Abstract
Fatty acid amide hydrolase (FAAH) terminates the endocannabinoid signaling pathway that regulates numerous neurobehavioral processes in animals by hydrolyzing N-acylethanolamines (NAEs). Recently, an Arabidopsis FAAH homologue (AtFAAH) was identified, and several studies, especially those using AtFAAH overexpressing and knock-out lines, have suggested an in vivo role for FAAH in the catabolism of NAEs in plants. We previously reported that overexpression of AtFAAH in Arabidopsis resulted in accelerated seedling growth, and in seedlings that were insensitive to exogenous NAEs but hypersensitive to abscisic acid (ABA) and hypersusceptible to nonhost pathogens. Here we show that whereas the enhanced growth and NAE tolerance of the AtFAAH overexpressing seedlings depend on the catalytic activity of AtFAAH, hypersensitivity to ABA and hypersusceptibility to nonhost pathogens are independent of its enzymatic activity. Five amino acids known to be critical for rat FAAH activity are also conserved in AtFAAH (Lys-205, Ser-281, Ser-282, Ser-305, and Arg-307). Site-directed mutation of each of these conserved residues in AtFAAH abolished its hydrolytic activity when expressed in Escherichia coli, supporting a common catalytic mechanism in animal and plant FAAH enzymes. Overexpression of these inactive AtFAAH mutants in Arabidopsis showed no growth enhancement and no NAE tolerance, but still rendered the seedlings hypersensitive to ABA and hypersusceptible to nonhost pathogens to a degree similar to the overexpression of the native AtFAAH. Taken together, our findings suggest that the AtFAAH influences plant growth and interacts with ABA signaling and plant defense through distinctly different mechanisms.
Introduction
Fatty acid amide hydrolase (FAAH)3 catalyzes the hydrolysis of acylethanolamides, such as N-acylethanolamines (NAEs) (1–4), as well as fatty acid primary amides (5–7). FAAH is known to terminate the “endocannabinoid” signaling pathway that regulates a variety of neurobehavioral processes in animals (reviewed in Refs. 8–10). This membrane-bound protein is a member of an enzyme superfamily termed the “amidase signature” (AS) family (11–12). Members of the AS family (more than 80 amidases) are characterized by a highly conserved region that consists of ∼130 amino acids rich in serine, glycine, and alanine residues (11–16). The x-ray crystal structure of rat FAAH revealed that the core catalytic machinery of FAAH, in contrast to the Ser-His-Asp triad typical of most serine hydrolases, consists of a novel Ser-Ser-Lys catalytic triad (12, 17–19). faah (−/−) knock-out mice had higher endogenous levels of NAEs compared with wild-type mice, and exhibited a variety of physiological and behavioral abnormalities in response to endocannabinoids, such as hypomotility, analgesia, catalepsy, and hypothermia (20–23). These observations suggested that FAAH was a key enzyme involved in the catabolism of NAEs in vivo and was responsible for termination of the endocannabinoid signaling.
In plants, FAAH homologues were identified and characterized recently at the biochemical level (24–25), but much remains to be learned regarding the precise cellular function and physiological significance of this enzyme in plants. An NAE hydrolase activity was first detected in vitro in homogenates of tobacco cells (26), and was demonstrated both in vivo and in vitro in imbibed cotton seeds through radiolabeling approaches (27). An Arabidopsis FAAH homologue (AtFAAH; locus At5g64440) was identified that encodes a protein of 607 amino acids with 37% identity to rat FAAH within the AS domain (24). Catalytic residues (Lys-205, Ser-281, and Ser-305) were absolutely conserved and a single transmembrane domain, like rat FAAH, was predicted to be present near the N terminus of the protein (24). Recombinant protein, expressed in Escherichia coli, was indeed active in hydrolyzing a variety of naturally occurring fatty acid amides (24). Functional FAAH homologues have been identified and characterized in diverse plant species (25). Homology modeling of the AS region of the plant FAAH revealed a highly conserved active site organization with the catalytic triad positioned in the substrate-binding site (25).
Several lines of evidence, especially those using AtFAAH- overexpressing and T-DNA insertion mutant plants, clearly support a role for FAAH in vivo in the catabolism of NAEs in plants. Exogenous NAE at low micromolar concentrations exhibited a dose-dependent reduction of Arabidopsis seedling growth (28), suggesting that hydrolysis of endogenous NAEs by AtFAAH might be important for normal development. Indeed, AtFAAH overexpressors displayed enhanced seedling growth and increased cell/organ size (29). Seeds of AtFAAH overexpressors had lower endogenous NAE content, and their seedling growth was less sensitive to exogenous NAE, whereas Atfaah knock-out seeds had elevated levels of endogenous NAEs in desiccated seeds, and their seedlings were hypersensitive to exogenous NAE (29). These results suggested that FAAH is a modulator of endogenous NAE levels in plants and that NAE turnover by the action of FAAH likely participates in the regulation of plant growth.
Recently, AtFAAH overexpressors have exhibited several additional intriguing phenotypes. Overexpression of AtFAAH resulted in seedlings that were hypersensitive to the growth inhibitory effects of a plant hormone abscisic acid (ABA) (30). AtFAAH overexpressors were also found to be hypersusceptible to several bacterial pathogens and nonhost pathogens compared with wild-type plants, and this was attributed, in part to alterations in phytohormone accumulation and signaling (31). Interestingly, however, these phenotypic effects did not seem to be directly attributed to NAE turnover by AtFAAH. Even though AtFAAH overexpressors were compromised in innate immunity compared with wild-type plants, their NAE content and compositions in mature leaves were similar to those of wild-type plants (31). Likewise, application of ABA on AtFAAH overexpressors resulted in a marked reduction in growth despite little difference in NAE content or composition between ABA-treated and untreated seedlings (30).
