-identified as a cysteine protease due to its inhibition by thiol-directed alkylation by N-ethylmaleimide with Cys-819 identified as the residue primarily responsible for the loss of activity (13, 21). Interestingly, similar to NEM alkylation, Cys-819 is also susceptible to oxidation and nitrosylation (13), resulting in the inactivation of IDE (13, 22). However, IDE behaves differently in the presence of oxidizing or nitrosylating agents as compared with the alkylative agent, NEM, indicating that a different mechanism is likely involved in these physiologically relevant regulations (13). The molecular basis of IDE regulation by oxidation and nitrosylation has not been explored. Additionally, whether an oxidative burst can inhibit endogenous IDE in cell-culture based conditions has not been examined.
To better understand the effect and underpinning mechanism of oxidative and nitrosylative modification of IDE, we investigated whether an oxidative burst of the microglial cell line, BV-2, can specifically inhibit the activity of secreted IDE. We also examined the biochemical effect of the ROS/RNS modification of the enzyme by fully characterizing the kinetics and the biophysical properties of the modified form of IDE. We then used mutational and proteomic approaches to define the roles of five specific cysteines that are involved in the modulation of the structure and the catalytic activity of IDE.
EXPERIMENTAL PROCEDURES
Materials
H2O2, S-nitrosoglutathione (GSNO), NEM, trypsin singles (proteomics grade), endoproteinase Lys-C, ampicillin, reduced glutathione, 5,5′-dithiobis(2-nitrobenzoate) (DTNB), β-mercaptoethanol, phenylmethanesulfonyl fluoride, lipopolysaccharide (LPS-O55:B5), 1,10-phenanthroline monohydrate, and ammonium bicarbonate were all obtained from Sigma. Fluorogenic peptide substrate V and amyloid-β were obtained from EZBiolab. Fluorescein-Aβ-(1–40)-Lys-biotin (FAβB) was purchased from AnaSpec. Neutravidin was obtained from Pierce. Oligonucleotides for mutagenesis, primers for DNA sequencing, and Griess reagent were purchased from Invitrogen. Reagents used for mutagenesis were from Stratagene. The kits used for plasmid extraction were from Qiagen, Inc., and Promega. Nickel-nitrilotriacetic acid-agarose was purchased from Qiagen, Inc. Source-15Q and Superdex 200 resins were obtained from GE Healthcare. Dye reagent concentrate was purchased from Bio-Rad. All other chemicals were of reagent grade.
Expression and Purification of IDE
All His tag wild-type and mutant enzymes (see Figs. 3–5 and Tables 1–3) were expressed in Rosetta (DE3) Escherichia coli cells. In each case, the cell cultures were grown at 37 °C in T7 medium containing 0.1 mg/ml ampicillin and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 25 °C for ∼18 h. The cell cultures were then centrifuged and collected, as described previously (12). In this method, the cell pellet of wild-type or mutant enzyme was sonicated, and the supernatant was applied to nitrilotriacetic acid-agarose, and IDE was eluted with 150 mm imidazole (11, 13, 23). Enzymes collected at this step were considered only partially pure (∼80%) and were utilized for preliminary analyses. The enzymes were further separated from the other proteins by elution through an anion exchange column (Source-Q) followed by one or two rounds of size-exclusion chromatography (Superdex 200) (11, 13, 23). The fractions with the fully purified IDE were pooled and concentrated using Amicon 30 (30 kDa) and stored at −80 °C.
FIGURE 3.
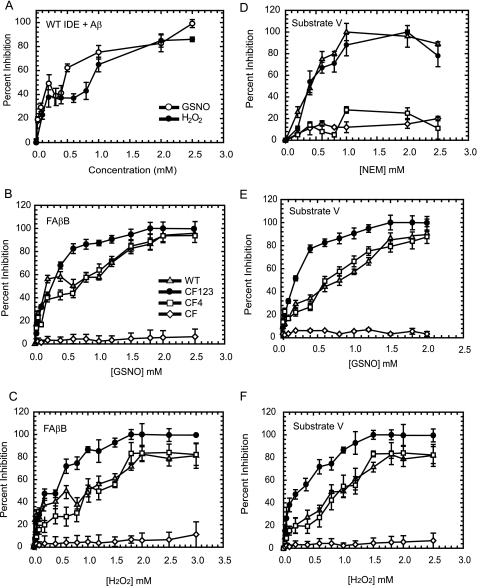
Concentration dependence for the inhibition of IDE by H2O2, GSNO, and NEM. IDE-WT (1.0 mg/ml) was incubated for 1 h with a range of concentrations of H2O2 (●) and GSNO (○), and the inhibition of IDE was measured by monitoring the degradation of 100 μm amyloid-β (A). In addition to WT IDE (▴), concentration-dependent inhibition was also evaluated for IDE-CF123 (●), IDE-CF4 (□), and IDE-CF (224) by GSNO (B) and H2O2 (C) with respect to 1.5 μm FAβB as a substrate. For comparison, concentration-dependent inhibition of IDE by NEM (D), GSNO (E), and H2O2 (F) was also measured with respect to 0.5 μm fluorogenic substrate V as a substrate.
FIGURE 4.
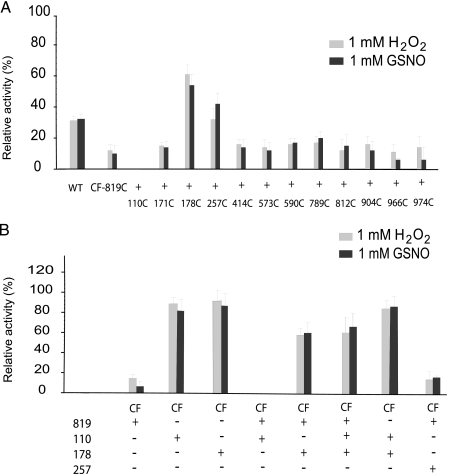
H2O2 and GSNO sensitivity of IDE. A single cysteine was restored from domains 1 to 4 into the IDE-CF-819C background, and activities were measured for partially purified samples upon incubation with H2O2 and GSNO. IDE-WT and IDE-CF-A819C were also analyzed for comparison (A). Relevant cysteines identified in B were introduced back into IDE-CF background, fully purified to exclude effects of contaminants from less purified samples, and assayed for activity after 1 h of incubation with H2O2 and GSNO (B). Relative activities are reported assuming as 100% the control reaction where each sample was incubated with buffer only under the same conditions. Activity was monitored by measuring the fluorescence resulting from the cleavage of 1.5 μm FAβB.
FIGURE 5.
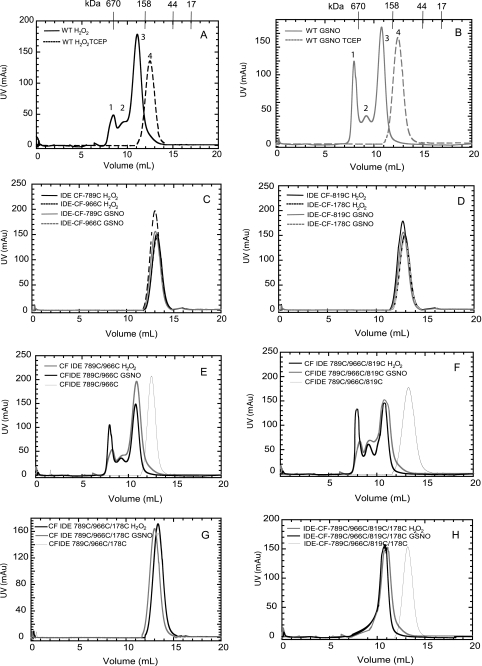
Gel filtration elution profiles of WT and mutant enzymes at 4 °C. These elution profiles were obtained by applying 1 ml of each protein (∼1 mg) to a Superdex 200 column equilibrated with 20 mm Tris-Cl buffer, pH 7.0, containing 50 mm NaCl. The molecular mass standards used are thyroglobulin, 670 kDa; γ-globulin, 158 kDa; ovalbumin, 44 kDa; and myoglobin, 17 kDa. The elution profiles of WT IDE were treated with H2O2 (A) and GSNO (B). The elution profiles of IDE-CF-789C, IDE-CF-966C (C), IDE-CF-819C, IDE-CF-178C (D), IDE-CF-789C/966C (E), IDE-CF-789C/966C/819C (F), IDE-CF-789C/966C/178C (G), and IDE-CF-819C/789C/966C/178C (H) enzymes were treated with H2O2 or GSNO as indicated. mAU, milli-absorbance units.
TABLE 1.
