Abstract
The low density lipoprotein receptor-related protein 1 (LRP1) is a multi-ligand receptor abundantly expressed in neurons. Previous work has shown that brain LRP1 levels are decreased during aging and in Alzheimer disease. Although mounting evidence has demonstrated a role for LRP1 in the metabolism of apolipoprotein E/lipoprotein and amyloid-β peptide, whether LRP1 also plays a direct role in neuronal survival is not clear. Here, we show that LRP1 expression is critical for the survival of primary neurons under stress conditions including trophic withdrawal, the presence of apoptosis inducers, or amyloid-β-induced neurotoxicity. Using lentiviral short hairpin RNA to knock down endogenous LRP1 expression, we showed that a depletion of LRP1 leads to an activation of caspase-3 and increased neuronal apoptosis, an effect that was rescued by a caspase-3 inhibitor. A correlation between decreased Akt phosphorylation and the activation of caspase-3 was demonstrated in LRP1 knocked down neurons. Notably, LRP1 knockdown decreased insulin receptor levels in primary neurons, suggesting that decreased neuronal survival might be a consequence of an impaired insulin receptor signaling pathway. Correspondingly, both insulin receptor and phospho-Akt levels were decreased in LRP1 forebrain knock-out mice. These results demonstrate that LRP1 mediates anti-apoptotic function in neurons by regulating insulin receptor and the Akt survival pathway and suggest that restoring LRP1 expression in Alzheimer disease brain might be beneficial to inhibiting neurodegeneration.
Introduction
Originally described as a clearance receptor for lipoproteins in the liver, the low density lipoprotein receptor-related protein 1 (LRP1)2 plays essential roles in development and mediates important tissue-specific functions throughout adulthood. These include regulation of glutamatergic synaptic transmission in the nervous system, prevention of vascular smooth muscle cell proliferation and atherosclerosis development, catabolism of activated coagulation factor VIII in hepatocytes, and regulation of body energy by adipocytes (1, 2). Mounting evidence suggests an important role for LRP1 in the pathogenesis of Alzheimer disease (AD). LRP1 regulates the production and clearance of amyloid-β (Aβ) peptide and the metabolism of several AD-associated ligands, including apolipoprotein E (apoE), which has three isoforms in humans with apoE4 being a major risk factor for late onset AD (3).
LRP1 is abundantly expressed in the cell body and proximal processes of cortical and hippocampal neurons in the brain (4–6). It consists of an extracellular 515-kDa subunit (LRP1–515) that binds more than 30 different ligands and a transmembrane 85-kDa chain (LRP1–85) that interacts with several adaptor proteins for efficient endocytic trafficking and signaling (7). LRP1 binds to Aβ either directly or via Aβ chaperones, such as apoE, to mediate Aβ clearance (3, 8, 9). In AD patients and in the elderly, brain LRP1 levels are significantly decreased and inversely correlate with the age of onset of AD, suggesting that a decrease in LRP1 functions might contribute to cognitive decline (10).
Both active caspase-3 and DNA breaks have been detected in neurons that are associated with AD plaques (11) and in tangle-bearing neurons (12, 13). Although the involvement of apoptosis in AD is still a matter of debate (14), in vitro evidence implicates caspase-3 and caspase-6 in the cell death mechanism of cultured human neurons (15) and their distinct roles in neuronal survival and axonal degeneration induced by trophic withdrawal and soluble amyloid precursor protein (16). Additionally, it has been demonstrated that caspase-3-mediated cleavage of GGA3 (17), amyloid precursor protein (18), and Tau (19) contributes to increased neuronal cell death in animal models of AD.
Aβ-induced neuronal apoptosis can be reverted by several neuroprotective factors including neurotrophin-3 (20), insulin-like growth factor I (21), and α7 nicotinic receptor agonists (22), which activate the serine/threonine protein kinase Akt. Recent evidence supports the hypothesis that the Akt signaling pathway might be deregulated in AD. In the hippocampus of AD patients, cytoplasmic phospho-Akt Ser473 levels decrease (23). Presenilins bearing familial AD mutations increase neuronal susceptibility to cell death by down-regulating the pro-survival Akt pathway (24, 25). Additionally, Aβ42 oligomers bind to the insulin receptor (26, 27) and the brain-derived neurotrophic factor receptor (28) and modify their signaling properties, suggesting that Akt deregulation might be central to AD. In this work, we demonstrate that LRP1 inhibits apoptosis in primary neurons under stress conditions by regulating insulin receptor and Akt signaling pathway.
EXPERIMENTAL PROCEDURES
Reagents
LRP1 shRNA plasmids TRCN119624 (target 1) and TRCN119625 (target 2) and the empty pLKO.1 vector were obtained from Sigma. Antibodies to cleaved (Asp175) caspase-3, caspase-3, phospho-Akt Ser473, total Akt, phospho-GSK-3β, and total GSK-3β were purchased from Cell Transduction Laboratories (Danvers, MA). p85 and MAP2 antibodies were from Millipore (Bedford, MA). Insulin receptor β subunit antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-LRP1 were in-house produced rabbit polyclonal antibodies. DAPI was from Millipore (Bedford, MA). Laminin was from BD Biosciences (Franklin Lakes, NJ). All of the other reagents not specified were from Sigma.
Lentivirus Preparation
Lentiviruses were produced by the viral core facility at Washington University School of Medicine. Briefly, 293T cells were transfected with pLKO.1 derived constructs together with the pHR'8.2ΔR and pCMV-VSV-G packaging systems as described (29). Conditioned medium was concentrated by ultracentrifugation, titrated against HT1080 cells, and kept frozen in 20-μl aliquots at −80 °C until use.
Primary Neuronal Culture, Lentiviral Infection, and Induction of Apoptosis
Primary mixed cortical and hippocampal neurons were obtained from 16–18-day-old embryos of wild type Swiss Webster mice (Taconic, Hudson, NY) and grown in neurobasal medium supplemented with 2% B27 and 0.5 mm l-glutamine (Invitrogen) on plastic multiwell plates or acid-washed (oven-sterilized) coverslips, all precoated with poly-d-lysine (100 μg/ml) plus laminin (4 μg/ml) solution overnight. The neurons were treated with 2 μm AraC after 2 days in vitro and infected the next day with equivalent transformation units of pLKO.1 and different LRP1 shRNA viruses. 3 days after the lentiviral infection (6 days in vitro), the cells were depleted of B27 supplements for the induction of neuronal apoptosis by replacing the growth media with neurobasal medium containing only 0.5 mm l-glutamine. The cultures were then processed for Western blot, quantitative PCR, or TUNEL and MTT assays 12 or 18 h after trophic withdrawal.