Because FAAH is able to hydrolyze other types of lipid substrates in vitro (like monoacylglycerols (32–34) and fatty acid primary amides (5–7, 34)), it is possible that AtFAAH-mediated hydrolysis of other endogenous substrates, yet to be identified, may explain the impact on interaction with plant defense and ABA signaling for this enzyme, separate from its role in NAE catabolism. Here we test this possibility by ectopic overexpression of catalytically inactive, site-directed mutant forms of AtFAAH in Arabidopsis and examining the resulting effects on growth, ABA sensitivity, and innate immunity. As expected, overexpression of the AtFAAH variants without catalytic activities led to no growth enhancement and no NAE tolerance. Interestingly the transgenic AtFAAH variant lines remained hypersensitive to ABA and hypersusceptible to nonhost pathogens despite lack of enzymatic activity. Consequently, our findings suggest that AtFAAH possesses at least two co-existing activities. It influences plant growth through its amidase activity toward NAEs, whereas interacting with plant defense and ABA signaling through other, unknown mechanisms independent of its catalytic activity.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
[1-14C]Lauric acid was purchased from Amersham Biosciences (Alameda, CA). [1-14C]Palmitic acid and [1-14C]linoleic acid were from PerkinElmer Life Sciences. Dimethyl sulfoxide (DMSO), isopropyl β-d-1-thiogalactopyranoside, ethanolamine, cis-9-octadecenamide, and ABA were from Sigma. Silica Gel G (60 Å)-coated glass plates (10 × 20 cm or 20 × 20 cm, 0.25 mm thickness) were from Whatman. n-Dodecyl β-d-maltoside was from Calbiochem (La Jolla, CA). All organic solvents (isopropyl alcohol, chloroform, hexane, ethyl acetate, and methanol) were from Fisher. Polyvinylidene fluoride membrane (0.2 μm) and goat anti-mouse (or anti-rabbit) IgG conjugated to horseradish peroxidase were from Bio-Rad. Anti-c-Myc monoclonal antibody was from Abgent (San Diego, CA). Anti-AtFAAH polyclonal antibody was generated in Biosynthesis (Lewisville, TX). N-Lauroyl ethanolamide, N-palmitoyl ethanolamide, N-linoleoyl ethanolamide, and sn-2-arachidonoyl glycerol were from Cayman Chemical (Ann Arbor, MI).
Plant Materials and Growth Measurements
AtFAAH T-DNA insertional mutant (SALK_095108) was originally obtained from the Arabidopsis Biological Resource Stock Center (Ohio State University, Columbus, OH) and was characterized previously (29). Transgenic Arabidopsis lines overexpressing native AtFAAH proteins under the control of the cauliflower mosaic virus 35S-promoter were previously described (29). Plants were screened for zygosity using the REDExtract-N-Amp Plant PCR kit (Sigma). Plants were propagated in soil for seed production. For growth assay, seeds were first surface-sterilized with 95% ethanol, 30% bleach containing 0.1% Tween 20 and deionized water, and stratified for 3 days at 4 °C in the dark. Seeds were grown for 10 days in nutrient media (0.5× Murashige and Skoog salts containing 1% sucrose) in a controlled environment room with a 16-h light/8-h dark cycle at 20 °C. For detailed growth measurements, seedlings grown on agar plates were tilted at an ∼60° angle to facilitate reproducible measurements of root elongation. Cotyledon area and primary root length were measured from captured images of the seedlings. For fresh weight measurements, seedlings were grown in liquid media with shaking at 75 rpm, harvested by filtration, dried, and quantified in terms of seedling mass (mg) normalized to mass of seeds sown (mg). ABA or NAE, both dissolved in DMSO, were added to the appropriate final concentrations, and untreated controls contained equivalent amounts of DMSO alone (always less than 0.05% by volume). Concentrations of exogenous ABA were calculated based on the active cis-isomer. For pathogen assays, the plants were grown in short day conditions (10-h light/14-h dark cycle), with day temperature of 21 °C and night temperature of 19 °C at a relative humidity of 70%.
Site-directed Mutagenesis of AtFAAH
The original construct pCAMBIA1390-ATFAAH used to generate overexpressor lines (29) was used as template in reactions of site-directed mutagenesis by using the QuikChange® II XL site-directed mutagenesis kit according to the manufacturer's recommendations (Stratagene, La Jolla, CA). In short, 50 ng of the template DNA was used in PCR with primers containing nucleotide corresponding to amino acid change. The PCR program includes the following steps: 95 °C for 1 min (95 °C for 50 s, 60 °C for 50 s, 68 °C for 12 min) repeat 17 more cycles, and 68 °C for 7 min. The reaction mixture was digested with DpnI at 37 °C for 2 h to remove parent plasmids. Then the DNA was precipitated and used to transform XL10-Gold competent cells. Mutations were confirmed by sequencing. The constructs were transformed into Agrobacterium strain GV3101 and used to transform Arabidopsis wild-type (Col-0) and Atfaah knock-out plants by floral dipping (35). Transgenic plants resistant to hygromycin (15 mg/liter) were selected from MS medium. Putative transgenic plants were further confirmed with sequencing and reverse transcription-PCR with construct specific primers.
Recombinant Protein Expression and Purification
For proteins expressed in E. coli, AtFAAH cDNAs with the site-directed mutations were PCR-amplified, agarose gel-purified, cloned into pTrcHis2 vector (Invitrogen), and transformed into E. coli TOP10 cells. Selected transformants were grown in LB medium at 37 °C with shaking at 250 rpm to an A600 of 0.6, and incubated with 1 mm isopropyl β-d-1-thiogalactopyranoside for 4 h. Recombinant proteins expressed in-frame with the His6 tag were nickel-nitrilotriacetic acid affinity purified using the QIAexpress protein purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Eluted proteins were concentrated, and imidazole was removed with 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, and 0.2 mm dodecylmaltoside by filtration centrifugation using Centricon YM-30 (Millipore, Bedford, MA). Protein concentrations were determined by the Bradford assay using bovine serum albumin as a standard. For proteins expressed in Arabidopsis, seedlings grown in liquid media were flash frozen and powdered in liquid nitrogen using a mortar and pestle, and suspended in homogenization buffer (100 mm potassium phosphate, pH 7.2, 400 mm sucrose, 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 5 mm MgCl2). After incubation on ice for 30 min, homogenates were centrifuged at 12,000 × g for 15 min, and the resulting supernatants were used for further experiments.
FAAH Enzyme Activity Assays
14C radiolabeled NAEs were synthesized from corresponding free fatty acids and ethanolamine (27), and combined with non-radiolabeled NAEs to achieve the desired final concentration. Enzyme activity was determined based on radiospecific activity. Protein samples were incubated with 100 μm (∼12,000 cpm) NAEs (12:0, 16:0, or 18:2) or 100 μm 2-arachidonoyl glycerol in 50 mm BisTris buffer (pH 9.0) in a final volume of 0.4 ml at 30 °C for 30 min with shaking at 120 rpm. Reactions were terminated by the addition of boiling isopropyl alcohol (70 °C) for 30 min. Total lipids were extracted into chloroform, washed twice with 1 m KCl and once with water, and separated by Silica Gel-thin layer chromatography (TLC) using an organic solvent mixture of hexane, ethyl acetate, and methanol (60:40:5, v/v/v). Distribution of unreacted substrates and products formed was evaluated either by radiometric scanning (AR-2000 Imaging Scanner, Bioscan, NW Washington, DC) of the TLC plate for amidase activity assays or by exposure of the plate to iodine vapors for monoacyl esterase activity assays.