Summary of the H2O2 and GSNO IC50 for wild-type and various mutants of IDE
The enzymes were incubated for 1 h with a range of concentrations (10–5000 μm) of H2O2 or GSNO, and the residual activity of IDE was measured by monitoring the degradation of 1.5 μm FAβB as described under “Experimental Procedures.” IC50 values ± S.D. were calculated from mean inhibition dose-response curves obtained from an average of two experiments for each enzyme.
| Enzyme | IC50 |
|
|---|---|---|
| H2O2 | GSNO | |
| μm | ||
| IDE-WT | 900 ± 100 | 1200 ± 200 |
| IDE-CF123 | 220 ± 30 | 330 ± 30 |
| IDE-CF4 | 700 ± 100 | 1000 ± 100 |
| IDE-CF | >5000 | >5000 |
| IDE-CF-110C | >5000 | >5000 |
| IDE-CF-178C | >5000 | >5000 |
| IDE-CF-110C/178C | >5000 | >5000 |
| IDE-CF-966C | >5000 | >5000 |
| IDE-CF-819C | 190 ± 30 | 250 ± 50 |
| IDE-CF-819C/110C | 30 ± 10 | 30 ± 10 |
| IDE-CF-819C/178C | 900 ± 200 | 1000 ± 100 |
| IDE-CF-819C/178C/110C | 800 ± 100 | 1300 ± 100 |
TABLE 2.
Free sulfhydryls present in IDE after incubation with H2O2 or GSNO
Incubations were performed by mixing 1 mg/ml purified IDE samples with 1 mm H2O2 or 1 mm GSNO in 20 mm Tris-Cl, pH 8.0, containing 50 mm NaCl. The excess reagent was separated from the modified enzyme by gel filtration, and each sample was subsequently assayed with 5,5′-dithiobis(2-nitrobenzoate) as described under “Experimental Procedures.”
| Enzyme | Control(–SH/subunit) | +H2O2 (–SH/subunit) | +GSNO(–SH/subunit) |
|---|---|---|---|
| IDE-WT | 13.1 | 9.2 | 8.8 |
| IDE-CF | 0 | 0 | 0 |
| IDE-CF-819C | 1.1 | 0 | 0 |
| IDE-CF-178C | 1.0 | 0.1 | 0.3 |
| IDE-CF-110C | 1.2 | 1.0 | 0.9 |
| IDE-CF-819C/110C | 2.2 | 0.2 | 0.1 |
| IDE-CF-819C/178C | 2.3 | 0.1 | 0 |
| IDE-CF-819C/110C/178C | 3.2 | 1.3 | 1.1 |
| GSH | 1.0 |
TABLE 3.
NO-modified peptides identified by LTQ FT-ICR MS/MS in IDE
Modified cysteines are shown in parentheses.
Reaction of H2O2 and GSNO with IDE
IDE enzymes (1.0 mg/ml) were incubated in the dark in 20 mm Tris-Cl buffer, pH 7.2, containing 50 mm NaCl at 25 °C with various concentrations (0.01–5 mm) of H2O2 or GSNO by the addition of appropriate stock solutions of H2O2 or GSNO in water. In control experiments, the enzyme was incubated under the same conditions, including water but excluding H2O2 or GSNO. In every case, aliquots of the reaction mixture were removed at specified times and assayed for enzymatic activities using either 0.5 μm fluorogenic substrate V (24) or 1.5 μm FAβB (25) as substrate. In the preparation of modified and control enzyme (incubated for 1 h with 1 mm H2O2 or 1 mm GSNO), excess reagent was separated from the enzyme by gel filtration in which aliquots (1.0 ml) of the reaction mixture were applied to a column of Superdex-200 equilibrated with 20 mm Tris-Cl buffer, pH 7.2, containing 50 mm NaCl. The protein concentration was then determined using the Bio-Rad protein reagent, which is based on the dye-binding method of Bradford (26). Bovine serum albumin was used as the protein standard, and absorbance at 600 nm was measured using a Bio-Rad model 2550 RIA reader.
Thermostability of Native and Modified IDE
IDE-WT (1 mg/ml) was incubated for 1 h in the absence or presence of 1 mm H2O2 or GSNO under standard reaction conditions. Excess reagents were removed by gel filtration columns, and the protein concentration was determined by the Bio-Rad method, as described above. The recovered enzymes were subsequently incubated at 1 mg/ml at 37 °C, and aliquots were assayed for activity using FAβB as a substrate (25) every 15 min for the 1st h and every 30 min for the next 3 h.
Sulfhydryl Determinations
In the preparation of the enzymes for the DTNB assay, excess H2O2 and GSNO was separated from the proteins by gel filtration as described above. Aliquots of each enzyme (0.2 ml of ∼1 mg/ml in 20 mm Tris-HCl buffer, pH 8.0, containing 50 mm NaCl) were added to 0.6 ml of 20 mm Tris-HCl buffer, pH 8.0, containing 50 mm NaCl, and SDS (0.1 ml, 10% (w/v)) was added to denature the protein. The absorbance of the solution was read at 412 nm against a protein-free buffer blank that was identical to the test solution. A freshly prepared 10 mm DTNB solution (0.1 ml) was added to the reaction mixture and the protein-free buffer blank. The reaction was complete after 5 min. The concentration of free –SH groups in the enzyme was calculated from the change in absorbance, using a molar extinction coefficient of 13,600 m−1 cm−1 at 412 nm for thionitrobenzoate (27).
Determination of the Rate of Degradation of Aβ by Modified IDE
Kinetic analysis of Aβ degradation was conducted and analyzed as described previously (12, 25).
BV-2 Maintenance and Stimulation of ROS/RNS
BV-2 cells (immortalized mouse microglia), developed by Dr. Elisabetta Blasi (28), have been shown to secrete IDE3 (29). The BV-2 cells were maintained in a 1:1 ratio of Dulbecco's modified Eagle's medium and F-12 media with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, plated at 2 × 105 cells/ml, and passaged twice a week. The conditioned media were collected, filtered, and centrifuged at 100,000 × g for 15 min to remove cell debris and membranes. Aliquots of the supernatant were stored at −80 °C without added protease inhibitors. BV-2 cells have been shown to respond to classical stimulation paradigms involving interferon-γ priming and exposure to LPS (30–32), resulting in an inflammatory response that includes NO production. Thus, we explored stimulation of the BV-2 cells with priming by interferon-γ (10 or 100 ng/ml) for 24 h before subsequent LPS exposure (1, 10, or 50 μg/ml). Media were collected every 24 h for 3 days to determine optimal doses of interferon-γ and LPS for NO production. As a measure of NO synthesis during the culture period, nitrite production, which accumulated in the culture media, was measured by a spectrophotometric assay based on the Griess reaction as described previously (33, 34).
Western Blotting of IDE from BV-2 Cell Media
Equal volumes of BV-2 cell media (treated and untreated with LPS) (concentrated 10-fold by Amicon-30 filter) were electrophoresed on 7% SDS-polyacrylamide gels and transferred to polyvinylidine difluoride membrane. An anti-IDE polyclonal antibody (Invitrogen) was used at 1:1000 dilution.
MS Analysis for Identifying S-Nitrosylated Sites of IDE
BV-2 IDE (in the absence or presence of LPS treatment) and GSNO-treated (and untreated) IDE were digested with Lys-C enzyme (final enzyme/protein ratio was 1:20) in 50 mm ammonium carbonate buffer (pH 8) at 37 °C for 12 h, after which modified trypsin was added to the sample (final enzyme/protein ratio was 1:10) and incubated at 37 °C for another 12 h. For BV-2-secreted IDE, the tryptic digestion of in-gel IDE was performed as described (35). Aliquots of digested IDE were loaded onto C18 ZipTip columns (Millipore). Peptides were eluted with 15 μl of a 4:1 acetonitrile/water solution in 0.1% formic acid and then diluted 1:1 with 0.1% formic acid.
The proteolytic digests of IDE were separated by nano-liquid chromatography on a 15-cm × 75-μm inner diameter reverse-phase C8 column with a linear gradient of 5–95% acetonitrile in 0.1% formic acid at a flow rate of 200 nl/min. FT-ICR MS analysis was performed on a linear quadrupole ion trap (LTQ) FT-ICR hybrid mass spectrometer. Eluted tryptic and Lys-C peptides were electrosprayed at 2 kV. Peptide fragmentation was induced by collision-induced dissociation in the ion trap, and fragment ions were also analyzed in the ion trap. The LTQ FT mass spectrometer was operated in a “top three” data-dependent acquisition mode. The mass spectrometer was set to switch between an FT-ICR MS full scan (200–2,000 m/z) followed by successive FT-ICR MS single-ion monitoring scans and LTQ MS/MS scans of the three most abundant precursor ions in the FT-ICR MS full scan as determined by the Xcalibur software (Thermo Electron Corp.).
The liquid chromatography-electrospray ionization LTQ MS/MS data were searched against a custom FASTA sequence data base containing IDE, as well as unrelated human proteins as a negative control, by use of the programs MASCOT and BioWorks. Monoisotopic precursor and fragment ion masses were searched with a mass tolerance of 10 and 5 ppm, respectively. For identification of any oxidized/nitrosylated residue, the MASCOT/BioWorks searches were amended to search for the mass addition of all known modifications caused by oxidation and nitrosylation of any IDE residue. Additionally, FT-ICR MS spectra were extracted out of each sample data set for manual identification of modifications based on high mass accuracy. The modified peptides were manually validated by their absence in the unmodified FT-ICR MS spectra; a mass accuracy cut-off of 10 ppm was used.
