Abstract
We investigated the regulatory effects of GRK2 on D2 dopamine receptor signaling and found that this kinase inhibits both receptor expression and functional signaling in a phosphorylation-independent manner, apparently through different mechanisms. Overexpression of GRK2 was found to suppress receptor expression at the cell surface and enhance agonist-induced internalization, whereas short interfering RNA knockdown of endogenous GRK2 led to an increase in cell surface receptor expression and decreased agonist-mediated endocytosis. These effects were not due to GRK2-mediated phosphorylation of the D2 receptor as a phosphorylation-null receptor mutant was regulated similarly, and overexpression of a catalytically inactive mutant of GRK2 produced the same effects. The suppression of receptor expression is correlated with constitutive association of GRK2 with the receptor complex as we found that GRK2 and several of its mutants were able to co-immunoprecipitate with the D2 receptor. Agonist pretreatment did not enhance the ability of GRK2 to co-immunoprecipitate with the receptor. We also found that overexpression of GRK2 attenuated the functional coupling of the D2 receptor and that this activity required the kinase activity of GRK2 but did not involve receptor phosphorylation, thus suggesting the involvement of an additional GRK2 substrate. Interestingly, we found that the suppression of functional signaling also required the Gβγ binding activity of GRK2 but did not involve the GRK2 N-terminal RH domain. Our results suggest a novel mechanism by which GRK2 negatively regulates G protein-coupled receptor signaling in a manner that is independent of receptor phosphorylation.
Introduction
Dopamine receptors (DARs)3 belong to the seven-transmembrane-spanning domain GPCR family and are encoded by five distinct genes (1, 2). The five DARs consist of two subfamilies, which are defined by their structural, pharmacological, and transductional properties (3). The “D1-like” receptors consist of the D1 and D5 DARs, which activate the G proteins GS or GOLF to stimulate adenylate cyclase activity. The D2-like receptors include the D2, D3, and D4 DARs and couple to Gi/o proteins to inhibit adenylate cyclase and to also modulate voltage-gated K+ or Ca2+ channels. All five DAR subtypes are known to regulate critical functions within the central nervous system, including movement, learning and memory, reward and addiction, and cognition. More importantly, many of the DARs are central in the therapy of a number of neuropsychiatric disorders. Indeed, the D2 receptor is one of the most highly validated drug targets in neurology and psychiatry. For instance, most antiparkinsonian drugs work by stimulating the D2 DAR (4), whereas all clinically used antipsychotics are antagonists of this receptor (5, 6). As such, more knowledge concerning the regulation of the D2 DAR might be helpful in devising new treatment strategies for D2 DAR-related diseases.
Most cells maintain homeostatic control of their responsiveness through regulating the expression and functional activity of their cell surface GPCRs. One widely studied form of GPCR regulation is that of desensitization. In this process, activation of the GPCR by an agonist also triggers a sequence of events that result in the dampening of the receptor-mediated signal. The mechanisms associated with this form of desensitization have been widely investigated, resulting in the following canonical paradigm (for reviews, see Refs. 7 and 8). Agonist occupancy of the receptor promotes its phosphorylation by a member of the G protein-coupled receptor kinase (GRK) family, leading to the binding of an arrestin protein and ultimately resulting in uncoupling of the receptor from its cognate G protein and decreased functional signaling. The binding of arrestin also promotes internalization of the receptor through clathrin-coated pits into endosomal compartments. Once internalized, GPCRs can be sorted for recycling to the plasma membrane or targeted for degradation (9–11). Although this desensitization paradigm has been shown to be operative for many GPCRs, recent studies have suggested that there may be exceptions and significant variations to this general scheme.
Indeed, recent evidence has accumulated for a number of GPCRs suggesting that receptor phosphorylation is not always required for their desensitization or internalization (12–17). This appears to be particularly true for Gq/11-linked GPCRs, which can be regulated by GRK2-mediated mechanisms that are independent of receptor phosphorylation. Ferguson and co-worker (for reviews, see Refs. 12 and 13) have investigated this extensively using the Gq-linked metabotropic glutamate receptor-1 (mGluR1) as a model. These investigators found that truncation of the C terminus of mGluR1 prevented GRK2-mediated phosphorylation but not desensitization of the receptor. In addition, overexpression of a catalytically inactive (kinase-dead) mutant of GRK2 was found to significantly suppress mGluR1 signaling. Further studies revealed that the inhibitory effect of GRK2 was due to its N-terminal regulator of G-protein signaling (RGS) homology (RH) domain simultaneously interacting with both Gαq and mGluR1, thus interdicting receptor activation of Gq. Indeed, overexpression of a truncated GRK2 RH domain was found to produce an inhibitory effect similar to that of the wild-type GRK2. Similar results have also been observed for a variety of Gq-coupled receptors (12, 13, 16, 17). In those systems that have been examined, catalytically inactive mutants of GRK2 were found to be effective at attenuating Gq-mediated signaling, as shown for the mGluR1.
Recently, we have investigated the role of GRK-mediated phosphorylation in agonist-induced regulation of the Gi/o-linked D2 DAR (15). Using in situ phosphorylation assays and site-directed mutagenesis approaches, we were able to identify all of the GRK phosphorylation sites and create a receptor construct that is completely refractory to GRK-mediated phosphorylation. Our use of this GRK phosphorylation-null construct resulted in the surprising finding that GRK-mediated receptor phosphorylation is not required for agonist-induced desensitization, β-arrestin association, or endocytosis of the D2 DAR. Rather, we found that the GRK phosphorylation-null receptor is impaired in its ability to recycle to the cell surface subsequent to internalization and is degraded to a greater extent in comparison with the wild-type receptor. These results suggested that GRK-mediated phosphorylation of the D2 DAR regulates its intracellular trafficking or sorting once internalized. During the course of this investigation, however, we observed that overexpression of GRK2 enhanced agonist-induced internalization of the phosphorylation-null receptor, suggesting that GRK2 was capable of regulating D2 DAR activity in the absence of receptor phosphorylation.
In the current study, we further investigated the regulatory effects of GRK2 on D2 DAR signaling and found that GRK2 can negatively suppress both receptor expression and functional coupling, potentially through different mechanisms. Our results suggest novel mechanisms for how GRK2 can negatively regulate GPCR signaling in a manner that is independent from receptor phosphorylation and unlike that described for Gq-coupled receptors.
EXPERIMENTAL PROCEDURES
Materials
HEK293-tsa201 (HEK293T) cells were a gift from Dr. Vanitha Ramakrishnan. [3H]Sulpiride (70–87 Ci/mmol), [3H]methylspiperone (80–85 Ci/mmol), [32P]orthophosphate (carrier-free), and [3H]cAMP (25–40 Ci/mmol) were purchased from PerkinElmer Life Sciences. Dulbecco's modified Eagle's medium (DMEM) was from Cellgro® Mediatech, Inc. (Herndon, VA). Fetal calf serum and other cell culture reagents were purchased from Invitrogen. Calcium phosphate transfection kits were from Clontech. MiniComplete™ protease inhibitor mixture was purchased from Roche Applied Science. Anti-FLAG M2 affinity gel and all other reagents were purchased from Sigma. Anti-GRK2 antibodies (C-9 and C-15) and anti-hemagglutinin antibodies (Y-11 and F-7) were from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids and siRNAs
Rat D2L cDNA (18) in pcDNA was used for this study. For phosphorylation and immunoprecipitation experiments, FLAG-tagged rat D2L construct was used as described previously (19). Expression constructs for GRK2, GRK3, GRK5, GRK6, GRK2-K220R, GRK2-D110A, GRK2-R587Q, and GRK2-(495–689) were a kind gift from Dr. Jeffrey L. Benovic. The HA-GRK2-(45–185) construct was a gift from Dr. Stephen S. G. Ferguson. The control siRNA (siGENOME non-targeting siRNA 3) and siRNA directed against human GRK2 were purchased from Dharmacon RNA Technology (Lafayette, CO). GRK2 siRNA was designed by using Dharmacon's siDesign® center software (the siRNA sequence is GCA AGA AAG CCA AGA ACA A).
