Abstract
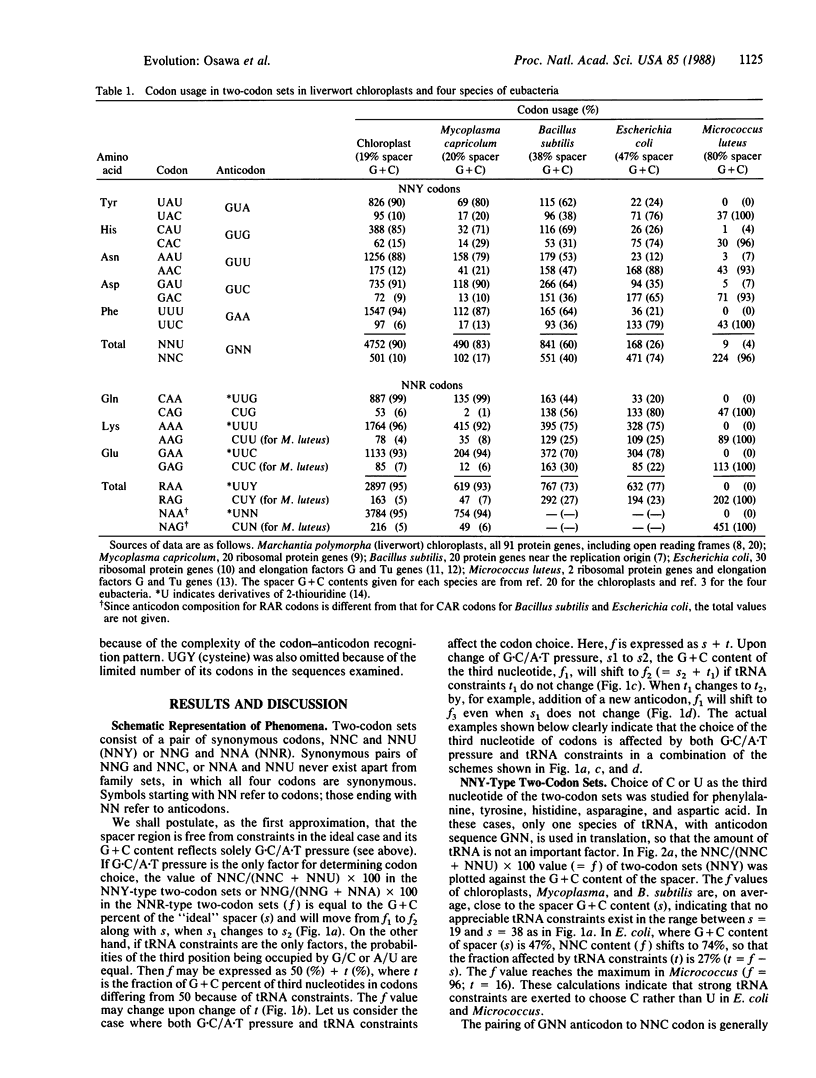
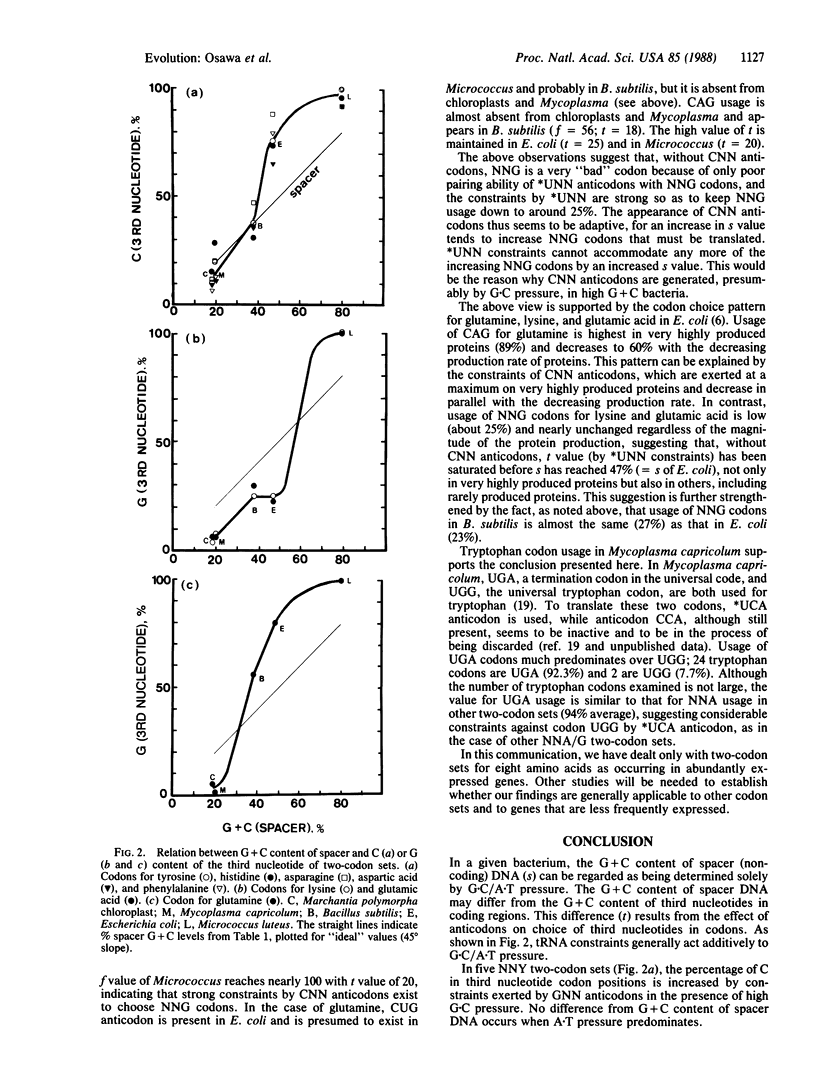
Bacterial species have diverged into a series of families, some with high G + C content in their DNA, and other with high A + T content, resulting, respectively, from G.C- and A.T-directional mutation pressures. Such mutation pressure (G.C/A.T pressure) may be an important determinant for codon usage. It has also been suggested that tRNA acts as a selective constraint for determining codon usage. We have studied the relation between G.C/A.T pressure and tRNA constraints in determining choice of the third nucleotide of eight two-codon sets, using codon usage data obtained from protein genes in four bacterial species, Mycoplasma capricolum, Bacillus subtilis, Escherichia coli, and Micrococcus luteus, and in liverwort (Marchantia polymorpha) chloroplasts. The genomic G + C contents of these range from 25% to 74%. The results demonstrate that tRNA levels act additively to A.T and G.C pressure in affecting contents of A (pairing with *UNN anticodons, in which *U indicates a 2-thiouridine derivative) and C (pairing with GNN anticodons) or G (pairing with CNN anticodons), respectively, in third nucleotide positions of codons.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Grosjean H., Sankoff D., Jou W. M., Fiers W., Cedergren R. J. Bacteriophage MS2 RNA: a correlation between the stability of the codon: anticodon interaction and the choice of code words. J Mol Evol. 1978 Dec 29;12(2):113–119. doi: 10.1007/BF01733262. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Bhushan V. Silent nucleotide substitutions and G + C content of some mitochondrial and bacterial genes. J Mol Evol. 1986;24(1-2):39–44. doi: 10.1007/BF02099949. [DOI] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N. Markedly unbiased codon usage in Bacillus subtilis. Gene. 1985;40(1):145–150. doi: 10.1016/0378-1119(85)90035-6. [DOI] [PubMed] [Google Scholar]
- Ohama T., Yamao F., Muto A., Osawa S. Organization and codon usage of the streptomycin operon in Micrococcus luteus, a bacterium with a high genomic G + C content. J Bacteriol. 1987 Oct;169(10):4770–4777. doi: 10.1128/jb.169.10.4770-4777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S., Muto A., Kawauchi Y., Yamao F., Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet. 1987 Dec;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- SUEOKA N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci U S A. 1962 Apr 15;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier J., Kitakawa M., Isono K. The nucleotide sequence of an Escherichia coli chromosomal region containing the genes for ribosomal proteins S6, S18, L9 and an open reading frame. Mol Gen Genet. 1986 Jul;204(1):126–132. doi: 10.1007/BF00330199. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T., Sugisaki H., Takanami M., Kaziro Y. The nucleotide sequence of the cloned tufA gene of Escherichia coli. Gene. 1980 Dec;12(1-2):25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Watanabe T., Murao K., Ishikura H., Yamaizumi Z., Nishimura S., Miyazawa T. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4905–4909. doi: 10.1073/pnas.82.15.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel J. M., Archer R. H., Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984 Feb 24;12(4):2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]