RESULTS
Catalytic Activity of IDE Is Reduced under Oxidative Stress in Cell Culture-based Conditions
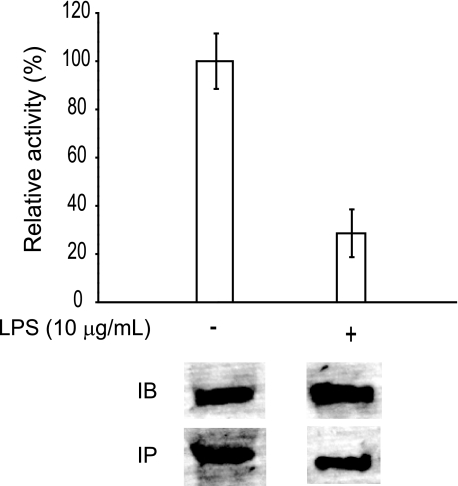
The vulnerability of endogenously produced IDE to oxidative stress under biologically relevant conditions has not been demonstrated. The immortalized mouse microglial cell line, BV-2, naturally secretes high levels of IDE3 (29, 36). To evaluate the effect of biological oxidant production on the catalytic activity of secreted IDE by BV-2 cells, we treated these cells with bacterial LPS, which are known to elicit a surge in oxidative activity. Indeed, a 24-h treatment of BV-2 cells with LPS results in an increase in nitrite levels from less than 5 to ∼80 μm. Thus, BV-2 cells were stimulated with LPS for 24 h, and the residual activity of the secreted IDE was measured using the FAβB degrading assay. LPS-stimulated BV-2 cells only had ∼30% of the FAβB degrading activity of the unstimulated cells (Fig. 1). Immunodepletion of the BV-2 medium with an IDE-specific polyclonal antibody removed ∼95% of the FAβB degrading activity, suggesting that the activity measured is indeed that of IDE.
FIGURE 1.
Relative activity of endogenous IDE from microglial BV-2 cells upon LPS stimulation. Relative activities of endogenous IDE in the conditioned media of mouse BV-2 cells in the absence (−) or presence (+) of LPS treatment. The IDE content was then analyzed directly after electrophoresis by immunoblot (IB) or after immunoprecipitation (IP).
This reduction of activity could be due to decreased levels of IDE upon LPS treatment. To address this, we performed an immunoblot of BV-2 media (Fig. 1, lower panel labeled IB) and found comparable levels of secreted IDE pre- and post-LPS treatment. We then immunoprecipitated IDE from the cultured media and immunoblotted the resulting pellet, confirming that the 110-kDa band is indeed IDE and that its production is comparable in the absence or presence of LPS treatment (Fig. 1, lower panel labeled IP). Therefore, we conclude that the activity, and not the production/degradation of the secreted IDE from BV-2 cells, is inhibited by oxidative/nitrosative stress.
Effects of H2O2 and GSNO Treatments on Half-life, Enzyme Kinetics, Oligomerization, and Thermostability of IDE
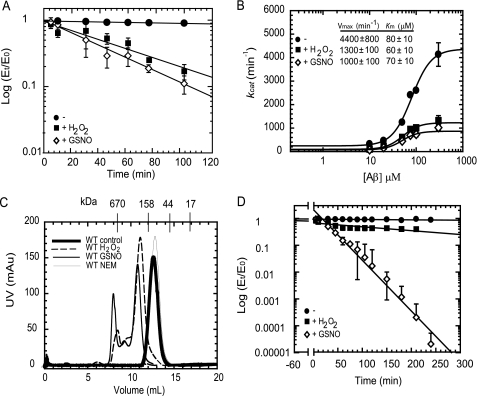
To understand the biochemical and biophysical effects of H2O2 and GSNO on IDE, the recombinant human IDE purified from E. coli (1 mg/ml) was treated with 1 mm H2O2 or 1 mm GSNO, representatives of physiologically relevant donors of reactive oxygen and nitrogen species. This results in a time-dependent inactivation (when assayed with FAβB as a substrate) with a rate constant of 0.022 ± 0.007 min−1, t½ = ∼32 min, and 0.018 ± 0.005 min−1, t½1= ∼39 min for the GSNO-treated and H2O2-treated enzymes, respectively (Fig. 2A). Control enzyme, incubated under the same conditions but in the absence of the reagents, showed no change in activity during this period. These results suggest that under our incubation conditions, both GSNO and H2O2 decrease the half-life of this enzyme.
FIGURE 2.
Biophysical characterization of wild-type IDE treated with H2O2 or GSNO. A, time-dependent inactivation plots (Et/E0 versus time) of native (●) and H2O2-modified (■) or GSNO-modified (224) IDE. B, kinetic analysis of Aβ degradation by native (●) and H2O2-modified (■) or GSNO-modified (224) IDE. Specific activities (min−1) of IDE samples were determined for the indicated Aβ-(1–40) concentrations using 0.25 μm FAβB as the tracer. The activities of the samples were measured at 37 °C for 5 and 10 min. C, gel filtration elution profiles of native (thick line) and H2O2-modified (dashed line), GSNO-modified (thin black line), and NEM-modified (thin gray line) enzymes at 4 °C. These elution profiles were obtained by applying 1 ml of each protein (∼1 mg) to a Superdex 200 column equilibrated with 20 mm Tris-Cl buffer, pH 7.0, containing 50 mm NaCl. mAU, milli-absorbance units. D, time-dependent thermal inactivation plots (Et/E0 versus time) of native (●) and H2O2-modified (■) or GSNO-modified (224) IDE at 37 °C.
The sensitivity of IDE to H2O2 and GSNO may be the result of an alteration in the affinity of the enzyme for substrates. Thus, the apparent Km values of treated and control enzymes were determined for Aβ as a substrate. As shown in Fig. 2B, the treated enzyme does not appreciably affect the apparent Km value for Aβ. In contrast, the velocity of the reaction decreased to ∼35% upon treatment of IDE with H2O2 and GSNO. Thus, the inhibition by H2O2 and GSNO is likely due to the loss of enzymatic activity in a chemically modified fraction of the enzyme pool. We also examined the ability of purified IDE treated with H2O2 or GSNO to recognize and degrade amyloid-β naturally produced by APP-overexpressing human embryonic kidney cells that harbor amyloid precursor protein containing the familial Alzheimer disease-linked “Swedish mutation.” The amyloid-β (4-kDa band) in the conditioned medium could be rapidly degraded by untreated IDE, but not by IDE treated with H2O2 and GSNO (supplemental Fig. S1). These results demonstrate that the H2O2- or GSNO-treated IDE has reduced efficiency to degrade naturally produced Aβ, heterogeneous in size and modifications, supporting our kinetic results.
To determine whether the altered kinetic parameters of modified IDE are attributed to the loss of quaternary structure of the enzyme upon modification, we analyzed these samples by size exclusion chromatography. The elution profile of the control enzyme from a Superdex-200 gel filtration column (Fig. 2C) indicates that in solution this enzyme exists as a monomer/dimer mixture (∼150 ± 20 kDa). The elution profiles of the enzymes modified with H2O2 or GSNO are also displayed in Fig. 2C. The major peak present in enzymes treated with H2O2 and GSNO have maxima at ∼280 ± 30 and 290 ± 40 kDa, respectively, suggesting that this peak has IDE that exists predominantly as dimer with small amounts of higher oligomers; the presence of higher molecular weight peaks indicates that these modified enzymes exist as mixtures of dimers, oligomers, and soluble aggregates. In contrast to H2O2/GSNO-treated IDE, NEM-modified IDE exhibits an elution profile similar to untreated IDE (Fig. 2C).
Modified IDE may possess intrinsically different kinetic parameters or may be particularly unstable. Therefore, the stability of these chemically modified enzymes was evaluated at 37 °C. The time-dependent thermal inactivation plots are shown in Fig. 2D. The enzymes were preincubated for 1 h with 1 mm H2O2 or GSNO (illustrated by the −60- to 0-min time points), and the excess reagent was subsequently removed by gel filtration. It is evident that within the first 4 h of incubation at 37 °C, the control and H2O2-treated enzyme does not lose any relative activity. On the other hand, the GSNO-treated protein lost most of its activity within about 1 h of incubation at 37 °C, suggesting that nitrosylation renders the enzyme unstable at the physiologically relevant temperature.
Concentration-dependent Inactivation of IDE by H2O2 and GSNO
In vivo, the concentration of ROS and RNS is in the nanomolar range during normal conditions to as high as 100 μm during stress conditions (9, 37). Therefore, we evaluated the concentration-dependent effects of H2O2 and GSNO on Aβ degradation by IDE-WT (Fig. 3A). The salient observations are the following: the cleavage of Aβ by IDE is significantly compromised at mid-micromolar concentrations of H2O2 and GSNO. We observed greater than 20% inhibition of IDE-WT with equal or above 50 μm GSNO treatment and up to 60% inhibition at 200 μm GSNO. Similar sensitivity was found upon the treatment of IDE by H2O2. Also, the concentration dependence of both H2O2 and GSNO inhibition is biphasic with one phase in the micromolar range and the second in the millimolar range (Fig. 3, A–C). The activity of human IDE is also sensitive to inhibition by a thiol-reactive alkylation agent, NEM (Fig. 3D) (13, 21). Interestingly, in contrast to the biphasic inhibitory curves for the treatment by H2O2 and GSNO, the dose-dependent inhibition curve of IDE by NEM is monophasic.