Cell Culture and Transfections
HEK293T cells were cultured in DMEM supplemented with 10% fetal calf serum, 1 mm sodium pyruvate, 50 units/ml penicillin, and 50 μg/ml streptomycin (DMEM complete). Cells were grown at 37 °C in 5% CO2 and 90% humidity. HEK293T cells were transfected using the calcium phosphate precipitation method (Clontech). Cells were seeded in 100- or 150-mm plates, and transfection was carried out at ∼50% confluency according to the manufacturer's instructions. After 18 h of transfection, the medium was replaced, and the cells were divided for subsequent experiments. For RNA knockdown experiments, HEK293T cells were seeded at a density of 1.5 × 106 cells/100-mm dish 1 day before siRNA transfection. The medium was changed to antibiotics-free DMEM complete before transfection, and then the cells were transfected with either control siRNA (100 nm) or GRK2 siRNA (100 nm) using Dharmafect I (Dharmacon) transfection reagent, according to the manufacturer's instructions. The following day, the medium was changed to fresh antibiotics-free DMEM complete, and then the cells were transfected with D2L DAR expression plasmid using FuGENE 6 transfection reagent (Roche Applied Science), according to the manufacturer's instructions. All assays were performed 3 days after siRNA transfection.
Determination of cAMP Production
HEK293T cells were seeded into poly-d-lysine-coated 24-well plates 1 day before the assay at a density of 2 × 105 cells/well. The cells were washed one time with prewarmed EBSS, and then the cells were further incubated with various concentrations of dopamine in a total volume of 0.4 ml at 37 °C for 10 min in the presence of 3 μm forskolin, 30 μm Ro-20-1724 (phosphodiesterase inhibitor), 0.2 mm sodium metabisulfite (to prevent oxidation of dopamine), and 10 μm propranolol (to block endogenous β-adrenergic receptors) in 20 mm HEPES-buffered DMEM. For the sensitization experiments, the cells were incubated in the presence of either 0.2 mm sodium metabisulfite (control) or 0.2 mm sodium metabisulfite plus 10 μm dopamine in DMEM-H (DMEM with 20 mm HEPES) for 4 h and then washed three times with prewarmed EBSS before forskolin stimulation. To terminate the reaction, the supernatant was aspirated, and 3% perchloric acid (200 μl/well) was added. After incubating for 30 min on ice, 80 μl of 15% KHCO3 was then added to neutralize the acid. The plates remained on ice for an additional 10 min and were then centrifuged at 1,300 × g for 20 min. The accumulation of cAMP was measured by a competitive binding assay described previously (20) with modifications. Briefly, 50 μl of the supernatant from each well was transferred to a 1.2-ml reaction tube containing 50 μl of cAMP-binding protein (cAMP-dependent protein kinase lysate prepared from bovine adrenals), 50 μl of [3H]cAMP (∼0.3–0.4 pmol), and 150 μl of Tris-EDTA buffer. The reaction was incubated for at least 90 min at 4 °C. After the incubation, 250 μl of charcoal solution (2% carbon and 0.5% bovine serum albumin) was added to each tube and vortexed gently. Tubes were then incubated at 4 °C for 10 min followed by centrifugation (1,300 × g) for 20 min. Radioactivity in the supernatant was then quantified by liquid scintillation spectroscopy at a counting efficiency of 58%. The cAMP concentrations were determined using a standard curve from 0.1 to 27 pmol of cAMP.
[35S]GTPγS Binding Assays
The [35S]GTPγS binding assays were carried out as described by Gardner et al. (21) with modifications. Briefly, HEK293T cells cultured in 100-mm dishes were washed twice with ice-cold membrane preparation buffer (50 mm Tris, 10 mm sodium pyrophosphate, 10 mm NaF, and 5 mm EDTA, pH 7.4). The membrane preparation buffer was removed and replaced with 8 ml of membrane preparation buffer. The cells were then scraped and homogenized with a Dounce homogenizer followed by centrifugation at 34,000 × g for 30 min. After removing the supernatant, 10 ml of ice-cold HEPES buffer (20 mm HEPES, 6 mm MgCl2, and 100 mm NaCl, pH 7.4) was added, and the pellet was again centrifuged at 34,000 × g for 30 min at 4 °C. The resulting pellet was stored at −80 °C until use. The frozen membrane pellet was thawed and resuspended with 4.5 ml of ice-cold HEPES buffer. The membrane suspension (∼20–40 μg of protein) was incubated with HEPES buffer with 0.1 mm dithiothreitol, 0.2 mm sodium metabisulfite, 5 μm GDP, 0.1 nm [35S]GTPγS, and various concentrations of dopamine at 30 °C for 30 min in a final volume of 1 ml. Basal binding was determined in the absence of agonist, and nonspecific binding was determined in the presence of 10 μm non-radioactive GTPγS. The reaction was terminated by rapid filtration through GF/C filters with four washes of 4 ml of ice-cold 50 mm Tris·HCl, pH 7.4. Radioactivity bound to the filters was quantified by liquid scintillation spectroscopy. Free [35S]GTPγS (0.1 fmol) was counted to calculate cpm to fmol conversion. The protein amount was measured using the bicinchoninic acid protein assay (Pierce). DA-stimulated [35S]GTPγS binding was calculated by subtracting the basal binding and normalized with the amount of membrane protein.
Intact Cell [3H]Sulpiride Binding Assays
HEK293T cells were seeded into poly-d-lysine-coated 24-well plates 1 day before the assay at a density of 2 × 105 cells/well. The cells were incubated in the presence of either 0.2 mm sodium metabisulfite (control) or 0.2 mm sodium metabisulfite plus 10 μm dopamine in DMEM-H for 1 h. Stimulation was terminated by quickly cooling the plates on ice and washing the cells three times with ice-cold EBSS. Then the cells were incubated with 0.5 ml of [3H]sulpiride in EBSS (final concentration, 6.4 nm) at 4 °C for 3.5 h. Nonspecific binding was determined in the presence of 5 μm (+)-butaclamol. Cells were washed three times with ice-cold EBSS, and ∼0.5 ml of 1% Triton X-100 and 5 mm EDTA was added. Samples were mixed with 4.5 ml of liquid scintillation mixture and counted with a Beckman LS6500 scintillation counter. The cells for measuring protein concentration were incubated with EBSS without [3H]sulpiride. After final washes, the protein concentration was determined by directly adding 2 ml of bicinchoninic acid (BCA) protein assay solution (Pierce) into each well. After incubation at room temperature for at least 30 min, the absorbance was read at 562 nm.
Membrane [3H]Methylspiperone Binding Assays
HEK293T cells were harvested by 10-min incubation with 5 mm EDTA in EBSS (without Ca2+ and Mg2+) at 37 °C, diluted 3-fold in EBSS, and collected by centrifugation at 300 × g for 10 min. The cells were resuspended in lysis buffer (5 mm Tris, pH 7.4 at 4 °C and 5 mm MgCl2) and were disrupted using a Dounce homogenizer followed by centrifugation at 34,000 × g for 30 min. The resulting membrane pellet was resuspended in binding buffer (50 mm Tris, pH 7.4) by homogenization. The membrane suspension was then added to assay tubes containing [3H]methylspiperone in a final volume of 1.0 ml. (+)-Butaclamol was added at a final concentration of 3 μm to determine nonspecific binding. The assay tubes were incubated at room temperature for 1.5 h, and the reaction was terminated by rapid filtration through GF/C filters pretreated with 0.3% polyethyleneimine. Radioactivity bound to the filters was quantitated by liquid scintillation spectroscopy. Protein concentrations were determined using the BCA protein assay from Pierce.
Co-immunoprecipitation Assays
HEK293T cells in 100-mm dishes were transfected with FLAG-tagged D2L DAR along with pcDNA (mock) or plasmid containing GRK2 constructs. The cells were harvested by 10-min incubation with 5 mm EDTA in EBSS (without Ca2+ and Mg2+), diluted 3-fold in EBSS, and collected by centrifugation at 300 × g for 10 min. Cells were solubilized for 1 h at 4 °C in 1 ml of solubilization buffer (50 mm HEPES, 1 mm EDTA, 10% glycerol, 1% Triton X-100, pH 7.4, 50 mm NaF, 40 mm sodium pyrophosphate, and 150 mm NaCl) supplemented with Complete protease inhibitor mixture. The samples were cleared by centrifugation, and the protein concentration was determined using the BCA protein assay kit from Pierce. The level of D2 DAR expression for each transfection was quantified via radioligand binding assays using the cells from the same transfection. After receptor/protein quantification, equal amounts of receptor protein (∼0.5–1 mg of total protein) were then transferred to fresh tubes with 40 μl of washed M2-agarose and incubated overnight with mixing at 4 °C. The samples were then washed once with solubilization buffer and 500 mm NaCl, once with solubilization buffer and 150 mm NaCl, and once with Tris-EDTA, pH 7.4 at 4 °C. Samples were then incubated in 2× SDS-PAGE sample buffer for 1 h at 37 °C followed by SDS-PAGE and immunoblotting to detect co-immunoprecipitated GRK2 or its mutants.