Western Blot Analysis
Protein samples were separated on 10% polyacrylamide SDS gels and electrophoretically transferred onto polyvinylidene fluoride membranes in a Semidry Trans-Blot apparatus (Bio-Rad) for 30 min at constant 14 V. The membranes were blocked in 5% nonfat milk in Tris-buffered saline (20 mm Tris-HCl, pH 7.5, and 500 mm NaCl) containing 0.1% Tween 20. Affinity-purified proteins expressed in E. coli as in-frame c-Myc-epitope fusions and proteins expressed in Arabidopsis were localized by overnight incubation at room temperature with mouse monoclonal anti-c-Myc antibodies (Abgent, San Diego, CA) or rabbit polyclonal anti-AtFAAH antibodies, respectively. Immunolocalized proteins were detected by chemiluminescence following incubation for 1 h at room temperature with either goat anti-mouse IgG or goat anti-rabbit IgG (Bio-Rad), both conjugated to horseradish peroxidase, according to the manufacturer's instructions.
Pathogen Assay
The pathogen growth assay was performed as previously described (31) whereby, Pseudomonas syringae pv. tabaci was grown in Kings B agar medium with appropriate antibiotics for 16 h at 30 °C. The cells were pelleted by centrifugation at 3000 × g, washed three times, and resuspended in sterile water for inoculation. Bacterial suspension was syringe-infiltrated into leaves at a concentration of 5 × 107 colony-forming units/ml for symptom development, and vacuum-infiltrated with a concentration of 107 colony-forming units/ml for growth assays. Leaf disks of ∼1 cm2 were taken from inoculated leaves, homogenized in sterile water, and the serial dilutions of the samples were plated onto Kings B agar plates.
RESULTS
Site-directed Mutagenesis of AtFAAH
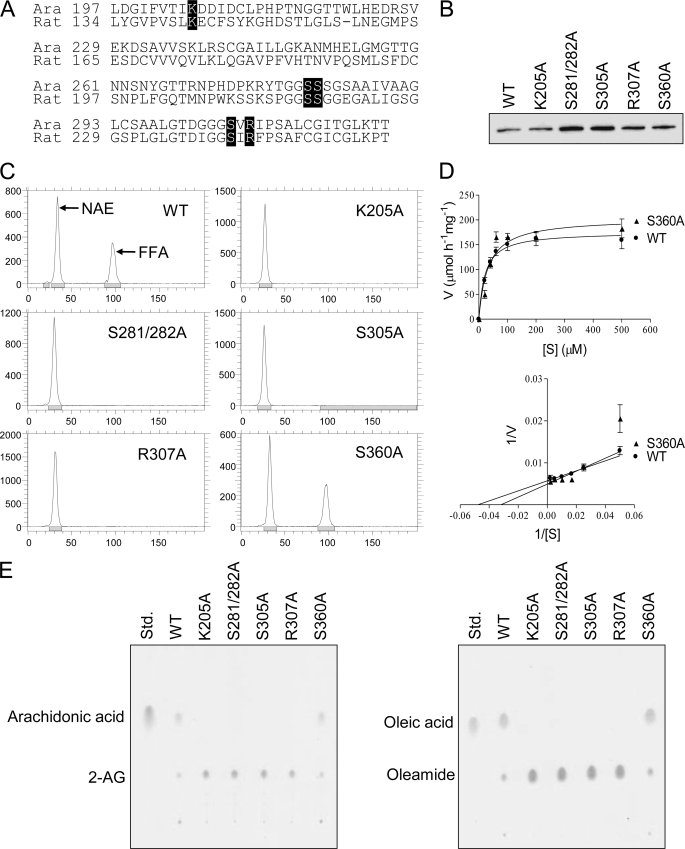
Site-directed mutagenesis studies of rat FAAH identified five amino acids (Lys-142, Ser-217, Ser-218, Ser-241, and Arg-243), including and nearby the catalytic triad, where substitution to alanine significantly decreased catalytic activity of the protein (19). Amino acid sequence alignment between rat and Arabidopsis FAAH proteins over the entire AS domain showed that the residues critical for the rat FAAH activity were absolutely conserved in the AtFAAH sequence (Lys-205, Ser-281, Ser-282, Ser-305, and Arg-307; Fig. 1). Therefore, the corresponding residues in AtFAAH could also be critical for its amidase activity.
FIGURE 1.
Expression of site-directed AtFAAH mutants in E. coli and evaluation of their enzyme activities. A, conserved catalytic residues in the amidase signature (AS) sequence between rat and Arabidopsis FAAH proteins. Full sequences of rat and Arabidopsis FAAH (Ara) were aligned using T-coffee software (Swiss Institute of Bioinformatics). AS regions that consist of 125 amino acids are shown here. Five residues (Lys-142, Ser-217, Ser-218, Ser-241, and Arg-243) known to be critical for rat FAAH activity are highlighted in black boxes. These residues are also conserved in the Arabidopsis sequence as shown (Lys-205, Ser-281, Ser-282, Ser-305, and Arg-307). B, Western blot analysis. Wild-type AtFAAH protein without mutation (WT) and the proteins with site-directed mutation (indicated by their corresponding residues) were expressed in E. coli, and affinity purified proteins (1 μg) were analyzed by Western blot. All mutated AtFAAH proteins were immunolocalized at the positions expected (∼69.4 kDa including epitope tags). C, representative radiochromatograms for NAE hydrolase assay. Equal amounts of affinity purified proteins were reacted with [1-14C]NAE16:0. Total lipids extracted from reaction mixtures were separated by thin layer chromatography and analyzed by radiometric scanning. Each radiochromatogram shows the distance (mm) on the x axis that lipids migrated on the TLC plate and their radioactivity (cpm) on y axis. Picks that represent NAE and FFA (free fatty acid) are indicated. Wild-type (WT) and S360A mutant show a significant production of free fatty acid (peak at ∼97 mm), whereas all mutants did not produce any detectable products. Enzyme activities for mutants compared with wild-type are summarized in Table 1. D, kinetic comparison of wild-type and S360A mutant. Initial velocities (for 10 min) were measured at increasing concentrations of [1-14C]NAE16:0. Michaelis-Menten and Lineweaver-Burk plots are shown for comparison. Plots were generated with Prism software version 3.0 (GraphPad Software, San Diego, CA). Data points represent mean ± S.D. of triplicate assays. Kinetic parameters for wild-type and S360A mutant are summarized in Table 2. E, lack of enzyme activity of the mutants toward monoacylester and primary amide substrates. 1 μg of purified wild-type and S360A proteins and 10 μg of all other mutant proteins were reacted with 2-arachidonoyl glycerol (2-AG; left plate) and 9-octadecenamide (oleamide; right plate). Total lipids from the reactions were separated by TLC, and visualized by iodine vapors. Positions for substrates (2-AG and oleamide) and products (arachidonic acid and oleic acid) are indicated on the left. Wild-type and S360A mutant show a significant formation of free fatty acid products, whereas all mutants display essentially no product formation. Arachidonic acid and oleic acid standards (Std.) are also included for position comparison.