IDE exhibits substantially different enzyme kinetic parameters with the longer peptide Aβ as compared with a shorter peptide substrate, the bradykinin-mimetic substrate V (13). The kcat for bradykinin (2000 s−1) is ∼200-fold higher than that for Aβ (70 s−1) (Fig. 2B). However, IDE has much higher affinity for Aβ (80 μm) as compared with bradykinin (4 mm). Therefore, we also examined the dose-dependent inhibition of IDE using substrate V (Fig. 3, E and F). Interestingly, we observed a monophasic response to the inhibition by GSNO and H2O2 when assaying the activity using substrate V.
It is well known that the nucleophilic nature of cysteines makes this residue a primary target of ROS/RNS. We have constructed a catalytically active human IDE that is devoid of all 13 cysteines, IDE-CF (13). We utilized IDE-CF to assess whether any of the 13 cysteine residues are indeed involved in the oxidative/nitrosative inactivation of IDE using FAβB (Fig. 3, B and C) and substrate V (Fig. 3, D–F) as substrates. We found that IDE-CF was relatively resistant to the inactivation by H2O2 and GSNO. This finding confirms that indeed cysteine residues are primarily responsible for the in vitro inactivation of IDE in the presence of H2O2 and GSNO.
IDE has four homologous domains known as domains 1–4, and the 13 cysteines are distributed throughout all four domains. We have previously constructed IDE-CF123, a mutant free of all cysteines in domains 1–3, and IDE-CF4, an IDE mutant free of all cysteines in domain 4 (13); both constructs are catalytically active. We examined the dose-dependent inhibition of these two mutants by H2O2, GSNO, and NEM. Previously, we found that although IDE-WT and IDE-CF123 were highly sensitive to the inhibition by NEM, IDE-CF4 was not affected by the treatment (Fig. 3D). From the systematic mutational study, we identified Cys-819 as the residue responsible for the inhibition by NEM (13). Here, we discovered that the treatment of IDE-CF123 and IDE-CF4 with H2O2 and GSNO results in completely different effects compared with the treatment with NEM (Fig. 3, B and C and E and F).
Using Aβ as the substrate, we found that at the high micromolar concentration ranges of H2O2 and GSNO (0.1–0.6 mm), IDE-CF4 became less sensitive to inhibition than wild-type IDE (IDE-WT) (Fig. 3, B and C). However, we observed no difference at the concentrations of H2O2 and GSNO above 0.6 mm. Additionally, we observed no difference between IDE-WT and IDE-CF4 in their sensitivity to H2O2 and GSNO when substrate V was used to assess catalytic activity (Fig. 3, E and F). Together, our data suggest that cysteine residue(s) in domain 4 of IDE are involved in the inhibitory effect at the low concentrations of H2O2 and GSNO and that domains 1–3 contain cysteine(s) that serves a modulatory or protective role against the inactivation of IDE by H2O2 and GSNO. We also suggest that the S-alkylation by NEM is not a suitable model to study thiol-directed post-translational modifications of enzymes possibly due to the very different chemical properties of the maleimidyl moiety of NEM.
Identification of Cysteine Residues Crucial for the Modulation of IDE Activity
Previously, we identified Cys-819 as a modification site for NEM, H2O2, and GSNO (supplemental Fig. S2) (13). However, for H2O2 and GSNO, the cysteine-free mutant containing only the Cys-819 has higher sensitivity to the treatments with H2O2 and GSNO, as compared with IDE-WT. This finding led us to hypothesize that there is a “protective” cysteine residue(s) in IDE that restores the activity in IDE-WT. Therefore, we individually reintroduced each of the 12 cysteines into the IDE-CF-A819C to identify the other cysteine(s) involved in the sensitivity observed in the higher H2O2 and GSNO concentrations as well as the cysteine(s) that serve to protect the catalytic activity of IDE. Initial characterization experiments were conducted on partially purified samples (Fig. 4A). We observed that IDE-CF-819C/110C had a striking loss of activity upon H2O2 or GSNO treatment. In contrast, IDE-CF-819C/257C and IDE-CF-819C/178C had partial resistance to inactivation comparable with IDE-WT when treated with the same reagents (Fig. 4A). These findings indicate that under these conditions, Cys-110 contributes to the further loss of IDE activity after treatment with H2O2 and GSNO, whereas Cys-178 and Cys-257 confer resistance to oxidative/nitrosative damage by H2O2 and GSNO.
To understand the role of each of these residues, we constructed and characterized a set of eight fully purified IDE mutants that have variable combinations of Cys-819, Cys-110, and Cys-178 as well as that with 819C and 257C in the IDE-CF background (Fig. 4B). As expected, IDE-CF-819C was sensitive to treatment with H2O2 and GSNO. Interestingly, both IDE-CF-110C and IDE-CF-178C, which have a single cysteine, were resistant to inactivation by either reagent. In contrast, the combination of Cys-819 with Cys-110 confers higher sensitivity toward both reagents as compared with either mutant alone. These results indicate that the modification of Cys-819 either increases the exposure of the reaction site of Cys-110 or, conversely, that modification of Cys-110 results in greater vulnerability of Cys-819. As observed previously, the combination of Cys-819 with Cys-178 affords protection against oxidative/nitrosative inactivation. Moreover, the inclusion of Cys-819, Cys-110, and Cys-178 in the IDE-CF background also provided considerable protection against inactivation caused by H2O2 and GSNO. These results confirm that the presence of Cys-178 is crucial for the protection of IDE against complete inactivation. Because the combination of Cys-819 and Cys-110 increases the sensitivity of IDE by H2O2 and GSNO, we evaluated if the combination of Cys-178 with Cys-110 has a similar effect. We found that IDE-CF-110C/178C is relatively unaffected by H2O2 and GSNO (Fig. 4B).
Our results in Fig. 4A suggested that the presence of both Cys-257 and Cys-819 results in a marked decrease in the inhibition caused by H2O2 and GSNO. However, when the same sample was purified to homogeneity, Cys-257 did not offer any protection against inactivation by the chemical reagents (Fig. 4B). Thus, the preliminary observations that directed us to Cys-257 are likely an artifact from impurities present in the partially purified samples.
We sought to screen relevant variants of IDE for their sensitivity to H2O2 and GSNO with sensitivities expressed as IC50 values using FAβB as a substrate (Table 1). For IDE-WT, in the presence of H2O2 and GSNO, the enzyme displays an IC50 of ∼1 mm, whereas the IC50 value is ∼4–5-fold less for IDE-CF123, which contains all of the cysteines in domain 4, including Cys-819. In contrast, IDE-CF4, with cysteines present in domains 1–3, has an IC50 value comparable with the wild-type enzyme. The absence of Cys-819 thus confers a 4-fold increase in the resistance to inactivation, as compared with IDE-CF123.
As expected, the IC50 value for IDE-CF, free of all cysteines, is greater than 5 mm. Therefore, any other non-cysteine residues that may be modified have little impact on the activity of the enzyme. Similarly, IDE-CF-110C, IDE-CF-178C, and IDE-CF-110C/178C displayed IC50 values greater than 5 mm. Thus, in the absence of Cys-819, H2O2 and GSNO have no effect on Cys-178 or Cys-110 of IDE. For IDE-CF-819C, the IC50 is reduced to ∼200 μm, consistent with the sensitivity observed for IDE-CF123 and confirming that the modification of Cys-819 in domain 4 affects the enzyme activity. Moreover, IDE-CF-819C/110C exhibits a significant decrease in the IC50 value for both H2O2 and GSNO (to ∼30 μm), supporting the proposal that only in the presence of Cys-819 does residue Cys-110 becomes vulnerable to modification, leading to greater loss of catalytic activity. Conversely, IDE-CF-819C/178C and IDE-CF-819C/178C/110C display less sensitivity to GSNO and H2O2, with IC50 values increased to ∼1 mm.
Loss of Thiol-specific Labeling to Quantify the Sites of Oxidation and Nitrosylation of IDE
We determined the free thiol content of the individual enzymes after the complete removal of H2O2 and GSNO, by the reaction of each protein sample with DTNB under denaturing conditions (Table 2) (27). As a quality control of our thiol count assay, the number of thiols was determined for reduced glutathione (1 –SH/molecule) (Table 2). As expected, the IDE mutant devoid of all cysteines has no free thiols present in the absence or presence of H2O2 and GSNO (Table 2). After complete removal of H2O2 or GSNO, the intact IDE-WT enzyme exhibited an average of 9.2 and 8.8 sulfhydryl groups, respectively. This indicates that four cysteine residues in IDE can be modified by H2O2 or GSNO. This number is higher than the three cysteines implicated in the inactivation of IDE, suggesting that at least one cysteine can be modified by these reagents without perturbing the catalytic activity of the enzyme.