Western Blotting
Cells were harvested and solubilized as described above. After clearing by centrifugation, cell lysates were resolved by 4–12% SDS-PAGE and transferred to nitrocellulose membrane. Blots were incubated with membrane blocking buffer (Zymed Laboratories Inc.) for 1 h at room temperature. For detection of GRK2 and its mutants, the blots were incubated with anti-GRK2 antibody or anti-hemagglutinin antibody for 1 h at room temperature. Membranes were washed with TBST (1× Tris-buffered saline and 0.1% Tween 20) and then incubated with secondary antibodies coupled to horseradish peroxidase in TBST for 30 min at room temperature. Membranes were washed, and immunoreactive proteins were visualized by chemiluminescence (SuperSignal West Pico chemiluminescent substrate, Pierce). To detect endogenous GRK2, SuperSignal West Dura substrate (Pierce) was used.
Whole Cell Phosphorylation Assays
Metabolic labeling of cells and subsequent immunoprecipitation of the FLAG-tagged D2L DAR were carried out as described previously (19). Briefly, 1 day after transfection, cells were seeded at 1–1.5 × 106/well of a poly-d-lysine-coated 6-well plate for phosphorylation assays and ∼2 × 106 cells in a 100-mm dish for radioligand binding assays to quantify the level of receptor expression. The next day, the cells were washed once with EBSS and incubated for 1 h in phosphate-free DMEM with 10% fetal calf serum. Medium was removed and replaced with 1 ml of fresh medium supplemented with 200 μCi/ml [32P]H3PO4. After 45 min at 37 °C, the cells were then challenged with 10 μm DA in medium supplemented with 0.2 mm sodium metabisulfite. Cells were then transferred to ice, washed twice with ice-cold EBSS, and solubilized for 1 h at 4 °C in 1 ml of solubilization buffer (50 mm HEPES, 1 mm EDTA, 10% glycerol, and 1% Triton X-100, pH 7.4 at 4 °C) + 150 mm NaCl supplemented with Complete protease inhibitor mixture and phosphatase inhibitors (40 mm sodium pyrophosphate and 50 mm NaF). The samples were cleared by centrifugation in a Microfuge, and the protein concentration was determined by the bicinchoninic acid protein assay (Pierce). The level of D2 DAR expression for each transfection was quantified via radioligand binding assays using the cells from the same transfection. After receptor/protein quantification, equal amounts of receptor protein were then transferred to fresh tubes with 40 μl of washed M2-agarose and incubated overnight with mixing at 4 °C. The samples were then washed once with solubilization buffer and 500 mm NaCl, once with solubilization buffer and 150 mm NaCl, and once with Tris-EDTA, pH 7.4 at 4 °C. Samples were then incubated in 2× SDS-PAGE sample buffer for 1 h at 37 °C before being resolved by 4–12% SDS-PAGE. The gels were dried and subjected to autoradiography. After developing, the band intensities were quantitated by LabWorks™ software (UVP Inc., Upland, CA).
Bioluminescence Resonance Energy Transfer (BRET) Assays
HEK293T cells (8 × 105 cells/ml) were transiently transfected using polyethyleneimine in a 1:3 ratio (Polysciences Inc.) with a constant amount of FLAG-D2L-RLuc8 as donor and increasing amounts of GRK2-mVenus as the acceptor for the titration experiments and with a fixed amount of donor and different acceptors (GRK2-mVenus, GRK2-GFP2, or GRK5-GFP2 (GFP2 fusions were kind gifts from C. E. Elling, 7TM Pharma A/S, Horsholm, Denmark)) for the concentration-effect studies. BRET1 or BRET2 experiments were performed 48 h after transfection. The cells were harvested, washed, and resuspended in phosphate-buffered saline. Approximately 200,000 cells/well, in triplicate, were distributed in 96-well plates, and the total fluorescence (excitation at 480 nm and emission at 530 nm for GFP2 or excitation at 500 nm and emission at 540 nm for mVenus; 1-s recordings for both) and luminescence in the presence of 5 μm Deep Blue coelenterazine for BRET2 or coelenterazine h for BRET1 (no filters and 1-s recording) were quantified (Polarstar and Pherastar, BMG Labtech). BRET titration curves were performed in the presence or absence of dopamine (final concentration, 10 μm) for 5 min, and the BRET signal was determined by calculating the ratio of the light emitted by GFP2 (515–530 nm) over that emitted by RLuc8 (410–480 nm) for BRET2 and the ratio of the light emitted by mVenus (510–540 nm) over that emitted by RLuc8 (485 nm) for BRET1. The net BRET values were obtained by subtracting the background from cells expressing RLuc8 alone.
Data Analyses
All binding assays were routinely performed in triplicate and were repeated three to four times. Cyclic AMP experiments were performed in duplicate and were repeated three to four times. Estimations of the IC50 values for dopamine inhibition of cAMP accumulation were calculated using the GraphPad Prizm curve fitting program. The curves presented throughout this study, representing the best fits to the data, were generated using this software program as well.
RESULTS
GRK2 Constitutively Suppresses D2 DAR-mediated Inhibition of cAMP Accumulation
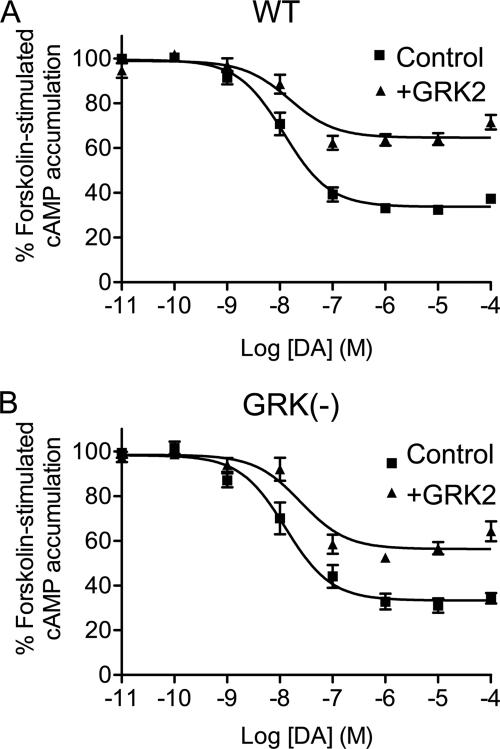
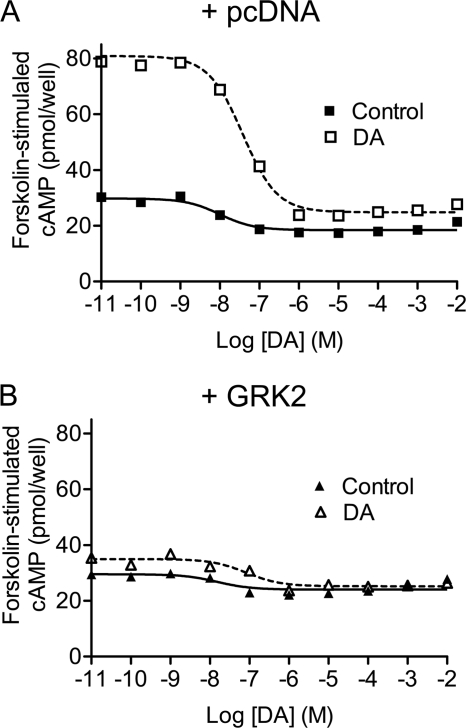
We initially investigated the effects of overexpressing GRK2 on D2 DAR-mediated signaling in HEK293T cells. Fig. 1A shows that in wild-type (WT) D2 DAR receptor-transfected cells DA inhibited forskolin-stimulated cAMP accumulation by ∼65% (see also Table 1). In contrast, in GRK2-cotransfected cells, the DA inhibition of cAMP accumulation was reduced to ∼35%. As overexpression of GRK2 is known to be associated with increased D2 DAR phosphorylation (Namkung et al. (15) and see below), we repeated this experiment using a D2 DAR receptor construct in which all of the GRK2 phosphorylation sites were mutated (GRK(−)). The GRK(−) receptor was found to inhibit cAMP accumulation to the same extent as the WT receptor (Fig. 1B) as we described previously (15). As with the WT D2 DAR, overexpression of GRK2 significantly diminished the ability of the GRK(−) receptor to inhibit cAMP accumulation, although the extent of this suppression was somewhat less than that seen with the WT receptor (Table 1). In separate experiments, we determined that overexpression of GRK2 also attenuated DA-induced [35S]GTPγS binding mediated by the WT and GRK(−) D2 DARs (supplemental Fig. 1). Taken together, these data suggest that GRK2 can constitutively attenuate D2 DAR signaling through a mechanism that is largely independent of receptor phosphorylation.