We performed a systematic mutational analysis for each of the five conserved residues of AtFAAH by converting them to alanine to evaluate their importance for catalytic activity and biological functions in development and stress responses. Because Ser-281 and Ser-282 are located adjacent to each other on the protein molecule, a S281A/S282A double mutant was generated to rule out the possibility that mutation of one of the two serine residues might compromise catalytic activity by structurally impacting the other residue. As a control, a S360A mutation was also generated because it is outside the amidase signature sequence and should have no impact on catalysis. Thus, the following mutants were generated for this study and expressed as recombinant proteins for functional analysis in E. coli: K205A, S281A/S282A, S305A, R307A, and S360A. Western blot analysis using affinity purified proteins with the mutations showed that the proteins are normally expressed in E. coli in roughly similar levels (Fig. 1B).
Mutations in the Amidase Domain of AtFAAH Abolished Enzyme Activity
When NAE hydrolase assays were performed with equal amounts of affinity purified, mutated proteins using NAE16:0 as a substrate, no detectable amount of the product (free fatty acid 16:0) was found in K205A, S281A/S282A, S305A, and R307A mutants, whereas the S360A mutant exhibited fairly similar levels of product formation to wild-type (native) protein (Fig. 1C), providing a negative control for comparison. Based on the assay results, specific activities for the mutants and their relative activities to wild-type protein were calculated and summarized in Table 1. Kinetic comparisons between wild-type AtFAAH and S360A mutant showed that the mutation outside of the FAAH active site had similar kinetic properties and catalytic efficiencies (Kcat/Km) toward NAE substrates as the native FAAH (Fig. 1D and Table 2).
TABLE 1.
NAE hydrolysis activity of mutant proteins expressed in E. coli
Proteins affinity purified from E. coli were reacted with 14C radiolabeled NAE12:0, NAE16:0, or NAE18:0. Specific activities were calculated based on radioactivity of the products formed. Values are shown as mean ± S.D. of triplicate measurements. For each NAE substrate, enzyme activities relative to wild-type AtFAAH without mutation (100%; WT) are indicated under “relative activity.”
| Enzymes | Specific activity |
Relative activity |
||||
|---|---|---|---|---|---|---|
| NAE12:0 | NAE16:0 | NAE18:2 | NAE12:0 | NAE16:0 | NAE18:2 | |
| μmol h−1mg−1 | % | |||||
| WT | 117 ± 8 | 134 ± 10 | 139 ± 8 | 100 | 100 | 100 |
| K205A | 0.06 ± 0.01 | 0.07 ± 0.02 | 0.10 ± 0.02 | 0.05 | 0.05 | 0.07 |
| S281A/ | 0.08 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.01 | 0.07 | 0.06 | 0.06 |
| S282A | ||||||
| S305A | 0.09 ± 0.02 | 0.11 ± 0.02 | 0.15 ± 0.01 | 0.08 | 0.08 | 0.11 |
| R307A | 0.16 ± 0.03 | 0.21 ± 0.03 | 0.34 ± 0.05 | 0.14 | 0.16 | 0.24 |
| S360A | 119 ± 7 | 145 ± 12 | 152 ± 11 | 102 | 108 | 109 |
TABLE 2.
Apparent kinetic parameters of wild-type AtFAAH and the S360A mutant
Initial velocities (for 10 min) of affinity purified wild-type (WT) and S360A proteins were measured at increasing concentrations of [1-14C]NAE16:0. Values were estimated with Prism software version 3.0 (GraphPad Software) from triplicate measurements.
| Enzyme | Vmax | Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| μmol h−1mg−1 | μm | s−1 | μm−1s−1 | |
| WT | 181.7 | 23.0 | 3.33 | 0.145 |
| S360A | 205.4 | 33.1 | 3.77 | 0.114 |
Enzyme activity assays were also conducted with NAE12:0, NAE18:2, a fatty acid primary amide, 9-octadecenamide (oleamide), and a monoacylglycerol, 2-arachidonylglycerol, to test the mutant enzymes against a broad range of substrate types including long- and short-chain acylamides, long-chain polyunsaturated acylamides, primary amides, and monoacylesters (see Fig. 1). These substrates were all hydrolyzed well by wild-type AtFAAH, but were hydrolyzed to a barely detectable degree by the site-directed mutants, except for the S360A mutant, which exhibited similar activity to wild-type protein for all the substrates tested (see Fig. 1E and Table 1). Overall, all AtFAAH mutants displayed slightly better activities toward long-chain polyunsaturated NAEs, and the R307A mutant hydrolyzed all the NAE substrates better than the other mutants (see Table 1). These results suggest that the five residues reported to be important for activity of rat FAAH (19) are also essential for the hydrolase activity of AtFAAH, and support a common catalytic mechanism of animal and plant FAAH enzymes.
Overexpression of the Site-directed Mutant AtFAAH Proteins in Arabidopsis
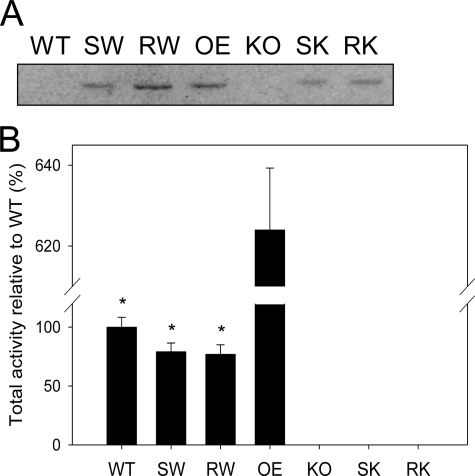
To test the impact of NAE turnover, or of hydrolysis of unknown lipid substrates by AtFAAH overexpressors, catalytically “dead” enzymes were overexpressed in wild-type and Atfaah knock-out backgrounds. Atfaah cDNAs with K205A, S281A/S282A, S305A, and R307A cloned in the pCAMBIA-1390 vector were used to transform Arabidopsis wild-type (Col-0) and the Atfaah knock-out mutant using floral dip transformation (35). Transgenic plants were successfully obtained for S281A/S282A and R307A mutants. Western blot analysis using homogenates of 10-day-old seedlings showed that S281A/S282A and R307A mutant lines overexpressed their respective mutated proteins (Fig. 2A). The two backgrounds for transformation allowed for the endogenous AtFAAH to be accounted. AtFAAH protein was essentially undetectable in homogenates of wild-type and Atfaah knock-out plants; the protein exists in very low abundance in wild-type, and it was barely detected in immunoblots of isolated microsomes prepared from homogenates of wild-type plants (not shown).