The IDE-CF-819C enzyme has no free thiols present after reaction with either H2O2 or GSNO; this result is consistent with Cys-819 being modified by this reaction. Similarly, IDE-CF-178C loses its free sulfhydryl upon reaction with H2O2 and GSNO; however, catalytic activity is not affected. In contrast, IDE-CF-110C retains its free sulfhydryl after the standard 1-h incubation with the chemical reagents, indicating that Cys-110 is inaccessible for oxidation and nitrosylation unless IDE is denatured. In contrast, IDE-CF-819C/110C contained no more than 0.2 mol of –SH/subunit of IDE (Table 2). This indicates that in the presence of Cys-819, Cys-110 becomes accessible to oxidation and nitrosylation by H2O2 and GSNO. This correlates with the total loss of activity of this mutant upon treatment with 1 mm H2O2 and GSNO. The chemically modified IDE-CF-819C/178C does retain appreciable activity and exhibited a similar decrease in the number of reduced cysteines, to no more than 0.1 mol of –SH/subunit of enzyme. This suggests that both Cys-819 and Cys-178 are modified by treatment with H2O2 or GSNO. The total number of free thiols of IDE-CF-819C/178C/110C following treatment with H2O2 or GSNO was reduced from 3 to 1 (Table 2), indicating that one of these cysteines is not vulnerable to chemical reaction by H2O2/GSNO when this combination of cysteines is present in IDE. However, the DTNB assay cannot identify which two cysteine residues are modified.
Identification of Nitrosylation Sites of IDE Mutants Using Liquid Chromatography-Electrospray Ionization LTQ FT-ICR MS and MS/MS
Although our IDE mutant analysis points to Cys-819, Cys-110, and Cys-178 as possible target sites of oxidation/nitrosylation, it cannot provide unequivocal information on the actual site of modification. FT-ICR MS provides high resolution and high mass accuracy and has become widely used in the characterization of post-translational modifications (38–43). To understand the molecular basis of oxidative/nitrosative modification of IDE, we used nitrosylation as a model system for our MS studies because only a single adduct results upon reaction with a residue.
We first examined the effect of GSNO treatment of IDE-CF-819C/110C and IDE-CF-819C/178C/110C. From our DTNB experiment, both mutants have an equivalent of two reactive cysteines. We hypothesized that the modification of 819C permits 110C to be modified, resulting in IDE-CF-819C/110C containing two nitrosylated cysteines because the thiol count is reduced from 2 to 0.2. Furthermore, the presence of modified 178C prevents 110C from being affected by the modification of 819C; thus, IDE-CF-819C/178C/110 also has only two nitrosylated cysteines following GSNO treatment, consistent with our finding that the free thiol count is reduced from 3 to ∼1.
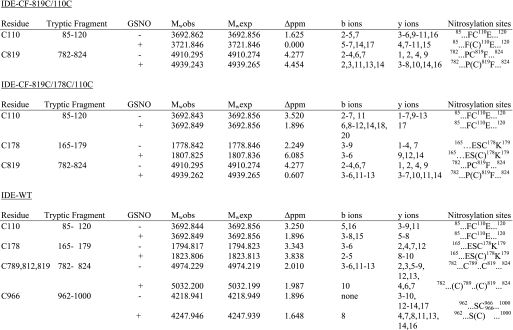
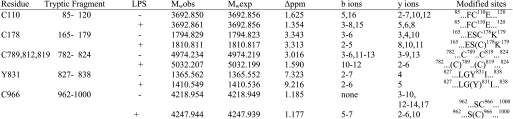
MS/MS can unambiguously map specific sites of post-translational modification, including amino acids modified by NO adduction. We used collision-induced dissociation for the gas phase fragmentation method to produce b- and y-type fragment ions for peptide identification. When IDE-CF-819C/110C was reacted with GSNO, two different sites of modification were localized with a mass accuracy within 5 ppm by MASCOT and BioWorks searches and manual inspection of the FT-ICR mass spectra (Table 3). The FT-ICR spectrum (data not shown) indicated that the ion species corresponded to the tryptic peptides 85SSAALDVHIGSLSDPPNIAGLSHFCEHMLFLGTKK119 and 782NEVHNNAGIEIYYQTDMQSTSENMFLELFAQIISEPCFNTLR823 both with one NO adduct (+29 Da), indicating that both peptides were modified (Table 3). Inspection of the LTQ MS/MS spectrum confirmed the peptide sequence with the addition of the NO moiety in Cys-110 and Cys-819, respectively. Together with our enzymatic assay, our data indicate that S-nitrosylation of Cys-110 and Cys-819 inactivates this enzyme.
We next examined the S-nitrosylation sites for IDE-CF-819C/178C/110C, which retains ∼60% of activity following incubation with GSNO. This is in sharp contrast with the activity of GSNO-treated IDE-CF-819C/110C, which is completely inactive. Again, MS analysis was performed to examine the protective effect of S-nitrosylation of Cys-178. Two peptide regions (165–179 and 782–824) registered a difference of 29 mass units, the characteristic peptide mass increase of an NO adduct (Table 3). The corresponding MS/MS data clearly assigned Cys-819 and Cys-178 as preferred S-nitrosylation sites. The tryptic peptide-(85–120) containing the third cysteine, 110, was only observed as an unmodified peptide following the GSNO treatment. This suggests that Cys-178 prevents Cys-110 from being modified, thereby protecting the enzyme from complete inactivation by S-nitrosylation. Together, our data clearly define Cys-819, Cys-110, and Cys-178 as targets of oxidation and nitrosylation. The modifications of Cys-819 and Cys-110 are responsible for the loss of enzymatic activity, whereas the modification of Cys-178 serves a protective role, preventing total inactivation of IDE.
Analysis of S-Nitrosylation Sites of IDE-WT and BV2-IDE
When GSNO was reacted with recombinant wild-type human IDE, which contains all of the 13 cysteines present, four different sites of modification were localized with a mass accuracy of better than 4 ppm by MASCOT and BioWorks searches and manual inspection of the FT-ICR mass spectra (Table 3). The tryptic peptide containing residues 85–120 had a molecular mass of 3692.844 Da equivalent to the unmodified form of this peptide; thus, Cys-110 is not a target site for nitrosylation in the WT enzyme. In contrast, the tryptic fragment containing the amino acids FAQFFLCPLFDESCK, corresponding to residues 165–179, had an atomic mass unit increase of 29 Da, corresponding to one NO adduct. This peptide, however, contains two cysteines, Cys-171 and Cys-178. Inspection of the LTQ MS/MS spectrum confirmed the peptide sequence with the addition of the NO moiety. From the N terminus, we only identified matches for b2–b5 ions, which is not sufficient to rule out Cys-171 as a target site, On the other hand, from the C terminus, the observed y ions (y8–y10), corresponding to KCSEDFLPy8Cy9Ly10, contain matches that correspond to all of the residues with a mass increase of 29 Da in y8–y10 ions, leading us to conclude that the single NO adduct is located at position Cys-178 and not Cys-171. The molecular mass of 5032.2 for the tryptic fragment 782NEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPCFNTLR823 is equivalent to this fragment plus two NO adducts; thus, two of the three cysteines present in this peptide (Cys-789, Cys-812, and Cys-819) are modified. From the N terminus, the b10 ion match contains the mass of residues 782–791 with a mass increase of 29 Da, corresponding to a single NO adduct on Cys-789. From the C terminus, the initial y4 ion corresponds to the first four amino acids, RLTN. The subsequent observed y ions (y6–y7) have a mass increase of 29 Da, corresponding to a nitrosyl adduct on Cys-819. This eliminates Cys-812 as a site of NO modification. Therefore, in this peptide the two sites of NO adducts are Cys-789 and Cys-819, but only the modification at Cys-819 affects IDE activity. We also identified a third tryptic fragment (962–1000) that has the molecular mass of the peptide and the NO adduct. This peptide includes a cysteine at position 966 and 974. The LTQ MS/MS spectrum confirmed the peptide sequence with the addition of a single NO adduct. Collision-induced dissociation product ions from both N- and C-terminal directions, b8 and y4–y16, confirm the expected sequence with the addition of 29 Da to Cys-966 but not Cys-974. Thus, our analysis unambiguously localizes four sites of NO modification in IDE-WT as follows: Cys-178, Cys-789, Cys-819, and Cys-966 (Table 3), confirming our previous thiol count of nine. As described above from our mutational analysis, nitrosylation of Cys-178 and Cys-819 is involved in the modulation of activity. The role of the modifications of Cys-789 and Cys-966 will be considered below.