FIGURE 1.
GRK2 regulation of D2 DAR-mediated inhibition of cAMP accumulation. DA inhibition of cAMP accumulation was measured using intact HEK293T cells transiently expressing WT (A) or GRK(−) (B) D2 DARs along with pcDNA (Control) or GRK2. Cells were incubated with various concentrations of DA for 10 min in the presence of 3 μm forskolin. cAMP accumulation was then assessed as described under “Experimental Procedures.” The data shown represent the mean ± S.E. values from at least six experiments and are expressed as a percentage of forskolin-stimulated cAMP accumulation in the absence of DA. The average estimated IC50 parameters (mean ± S.E.) are as follows: WT D2 DAR, 15.5 ± 4.5 nm for control and 25.3 ± 10.2 nm for GRK2 overexpression; GRK(−) mutant, 27.7 ± 10.4 nm for control and 36.0 ± 13.6 nm for GRK2 overexpression.
TABLE 1.
GRK2 regulation of D2 DAR-mediated inhibition of cAMP accumulation
DA inhibition of cAMP accumulation was measured using intact HEK293T cells transiently expressing WT or GRK(−) D2 DARs along with pcDNA (control), GRK2, GRK2-D110A (D110A), GRK2-K220R (K220R), GRK2-R587Q (R587Q), GRK2-(45–185), or GRK2-(495–689) expression constructs. Cells were incubated with various concentrations of DA for 10 min in the presence of 3 μm forskolin as described in Figs. 1–4. cAMP accumulation was then assessed as described under “Experimental Procedures.” The maximal inhibition of forskolin-stimulated cAMP accumulation was calculated as a percentage of the cAMP produced with 1 μm DA divided by that produced with forskolin only. Data are represented as mean ± S.E. from 3–10 independent experiments, indicated in parentheses. NT, not tested.
| Inhibition of forskolin-stimulated cAMP accumulation |
IC50 |
|||
|---|---|---|---|---|
| WT | GRK(−) | WT | GRK(−) | |
| % | nm | |||
| Control | 65.4 ± 2.6 (10) | 67.2 ± 9.4 (6) | 15.5 ± 4.5 (7) | 27.7 ± 10.4 (6) |
| +GRK2 | 36.7 ± 2.4a (8) | 47.5 ± 4.5b (6) | 25.3 ± 10.2 (6) | 36.0 ± 13.6 (6) |
| +K220R | 55.4 ± 3.6c,d (6) | 63.9 ± 6.3 (4) | 14.0 ± 6.3 (4) | 25.2 ± 12.4 (4) |
| +D110A | 32.1 ± 3.4a (5) | NT | 6.1 ± 2.9 (3) | NT |
| +R587Q | 63.2 ± 4.2 (7) | NT | 12.9 ± 3.2 (5) | NT |
| +(45–185) | 64.7 ± 2.9 (3) | NT | 13.6 ± 1.5 (3) | NT |
| +(495–689) | 69.0 ± 1.9 (4) | NT | 13.0 ± 2.4 (4) | NT |
ap < 0.001 versus control, unpaired Student's t test.
b p < 0.01 versus WT D2 + GRK2, unpaired Student's t test.
c p < 0.02 versus GRK2 or D110A, unpaired Student's t test.
d p < 0.05 versus control, unpaired Student's t test.
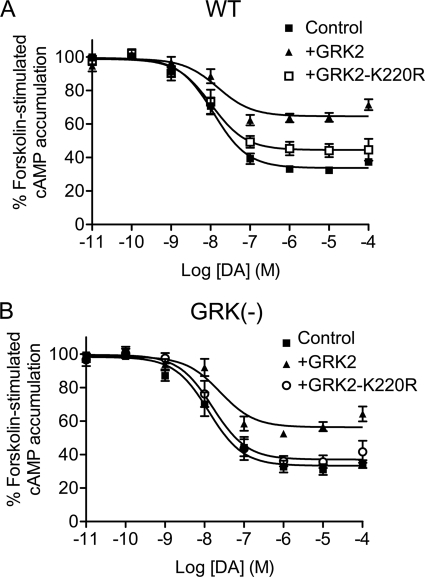
To investigate further GRK2-mediated regulation of the D2 DAR, we tested various GRK2 constructs with mutations in specific functional domains. Fig. 2 shows results using the K220R GRK2 mutant, which is completely devoid of kinase activity (22). In contrast to the WT GRK2, the constitutive effects of the K220R mutant on D2 DAR signaling were greatly attenuated with the WT D2 DAR (Fig. 2A) and completely absent with the GRK(−) receptor (Fig. 2B). The K220R mutant was able to slightly, but significantly (Fig. 2A and Table 1) diminish WT D2 DAR signaling, suggesting that whereas the inhibitory effects of GRK2 are largely kinase-dependent there is a minor kinase-independent component to this regulation (see below). This kinase-independent component was not observed with the GRK(−) receptor for reasons that are unclear, but it might be related to conformational alterations in the mutant that are independent from negating receptor phosphorylation. Overall, these results suggest that the kinase activity of GRK2 is an important determinant for its constitutive suppression of D2 DAR signaling but that this inhibitory effect does not require receptor phosphorylation.
FIGURE 2.
Effect of catalytically inactive GRK2 mutant (K220R) expression on D2 DAR-mediated inhibition of cAMP accumulation. DA inhibition of cAMP accumulation was measured using intact HEK293T cells transiently expressing WT (A) or GRK(−) (B) D2 DARs along with pcDNA (Control), GRK2, or GRK2-K220R. Cells were incubated with various concentrations of DA for 10 min in the presence of 3 μm forskolin. cAMP accumulation was then assessed as described under “Experimental Procedures.” The GRK2-K220R data shown represent the mean ± S.E. values from at least four experiments and are expressed as a percentage of forskolin-stimulated cAMP accumulation in the absence of DA. The average estimated IC50 parameters (mean ± S.E.) with GRK2-K220R expression are as follows: WT D2 DAR, 14.0 ± 6.3 nm; GRK(−) mutant, 25.2 ± 12.4 nm. The control and GRK2 (WT) data are replicated from Fig. 1 for comparative purposes.
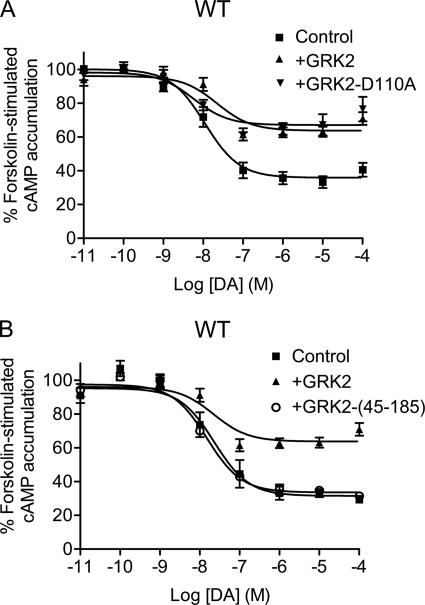
The recent elucidation of the GRK2 structure has shown that it contains three functional domains: an N-terminal RH domain, a central protein kinase domain, and a C-terminal Gβγ-binding pleckstrin homology (PH) domain (23). The RH domain of GRK2 has been shown to interact with some GPCRs and Gα subunits, particularly Gαq (23). Recently, a number of investigators have shown that GRK2 can constitutively suppress receptor-mediated signaling through Gq-mediated pathways by physically interrupting GPCR-Gq interactions (for reviews, see Refs. 12 and 13). This phosphorylation-independent GRK2-mediated inhibition of signaling is not observed with RH domain mutants that are Gαq binding-impaired (12, 13). We thus decided to explore this mechanism by examining the effects of a D110A GRK2 mutant, which lacks Gq interactions (24) (Fig. 3A and Table 1). As can be seen, the D110A mutant was as effective as the WT GRK2 in suppressing D2 DAR-mediated inhibition of cAMP accumulation. Because other studies have shown that expression of the GRK2 N-terminal RH domain alone is sufficient to attenuate signaling of some GPCRs, mainly through Gq interactions (for reviews, see Refs. 12 and 13), we overexpressed a GRK2 RH domain construct (residues 45–185), as shown in Fig. 3B. In this case, overexpression of the RH domain of GRK2 was ineffective in suppressing D2 DAR-mediated inhibition of cAMP accumulation. These results suggest that GRK2 suppresses D2 DAR-mediated signaling through a mechanism that is RH domain-independent and unlike that seen for some Gq-coupled GPCRs.
FIGURE 3.