FIGURE 2.
Expression of site-directed AtFAAH mutants in Arabidopsis and evaluation of their enzyme activities. A, Western blot analysis. Arabidopsis wild-type (WT) and transgenic lines (SW, RW, OE, KO, SK, and RK) were grown for 10 days as described under ”Experimental Procedures.“ Total proteins (200 μg) from each seedling homogenates were analyzed by Western blot. All AtFAAH proteins of overexpressors (SW, RW, OE, SK, and RK) were immunolocalized at the position expected (∼66.1 kDa). SW, S281A/S282A mutant expressed in wild-type background; RW, R307A mutant expressed in wild-type; OE, native AtFAAH overexpressor; KO, Atfaah T-DNA insertion knock-out; SK, S281A/S282A mutant expressed in Atfaah knock-out background; RK, R307A mutant expressed in Atfaah knock-out. B, NAE hydrolysis activities. Total proteins (50 μg) from each of 10-day-old seedling homogenates were reacted with [1-14C]NAE16:0. Total activities (micromole/h) were measured based on radioactivity of the product formed. All AtFAAH overexpressors with site-directed mutation (SW, RW, SK, and RK) exhibited significantly lower NAE hydrolase activity than the native AtFAAH overexpressor (OE). The error bars represent S.D. from triplicate measurements. Asterisks indicate a significant difference (p < 0.0001) compared with OE, which was determined by Student's t test.
When NAE hydrolase assays were performed with equal amounts of total proteins extracted from 10-day-old seedlings using NAE16:0 as a substrate, both S281A/S282A and R307A mutants transformed into the wild-type background exhibited slightly reduced activities compared with wild-type plants alone, presumably due to the competition for substrate binding between the active and inactive enzymes (Fig. 2B) (25). However, both of these two site-directed mutants exhibited significantly lower enzyme activity than overexpression of the native AtFAAH, clearly distinguishing the expression of site-directed mutant forms from overexpression of authentic AtFAAH. As expected, no measurable activity was found when either of the mutant forms were transformed into the Atfaah knock-out plants (Fig. 2B), despite protein accumulation detected on Western blots (Fig. 2A).
AtFAAH Enzyme Activity Is Required for Enhanced Seedling Growth Observed in AtFAAH Overexpressing Plants
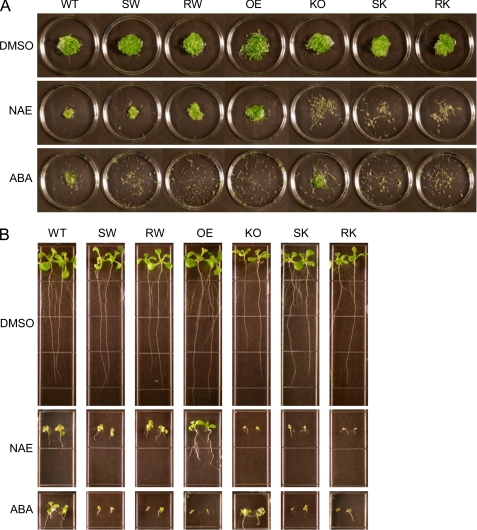
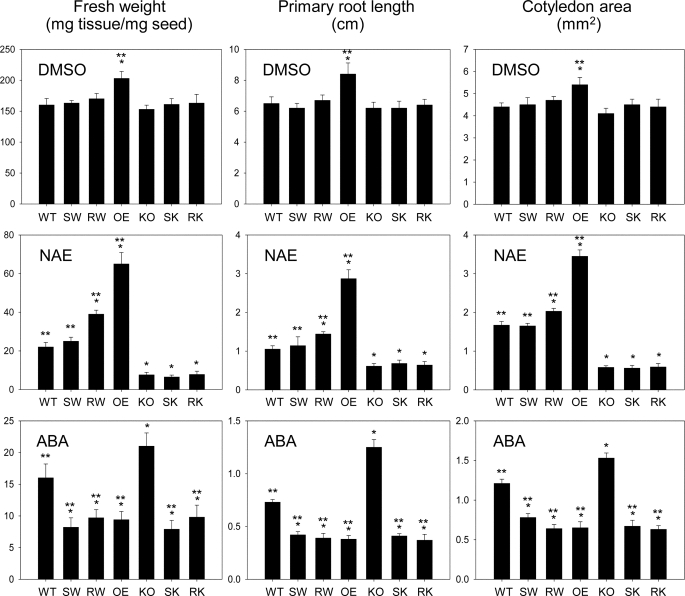
To ask whether NAE hydrolase activity of AtFAAH was required for enhanced seedling growth (29–30), growth phenotypes of 10-day-old seedlings of the site-directed mutant lines in response to NAE12:0 were determined by measuring their seedling fresh weights, primary root lengths, and cotyledon areas. Generally, both S281A/S282A and R307A mutants exhibited growth phenotypes essentially identical to those of their background lines (Col-0 and Atfaah knock-out) when treated with either solvent (DMSO) only or NAE12:0 (Figs. 3 and 4). Overexpression of the inactive AtFAAH proteins showed no statistically significant enhancement of growth, whereas the native AtFAAH overexpressors exhibited ∼30% increase in overall seedling growth when compared with wild-type plants. Moreover, unlike the native AtFAAH overexpressors, none of these site-directed mutant lines showed any significant tolerance to exogenous NAE except that the R307A mutant transformed in wild-type background appeared to grow slightly better than either wild-type or S281A/S282A mutant in the same background.
FIGURE 3.
Overall apparent growth phenotype of Arabidopsis seedlings expressing AtFAAH mutants in response to NAE and ABA. Seedlings were grown for 10 days in the presence of DMSO only, 30 μm NAE12:0, or 0.25 μm ABA (A). Growth phenotype in liquid media. Seedlings were grown in flasks, and then transferred onto Petri dishes to photograph. Representative images of triplicate experiments are shown here. All site-directed mutant lines (SW, RW, SK, and RK) display no growth enhancement and no NAE tolerance compared with native AtFAAH overexpressor (OE), whereas they all show ABA hypersensitivity compared with their background lines (WT and KO). B, growth phenotype in solid media. Representative images of 24 individual seedlings for each line are shown. Note that relative growths of the different lines are highly analogous to those in liquid medium (A). KO, knock-out; SW, S281A/S282A mutant expressed in wild-type background; RW, R307A mutant expressed in wild-type background; SK, S281A/S282A mutant expressed in Atfaah knock-out background; RK, R307A mutant expressed in Atfaah knock-out background.
FIGURE 4.