Immunofluorescence and TUNEL Assay
For immunostaining procedures, all of the solutions were prewarmed at 37 °C before use. The neurons were washed twice with PBS and fixed in PBS containing 4% paraformaldehyde and 4% sucrose for 20 min at room temperature. After four short washes, fixed neurons were permeablized with PBS containing 0.2% Triton X-100 for 10 min. After three washing steps, permeabilized neurons were blocked for 1 h with PBS containing 5% bovine serum albumin and were incubated with MAP2 (1:100) and anti-LRP1 antibodies (25 μg/ml) in PBS containing 1% bovine serum albumin for 30 min at 37 °C. After three washes, the cells were incubated with secondary antibodies, and a short DAPI incubation was included during the final wash before mounting the coverslips. For detection of apoptosis, the neurons were stained using the TUNEL apoptosis detection kit (Millipore, Bedford, MA) following the manufacturer's instructions. The cells were counterstained with DAPI to reveal normal and apoptotic nuclear morphology. The images were captured using the LSM5 Pascal software coupled to a Zeiss LSM Pascal Vario 2 RGB confocal system.
MTT and MTS Assay
After different treatments, neuronal viability was measured by using the cell proliferation kit I MTT (Roche Applied Science). The neurons were labeled for 45–60 min, and the reduced formazan product was solubilized overnight in lysis buffer and quantified as recommended by the manufacturer using a SynergyTM microplate reader (BIO-TEK Instrument, Winooski, VT). For the rescue experiments, 50 μm of the caspase-3 inhibitor z-DEVD-fmk (R & D Systems, Minneapolis, MN) or control Me2SO vehicle were incorporated into the trophic withdrawal treatment. For Aβ toxicity experiments, infected neurons were co-treated with freshly resuspended Aβ42 at indicated doses for 18 h in the presence of B27 supplements. After treatment, the neuronal viability was assessed by the MTS reduction assay (Promega, Madison, WI), and the reduced, soluble formazan product obtained in 3 h of incubation was quantified by directly reading of the plate.
Western Blotting
After treatment, the cells were immediately washed twice with cold PBS and lysed in PBS containing 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 1× Complete protease inhibitor mix (Roche Applied Science), 10 mm NaF, and 1 mm Na3VO4. Protein determination was performed in triplicate using the Bio-Rad Bradford reagent. Twenty μg of lysate were subjected to reducing SDS-polyacrylamide gel electrophoresis, the proteins were transferred overnight, and specific proteins were detected with antibodies following the manufacturer's recommendations. All of the determinations were performed with an enhanced chemiluminiscence kit (Pierce), and a Reblot Plus Strong stripping solution kit (Millipore, Bedford, MA) was used when needed. The images were obtained with the Chemidoc-XRS system, and densitometric analyses were performed using Quantity One software (Bio-Rad).
Assessment of Aβ42 Aggregation in the Conditioned Media of Primary Neuronal Cultures
Conditioned media from infected or control, uninfected primary neurons incubated with Aβ42 for 24 h were electrophoretically separated on 16% polyacrylamide Tris-Tricine gels, and transferred to a polyvinylidene difluoride membrane. Immunoblotting with 6E10 antibody against Aβ (0.5 μg/ml) was carried out overnight at 4 °C followed by incubation with fluorescent anti-mouse IgG (1:10,000 dilution) for 1 h at room temperature. Fluorescent signals were detected with an Odyssey imaging scanner (LI-COR, Lincoln, NE).
LRP1 Knockdown and Cell Death Studies in GT1-7 Cells
GT1-7 cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The cells were simultaneously seeded at a density of 30,000 cells/cm2 in 6-well plates (Western blot) or 48-well dishes (MTT) and transfected with 90 nm LRP1 siRNA (GGA GUC ACU UAC AUC AAU A, J-040764-07) or control siGENOME nontargeting siRNA 5 (Dharmacon, Lafayette, CO) using Lipofectamine 2000 reagent (Invitrogen). Seventy-two h after transfection, the cells were washed twice with Dulbecco's modified Eagle's medium and incubated with growing medium or with Dulbecco's modified Eagle's medium-only medium for additional 24 h. Total lysates were prepared and LRP1, Akt, Caspase-3, and Cleaved Caspase-3 levels were determined by Western blot. Cell viability was determined using MTT proliferation assay kit (Roche Applied Science).
Quantitative PCR
The cells were washed twice in PBS, and total RNA was extracted with the TRIzol reagent as recommended by the manufacturer (Invitrogen). Two μg RNA were subjected to first strand cDNA synthesis using the Superscript III reverse transcription kit with random hexamers as primers (Invitrogen). cDNAs were diluted five times, and 3 μl were used for each determination using the Platinum® SYBR® Green qPCR SuperMix-UDG kit (Invitrogen) and the iCycler iQ® single-color real time PCR detection system (MyiQTM real time PCR software; Bio-Rad). The primers utilized were: ATG GGC TTC GGG AGA GGA T (insulin receptor, sense), GGA TGT CCA TAC CAG GGC AC (insulin receptor, antisense), AGT GTG ACG TTG ACA TCC GTA (β-actin, sense), and GCC AGA GCA ATC TCC TTC T (β-actin, antisense).
Brain Lysates and Detection of Phospho-Tau in Cortical Brain Lysates
Brain cortices were dissected from flox-LRP1 transgenic mice or Cre-flox-LRP1 double transgenic mice and lysed in PBS containing 1% Triton X-100, 2 mm phenylmethylsulfonyl fluoride, 2× Complete protease inhibitor mix (Roche Applied Science), 10 mm NaF, and 1 mm Na3VO4. Protein determination was performed in triplicate using the Bio-Rad Bradford reagent (Bio-Rad), and 20 μg of lysate were subjected to reducing SDS-polyacrylamide gel electrophoresis. The proteins were transferred overnight to a polyvinylidene difluoride membrane, and specific proteins were detected with LRP1, phospho-Tau (AT270, proline-directed kinase-dependent), and total Tau (14/46) antibodies.
Statistical Methods
The data are presented as the means ± S.D. from three independent experiments or from indicated number of animals. Differences between and among groups were determined by ANOVA with Dunnett's (multiple comparison with a single control group) or Bonferroni's test (selected comparisons) or by unpaired Student's t test, as indicated in the figure legends. The values of p < 0.05 were considered statistically significant.