Effect of GRK2 RH domain mutant (D110A) or GRK2 RH domain fragment on D2 DAR-mediated inhibition of cAMP accumulation. DA inhibition of cAMP accumulation was measured using intact HEK293T cells transiently expressing WT D2 DAR along with pcDNA, GRK2, GRK2-D110A (A), or GRK2-(45–185) (B). Cells were incubated with various concentrations of DA for 10 min in the presence of 3 μm forskolin. cAMP accumulation was then assessed as described under “Experimental Procedures.” The data shown represent the mean ± S.E. values from at least three experiments and are expressed as a percentage of forskolin-stimulated cAMP accumulation in the absence of DA. The average estimated IC50 parameters (mean ± S.E.) with GRK2-D110A or GRK2-(45–185) expression are 6.1 ± 2.9 and 13.6 ± 1.5 nm, respectively. The control and GRK2 (WT) data are replicated from Fig. 1 for comparative purposes.
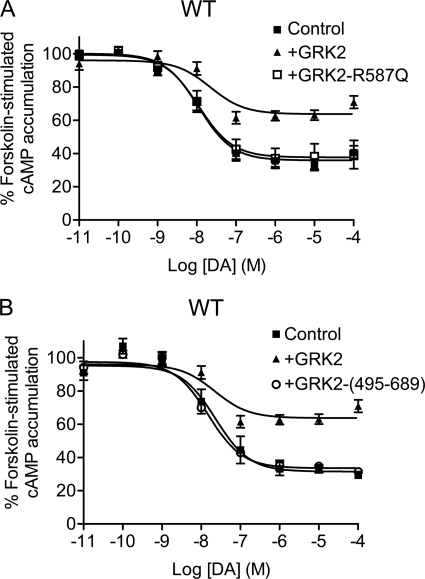
We next wished to evaluate the potential role of the GRK2 C-terminal Gβγ-binding PH domain. Fig. 4A shows that expression of a R587Q GRK2 mutant, which lacks Gβγ binding (25), was completely inactive in suppressing D2 DAR-mediated inhibition of cAMP accumulation. These results suggest that Gβγ interactions are necessary for GRK2-mediated suppression of D2 DAR signaling. We also overexpressed a construct encoding just the C-terminal fragment (residues 495–689) of GRK2, which is known to bind to and sequester Gβγ subunits (26). Fig. 4B shows that this construct was ineffective in suppressing D2 DAR-mediated regulation of cAMP accumulation (see also Table 1). These results suggest that GRK2 interactions with Gβγ are necessary but not sufficient to attenuate D2 DAR signaling. Overall, the data in Figs. 1–4 suggest that GRK2 can constitutively attenuate D2 DAR signaling in the absence of receptor phosphorylation, and this suppression mainly requires GRK2 kinase activity and GRK2-Gβγ interactions.
FIGURE 4.
Effect of GRK2 PH domain mutant (R587Q) or the GRK2 PH domain fragment on D2 DAR-mediated inhibition of cAMP accumulation. DA inhibition of cAMP accumulation was measured using intact HEK293T cells transiently expressing WT D2 DAR along with pcDNA, GRK2, GRK2-R587Q (A), or GRK2-(495–689) (B). Cells were incubated with various concentrations of DA for 10 min in the presence of forskolin. cAMP accumulation was then assessed as described under “Experimental Procedures.” The data shown represent the mean ± S.E. values from at least three experiments and are expressed as a percentage of forskolin-stimulated cAMP accumulation in the absence of DA. The average estimated IC50 parameters (mean ± S.E.) with GRK2-R587Q or GRK2-(495–689) overexpression are 12.9 ± 3.2 and 13.0 ± 2.4 nm, respectively. The control and GRK2 (WT) data are replicated from Fig. 1 for comparative purposes.
GRK2 Constitutively Suppresses D2 DAR-mediated Sensitization of Adenylate Cyclase
Because GRK2 overexpression constitutively suppresses D2 DAR signaling, we wondered whether this regulatory effect would have any impact on agonist-induced receptor desensitization. Fig. 5A shows an experiment in which WT D2 DAR-expressing cells were pretreated with DA for 6 h prior to assay. Interestingly, we found that this DA pretreatment led to a very significant “sensitization” of forskolin-stimulated cAMP accumulation, although DA was still able to inhibit this activity in a dose-dependent fashion. Sensitization of adenylate cyclase by agonist activation of Gαi/o-coupled receptors has been well described, including that for the D2 DAR (27). The mechanism(s) involved are not clear but may be cell type-specific and are thought to involve enhanced Gαs-adenylate cyclase interactions and/or isoform-specific adenylate cyclase phosphorylation. In any case, this sensitization of downstream coupling pathways renders it difficult to assess the functional activity of the receptor per se. Surprisingly, however, when GRK2 was expressed with the D2 DAR, there was a near complete suppression of the DA-induced adenylate cyclase sensitization (Fig. 5B). Similar results were observed with the GRK(−) D2 DAR (data not shown), suggesting that receptor phosphorylation is not involved.
FIGURE 5.
GRK2 regulation of D2 DAR-mediated adenylate cyclase sensitization. HEK293T cells were transfected with D2 DAR along with pcDNA (A) or WT GRK2 (B). Cells were preincubated in the absence (Control) or presence of DA (10 μm) for 6 h at 37 °C and then washed extensively. Cells were further incubated with various concentrations of DA for 10 min in the presence of 3 μm forskolin. cAMP accumulation was then assessed as described under “Experimental Procedures.” Data shown are from one of three representative experiments.
We explored this further in Fig. 6A by comparing the ability of other GRK isoforms to suppress agonist-induced sensitization of adenylate cyclase activity. Interestingly, only GRK2 and GRK3 were effective in this model. In contrast, overexpression of GRK5 or GRK6 had no impact on agonist-induced sensitization. It is of interest to note that a common feature of both GRK2 and GRK3 is their ability to bind Gβγ. In Fig. 6B, we examined the ability of various GRK2 mutants to block the adenylate cyclase sensitization response. Overexpression of the D110A mutant proved as effective as WT GRK2, whereas more graded responses were observed with the K220R and R587Q mutants. Interestingly, the GRK2 mutant with the least activity (R587Q) was deficient in Gβγ interactions as are GRK5 and GRK6. These results suggest that Gβγ binding may be important for GRK2-mediated attenuation of adenylate cyclase sensitization. Although it is not clear whether the GRK2 suppression of D2 DAR-mediated inhibition of cAMP accumulation (Figs. 1–4) is related to the GRK2 suppression of adenylate cyclase sensitization (Figs. 5 and 6), both data sets speak to the fact that GRK2 can constitutively attenuate D2 DAR-mediated functional responses in the absence of receptor phosphorylation.
FIGURE 6.
Effect of GRK isozymes and GRK2 functional mutants on D2 DAR-mediated adenylate cyclase sensitization. HEK293T cells were transfected with D2 DAR along with pcDNA or various GRK isozymes as indicated in A or GRK2 mutants as indicated in B. Cells were preincubated in the absence (control) or presence of 10 μm DA for 4 h at 37 °C and then stimulated with 3 μm forskolin for 10 min after extensive washing. The accumulated cAMP amount was measured as described under “Experimental Procedures.” Values are expressed as percentage of forskolin-stimulated cAMP accumulation in the absence of DA preincubation (control). Data shown are the mean ± S.E. from three to five experiments. *, p < 0.05; **, p < 0.001, compared with pcDNA, unpaired Student's t test. #, p < 0.01, compared with GRK2, unpaired Student's t test.
GRK2 Constitutively Suppresses Cell Surface Expression of D2 DAR
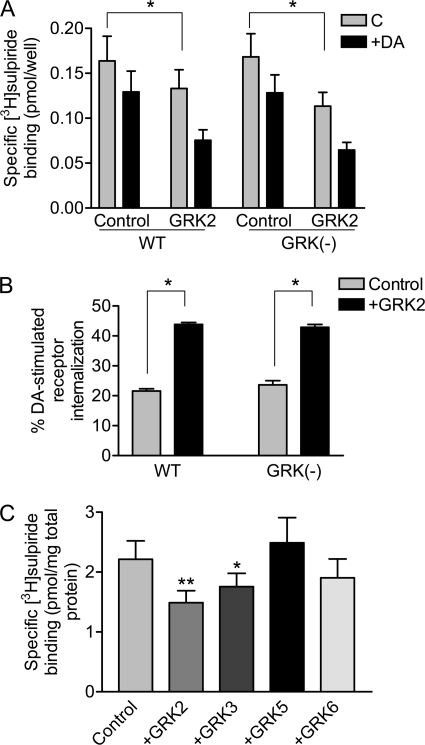
An important question given our functional data is whether or not GRK2 negatively modulates D2 DAR expression at the cell surface. We explored this in Fig. 7 using an intact cell binding assay and [3H]sulpiride, which is a hydrophilic antagonist that only labels receptors on the cell surface (15, 19). Overexpression of GRK2 indeed led to decreased receptor expression at the cell surface, and this was observed with both the WT and GRK(−) D2 DARs (Fig. 7A). GRK2 is thus capable of suppressing cell surface expression of the D2 DAR through a mechanism that does not involve receptor phosphorylation. Notably, acute treatment of the cells with DA promoted a further loss of receptors from the cell surface in a phosphorylation-independent manner as we described previously (15). Interestingly, the DA-induced receptor internalization was significantly greater in cells overexpressing GRK2 (Fig. 7B), which is true for both the WT and GRK(−) receptors. In Fig. 7C, we evaluated other GRK isoforms for their ability to suppress cell surface expression of the D2 DAR. Notably, only GRK2 and GRK3 were active in this regard; GRK5 and GRK6 were ineffective. These results agree well with our functional response data in that only GRK2 and GRK3 appear to constitutively regulate the D2 DAR.