IDE secreted from the mouse microglial cell line BV-2, has decreased activity upon oxidative stress induction; however, there is no direct evidence that the enzyme is modified. To characterize the oxidative modification of LPS-stimulated BV-2 IDE, gel bands corresponding to IDE (from the BV-2 media) were excised from the electrophoretic gel and subjected to tryptic in-gel digestion prior to FTI-ICR mass spectrometric analysis of the resulting mixture (Table 4). MS analysis of the BV-2 tryptic digests yielded peptides corresponding to ∼70% of the mouse IDE amino acid sequence. All four cysteines that we identified in IDE-WT treated with GSNO (Cys-178, Cys-789, Cys-819, and Cys-966) were also modified in LPS-treated BV2-IDE. Interestingly, although Cys-789, Cys-819, and Cys-966 were nitrosylated (+ 29 Da), Cys-178 had a mass difference of only 16 Da, indicating that this residue was modified to sulfenic acid (Cys-SOH). In addition to the four modified cysteines, we also observed a mass increase of 45 Da on Tyr-831, indicating that this is a nitrotyrosine. This modification was never observed in the mass spectrometric analysis of GSNO-treated IDE-WT. The importance of Tyr-831 was previously demonstrated by Shen et al. (11), showing that it is a crucial residue involved in substrate binding and catalysis; thus the nitrosylation of Tyr-831 likely affects catalytic activity. Overall, these results provide direct evidence for the oxidative modifications of IDE secreted by LPS-induced BV-2 cells, which in turn results in the inhibition of IDE.
TABLE 4.
Modified peptides identified by LTQ FT-ICR MS/MS in BV2-IDE
Modified residues are shown in parentheses.
Oligomerization of Modified IDE
As described in Fig. 2, IDE-WT treated with H2O2 or GSNO elutes in three peaks after size exclusion chromatography (Fig. 2C and Fig. 5, A and B). Peak 1 of ∼680 ± 50 kDa approximately corresponds to a hexamer, whereas peaks 2 and 3 with molecular masses of 440 ± 60 and 260 ± 40 kDa, respectively, represent mostly tetramer and dimer. In contrast, cysteine-free IDE, under the same treatment conditions, elutes as a single peak at ∼180 ± 30 kDa (data not shown). This suggests that the oligomerization resulting from H2O2 or GSNO treatment is likely mediated by cysteine residue(s). All three peaks were separately pooled and assayed for FAβB activity. Of the ∼60% relative activity that remains in IDE-WT after modification, no activity is seen in peak 1, whereas peaks 2 and 3 have ∼15 and ∼45% of the total activity, respectively. We also found that the addition of a reducing agent, tris(2-carboxyethyl)phosphine (1 mm), to the modified IDE-WT resulted in a complete conversion of the oligomeric forms of the enzyme to its natural monomer-dimer equilibrium state when in solution (Fig. 5, A and B, peak 4). Moreover, the FAβB activity was restored to ∼85% that of the control enzyme (under the same conditions but without chemical treatment). These data confirm that cysteines are involved in the oligomerization of oxidized IDE and that activity is influenced by the oligomeric state.
We then used a mutational approach to identify the cysteine residue(s) responsible for the oligomerization of oxidized IDE. In IDE-WT, Cys-178, Cys-789, Cys-819, and Cys-966 were modified by oxidation and nitrosylation. Although the modification of Cys-178 and Cys-819 affects the catalytic activity of IDE, the roles of the oxidative reaction of Cys-789 and -966 are not apparent. Because both Cys-789 and Cys-966 are exposed to the outer surface of the protease and are in close proximity, we hypothesized that they affect the oligomerization and stability of the enzyme. To evaluate this, IDE-CF variants containing Cys-789 and/or Cys-966 were subjected to gel filtration analysis after inducing oxidative/nitrosative modifications (Fig. 5, C–H). All of the IDE-CF mutants containing only one of the four identified cysteine residues (789, 966, 819, or 178) eluted as a single peak upon chemical treatment, at position 4. Thus, oxidation- or nitrosylation-induced oligomerization of IDE is a process that involves more than one cysteine (Fig. 5, C and D). After chemical treatment, IDE-CF containing both Cys-789 and Cys-966 showed a similar elution pattern (containing peaks 1–3) as that of modified IDE-WT (Fig. 5, A, B, and E), although the untreated enzyme eluted at position 4 (Fig. 5, E and F). Furthermore, the inclusion of Cys-819 along with Cys-789 and Cys-966 did not alter the elution profile of the chemically treated enzyme (Fig. 5F). However, the addition of Cys-178, which we identified as a key residue that protects the activity of IDE after oxidation/nitrosylation, also prevents the GSNO- and H2O2-induced oligomerization/aggregation of the enzyme (Fig. 5, G and H). It is worth noting that the protection against oligomerization, rendered by Cys-178 is not complete for IDE-CF-789C/966C/819C/178C because most of the enzyme eluted at position 3 and not 4 as in the control (Fig. 5H). These results indicate that in addition to serving a protective role against catalytic inactivation of IDE, Cys-178 also serves a protective role against oligomerization of IDE induced by oxidation and nitrosylation.
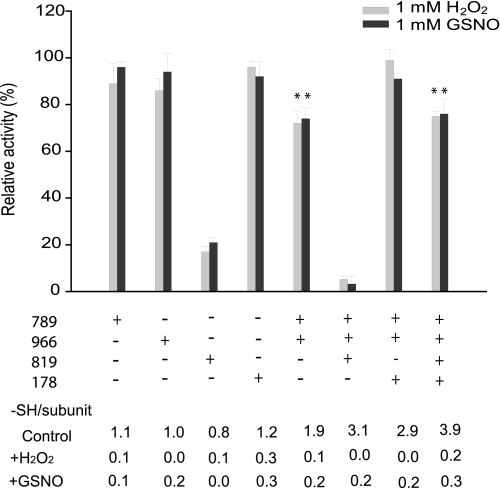
To study whether the oligomeric structure of modified IDE is a function of the status of some cysteine groups, we measured the free cysteine content of each mutant in Fig. 5 in the absence or presence of H2O2 or GSNO treatment (Fig. 6). Activity was determined in parallel using substrate V. For mutants containing a single cysteine (789, 966, 819, or 178), which eluted as a single peak at position 4 (Fig. 5, C and D), the thiol count decreased from ∼1 to no more than 0.3 mol of –SH/subunit of IDE (Fig. 6). Furthermore, despite the apparent modification of the thiol groups, each single cysteine mutant retained greater than 90% activity with the exception of IDE-CF-819C. As expected, both IDE-CF-789/966C and IDE-CF-789C/966C/819C, which oligomerize in solution upon chemical treatment (Fig. 5, E and F), exhibit a loss of ∼2 or 3 sulfhydryl groups, respectively (Fig. 6). Thus, all of these cysteines are modified upon treatment. This suggests the involvement of both Cys-789 and Cys-966 in the mechanism of oligomerization upon oxidative or nitrosative modification. We also found that IDE-CF-789C/966C has reduced catalytic activity (∼25%) compared with either IDE-CF-789C or IDE-CF-966C. Furthermore, the reduction in the activity of IDE-CF-789C/966C/819C (∼90%) is greater than that of IDE-CF-819C (∼70%). This is consistent with the notion that the oligomerization of IDE leads to decreased activity. The introduction of Cys-178 to either IDE-CF-789C/966C or IDE-CF-789C/819C/966C restores the catalytic activity to full or ∼75% activity, respectively, which is significantly higher than those of the original mutants (Fig. 6). Interestingly, the presence of Cys-178 did not increase the number of free thiols after H2O2/GSNO incubation. Thus, Cys-178 does not prevent the modification of Cys-789 or Cys-966. Together, our data show that Cys-178 can protect IDE against both loss of catalytic activity and oligomerization caused by oxidative or nitrosative attack.
FIGURE 6.
Analysis of activity and quantitation of sulfhydryls for IDE mutants involved in oxidation/nitrosylation-induced oligomerization. IDE mutants with the various cysteine combinations analyzed in Fig. 5 were assayed for residual activity and titrated with DTNB upon treatment with 1 mm H2O2 or GSNO. Relative activities are reported assuming as 100% the control reaction where each sample was incubated with buffer only under the same conditions. Activity was monitored by measuring the fluorescence resulting from the cleavage of 5 μm substrate V, and DTNB titrations were performed as described under “Experimental Procedures.” Data are means ± S.E. for two independent experiments with three to five replications per condition. Statistical significance reflects comparison with the activity of CF-IDE using the same substrate. **, p < 0.05.
DISCUSSION
Our study demonstrates that inactivation of naturally produced IDE occurs in a biologically relevant model of oxidative stress. This finding led us to evaluate the biochemical and biophysical properties of human IDE exposed to agents that promote oxidation (H2O2) and nitrosylation (GSNO). As a result, significant new properties of IDE were discovered. H2O2 and GSNO inactivate the catalytic activity of IDE through covalent modifications. Both chemical reagents inactivate the enzyme in a time-dependent manner, which is not reversed by gel filtration or dialysis. Furthermore, these chemical modifications caused a marked decrease in enzymatic activity toward Aβ hydrolysis and shifted the equilibrium of the enzyme toward higher average masses. GSNO-treated IDE also exhibited striking instability (∼10% residual activity after 1 h) relative to the untreated and H2O2-treated IDE, suggesting the higher magnitude of structural abnormality arising from the GSNO-mediated modification. Our dose-dependent studies document that exposure of IDE to 10–100 μm H2O2 or GSNO, equivalent to the range of ROS/RNS released under pathophysiological conditions, caused inhibition of Aβ hydrolysis. Furthermore, the inhibition response is substrate-dependent, suggesting that modified IDE retains substrate specificity.