RESULTS
Knockdown of LRP1 Expression in Primary Neurons Increases Cell Death
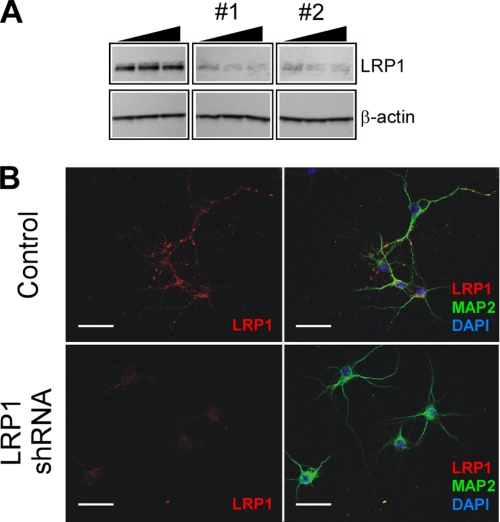
To test whether LRP1 regulates neuronal apoptosis, we decreased LRP1 levels in cultured primary neurons by lentivirus-mediated delivery of short hairpin RNA (shRNA). The cells were infected either with one of two LRP1 shRNAs that target different regions of LRP1 or with the empty pLKO.1 vector as a negative control, and LRP1 levels were determined by Western blot and indirect immunofluorescence. As shown in Fig. 1A, infection with either shRNA decreased LRP1 levels by about 80% in cultured neurons compared with control. We selected LRP1 shRNA 2 for the remainder of this study. It has been previously demonstrated that LRP1 subcellular localization is confined to the neuronal cell bodies and to the proximal processes in vivo (4, 6), and it is progressively restricted to a MAP2-positive somatodendritic region as neuronal cells differentiate in vitro (30). A decrease in LRP1 immunostaining was observed after 3 days of shRNA infection as confirmed by confocal microscopy (Fig. 1B). These results demonstrate that endogenous LRP1 levels can be efficiently knocked down in mouse primary neurons by lentivirus-delivered shRNA. Next, we determined the effects of LRP1 knockdown on neuronal survival. The cultures were infected with lentiviruses as described under “Experimental Procedures.” After 3 days of infection, neuronal viability was assessed with the MTT reduction assay. We were unable to detect any differences in neuronal viability under these conditions (Fig. 2A). It is possible that growth factors, antioxidants, and trophic factors in the B27 supplement might compensate for the effect of decreasing LRP1 expression in our experimental system. We therefore analyzed cell viability upon trophic support withdrawal as a paradigm for inducing neuronal injury. The cultures were similarly infected for 3 days, and the cells were incubated with Neurobasal-only medium for 18 h, and neuronal viability was assessed by MTT. Under these conditions, knockdown of LRP1 expression significantly increased neuronal cell death compared with cultures infected with control viruses (Fig. 2A). These results demonstrate that LRP1 is important for mediating neuronal survival under stress conditions.
FIGURE 1.
Knockdown of LRP1 in primary neurons. A, titration of LRP1 shRNA lentiviruses targeting two different regions of LRP1. The neurons were infected with 1 × 105, 2.5 × 105, or 1 × 106 transforming units of the indicated lentiviruses, and LRP1 and β-actin levels were analyzed by Western blot. A lentivirus prepared from pLKO.1 empty vector was used as a control. Both LRP1 shRNAs efficiently decreased LRP1 levels. LRP1 shRNA 2 was selected for the remainder of this study. B, triple staining for LRP1 (red), MAP2 (green), and DAPI (blue) in control and LRP1 shRNA-infected neurons. A robust LRP1 staining was detected in the soma and a spotted pattern in MAP2-positive processes and were significantly decreased upon infection with LRP1 shRNA. Scale bar, 50 μm.
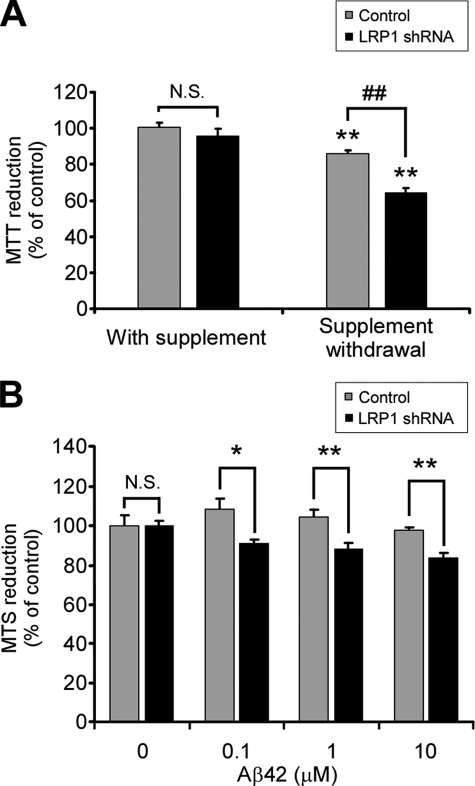
FIGURE 2.
LRP1 knockdown decreases cell viability in primary neurons upon trophic withdrawal and Aβ42-induced toxicity. A, effect of control or LRP1 shRNA on neuronal viability analyzed by the MTT reduction assay in the presence or absence of B27 supplement. Increased neuronal cell death was detected in LRP1 shRNA-infected neurons upon trophic support withdrawal. The mean differences were compared by ANOVA and Dunnett's test using control infection + B27 supplement cells as the reference group (**, p < 0.01) or by ANOVA and Bonferroni's test for selected groups (##, p < 0.01). B, LRP1 knockdown renders neurons susceptible to Aβ-induced toxicity. Primary neurons were infected with control or LRP1 shRNA and incubated with 0, 0.1, 1, and 10 μm Aβ42 for 18 h, and neuronal viability was assessed by reduction of the MTS redox dye. The mean differences were compared by unpaired Student's t test. *, p < 0.05; **, p < 0.01.
LRP1 Protects against Aβ42-induced Neurotoxicity
It has been well established that Aβ42 impairs neuronal survival in vitro and that the pro-survival Akt signaling is a target for Aβ42-induced neurotoxicity. However, it is not known whether LRP1 plays a role in protecting neurons against Aβ42-induced cell death. We exposed control and LRP1 knocked down neurons to Aβ42 in the presence of B27 supplement and evaluated cell viability with the MTS reduction assay, a more suitable tetrazolium redox dye for evaluating amyloid-induced toxicity (31, 32). In control neurons, no viability loss was observed upon 18 h of treatment of Aβ42 for all of the doses utilized. However, LRP1 knocked down neurons were susceptible to Aβ42 toxicity from concentrations above 100 nm, as evidenced by a statistically significant decrease in the reduction of the MTS reagent (Fig. 2B). The decrease in cell viability in LRP1 knocked down cells might result from a differential balance of Aβ42 species in the conditioned medium during the incubation period. To rule out such a possibility, we analyzed the aggregation state of Aβ42 at the end of the experiment. Immunoblotting revealed that only a small fraction of Aβ42 became SDS-stable oligomers under our experimental conditions (supplemental Fig. S1), indicating that neither the presence of lentiviruses nor a decreased expression of LRP1 affected Aβ aggregation and suggesting that the toxic effect was derived mostly from Aβ monomers rather than oligomers. These results further support an important role for LRP1 in neuronal survival under stress conditions such as trophic withdrawal and the neurotoxicity induced by Aβ42.