FIGURE 7.
Effect of GRK2 and other GRK isozymes on D2 DAR cell surface expression. A, HEK293T cells were transfected with either WT or GRK(−) D2 DARs along with pcDNA (Control) or GRK2. The cells were incubated in the absence (C) or presence of 10 μm DA for 1 h and then subjected to intact cell [3H]sulpiride binding assays as described under “Experimental Procedures.” The values shown represent the means ± S.E. of six independent experiments. *, p < 0.05, paired Student's t test. B, the DA-induced loss of receptors from the cell surface from A is replotted and shown as percent receptor internalization. C, HEK293T cells were transfected with the WT D2 DAR along with pcDNA (Control), GRK2, GRK3, GRK5, or GRK6 constructs. The cells were washed and then subjected to intact cell [3H]sulpiride binding assays as described under “Experimental Procedures.” The values shown represent the means ± S.E. of at least three independent experiments. *, p < 0.05; **, p < 0.01, compared with pcDNA, unpaired Student's t test.
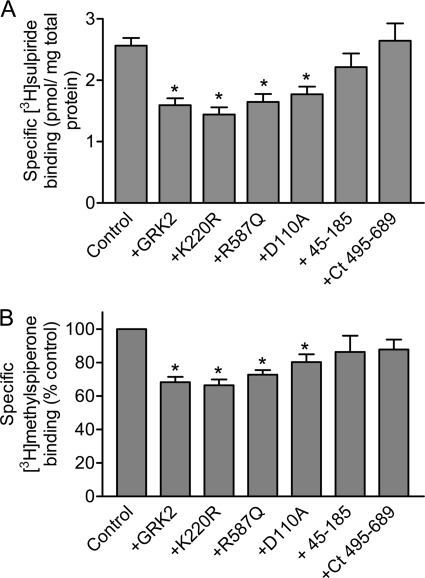
In Fig. 8, we overexpressed the GRK2 mutant and truncated constructs to evaluate their ability to regulate D2 DAR expression. Interestingly, all of the singly mutated GRK2 constructs behaved as the WT GRK2 in suppressing D2 DAR surface expression by ∼40% (Fig. 8A). In contrast, the N- or C-terminal GRK2 constructs had no effect on D2 DAR surface expression. To further investigate this regulatory response, we quantitated receptor binding in crude membranes using the hydrophobic antagonist [3H]methylspiperone as this technique will more closely reflect the total cellular expression levels of the D2 DAR. As with the intact cell binding assays, all of the singly mutated GRK2 constructs suppressed D2 DAR expression, as determined with [3H]methylspiperone binding, by 20–30% (Fig. 8B). This suggests that at least some, if not most, of the GRK2-induced loss of cell surface receptors is due to a decrease in total cellular expression of the D2 DAR.
FIGURE 8.
Effect of GRK2 and its mutants on D2 DAR expression. HEK293T cells were transfected with the D2 DAR with pcDNA (Control), GRK2, GRK2-K220R, GRK2-R587Q, GRK2-D100A, GRK2-(45–185), or GRK2-(495–689) constructs. A, the cells were subjected to intact cell [3H]sulpiride binding assays as described under “Experimental Procedures.” The values shown represent the means ± S.E. from 3 to 11 independent experiments. *, p < 0.01, compared with control, unpaired Student's t test. B, total cellular D2 DAR expression was measured by [3H]methylspiperone (2 nm) binding as described under “Experimental Procedures.” Data are normalized to the control binding values for each individual experiment and expressed as means ± S.E. of 3–10 experiments. Statistical analyses were performed before normalization. *, p < 0.05, compared with pcDNA (Control), paired Student's t test. Ct, C-terminal.
It was surprising to observe that both the K220R and R587Q GRK2 mutants moderately decreased total and cell surface receptor expression without significantly affecting receptor function (cf. Figs. 2 and 4), thus suggesting the existence of “receptor reserve” in our HEK293T cell expression system. We investigated this further by using HEK293 cells for these experiments rather than HEK293T cells as the HEK293 cells express ∼70% fewer receptors when transfected compared with the HEK293T cells. supplemental Fig. 2 shows that, when using the HEK293 cells for expression, we did indeed observe a small, but significant reduction in functional receptor signaling when expressing the GRK2-K220R and GRK-R587Q constructs as opposed to that observed when using the HEK293T cells (cf. Figs. 2 and 4). Taken together, these observations suggest that GRK2 negatively modulates D2 DAR signaling through more than a single mechanism as will be discussed below.
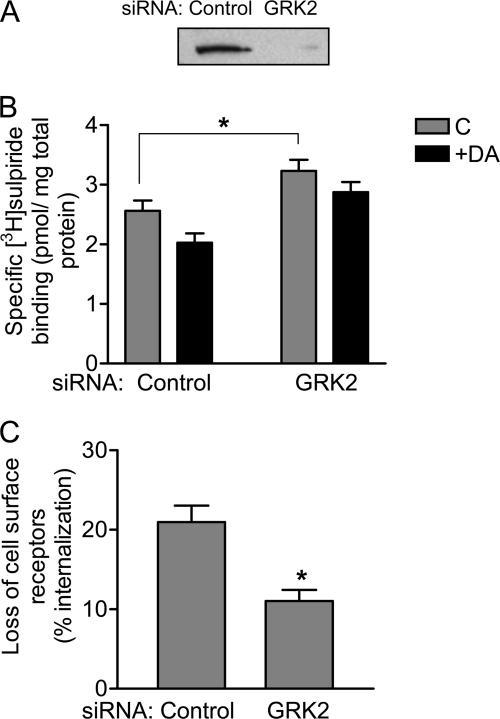
Although HEK293 cells endogenously express relatively low levels of GRK2, we thought it would be important to try to “knock down” GRK2 expression using siRNA techniques to see what effect this would have on D2 DAR expression. Fig. 9A shows a GRK2 immunoblot of lysates from cells treated with either a control siRNA or an siRNA directed against GRK2. As can be seen, treatment with the GRK2 siRNA effectively decreased its cellular expression. Interestingly, in Fig. 9B, we show that siRNA knockdown of GRK2 significantly increased cell surface expression of the D2 DAR and also impaired agonist-promoted receptor internalization. This latter point is illustrated better in Fig. 9C. GRK2 thus appears to enhance agonist-induced receptor sequestration, although not through receptor phosphorylation (cf. Fig. 7). Taken together, the data in Figs. 7–9 indicate that the cell surface expression of the D2 DAR is inversely related to the expression levels of GRK2.
FIGURE 9.
Effect of siRNA-mediated suppression of endogenous GRK2 expression on D2 DAR surface expression and internalization. HEK293T cells were transfected with either control siRNA or GRK2 siRNA followed by transfection with the D2 DAR. A, endogenous GRK2 expression level was assessed by Western blotting using an anti-GRK2 antibody as described under “Experimental Procedures.” Representative data from four independent experiments are shown. B, the cells were incubated in the absence (C) or presence of 10 μm DA for 1 h and then subjected to intact cell [3H]sulpiride binding assays as described under “Experimental Procedures.” C, the internalization of the D2 DAR was assessed by measuring the decrease in cell surface [3H]sulpiride binding sites, and the data are expressed as the percent loss of cell surface receptors. The values shown represent the means ± S.E. of four independent experiments. *, p < 0.05, unpaired Student's t test.