All of these notable biochemical deviations of the modified enzyme prompted us to characterize the mechanism of modification at the molecular level by mass spectrometry to identify and localize sites of modification. We show that modification of Cys-819 by H2O2 and GSNO leads to the accessibility of Cys-110 for oxidation or nitrosylation, which leads to a completely inactivated enzyme. The most significant finding in this study is that the modification of Cys-178 protects the active site Cys-110 from subsequent oxidation/nitrosylation. This role is completely different from the assigned inhibitory role of Cys-178 revealed by using NEM as the thiol alkylation agent (21).
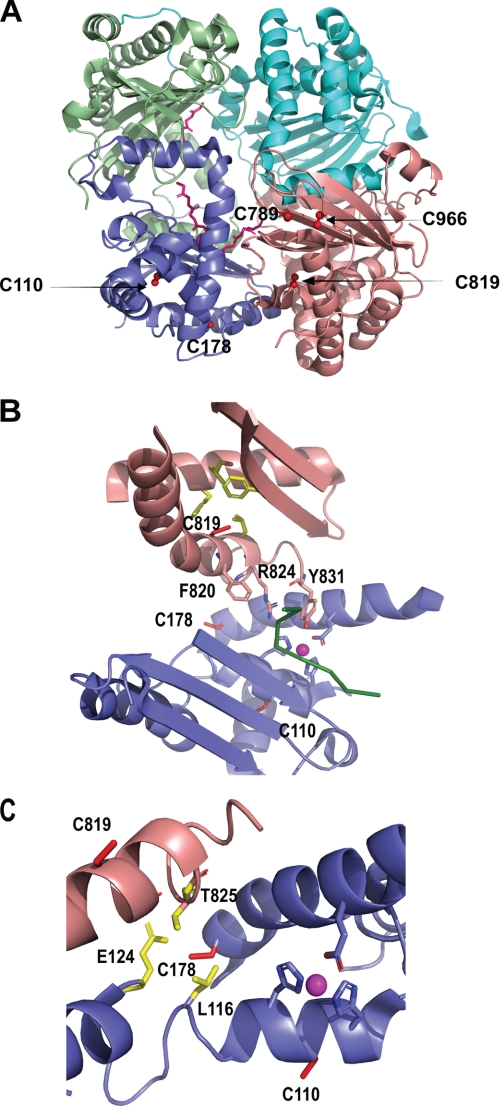
Examination of the crystal structure of human IDE allows us to better understand our experimental results (11). IDE consists of the four homologous domains 1–4, and the five cysteines that can be nitrosylated are distributed in domain 1 and 4 (Fig. 7A). Cys-819 is partially buried in a hydrophobic pocket, and we postulate that the nitrosylation or oxidation of this residue perturbs the local structure and residues crucial for substrate binding (Fig. 7B) (13). In this study, we found that the modification of Cys-819 must precede the modification of Cys-110 to fully inactivate IDE. At first glance, the location of Cys-110 does not immediately suggest it as an ideal candidate for reaction with ROS/RNS because it appears to be buried in the structure within a mixture of hydrophobic and hydrophilic residues (Fig. 7B). Indeed, the lack of reaction of CF-IDE-110C with H2O2 and GSNO support this notion. Thus, we hypothesize that the modification of Cys-819 induces a conformational change that destabilizes the active site motif, resulting in the exposure of Cys-110; thus, making it accessible for reaction with ROS/RNS. The modification of Cys-110 alters the catalytic pocket formed by the conserved inverted HFCEH motif to HFC-SNO-EH, potentially blocking substrate access to the active site residues specifically or completely blocking access to the binding pocket.
FIGURE 7.
A, schematic representation of IDE highlighting the position of Cys-110, Cys-178, Cys-789, Cys-819, and Cys-966 (red ball and sticks) (Protein Data Bank 2G47). IDE domains 1–4 are depicted as slate blue, pale green, cyan, and salmon pink schematics, respectively. Aβ-(1–40) is shown as pink sticks. B, schematic representation of the relative location of the three cysteines involved in the modulation of activity upon oxidation and nitrosylation. Side chains of Cys-110, Cys-178, and Cys-819 are shown as red sticks. The hydrophobic residues near Cys-819 are depicted as yellow sticks, and residues involved in substrate binding are highlighted as salmon pink sticks. The buried Cys-110 residue is located near the catalytic residues highlighted as slate blue sticks and near the Zn2+, shown as a magenta sphere. Aβ-(1–40) is shown as a green ribbon. C, zoom of the interplay between Cys-819, Cys-178, and Cys-110, all shown as red sticks. The side chains of Leu-116, Glu-124, and Thr-825 are shown as yellow sticks. The catalytic site is also shown (blue sticks and magenta sphere).
We also found that modification of Cys-178 of IDE compensates for the distortion of the active site network caused by modification of Cys-819. This is supported by experimental evidence, including the finding that both CF-IDE-819C/178C and CF-IDE-819C/178C/110C retain ∼60% residual activity upon reaction with 1 mm H2O2 and GSNO as compared with ∼20% activity and no activity upon reaction with CF-IDE-819C and CF-IDE-IDE-819C/110C, respectively. Cys-178 faces the outer surface of the enzyme, and similar to Cys-819, it is positioned in an α-helix located near the interface between the N- and C-terminal halves of the protease (Fig. 7C). Because of this strategic location, it could likely reduce the structural perturbations, resulting from the modification of Cys-819, by two possible mechanisms. The first mechanism involves the stabilization of the active site network. Glu-189, a key Zn2+-binding residue, is located in the same helix as Cys-178. Furthermore, the –SH group of Cys-178 is positioned in very close proximity to Leu-116 and Glu-124, both of which lie in the α-helix containing the remaining three active site residues (Glu-111, His-108, and His-112) (Fig. 7C). Therefore, oxidation or nitrosylation of Cys-178 may reposition the network of active site residues, compensating for the distortion that is caused by the modification of Cys-819. The modification of Cys-178 may additionally preserve proper substrate binding because Cys-178 and Thr-825 are in close proximity (Fig. 7B), so that the reaction of Cys-178 likely readjusts the C-terminal residues that participate in substrate binding (Phe-820, Tyr-821, and Arg-824) that are shifted out of position by the modification of Cys-819. Interestingly, of the three cysteines, Cys-178 of IDE is the least conserved, present only in higher vertebrates, which have evolved to have three different types of NO synthases and are in most pathogenic fungi (supplemental Fig. S3). This suggests that the evolution of this residue might contribute to the generation of a more stable and efficient IDE that is amendable to fine regulation in an ROS/RNS-rich environment.
Our study also reveals a surprising finding on the role of cysteine modification on the oligomerization of IDE. The modification of 789 and 966 does not directly affect catalysis. Instead, these modifications influence enzyme structure at the quaternary level, ultimately resulting in enzyme instability (i.e. aggregation and/or thermal instability). Interestingly, the enzyme aggregation only occurs when both Cys-789 and Cys-966 cysteines are nitrosylated or oxidized. The sulfur atom of Cys-789 is in close proximity to that of Cys-966 (6.5 Å) (Fig. 7A). The modification of these two residues together likely results in steric and electrostatic clashes, causing a local conformational change that leads to aggregation of IDE followed by a reduction of catalytic activity. Surprisingly, in addition to protecting Cys-110 from chemical modification, our studies indicate that Cys-178 modification also protects the enzyme from aggregation because its presence led to a striking decrease in oligomerization induced by ROS/RNS attack.
Our study suggests that the cysteines of IDE act as sensors of environmental ROS/RNS levels through subtle alterations of the protein structure. We uncovered two means by which ROS/RNS can affect the activity of IDE as follows: first by affecting the catalytic activity and second by promoting the oligomerization. Cys-178 serves as a buffer to refine the effects of ROS/RNS. We also found that the oxidation or nitrosylation of IDE can be reversed by a reducing agent. Thus, the spatial, temporal, and concentration-dependent modifications of these cysteines could fine-tune IDE activity in response to the environmental oxidative and nitrosative insults. Furthermore, we also found that the inhibitory response of IDE by oxidation or nitrosylation is substrate-dependent, biphasic for Aβ degradation and monophasic for a shorter bradykinin-mimetic substrate, adding another layer of complexity to this intricate regulation. It is worth noting that S-nitrosylation of Cys-215 in tyrosine phosphatase 1B protects the enzyme from further oxidative damage, suggesting that the dynamic regulation of enzymatic activity via nitrosylation or oxidation is a common phenomenon (44).