Quantitative growth measurements of Arabidopsis seedlings expressing AtFAAH mutants in response to NAE and ABA. 10-Day-old seedlings were quantified in terms of seedling fresh weight (milligrams of seedling tissue/mg of seed sown), primary root length (cm), and cotyledon area (mm2). Values for fresh weight represent mean ± S.D. of triplicate measurements. Values for primary root length and cotyledon areas represent mean ± S.D. of 10 individual seedlings. Single and double asterisks indicate a significant difference (p < 0.0001) compared with wild-type (WT) and Atfaah knock-out (KO), respectively, which was determined by Student's t test. OE, overexpressor; SW, S281A/S282A mutant expressed in wild-type background; RW, R307A mutant expressed in wild-type background; SK, S281A/S282A mutant expressed in Atfaah knock-out background; RK, R307A mutant expressed in Atfaah knock-out background.
AtFAAH Enzyme Activity Is Not Required for Hypersensitivity to ABA and Hypersusceptibility to Nonhost Pathogens
We have previously shown that transcript levels of ABA-insensitive 3 (ABI3), a key transcription factor for ABA-responsive genes, are inversely associated with AtFAAH expression levels in Arabidopsis (30). Inconsistent with this observation, overexpression of the AtFAAH protein resulted in seedlings that were hypersensitive to ABA despite lower transcript levels of ABI3 (30). To determine whether NAE hydrolase activity of AtFAAH was required for ABA hypersensitivity observed in AtFAAH overexpressing plants (30), the plants overexpressing the mutant forms of AtFAAH were grown for 10 days in the presence of ABA and their growth phenotypes were determined in detail as described above. Surprisingly, all seedlings expressing S281A/S282A or R307A mutants, regardless of their backgrounds, still exhibited severe hypersensitivity to ABA to a very similar degree observed for the native AtFAAH overexpressors (Figs. 3 and 4), suggesting that AtFAAH enzyme activity and NAE turnover by the enzyme are not required for the ABA hypersensitivity.
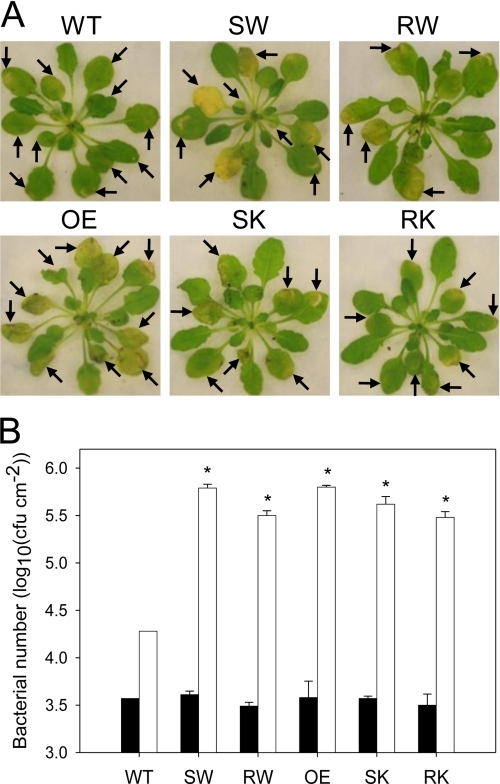
Previous studies showed that the AtFAAH overexpressors had compromised basal resistance and were susceptible to certain nonhost pathogens (31). However, it was not established if the enzyme activity was involved in causing the susceptibility. To check this, plants expressing S281A/S282A or R307A mutants in both wild-type and Atfaah knock-out backgrounds were inoculated with nonhost pathogen as described (31). Development of symptoms on the inoculated leaves was monitored and the bacterial growth was quantified as described under “Experimental Procedures.” Consistent with earlier observations (31), at 5 days post-inoculation with nonhost pathogen, P. syringae pv. tabaci, the native AtFAAH overexpressors had chlorotic lesions in most of the infiltrated leaves as compared with Col-0 (Fig. 5A). Interestingly, transgenic plants expressing S281A/S282A or R307A mutants in both Col-0 and Atfaah knock-out backgrounds showed more chlorotic lesions, similar to those of native AtFAAH overexpressors, in most of the infiltrated leaves as compared with Col-0 (Fig. 5A). Consistent with the disease symptoms, there was an increase in bacterial growth at 3 days post-inoculation in transgenic plants expressing either native AtFAAH or AtFAAH mutants (S281A/S282A or R307A) when compared with Col-0 (Fig. 5B).
FIGURE 5.
Enhanced growth of nonhost pathogen P. syringae pv. tabaci in Arabidopsis expressing AtFAAH mutants. A, leaves were syringe-infiltrated with bacteria of a concentration of 5 × 107 colony-forming units/ml for symptom development. The disease symptoms were photographed at 5 days post-inoculation. Inoculated leaves are indicated by arrows. B, plants were vacuum-infiltrated with bacteria of a concentration of 107 colony-forming units/ml for growth assays. Bacterial growth was quantified at 3 (white bars) and 0 days post-inoculation (black bars) for comparison. The error bars represent S.D. from triplicate measurements. Asterisks indicate a significant difference (p < 0.0001) compared with wild-type (WT; 3 days post-inoculation), which was determined by Student's t test. OE, overexpressor; SW, S281A/S282A mutant expressed in wild-type background; RW, R307A mutant expressed in wild-type background; SK, S281A/S282A mutant expressed in Atfaah knock-out background; RK, R307A mutant expressed in Atfaah knock-out background.
Collectively, these results indicate that the enhanced growth and the tolerance to exogenous NAE of the native AtFAAH-overexpressing seedlings are attributable to elevated NAE hydrolase activity of the AtFAAH protein, whereas the hypersensitivity to ABA and hypersusceptibility to nonhost pathogens of the AtFAAH overexpressors are independent of the catalytic activity of the enzyme but rather dependent on the presence of higher amounts of the protein only.
DISCUSSION
After “oleamide hydrolase” activity was first affinity purified from rat liver membranes and the same enzyme was found to display high levels of “anandamide hydrolase” activity (7), FAAH has been intensely investigated in animal systems to uncover its functions in regulating the endocannabinoid signaling system and to develop new therapeutics for the treatment of human disorders (reviewed in Refs. 9–10 and 36). NAEs in plants are hydrolyzed by a membrane-associated hydrolase functionally analogous to the mammalian FAAH (24–25, 27, 29), and an Arabidopsis FAAH homologue was identified (24), suggesting that a FAAH-mediated pathway exists in plants as well for the metabolism of endogenous NAEs (reviewed in Ref. 37–39). However, because plant FAAH homologues have been studied only recently, our knowledge on functions of this enzyme in plants is fragmentary and many questions remain to be addressed.