GRK2 Is Constitutively Associated with a D2 DAR Complex
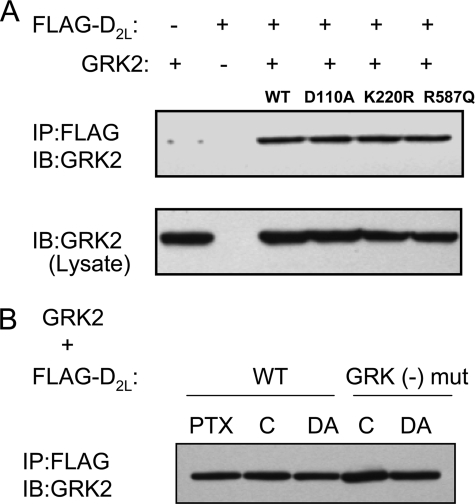
Given the constitutive nature of the regulatory effects of GRK2 on D2 DAR signaling, we wondered whether GRK2 might be constitutively associated in some way with the D2 DAR. In Fig. 10, we investigated this possibility using co-immunoprecipitation analyses. Fig. 10A shows the results of co-expressing GRK2 constructs with the D2 DAR followed by immunoprecipitation of the receptor and then immunoblotting against GRK2 to detect co-immunoprecipitation. Interestingly, the WT GRK2 and all of the singly mutated GRK2 constructs were found to co-immunoprecipitate with the D2 DAR. When normalized to the amount of D2 DAR that was immunoprecipitated, as determined by blotting the gels with an anti-FLAG antibody, there was no significant difference in the ability of the various GRK2 constructs to co-immunoprecipitate with the D2 DAR (supplemental Fig. 3A). Notably, these D2 DAR-GRK2 interactions appear to occur in the plasma membrane as the D2 receptor is almost exclusively localized at the cell surface, even with GRK2 overexpression, as detected by expressing a D2 DAR-YFP construct and visualization with confocal fluorescence microscopy (data not shown).
FIGURE 10.
Co-immunoprecipitation of GRK2 and its mutants with D2 DAR. A, HEK293T cells were transfected with FLAG-tagged D2 DAR plus WT GRK2 or various GRK2 mutants as indicated. Solubilized cell lysates were immunoprecipitated (IP) using anti-FLAG-agarose, separated by SDS-PAGE, and immunoblotted (IB) with an anti-GRK2 antibody as described under “Experimental Procedures.” A representative experiment, which was performed twice with identical results, is shown. B, HEK293T cells were transfected with either FLAG-tagged WT D2 DAR or FLAG-tagged GRK(−) D2 DAR plus GRK2. Cells were treated with 200 ng/ml pertussis toxin (PTX) overnight or incubated in the absence (C) or presence of 10 μm DA for 30 min before harvesting. Solubilized cell lysates were prepared and subjected to immunoprecipitation using anti-FLAG-agarose as described in A. A representative experiment, which was performed three times with identical results, is shown. mut, mutant.
As agonist occupancy leads to an increase in D2 DAR phosphorylation by GRK2 (Namkung et al. (15) and see below), we evaluated whether or not agonist treatment would affect GRK2 association with the D2 DAR using both the WT and GRK(−) receptors. Surprisingly, pretreatment of the cells with DA, prior to solubilization, had no effect on the ability of GRK2 to co-immunoprecipitate with the D2 DAR (Fig. 10B). Normalization of the amount of GRK2 co-immunoprecipitated to that of the D2 DAR confirmed this observation (supplemental Fig. 3B). GRK2 thus appears to constitutively associate with the D2 DAR complex, even if the receptor is not actually phosphorylated by GRK2, as suggested by co-immunoprecipitation experiments using the GRK(−) receptor (Fig. 10B and supplemental Fig. 3B).
Previously, we showed that disrupting the D2 DAR-Gαi/o interactions, through treatment of the cells with pertussis toxin, had no effect on agonist-promoted GRK2-mediated phosphorylation of the receptor (15). Similarly, we found that pertussis toxin treatment, which completely inhibited D2 DAR-mediated inhibition of adenylate cyclase (data not shown), had no effect on the ability of GRK2 to co-immunoprecipitate with the D2 DAR (Fig. 10B).
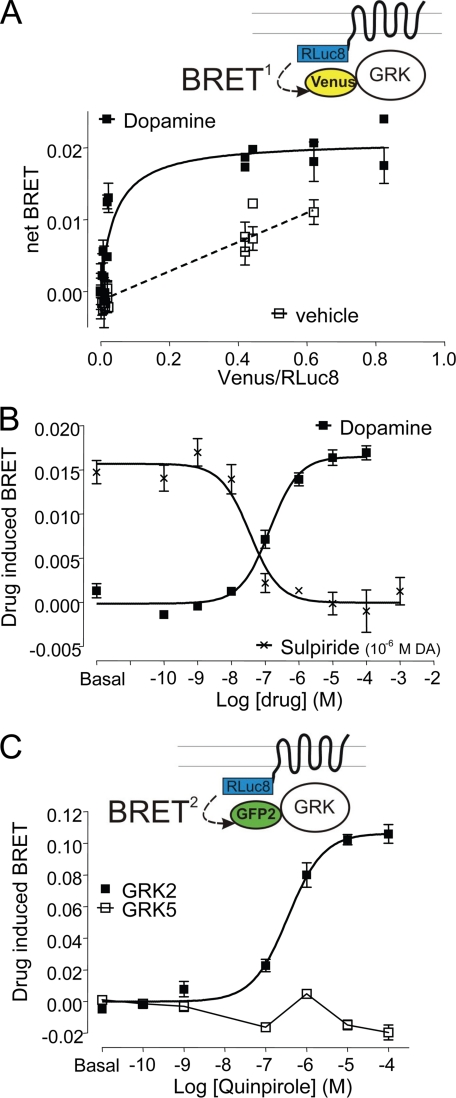
To assess the proximity of the D2 DAR and GRK2 using a parallel approach, we developed a BRET-based assay. Dopamine led to a substantial increase in BRET measured between the D2 DAR fused to RLuc8 and GRK2 fused to mVenus (Fig. 11A). This agonist-mediated modulation was concentration-dependent and specific, as it was blocked by the selective D2 DAR antagonist sulpiride (Fig. 11B), and also was seen with the D2 DAR agonist quinpirole (data not shown). In contrast, consistent with the ability of GRK2, but not GRK5, to phosphorylate and regulate D2 DAR expression and function (Ref. 15 and see below), dopamine had no effect on BRET between D2 DAR and GRK5 fused to GFP2 (Fig. 11C). Notably, BRET could not distinguish conformational changes that increase the efficiency of energy transfer from differences in the association-dissociation dynamics between the proteins of interest. Thus, in light of the co-immunoprecipitation data shown above (Fig. 10), the BRET changes observed upon incubation with the agonist may be the result of a conformational rearrangement within a pre-existing constitutive complex.
FIGURE 11.
Direct assessment of D2 DAR-GRK2 association using BRET. A, the molecular proximity between GRK2 and D2 DARs was studied by a newly developed BRET-based biosensor. Titration experiments were performed in HEK293T cells in the presence (filled symbols and solid lines) and absence (open symbols and dashed lines) of 10 μm dopamine for 5 min as described under “Experimental Procedures.” Values are expressed as means ± S.E. of three independent experiments. B and C, constant amounts of D2L-RLuc8 fusion and GRK2-mVenus, GRK2-GFP2, or GRK5-GFP2 were expressed in HEK293T cells, and BRET was recorded as described under “Experimental Procedures” after incubation with the indicated concentration of dopamine or quinpirole for 5 min. Sulpiride inhibition curves were done using a 15-min preincubation of the indicated concentration of antagonist at 37 °C and subsequent addition of 10 μm dopamine. BRET signals are shown as means ± S.E. of representative experiments, which were performed three times independently.
Overall, these results suggest that GRK2 is constitutively associated with the D2 DAR but most likely within a multiprotein complex given the BRET data. Furthermore, GRK2 association with this protein complex is not regulated by agonist activation of the receptor or liberation of Gβγ from activated Gi/o.
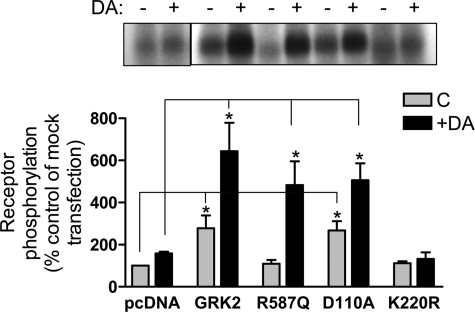
Although we showed that the GRK2 mutants are constitutively associated with the D2 DAR complex, an important question is whether or not this association leads to enhanced receptor phosphorylation as we have shown for the WT GRK2 (15). Fig. 12 shows in situ phosphorylation assays for the D2 DAR. As can be seen, overexpression of the WT and D110A GRK2 constructs led to an increase in both basal and agonist-stimulated receptor phosphorylation. Interestingly, overexpression of the R587Q mutant had little effect on basal phosphorylation but led to an enhancement of agonist-stimulated phosphorylation. In contrast, overexpression of the kinase-dead K220R mutant appeared to block agonist-stimulated phosphorylation consistent with a dominant-negative mechanism. Overall, the results confirm that GRK2 and its mutant constructs are effectively expressed, constitutively associated with the D2 DAR, and functional with respect to modulating receptor phosphorylation.