LRP1 Mediates Anti-apoptotic Function in Primary Neurons
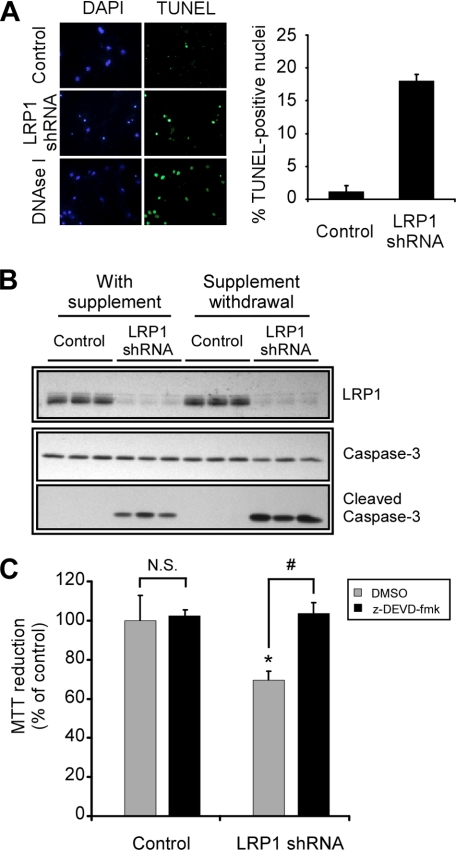
To determine whether apoptosis was responsible for the cell death observed in neurons with decreased LRP1 expression, we analyzed DNA breaks by TUNEL assay in control and LRP1 shRNA-infected neurons upon trophic withdrawal. The percentage of neurons containing positive TUNEL-stained nuclei increased from 1.2 ± 0.9 to 18.0 ± 1.0 when LRP1 was knocked down (Fig. 3A). To confirm these results, we performed similar experiments, and the levels of activated caspase-3 were determined by Western blot. Caspase-3 is an executioner caspase that mediates apoptosis downstream of both mitochondrial and extramitochondrial pathways by proteolytic cleavage. LRP1 knockdown increased the levels of cleaved caspase-3 and decreased the levels of procaspase-3 in neurons deprived of trophic support (Fig. 3B). Increased activation of caspase-3 was also detected in LRP1 knocked down neurons treated with staurosporine, indicating that LRP1 protects neurons from apoptosis under different stress conditions (supplemental Fig. S2). To confirm that the activation of caspase-3 is the pathway affected by decreased LRP1 levels, we determined the effects of caspase-3 inhibition on cell survival in neurons infected with control or LRP1 shRNA and subjected to trophic withdrawal. Primary neurons were infected as before and subjected to trophic withdrawal in the presence or absence of the caspase-3 inhibitor z-DEVD-fmk. Caspase-3 inhibition decreased cell death in neurons with LRP1 knockdown, indicating that the effects of decreased LRP1 levels on neuronal death were through the induction of caspase-3-dependent apoptosis (Fig. 3C). Together, these results demonstrate that LRP1 mediates an anti-apoptotic function in primary neurons by inhibiting caspase-3 activation.
FIGURE 3.
LRP1 regulates apoptosis in primary neurons. A, neurons were infected with control or LRP1 shRNA, and apoptosis was analyzed by TUNEL assay in neurons treated for 18 h with neurobasal-only medium. Left panel, representative images showing nuclear staining of DNA (DAPI) or single and double strand DNA breaks (TUNEL). Positive controls from an untreated culture incubated with DNase-I are shown. LRP1 knockdown increases apoptosis in primary neurons. Right panel, quantification of TUNEL staining. TUNEL-positive neurons were counted blindly from at least 500 neurons from two fields of three independent experiments. B, primary neurons were infected with control or LRP1 shRNA and cleaved caspase-3, full-length caspase-3, LRP1, and β-actin levels were analyzed by Western blot upon 18 h of treatment in neurobasal-only media. Active caspase-3 is increased in LRP1 knocked down neurons. Each lane represents the result obtained from an independent infection. C, primary neurons were infected with control or LRP1 shRNA and were subjected to trophic withdrawal in the presence of vehicle control (dimethyl sulfoxide, DMSO) or the caspase-3 inhibitor z-DEVD-fmk for 18 h, and cell viability was analyzed by MTT assay. Caspase-3 inhibition decreases cell death in LRP1 knocked down neurons. The mean differences were compared by ANOVA and Dunnett's test using control infection + Me2SO-treated cells as the reference group (*, p < 0.05) or by ANOVA and Bonferroni's test for selected groups (#, p < 0.05). The effects of LRP1 knockdown on neuronal cell death were rescued in the presence of the caspase-3 inhibitor.
LRP1 Activates Akt in Primary Neurons and in Mouse Brain
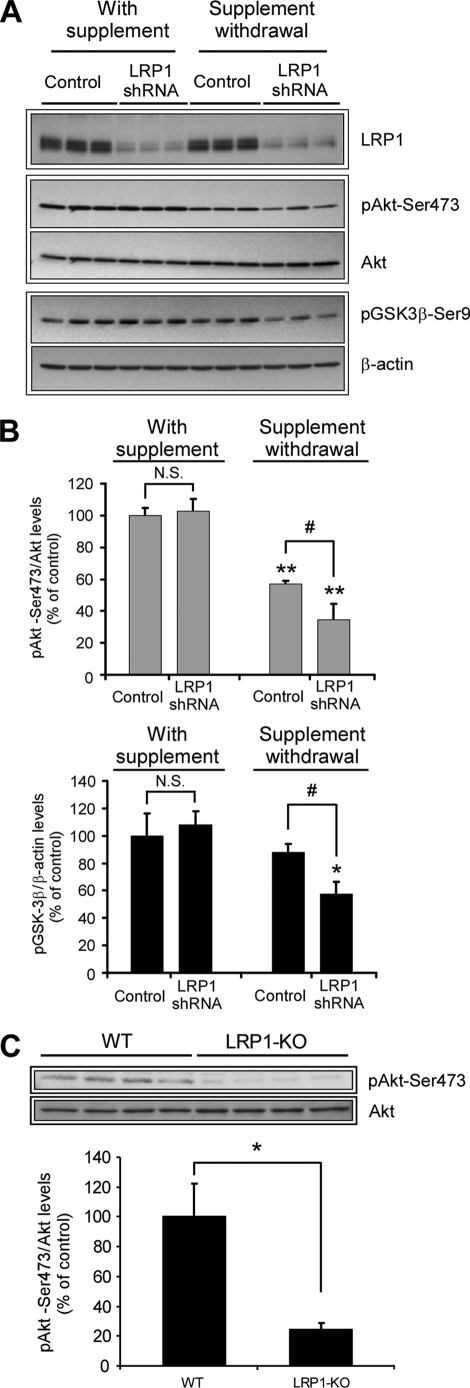
It has been well documented that the PI3K/Akt signaling pathway is essential for neuronal survival and for the neuroprotective action of growth factors in cortical neurons in vitro. Moreover, it has been demonstrated that this signaling pathway is a target of different types of neuronal injuries such as ischemia, seizure, and Aβ toxicity in vivo. Phosphorylation at serine residue 473 is required for full activation of Akt, for growth factor signal transduction, and also for neuronal survival against Aβ-induced toxicity (21, 33). We therefore analyzed the effects of LRP1 knockdown on phospho-Akt Ser473 levels in both normal and trophic deprived neurons. In control neurons, trophic withdrawal substantially decreased phospho-Akt Ser473 levels (Fig. 4A). This is likely due to the removal of growth factors (e.g. brain-derived neurotrophic factor, insulin) present in the B27 supplement already known to activate the Akt pathway. However, a further decrease in phospho-Akt levels was detected in LRP1 knocked down neurons compared with control neurons after removal of trophic support (Fig. 4, A and B), suggesting that LRP1 is important for activating an endogenous signaling pathway that leads to Akt activation in the absence of growth factors. To further examine the effects of LRP1 on Akt signaling, we analyzed the status of GSK-3β phosphorylation, a downstream target of Akt (Fig. 4, A and B). A decrease in GSK-3β phosphorylation was detected in LRP1 knocked down neurons after trophic withdrawal, which parallels the decrease in Akt activation. These results demonstrate that LRP1 activates Akt signaling and its downstream effector GSK-3β in primary neurons.