In summary, we have characterized the molecular basis for oxidation and nitrosylation of IDE. Because it is well documented that diabetes, Alzheimer disease, and cardiovascular disease lead to increased production of ROS and RNS, these studies may provide the knowledge basis for the development of novel methods that will effectively preserve the activity of this enzyme in the presence of oxidative/nitrosative stress conditions. Furthermore, these studies demonstrate that whereas thiol-alkylating compounds, such as NEM and iodoacetamide, are useful tools in enzymology, they do not provide an accurate view of the consequences of physiologically relevant modifications of enzymes.
Supplementary Material
Acknowledgments
We thank Dr. Sangram S. Sisodia (University of Chicago) for providing conditioned media from HEK293APPswe.3 cells, Dr. Linda Van Eldik (Northwestern University) for providing BV-2 cells, and Raymond Hulse for technical assistance and helpful discussion on studies related to BV-2 cells.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 81539 (to W. J. T.) and Fellowship F32 GM 87093 (to L. A. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
The amino acid sequences of the human and mouse IDE are 94% identical plus 3% similar, justifying the use of mouse IDE from the BV-2 immortalized microglial cell line.
- ROS
- reactive oxygen species
- IDE
- insulin-degrading enzyme
- WT
- wild-type enzyme
- RNS
- reactive nitrogen species
- H2O2
- hydrogen peroxide
- GSNO
- S-nitrosoglutathione
- NEM
- N-ethylmaleimide
- Aβ
- amyloid-β
- FAβB
- fluorescein-Aβ-(1–40)-Lys-biotin
- NO
- nitric oxide
- LPS
- lipopolysaccharides
- DTNB
- 5-5′-dithiobis(2-nitrobenzoic acid)
- FT-ICR MS
- Fourier transform-ion cyclotron resonance mass spectrometer
- LTQ
- linear quadrupole ion trap
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1.Finkel T., Holbrook N. J. (2000) Nature 408, 239–247 [DOI] [PubMed] [Google Scholar]
- 2.Lambeth J. D. (2004) Nat. Rev. Immunol. 4, 181–189 [DOI] [PubMed] [Google Scholar]
- 3.Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 4.Derakhshan B., Hao G., Gross S. S. (2007) Cardiovasc Res. 75, 210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashan N., Kovsan J., Kachko I., Ovadia H., Rudich A. (2009) Physiol. Rev. 89, 27–71 [DOI] [PubMed] [Google Scholar]
- 6.Foster M. W., Hess D. T., Stamler J. S. (2009) Trends Mol. Med. 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hultqvist M., Olsson L. M., Gelderman K. A., Holmdahl R. (2009) Trends Immunol. 30, 201–208 [DOI] [PubMed] [Google Scholar]
- 8.Owusu-Ansah E., Banerjee U. (2009) Nature 461, 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dröge W. (2002) Physiol. Rev. 82, 47–95 [DOI] [PubMed] [Google Scholar]
- 10.Malito E., Hulse R. E., Tang W. J. (2008) Cell. Mol. Life Sci. 65, 2574–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y., Joachimiak A., Rosner M. R., Tang W. J. (2006) Nature 443, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Im H., Manolopoulou M., Malito E., Shen Y., Zhao J., Neant-Fery M., Sun C. Y., Meredith S. C., Sisodia S. S., Leissring M. A., Tang W. J. (2007) J. Biol. Chem. 282, 25453–25463 [DOI] [PubMed] [Google Scholar]
- 13.Malito E., Ralat L. A., Manolopoulou M., Tsay J. L., Wadlington N. L., Tang W. J. (2008) Biochemistry 48, 12822–12834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurochkin I. V. (2001) Trends Biochem. Sci. 26, 421–425 [DOI] [PubMed] [Google Scholar]
- 15.Duckworth W. C., Bennett R. G., Hamel F. G. (1998) Endocr. Rev. 19, 608–624 [DOI] [PubMed] [Google Scholar]
- 16.Hersh L. B. (2006) Cell. Mol. Life Sci. 63, 2432–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirsky I. A., Broh-Kahn R. H. (1949) Arch Biochem. 20, 1–9 [PubMed] [Google Scholar]
- 18.Kurochkin I. V., Goto S. (1994) FEBS Lett. 345, 33–37 [DOI] [PubMed] [Google Scholar]
- 19.Selkoe D. J. (2001) Neuron 32, 177–180 [DOI] [PubMed] [Google Scholar]
- 20.Müller D., Baumeister H., Buck F., Richter D. (1991) Eur. J. Biochem. 202, 285–292 [DOI] [PubMed] [Google Scholar]
- 21.Neant-Fery M., Garcia-Ordoñez R. D., Logan T. P., Selkoe D. J., Li L., Reinstatler L., Leissring M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9582–9587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cordes C. M., Bennett R. G., Siford G. L., Hamel F. G. (2009) Biochem. Pharmacol. 77, 1064–1073 [DOI] [PubMed] [Google Scholar]
- 23.Manolopoulou M., Guo Q., Malito E., Schilling A. B., Tang W. J. (2009) J. Biol. Chem. 284, 14177–14188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson G. D., Ahn K. (2000) Anal. Biochem. 286, 112–118 [DOI] [PubMed] [Google Scholar]
- 25.Leissring M. A., Lu A., Condron M. M., Teplow D. B., Stein R. L., Farris W., Selkoe D. J. (2003) J. Biol. Chem. 278, 37314–37320 [DOI] [PubMed] [Google Scholar]
- 26.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27.Lundblad R. L., Noyes C. M. (1984) Chemical Reagents for Protein Modification, Vol. 1, p. 72, CRC Press, Boca Raton, FL [Google Scholar]
- 28.Blasi E., Barluzzi R., Bocchini V., Mazzolla R., Bistoni F. (1990) J. Neuroimmunol. 27, 229–237 [DOI] [PubMed] [Google Scholar]
- 29.Qiu W. Q., Walsh D. M., Ye Z., Vekrellis K., Zhang J., Podlisny M. B., Rosner M. R., Safavi A., Hersh L. B., Selkoe D. J. (1998) J. Biol. Chem. 273, 32730–32738 [DOI] [PubMed] [Google Scholar]
- 30.Gibbons H. M., Dragunow M. (2006) Brain Res. 1084, 1–15 [DOI] [PubMed] [Google Scholar]
- 31.Horvath R. J., Nutile-McMenemy N., Alkaitis M. S., Deleo J. A. (2008) J. Neurochem. 107, 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehlhase J., Gieche J., Ullrich O., Sitte N., Grune T. (2000) IUBMB Life 50, 331–335 [DOI] [PubMed] [Google Scholar]
- 33.Hey C., Wessler I., Racké K. (1995) Naunyn-Schmiedebergs Arch. Pharmacol 351, 651–659 [DOI] [PubMed] [Google Scholar]
- 34.Green L. C., Wagner D. A., Glogowski J., Skipper P. L., Wishnok J. S., Tannenbaum S. R. (1982) Anal. Biochem. 126, 131–138 [DOI] [PubMed] [Google Scholar]
- 35.Huynh M. L., Russell P., Walsh B. (2009) Methods Mol. Biol. 519, 507–513 [DOI] [PubMed] [Google Scholar]
- 36.Qiu W. Q., Ye Z., Kholodenko D., Seubert P., Selkoe D. J. (1997) J. Biol. Chem. 272, 6641–6646 [DOI] [PubMed] [Google Scholar]
- 37.Hanafy K. A., Krumenacker J. S., Murad F. (2001) Med. Sci. Monit. 7, 801–819 [PubMed] [Google Scholar]
- 38.Chalmers M. J., Håkansson K., Johnson R., Smith R., Shen J., Emmett M. R., Marshall A. G. (2004) Proteomics 4, 970–981 [DOI] [PubMed] [Google Scholar]
- 39.Chalmers M. J., Quinn J. P., Blakney G. T., Emmett M. R., Mischak H., Gaskell S. J., Marshall A. G. (2003) J. Proteome Res. 2, 373–382 [DOI] [PubMed] [Google Scholar]
- 40.Renfrow M. B., Cooper H. J., Tomana M., Kulhavy R., Hiki Y., Toma K., Emmett M. R., Mestecky J., Marshall A. G., Novak J. (2005) J. Biol. Chem. 280, 19136–19145 [DOI] [PubMed] [Google Scholar]
- 41.Dalpathado D. S., Irungu J., Go E. P., Butnev V. Y., Norton K., Bousfield G. R., Desaire H. (2006) Biochemistry 45, 8665–8673 [DOI] [PubMed] [Google Scholar]
- 42.Cooper H. J., Tatham M. H., Jaffray E., Heath J. K., Lam T. T., Marshall A. G., Hay R. T. (2005) Anal Chem 77, 6310–6319 [DOI] [PubMed] [Google Scholar]
- 43.Håkansson K., Cooper H. J., Emmett M. R., Costello C. E., Marshall A. G., Nilsson C. L. (2001) Anal. Chem. 73, 4530–4536 [DOI] [PubMed] [Google Scholar]
- 44.Chen Y. Y., Chu H. M., Pan K. T., Teng C. H., Wang D. L., Wang A. H., Khoo K. H., Meng T. C. (2008) J. Biol. Chem. 283, 35265–35272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.