Structure-function relationships for AtFAAH were predicted by homology-based modeling of the plant FAAH AS domain using the rat FAAH three-dimensional structure as a template (25). However no direct experimental evidence other than inhibitor studies has suggested that the plant and animal enzymes operate by a conserved mechanism. And inhibition by serine hydrolase inhibitors cannot distinguish between the Ser-Ser-Lys and Ser-His-Asp catalytic triads. Here we show that site-directed mutagenesis of five residues conserved in the AS region abolished the amidase activity of AtFAAH, supporting a conserved Ser-Ser-Lys catalytic mechanism. All five residues were predicted to be located in the immediate vicinity of the active site pocket and the putative catalytic residues (Lys-205, Ser-281, and Ser-305) showed nearly direct overlap among all plant and rat FAAH proteins (25).
FAAH has an unusual catalytic feature in that, in addition to amidase activity, it possesses esterase activity at an equivalent rate (32). Among the five catalytically important residues of rat FAAH, the R243A mutant was reported to exhibit unaffected esterase activity despite severely compromised amidase activity (19). In contrast to this finding, the corresponding mutant of AtFAAH (R307A) displayed abolished esterase activity in a similar manner to other site-directed mutants tested (Fig. 1E). This noticeable difference between rat and Arabidopsis FAAH indicates that, unlike rat FAAH, the amidase and esterase efficiencies of AtFAAH are functionally and tightly coupled. Another differential catalytic property between animal and plant FAAH enzymes has been previously shown by tolerance of plant FAAH to URB597, a specific inhibitor of animal FAAH (25). Collectively, these findings suggest that although the catalytic mechanism is conserved between the plant and animal enzymes, there are likely subtle differences within the AtFAAH active site that remain to be resolved at the structural level.
In addition to the expected phenotypes of plants overexpressing AtFAAH (e.g. tolerance to exogenous NAEs), these plants exhibited several unexpected phenotypes unable to be explained by NAE hydrolysis, such as hypersensitivity to ABA (30) and enhanced susceptibility to several bacterial pathogens (31). Here we provide experimental evidence that AtFAAH influences Arabidopsis growth and responses to ABA and pathogens through distinctly different molecular mechanisms.
Our previous studies suggested that hydrolysis of endogenous NAEs by the amidase activity of AtFAAH were important for normal Arabidopsis seedling growth (28–29). Here we further support this hypothesis by observing growth phenotype of the plants that overexpress the inactive enzymes. Lack of both enhancement of growth and NAE tolerance by overexpressing inactive AtFAAH proteins reinforces our previous conclusions that FAAH is a modulator of endogenous NAE levels in plants and depletion of NAE by the action of FAAH is one of the key components that participate in the regulation of seedling growth.
Surprisingly, transgenic lines overexpressing S281A/S282A or R307A mutants of AtFAAH that produced inactive enzyme still exhibited ABA hypersensitivity and hypersusceptibility to nonhost pathogens to a degree similar to the native AtFAAH overexpressors, suggesting that AtFAAH-mediated ABA hypersensitivity and disease susceptibility are independent of catalytic activity of the enzyme toward acylamide or acylester substrates. The disease susceptibility and ABA hypersensitivity of the transgenic plants expressing mutant AtFAAH (S281A/S282A or R307A) in the Atfaah knock-out background is almost similar to overexpressors (see Figs. 3 and 5). This can be attributed to the presence of more mutant AtFAAH protein because the expression is constitutively driven by cauliflower mosaic virus 35S-promoter. Indeed, Western analysis clearly shows more accumulation of mutant proteins in the knock-out background (see Fig. 2).
We speculate that the AtFAAH protein itself might directly interact with other protein(s) involved in ABA and/or defense signaling. We identified two conserved domains near the C terminus of plant FAAH proteins outside of the catalytic site (25, 37), which perhaps could facilitate interactions with target proteins. A recent report indicated that mammalian FAAH can interact with a membrane protein, ERp57, in caveolin-rich membranes (40), but the physiological significance of this interaction remains unclear. Future efforts will be aimed at uncovering the mechanism(s) by which AtFAAH may exert its effects on ABA sensitivity and disease susceptibility by identifying the binding partner molecule(s) of AtFAAH in Arabidopsis cells and identifying domains responsible for interactions.
Alternatively, differential localization of overexpressed FAAH protein (mutant or otherwise) might be responsible for the ABA and pathogen sensitivity phenotypes. However, this explanation is not entirely satisfactory because in previous experiments, when FAAH-green fluorescent protein was overexpressed in the Atfaah knock-out background this overexpressed protein complemented the knock-out phenotype and conferred tolerance to exogenous NAE in a manner similar to overexpression of FAAH without green fluorescent protein (31). This suggested that the endoplasmic reticulum/plasma membrane localization of the FAAH-green fluorescent protein was at least partially reflective of the normal location of FAAH (to functionally restore the phenotype of knock-outs) and that this FAAH-green fluorescent protein overexpression was a reasonable reporter of overexpressed FAAH location because the NAE-tolerant growth was similar between plants overexpressing either FAAH protein (31). Nonetheless, an effect of FAAH location due to overabundance of active or inactive FAAH transgene product should not be entirely ruled out.
In conclusion, we have shown that the AtFAAH influences plant growth through its hydrolysis of acylethanolamides, but that interactions with ABA and defense signaling are independent of its hydrolytic activity. We proposed previously that NAE metabolism and its influence by FAAH resides at the balance between plant growth and the responses of plants to stress (38). The results presented here are consistent with this concept and offer bifurcating mechanisms that may mediate this physiological control. Although understanding the detailed mechanisms involved in these two processes will require further experimentation beyond the scope of this paper, the novel results presented here provide continued direction to functionally define the group of enzymes that metabolize NAEs in plants. Furthermore, this work suggests the future possibility to uncouple the remarkable increase in overall plant growth seen in AtFAAH overexpressors from the concomitant increased susceptibility to stress, which could have important applications in crop biotechnology.
This work was supported by the Samuel Roberts Noble Foundation and a grant from the United States Department of Energy Office of Science (BES) agreement number DE-FG02-05ER15647 (to E. B. B. and K. D. C.).