FIGURE 4.
LRP1 regulates Akt phosphorylation in vitro and in vivo. A, decreased phospho-Akt and phospho-GSK-3β levels in LRP1 knocked down neurons upon trophic withdrawal. Primary neurons were infected with control or LRP1 shRNA, and the levels of LRP1, phospho-Akt (Ser473), total Akt, and phospho-GSK-3β were analyzed by Western blot under both control conditions and upon 18 h of treatment with neurobasal-only media. The β-actin levels were determined as a loading control. The levels of phospho-Akt and phospho-GSK-3β were additionally decreased in LRP1 knocked down neurons upon trophic withdrawal. B, protein levels from experiments in A were determined by densitometry, and the corresponding phospho-Akt to total Akt and phospho-GSK-3β to β-actin ratios were calculated and plotted relative to control neurons. The mean differences were compared by ANOVA and Dunnett's test using control infection + B27 supplement treated cells as the reference group (**, p < 0.01; *, p < 0.05) or by ANOVA and Bonferroni's test for selected groups (#, p < 0.05). C, decreased phospho-Akt in LRP1 forebrain knock-out mice. Equal amounts of total brain homogenates (40 μg) from LRP1 forebrain knock-out mice and littermate controls were subjected to Western blot analysis, and both phospho-Akt (Ser473) and total Akt levels were determined (n = 4). Lower panel, densitometric analysis of phospho-Akt and total Akt levels determined from experiments as in A. The mean differences were compared by unpaired Student's t test, * p < 0.05. WT, wild type.
To verify these results in a cell culture system that is not subject to lentiviral infection, we performed siRNA knockdown of LRP1 expression in GT1-7 cells, a mouse central nervous system neuronal cell line previously used for studying cell death mechanisms in vitro, including trophic withdrawal- and Aβ42-induced cell death (34, 35). We transfected GT1-7 cells with control, nontargeting siRNA or LRP1 siRNA and analyzed the effect of serum deprivation on apoptosis activation and Akt phosphorylation (supplemental Fig. S3). We found that GT1-7 cells with decreased LRP1 levels are more susceptible to trophic withdrawal-induced apoptosis, as evidenced by a decrease in MTT reduction and an increase in the cleavage of Caspase-3 upon serum deprivation. A corresponding decrease in phospho-Akt levels was also detected. Together, these results demonstrate that LRP1 regulates Akt signaling and cell survival both in primary neurons and in neuronal cells (supplemental Fig. S3).
To examine whether Akt activation is also regulated by LRP1 in vivo, we analyzed the expression of phospho-Akt Ser473 in LRP1 forebrain knock-out mice and wild type controls (36). LRP1 forebrain knock-out mice were generated by crossing LRP1 floxP mice (1) with the CamKIIα-Cre mice (37), which allows LRP1 deletion only in mature forebrain neurons (36). Using immunoblot, we found that the phospho-Akt Ser473 level was significantly decreased in the forebrain of LRP1 knock-out mice (Fig. 4C). These results demonstrate that Akt activation is regulated by LRP1 in neurons both in vitro and in vivo and suggest that LRP1 regulates neuronal survival through the Akt signaling pathway. To determine whether a decrease in neuronal Akt signaling affects a downstream substrate of the Akt-GSK-3β cascade, we analyzed Tau phosphorylation in cortical lysates of LRP1 forebrain knock-out mice and controls. We selected the antibody AT270 that recognizes phospho-Thr181, a target residue sensitive to GSK-3β inhibition (38, 39). We did not detect any difference in Tau phosphorylation in LRP1 forebrain knock-out mice compared with control littermates (supplemental Fig. S4), suggesting that additional signals might be required for Tau phosphorylation after inhibition of the LRP1-Akt-GSK-3β cascade in vivo.
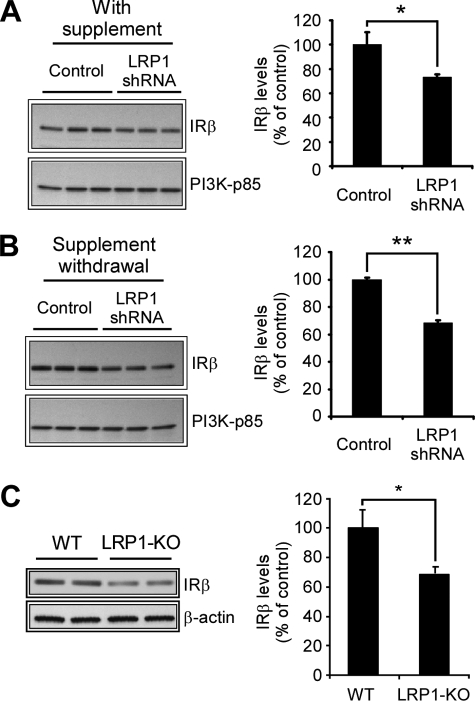
LRP1 Regulates Insulin Receptor Levels
Mounting evidence indicates that there is a correlation between diabetes and AD, and it has been hypothesized that impaired neuronal insulin receptor function and/or an insulin resistance condition might contribute to AD (40, 41). To further investigate the mechanism by which LRP1 activates Akt, we analyzed the levels of the Akt activator PI3K and insulin receptor in primary neurons infected with control or LRP1 shRNA. The PI3K kinase is a dimeric protein consisting of a 110-kDa catalytic subunit and an 85-kDa regulatory subunit (p85) whose levels decrease in AD and in an animal model for dietary-induced insulin resistance (42). We found that p85 levels remain unmodified upon LRP1 knockdown in primary neurons under both basal and trophic withdrawal conditions. However, a significant decrease in the levels of the β catalytic subunit of the insulin receptor was detected in LRP1 knocked down neurons under both basal (Fig. 5A) and trophic withdrawal (Fig. 5B) conditions. Similar results were obtained when a different LRP1 shRNA was used (supplemental Fig. S5). To determine whether LRP1 knockdown decreases insulin receptor mRNA levels, we conducted quantitative PCR experiments in similarly treated neurons (supplemental Fig. S6). We did not detect a decrease but a slight increase in the insulin receptor mRNA levels in neurons infected with LRP1 shRNA compared with controls under the conditions where decreased insulin receptor protein levels were detected (Fig. 5, A and B). These results indicate that LRP1 likely regulates insulin receptor levels through a post-translational mechanism. It is worthwhile to mention that the B27 supplement contains insulin and that the insulin receptor levels are negatively regulated by its ligand (43), which might explain the slight increase in insulin receptor levels upon trophic withdrawal observed in control infected cells (Fig. 5, A and B) and the decrease in phospho-Akt Ser473 levels upon trophic withdrawal (Fig. 4A). These results suggest that, in the absence of supplements, LRP1 knockdown decreases phospho-Akt levels by decreasing insulin receptor β-subunit levels. Next, we analyzed the expression of insulin receptor in the LRP1 forebrain knock-out mice. We detected a slight, yet significant, decrease in insulin receptor levels in the forebrain of LRP1 knock-out mice compared with wild type controls (Fig. 5C), suggesting that LRP1 regulates neuronal insulin receptor levels in vivo. Collectively, these findings demonstrate that LRP1 has an anti-apoptotic function in neurons and strongly suggest that LRP1 knockdown impairs the Akt signaling pathway by decreasing insulin receptor levels in primary neurons and in mouse brain.
FIGURE 5.
Decreased insulin receptor levels in LRP1 knocked down neurons and in LRP1 forebrain knock-out mice. Primary neurons were infected with control or LRP1 shRNA, and insulin receptor (IR) and PI3K-p85 levels were determined by Western blot under both control conditions (A) or upon 18 h of treatment with neurobasal-only media (B). Left panels, protein levels of the insulin receptor-β subunit from experiments in A and B were determined by densitometry and plotted relative to control neurons. The levels of the insulin receptor, but not of PI3K-p85, were decreased in LRP1 knocked down neurons under both basal and trophic withdrawal conditions. The mean differences were compared by unpaired Student's t test. *, p < 0.05; **, p < 0.01. C, decreased insulin receptor in LRP1 forebrain knock-out mice. Equal amounts of total brain homogenates (40 μg) from LRP1 forebrain knock-out mice and littermate controls were subjected to Western blot analysis, and the levels of both insulin receptor and β-actin were determined. Representative results from n = 4 are shown. Left panel, densitometric analysis showing the insulin receptor-to-β-actin ratio determined from experiments in C. The mean differences were compared by unpaired Student's t test. *, p < 0.05. WT, wild type.
DISCUSSION
Most published work studying LRP1 function in signal transduction relies on the use of LRP1 antagonist RAP (44). However, RAP also binds and antagonizes other LDL receptor family members, making it difficult to differentiate the role of LRP1 from other receptors. In the present work, we utilized lentivirus-delivered LRP1 shRNA to specifically knock down LRP1 expression in primary neurons. Our results show that decreasing LRP1 levels in primary neurons increases susceptibility to apoptosis induced by trophic withdrawal and renders neurons more vulnerable to Aβ42-induced toxicity. This increase in neuronal cell death correlates with a decrease in the phosphorylation status of the serine 473 residue of Akt. We additionally demonstrate that the insulin receptor levels were decreased in the LRP1 knocked down neurons, which is a likely mechanism responsible for decreased Akt phosphorylation. The important role of LRP1 in regulating insulin receptor and Akt survival pathways was also demonstrated in vivo in LRP1 forebrain knock-out mice. Together, our results reveal a novel mechanism by which lipoprotein receptor LRP1 promotes neuronal survival.
It is widely accepted that the PI3K/Akt signaling pathway is essential for neuronal survival during development of the nervous system and throughout adulthood (45). Deregulation of the Akt signaling pathway has been observed in neuropathological conditions such as schizophrenia (46), ischemia (47), and AD (see below). Our present results suggest that LRP1 mediates Akt activation in neurons and that decreased LRP1 levels may be important for neuronal survival when cells are exposed to injury or stress, such as a decrease in growth factor function and Aβ-induced toxicity. Previous work has shown that lipoproteins protect retinal ganglion neurons from trophic deprivation-induced cell death through LRP1 in vitro (48). In that study, however, lipoproteins activate protein kinase C and GSK-3β signaling, and neither a correlation between Akt phosphorylation and the neuronal cell death nor an essential role for Akt was observed. We speculate that different in vitro neuronal culture systems in our study and that by Hayashi et al. (48) might explain these differences.
Our finding that Akt is a target for LRP1 signaling in vitro is consistent with the observation that Akt deregulation also occurs in AD regions where LRP1 levels are decreased (10, 23). It has been shown that, depending on the severity of the disease, cytoplasmatic phospho-Akt Ser473 levels decrease in the hippocampus of AD brains. However, in the temporal cortex of AD patients, a mislocalization of active Akt into neuronal membranes as well as an increase in Akt activity have also been reported (23, 49, 50). Further experiments will be required to determine whether LRP1, insulin receptor, and phospho-Akt Ser473 levels are decreased in the hippocampus of AD brains from same specimens. Another physiological scenario for deregulated Akt signaling by LRP1 in AD might involve the uptake of Aβ by neurons. It has been demonstrated that intracellular accumulation of Aβ down-regulates Akt signaling in cultured neurons (51). It is known that the endocytic function of LRP1 is important for the uptake of Aβ in neuroblastoma and fibroblasts, where the formation of a complex between Aβ and α2M increases the uptake and degradation of Aβ (52, 53). We propose that LRP1-mediated uptake of Aβ in neuronal cells might interfere with LRP1 to Akt signaling and therefore may also contribute to the neurodegeneration seen in AD.
It has been demonstrated that insulin mediates the translocation of LRP1 to caveolae in adipocytes (54) and increases cell surface levels of LRP1 in adipocytes and hepatocytes (54, 55). Because insulin receptor association with lipid rafts is important for cell survival signaling in cells lacking caveolin (56) and because Akt activation requires intact lipid rafts (57), we hypothesize that LRP1, insulin receptor, and Akt activation might be linked by its association with lipid rafts and might be required for the transduction of the survival signal through Akt in primary neurons. It will be interesting to analyze the role of LRP1 lipid raft association in the regulation of Akt phosphorylation and signaling through the insulin receptor.
The decrease of insulin receptor levels by LRP1 knockdown strongly suggests that down-regulation of phospho-Akt Ser473 is likely mediated through decreased expression of this upstream receptor. Supporting this hypothesis is the in vivo evidence showing that levels of insulin receptor, insulin receptor substrate 2, PI3K-p85, and phospho-Akt Ser473 as well as LRP1 are significantly decreased in AD brains (41, 42). Together with our present in vitro and in vivo findings, we hypothesize that decreased LRP1 levels in neurons might evoke an insulin receptor resistance phenotype. It is worth mentioning that the insulin sensitizer drug and peroxisome proliferator-activated receptor γ agonist troglitazone induces the expression of LRP1 in adipocytes (58). It will be of great interest to test whether peroxisome proliferator-activated receptor γ agonists can rescue the apoptotic phenotype seen in LRP1 knocked down neurons.
Although impaired insulin receptor signaling might explain decreased phospho-Akt levels in LRP1 knocked down neurons, we cannot rule out additional mechanisms by which LRP1 activates Akt pathway. For example, it has been demonstrated that several cytosolic adaptors important for signal transduction bind to the LRP1 tail. Yeast two-hybrid experiments have demonstrated that Dab1 binds to the cytoplasmic tail of LRP1 and that this binding occurs specifically at the second NPXY motif (59, 60). A signaling pathway from reelin to neuronal Akt activation through the apoE receptor 2 (ApoER2) has been well characterized (61). When reelin binds to ApoER2, tyrosine phosphorylation of the cytoplasmic tail of ApoER2 by Src family members increases the binding of ApoER2 to Dab1, which in turn binds to the p85 regulatory subunit of PI3K. The latter binding activates Akt and leads to the reorganization of the actin cytoskeleton in a reelin- and ApoER2-dependent manner. A similar signaling pathway through LRP1 might also occur in neurons. Because α2 M binds to LRP1 and activates Akt in both Schwann and PC12 cells (62), LRP1 and Dab1 interaction might be important for α2M-induced Akt activation. On the other hand, it has been well established that LRP1 inhibits the signal transduction of platelet-derived growth factor to ERK kinase activation through the binding and modulation of the platelet-derived growth factor receptor trafficking in smooth muscle cells (63–65). Platelet-derived growth factor binding to its receptor mediates an Akt- and Shc-sensitive tyrosine phosphorylation of the LRP1 tail in caveolae that is required for ERK regulation (64). Additional experiments will be required to clarify the mechanism by which LRP1 activates Akt and the role of insulin receptor in this regulation.