- FAAH
- fatty acid amide hydrolase
- NAE
- N-acylethanolamine
- ABA
- abscisic acid
- AtFAAH
- Arabidopsis thaliana fatty acid amide hydrolase
- AS
- amidase signature
- ABI
- ABA-insensitive
- 2-AG
- sn-2-arachidonyl glycerol
- DMSO
- dimethyl sulfoxide
- URB597
- 3′-carbamoyl-biphenyl-3-yl-cyclohexylcarbamate
- ER
- endoplasmic reticulum
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Desarnaud F., Cadas H., Piomelli D. (1995) J. Biol. Chem. 270, 6030–6035 [DOI] [PubMed] [Google Scholar]
- 2.Deutsch D. G., Chin S. A. (1993) Biochem. Pharmacol. 46, 791–796 [DOI] [PubMed] [Google Scholar]
- 3.Schmid P. C., Zuzarte-Augustin M. L., Schmid H. H. (1985) J. Biol. Chem. 260, 14145–14149 [PubMed] [Google Scholar]
- 4.Ueda N., Kurahashi Y., Yamamoto S., Tokunaga T. (1995) J. Biol. Chem. 270, 23823–23827 [DOI] [PubMed] [Google Scholar]
- 5.Cravatt B. F., Prospero-Garcia O., Siuzdak G., Gilula N. B., Henriksen S. J., Boger D. L., Lerner R. A. (1995) Science 268, 1506–1509 [DOI] [PubMed] [Google Scholar]
- 6.Maurelli S., Bisogno T., De Petrocellis L., Di Luccia A., Marino G., Di Marzo V. (1995) FEBS Lett. 377, 82–86 [DOI] [PubMed] [Google Scholar]
- 7.Cravatt B. F., Giang D. K., Mayfield S. P., Boger D. L., Lerner R. A., Gilula N. B. (1996) Nature 384, 83–87 [DOI] [PubMed] [Google Scholar]
- 8.Fowler C. J. (2006) Fundam. Clin. Pharmacol. 20, 549–562 [DOI] [PubMed] [Google Scholar]
- 9.McKinney M. K., Cravatt B. F. (2005) Annu. Rev. Biochem. 74, 411–432 [DOI] [PubMed] [Google Scholar]
- 10.Fezza F., De Simone C., Amadio D., Maccarrone M. (2008) Subcell. Biochem. 49, 101–132 [DOI] [PubMed] [Google Scholar]
- 11.Chebrou H., Bigey F., Arnaud A., Galzy P. (1996) Biochim. Biophys. Acta 1298, 285–293 [DOI] [PubMed] [Google Scholar]
- 12.Patricelli M. P., Lovato M. A., Cravatt B. F. (1999) Biochemistry 38, 9804–9812 [DOI] [PubMed] [Google Scholar]
- 13.Cai G., Zhu S., Wang X., Jiang W. (2005) FEMS Microbiol. Lett. 249, 15–21 [DOI] [PubMed] [Google Scholar]
- 14.Gopalakrishna K. N., Stewart B. H., Kneen M. M., Andricopulo A. D., Kenyon G. L., McLeish M. J. (2004) Biochemistry 43, 7725–7735 [DOI] [PubMed] [Google Scholar]
- 15.Labahn J., Neumann S., Büldt G., Kula M. R., Granzin J. (2002) J. Mol. Biol. 322, 1053–1064 [DOI] [PubMed] [Google Scholar]
- 16.Neu D., Lehmann T., Elleuche S., Pollmann S. (2007) FEBS J. 274, 3440–3451 [DOI] [PubMed] [Google Scholar]
- 17.Bracey M. H., Hanson M. A., Masuda K. R., Stevens R. C., Cravatt B. F. (2002) Science 298, 1793–1796 [DOI] [PubMed] [Google Scholar]
- 18.McKinney M. K., Cravatt B. F. (2003) J. Biol. Chem. 278, 37393–37399 [DOI] [PubMed] [Google Scholar]
- 19.Patricelli M. P., Cravatt B. F. (2000) J. Biol. Chem. 275, 19177–19184 [DOI] [PubMed] [Google Scholar]
- 20.Cravatt B. F., Demarest K., Patricelli M. P., Bracey M. H., Giang D. K., Martin B. R., Lichtman A. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9371–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clement A. B., Hawkins E. G., Lichtman A. H., Cravatt B. F. (2003) J. Neurosci. 23, 3916–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cravatt B. F., Saghatelian A., Hawkins E. G., Clement A. B., Bracey M. H., Lichtman A. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10821–10826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichtman A. H., Shelton C. C., Advani T., Cravatt B. F. (2004) Pain 109, 319–327 [DOI] [PubMed] [Google Scholar]
- 24.Shrestha R., Dixon R. A., Chapman K. D. (2003) J. Biol. Chem. 278, 34990–34997 [DOI] [PubMed] [Google Scholar]
- 25.Shrestha R., Kim S. C., Dyer J. M., Dixon R. A., Chapman K. D. (2006) Biochim. Biophys. Acta 1761, 324–334 [DOI] [PubMed] [Google Scholar]
- 26.Chapman K. D., Tripathy S., Venables B., Desouza A. D. (1998) Plant Physiol. 116, 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shrestha R., Noordermeer M. A., van der Stelt M., Veldink G. A., Chapman K. D. (2002) Plant Physiol. 130, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blancaflor E. B., Hou G., Chapman K. D. (2003) Planta 217, 206–217 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y. S., Shrestha R., Kilaru A., Wiant W., Venables B. J., Chapman K. D., Blancaflor E. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12197–12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teaster N. D., Motes C. M., Tang Y., Wiant W. C., Cotter M. Q., Wang Y. S., Kilaru A., Venables B. J., Hasenstein K. H., Gonzalez G., Blancaflor E. B., Chapman K. D. (2007) Plant Cell 19, 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang L., Wang Y. S., Uppalapati S. R., Wang K., Tang Y., Vadapalli V., Venables B. J., Chapman K. D., Blancaflor E. B., Mysore K. S. (2008) Plant J. 56, 336–349 [DOI] [PubMed] [Google Scholar]
- 32.Patricelli M. P., Cravatt B. F. (1999) Biochemistry 38, 14125–14130 [DOI] [PubMed] [Google Scholar]
- 33.Ghafouri N., Tiger G., Razdan R. K., Mahadevan A., Pertwee R. G., Martin B. R., Fowler C. J. (2004) Br. J. Pharmacol. 143, 774–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fowler C. J., Jonsson K. O., Tiger G. (2001) Biochem. Pharmacol. 62, 517–526 [DOI] [PubMed] [Google Scholar]
- 35.Clough S. J., Bent A. F. (1998) Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 36.Labar G., Michaux C. (2007) Chem. Biodivers. 4, 1882–1902 [DOI] [PubMed] [Google Scholar]
- 37.Chapman K. D. (2004) Prog. Lipid Res. 43, 302–327 [DOI] [PubMed] [Google Scholar]
- 38.Kilaru A., Blancaflor E. B., Venables B. J., Tripathy S., Mysore K. S., Chapman K. D. (2007) Chem. Biodivers. 4, 1933–1955 [DOI] [PubMed] [Google Scholar]
- 39.Gertsch J. (2008) Planta Med. 74, 638–650 [DOI] [PubMed] [Google Scholar]
- 40.Yates M. L., Barker E. L. (2007) FASEB J. 21, 885.117197387 [Google Scholar]