In summary, our results demonstrate that LRP1 is important for mediating an anti-apoptotic function in primary neurons through a mechanism that involves Akt phosphorylation and insulin receptor regulation. Our results provide the first evidence linking LRP1 function to insulin receptor and Akt in differentiated neurons. These results further explain the roles of LRP1 in the pathogenesis of AD and suggest that increasing LRP1 levels in AD brains might be a viable approach in the treatment of this devastating disease.
Supplementary Material
Acknowledgments
We thank Dr. Mingjie Li and Nada Husic from the Viral Vectors Core at the Hope Center for Neurological Disorders (Washington University) for producing the lentiviruses and for technical assistance and to Dr. Paul Kotzbauer and Dr. Carlos Cruchaga for providing Tau antibodies. We also thank Julie Trausch-Azar for critical reading of the manuscript and to all members of the Bu and Dr. Alan Schwartz laboratories for valuable comments and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AG027924 and R01 AG031784. This work was also supported by a Zenith Fellows Award from the Alzheimer's Association (to G. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- LRP
- low density lipoprotein receptor-related protein
- AD
- Alzheimer disease
- apo
- apolipoprotein
- Aβ
- amyloid-β
- DAPI
- 4′,6-diamidino-2-phenylindole
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick end labeling
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- ANOVA
- analysis of variance
- shRNA
- short hairpin RNA
- PI3K
- phosphatidylinositol 3-kinase
- ERK
- extracellular signal-regulated kinase
- GSK
- glycogen synthase kinase
- Tricine
- N-[2-hydroxy-1,1- bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Herz J., Clouthier D. E., Hammer R. E. (1992) Cell 71, 411–421 [DOI] [PubMed] [Google Scholar]
- 2.Lillis A. P., Van Duyn L. B., Murphy-Ullrich J. E., Strickland D. K. (2008) Physiol. Rev. 88, 887–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bu G., Cam J., Zerbinatti C. (2006) Ann. N.Y. Acad. Sci. 1086, 35–53 [DOI] [PubMed] [Google Scholar]
- 4.Wolf B. B., Lopes M. B., VandenBerg S. R., Gonias S. L. (1992) Am J. Pathol. 141, 37–42 [PMC free article] [PubMed] [Google Scholar]
- 5.Bu G., Maksymovitch E. A., Nerbonne J. M., Schwartz A. L. (1994) J. Biol. Chem. 269, 18521–18528 [PubMed] [Google Scholar]
- 6.Tooyama I., Kawamata T., Akiyama H., Kimura H., Moestrup S. K., Gliemann J., Matsuo A., McGeer P. L. (1995) Brain Res. 691, 235–238 [DOI] [PubMed] [Google Scholar]
- 7.Lillis A. P., Mikhailenko I., Strickland D. K. (2005) J. Thromb. Haemostasis 3, 1884–1893 [DOI] [PubMed] [Google Scholar]
- 8.Van Uden E., Mallory M., Veinbergs I., Alford M., Rockenstein E., Masliah E. (2002) J. Neurosci. 22, 9298–9304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada K., Hashimoto T., Yabuki C., Nagae Y., Tachikawa M., Strickland D. K., Liu Q., Bu G., Basak J. M., Holtzman D. M., Ohtsuki S., Terasaki T., Iwatsubo T. (2008) J. Biol. Chem. 283, 34554–34562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang D. E., Pietrzik C. U., Baum L., Chevallier N., Merriam D. E., Kounnas M. Z., Wagner S. L., Troncoso J. C., Kawas C. H., Katzman R., Koo E. H. (2000) J. Clin. Invest. 106, 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaFerla F. M., Troncoso J. C., Strickland D. K., Kawas C. H., Jay G. (1997) J. Clin. Invest. 100, 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamblin T. C., Chen F., Zambrano A., Abraha A., Lagalwar S., Guillozet A. L., Lu M., Fu Y., Garcia-Sierra F., LaPointe N., Miller R., Berry R. W., Binder L. I., Cryns V. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10032–10037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohn T. T., Head E., Su J. H., Anderson A. J., Bahr B. A., Cotman C. W., Cribbs D. H. (2001) Am J. Pathol. 158, 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlanc A. C. (2005) Curr. Alzheimer Res. 2, 389–402 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Goodyer C., LeBlanc A. (2000) J. Neurosci. 20, 8384–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolaev A., McLaughlin T., O'Leary D. D., Tessier-Lavigne M. (2009) Nature 457, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Tesco G., Koh Y. H., Kang E. L., Cameron A. N., Das S., Sena-Esteves M., Hiltunen M., Yang S. H., Zhong Z., Shen Y., Simpkins J. W., Tanzi R. E. (2007) Neuron 54, 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvan V., Gorostiza O. F., Banwait S., Ataie M., Logvinova A. V., Sitaraman S., Carlson E., Sagi S. A., Chevallier N., Jin K., Greenberg D. A., Bredesen D. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7130–7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rissman R. A., Poon W. W., Blurton-Jones M., Oddo S., Torp R., Vitek M. P., LaFerla F. M., Rohn T. T., Cotman C. W. (2004) J. Clin. Invest. 114, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesné S., Gabriel C., Nelson D. A., White E., Mackenzie E. T., Vivien D., Buisson A. (2005) J. Biol. Chem. 280, 24941–24947 [DOI] [PubMed] [Google Scholar]
- 21.Wei W., Wang X., Kusiak J. W. (2002) J. Biol. Chem. 277, 17649–17656 [DOI] [PubMed] [Google Scholar]
- 22.Kihara T., Shimohama S., Sawada H., Honda K., Nakamizo T., Shibasaki H., Kume T., Akaike A. (2001) J. Biol. Chem. 276, 13541–13546 [DOI] [PubMed] [Google Scholar]
- 23.Griffin R. J., Moloney A., Kelliher M., Johnston J. A., Ravid R., Dockery P., O'Connor R., O'Neill C. (2005) J. Neurochem. 93, 105–117 [DOI] [PubMed] [Google Scholar]
- 24.Baki L., Neve R. L., Shao Z., Shioi J., Georgakopoulos A., Robakis N. K. (2008) J. Neurosci. 28, 483–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weihl C. C., Ghadge G. D., Kennedy S. G., Hay N., Miller R. J., Roos R. P. (1999) J. Neurosci. 19, 5360–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Townsend M., Mehta T., Selkoe D. J. (2007) J. Biol. Chem. 282, 33305–33312 [DOI] [PubMed] [Google Scholar]
- 27.Zhao W. Q., De Felice F. G., Fernandez S., Chen H., Lambert M. P., Quon M. J., Krafft G. A., Klein W. L. (2008) FASEB J. 22, 246–260 [DOI] [PubMed] [Google Scholar]
- 28.Tong L., Balazs R., Thornton P. L., Cotman C. W. (2004) J. Neurosci. 24, 6799–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart S. A., Dykxhoorn D. M., Palliser D., Mizuno H., Yu E. Y., An D. S., Sabatini D. M., Chen I. S., Hahn W. C., Sharp P. A., Weinberg R. A., Novina C. D. (2003) RNA 9, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown M. D., Banker G. A., Hussaini I. M., Gonias S. L., VandenBerg S. R. (1997) Brain Res. 747, 313–317 [DOI] [PubMed] [Google Scholar]
- 31.Wogulis M., Wright S., Cunningham D., Chilcote T., Powell K., Rydel R. E. (2005) J. Neurosci. 25, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shearman M. S., Hawtin S. R., Tailor V. J. (1995) J. Neurochem. 65, 218–227 [DOI] [PubMed] [Google Scholar]
- 33.van der Heide L. P., Ramakers G. M., Smidt M. P. (2006) Prog. Neurobiol. 79, 205–221 [DOI] [PubMed] [Google Scholar]
- 34.Dhandapani K. M., Hadman M., De Sevilla L., Wade M. F., Mahesh V. B., Brann D. W. (2003) J. Biol. Chem. 278, 43329–43339 [DOI] [PubMed] [Google Scholar]
- 35.Simakova O., Arispe N. J. (2007) J. Neurosci. 27, 13719–13729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q., Zerbinatti C. V., Zhang J., Hoe H. S., Wang B., Cole S. L., Herz J., Muglia L., Bu G. (2007) Neuron 56, 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohlmann A., Gotthardt M., Hammer R. E., Herz J. (1998) J. Clin. Invest. 101, 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong M., Chen D. C., Klein P. S., Lee V. M. (1997) J. Biol. Chem. 272, 25326–25332 [DOI] [PubMed] [Google Scholar]
- 39.Caccamo A., Oddo S., Tran L. X., LaFerla F. M. (2007) Am J. Pathol. 170, 1669–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole G. M., Frautschy S. A. (2007) Exp. Gerontol. 42, 10–21 [DOI] [PubMed] [Google Scholar]
- 41.Steen E., Terry B. M., Rivera E. J., Cannon J. L., Neely T. R., Tavares R., Xu X. J., Wands J. R., de la Monte S. M. (2005) J. Alzheimers Dis. 7, 63–80 [DOI] [PubMed] [Google Scholar]
- 42.Zhao L., Teter B., Morihara T., Lim G. P., Ambegaokar S. S., Ubeda O. J., Frautschy S. A., Cole G. M. (2004) J. Neurosci. 24, 11120–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carpentier J. L. (1989) Diabetologia 32, 627–635 [DOI] [PubMed] [Google Scholar]
- 44.Bu G. (2001) Int. Rev. Cytol. 209, 79–116 [DOI] [PubMed] [Google Scholar]
- 45.Brunet A., Datta S. R., Greenberg M. E. (2001) Curr. Opin. Neurobiol. 11, 297–305 [DOI] [PubMed] [Google Scholar]
- 46.Arguello P. A., Gogos J. A. (2006) Neuron 52, 179–196 [DOI] [PubMed] [Google Scholar]
- 47.Saito A., Narasimhan P., Hayashi T., Okuno S., Ferrand-Drake M., Chan P. H. (2004) J. Neurosci. 24, 1584–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi H., Campenot R. B., Vance D. E., Vance J. E. (2007) J. Neurosci. 27, 1933–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei J. J., Khatoon S., An W. L., Nordlinder M., Tanaka T., Braak H., Tsujio I., Takeda M., Alafuzoff I., Winblad B., Cowburn R. F., Grundke-Iqbal I., Iqbal K. (2003) Acta Neuropathol. 105, 381–392 [DOI] [PubMed] [Google Scholar]
- 50.Rickle A., Bogdanovic N., Volkman I., Winblad B., Ravid R., Cowburn R. F. (2004) Neuroreport 15, 955–959 [DOI] [PubMed] [Google Scholar]
- 51.Magrané J., Rosen K. M., Smith R. C., Walsh K., Gouras G. K., Querfurth H. W. (2005) J. Neurosci. 25, 10960–10969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narita M., Bu G., Holtzman D. M., Schwartz A. L. (1997) J. Neurochem. 68, 587–595 [DOI] [PubMed] [Google Scholar]
- 53.Zerbinatti C. V., Wahrle S. E., Kim H., Cam J. A., Bales K., Paul S. M., Holtzman D. M., Bu G. (2006) J. Biol. Chem. 281, 36180–36186 [DOI] [PubMed] [Google Scholar]
- 54.Ko K. W., Avramoglu R. K., McLeod R. S., Vukmirica J., Yao Z. (2001) Biochemistry 40, 752–759 [DOI] [PubMed] [Google Scholar]
- 55.Tamaki C., Ohtsuki S., Terasaki T. (2007) Mol. Pharmacol. 72, 850–855 [DOI] [PubMed] [Google Scholar]
- 56.Vainio S., Heino S., Mansson J. E., Fredman P., Kuismanen E., Vaarala O., Ikonen E. (2002) EMBO Rep. 3, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lasserre R., Guo X. J., Conchonaud F., Hamon Y., Hawchar O., Bernard A. M., Soudja S. M., Lenne P. F., Rigneault H., Olive D., Bismuth G., Nunès J. A., Payrastre B., Marguet D., He H. T. (2008) Nat. Chem. Biol. 4, 538–547 [DOI] [PubMed] [Google Scholar]
- 58.Gauthier A., Vassiliou G., Benoist F., McPherson R. (2003) J. Biol. Chem. 278, 11945–11953 [DOI] [PubMed] [Google Scholar]
- 59.Howell B. W., Lanier L. M., Frank R., Gertler F. B., Cooper J. A. (1999) Mol. Cell. Biol. 19, 5179–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trommsdorff M., Borg J. P., Margolis B., Herz J. (1998) J. Biol. Chem. 273, 33556–33560 [DOI] [PubMed] [Google Scholar]
- 61.Beffert U., Durudas A., Weeber E. J., Stolt P. C., Giehl K. M., Sweatt J. D., Hammer R. E., Herz J. (2006) J. Neurosci. 26, 2041–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mantuano E., Mukandala G., Li X., Campana W. M., Gonias S. L. (2008) J. Biol. Chem. 283, 19904–19911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boucher P., Gotthardt M., Li W. P., Anderson R. G., Herz J. (2003) Science 300, 329–332 [DOI] [PubMed] [Google Scholar]
- 64.Boucher P., Liu P., Gotthardt M., Hiesberger T., Anderson R. G., Herz J. (2002) J. Biol. Chem. 277, 15507–15513 [DOI] [PubMed] [Google Scholar]
- 65.Loukinova E., Ranganathan S., Kuznetsov S., Gorlatova N., Migliorini M. M., Loukinov D., Ulery P. G., Mikhailenko I., Lawrence D. A., Strickland D. K. (2002) J. Biol. Chem. 277, 15499–15506 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.