FIGURE 12.
Effect of overexpressing GRK2 WT and mutants on basal and DA-induced D2 DAR phosphorylation. HEK293T cells were transfected with FLAG-tagged D2 DAR along with pcDNA (control), WT GRK2, GRK2-R587Q, GRK2-D110A, or GRK2-K220R. Cells were metabolically labeled with [32P]H3PO4 for 45 min prior to stimulation with 10 μm DA for 20 min. The cells were then solubilized, and the samples were subjected to immunoprecipitation as described under “Experimental Procedures.” Receptors were quantified in each transfection, and equal amounts of receptor protein were loaded in each gel lane followed by SDS-PAGE resolution. The extent of receptor phosphorylation was visualized by autoradiography and quantified as described below. The upper panel shows a representative autoradiogram. In the lower panel, the receptor phosphorylation was quantified by scanning the autoradiograms followed by analysis with the LabWorks software. The data are presented as the percentage of the basal phosphorylation of the mock (pcDNA) transfection and expressed as the mean ± S.E. values from three independent experiments. Statistical analyses were performed before normalization. *, p < 0.05, compared with the control (C) or DA-stimulated values of the mock (pcDNA) transfection, respectively; paired Student's t test.
DISCUSSION
Contrary to earlier developed models based mostly on the β2-adrenergic receptor (for reviews, see Ferguson et al. (7) and Gainetdinov et al. (8)), GPCRs can undergo agonist-induced desensitization and other forms of regulation in the absence of GRK-mediated receptor phosphorylation (12–17). More recently, it has also been suggested that GRKs can negatively regulate GPCR function even in the absence of receptor phosphorylation. This appears to be particularly true for Gq/11-linked GPCRs where GRK2 has been shown to interact with both Gqα and the cognate receptor to prevent G protein activation and signaling (for reviews, see Refs. 12 and 13). Whether or not GRKs can regulate GPCRs linked to other G proteins in such a phosphorylation-independent manner has hitherto been unclear. Indeed, we are aware of only two previous investigations suggesting such phenomena. In one study, GRK2/5/6 were found to attenuate Gαs-coupled follicle-stimulating hormone signaling (28), whereas, in a second study, GRK4 was found to desensitize Gαi-linked γ-aminobutyric acid type B receptors in cerebellar granule cells (29). Importantly, in both of these investigations, catalytically inactive GRK mutants were found to mimic their wild-type counterparts in promoting attenuation of receptor signaling, thus suggesting phosphorylation-independent mechanism(s).
Recently, we showed that GRK-mediated receptor phosphorylation is not required for agonist-induced arrestin association, desensitization, or internalization of the D2 DAR (15). Rather, GRK2 phosphorylation appears to regulate the intracellular trafficking and recycling of the D2 DAR subsequent to agonist-induced endocytosis (15). In our current study, we showed that GRK2 is also involved in regulating cell surface receptor activity as its overexpression led to decreased expression of both the wild-type receptor and a phosphorylation-null mutant. In contrast, siRNA knockdown of endogenous GRK2 expression led to an increase in cell surface expression of the D2 DAR. Thus, the expression of the D2 DAR appears to be inversely correlated with the expression of GRK2. We also observed that overexpression of GRK2 led to enhanced agonist-induced receptor internalization, again in a phosphorylation-independent manner. GRK3 mimicked these effects of GRK2 on receptor expression, whereas GRK5/6 did not. In agreement with this, we previously observed that only GRK2/3 promoted D2 DAR phosphorylation (15), suggesting that only these GRKs interact with the D2 DAR. It will be important, however, to investigate GRK-mediated regulation of the D2 DAR in other cell types, such as neurons, that express endogenous levels of GRK isotypes and the D2 DAR.
Interestingly, overexpression of various functional domain mutants of GRK2, including the catalytically inactive K220R mutant, also resulted in decreased D2 DAR expression, whereas overexpression of the truncated N- and C-terminal GRK2 fragments did not. When we expressed the D2 DAR at lower levels such that there was no receptor reserve, the GRK2-induced attenuation of receptor expression was also found to correlate with decreased receptor signaling. This regulatory effect of GRK2 appears similar to those described for the follicle-stimulating hormone and γ-aminobutyric acid type B receptors (cited above) in that the GRK catalytic activity was not required to inhibit the receptor response. However, it should be noted that the follicle-stimulating hormone and γ-aminobutyric acid type B receptor expression levels were not investigated in those studies, thus leaving open the possibility for the involvement of different GRK-associated mechanisms.
Notably, we found that GRK2 and all associated functional domain mutants co-immunoprecipitated with the D2 DAR, suggesting an association under basal conditions. GRK2 was also found to co-immunoprecipitate with the phosphorylation-null D2 DAR construct, indicating that receptor phosphorylation is not required for this association. Surprisingly, pretreatment of the cells with DA prior to solubilization and immunoprecipitation had no effect on the ability of GRK2 and the D2 DAR to co-immunoprecipitate or on the apparent quantity of GRK2 associated with the receptor. In addition, we found that pretreatment of the cells with pertussis toxin, which negates the ability of the receptor to activate Gi/o proteins and release βγ subunits, had no effect on the co-immunoprecipitation of GRK2 and the receptor. These results contrast with the generally accepted model whereby receptor activation by agonists releases Gβγ subunits, thus promoting the translocation of GRK2 to the plasma membrane, leading to receptor phosphorylation (7, 8). Rather, GRK2 appears to be preassociated with the D2 DAR, although it should be noted that our co-immunoprecipitation data do not distinguish whether GRK2 and the D2 DAR are in direct physical contact under basal conditions or whether they are precipitating within a multiprotein complex. In fact, our BRET results actually favor the latter hypothesis. Nonetheless, given that all of the GRK2 mutants, especially the K220R catalytically inactive construct, are constitutively associated with the D2 DAR complex and that all cause a loss of receptor expression, it is tempting to speculate that this is, at least in part, the mechanism for GRK2-mediated down-regulation of the D2 DAR.
In this study, we also observed a constitutive suppressive effect of GRK2/3 on agonist-induced adenylyl cyclase sensitization. In contrast, this regulation was not observed with GRK5/6, which notably have no functional effects on the D2 DAR. The functional domain mutants of GRK2 produced somewhat graded responses in this paradigm where the Gβγ binding-defective R587Q mutant was the least active but still significantly suppressed the sensitization response. In contrast, the K220R catalytically inactive mutant was nearly as active as wild-type GRK2. Notably, the R587Q mutant was as effective as wild-type GRK2 in promoting receptor phosphorylation (Fig. 11), indicating that this construct is fully capable of functional interactions with the D2 DAR. These results thus suggest that GRK2 suppression of adenylyl cyclase sensitization is linked to constitutive association with the receptor but may be more complex in that Gβγ interactions may facilitate the suppressive effect. However, given that the mechanisms involved with Gi/o-linked GPCR sensitization of adenylyl cyclase activity are not known with certainty (27), it is difficult to speculate further.
Perhaps the most interesting finding of our current study is the observation that GRK2 can also attenuate the signaling of the D2 DAR through a mechanism that requires kinase activity and Gβγ binding but does not involve receptor phosphorylation. Interestingly, we were able to differentiate this regulatory effect from that involving a reduction in receptor expression by expressing the receptor at high levels, thus creating the existence of receptor reserve. Thus, a small reduction in receptor expression did not translate into a loss of functional response, which explains why the K220R and R587Q mutants did not attenuate D2 DAR-mediated regulation of adenylyl cyclase activity in HEK293T cells, despite their attenuation of receptor expression. The observation that GRK2 kinase activity is required for this additional regulatory response suggests that GRK2 must phosphorylate another protein that is involved in regulating D2 DAR signaling. At the moment, the identity of this protein is unclear, although GRK2 is known to phosphorylate multiple non-GPCR substrates (30–33). We investigated the potential role of some of these additional substrates including the protein ezrin, which has been shown to play a role in α1B-adrenergic receptor recycling (33); however, overexpression of ezrin did not affect D2 DAR expression or signaling (data not shown). Thus, the identity of the relevant phosphoprotein remains unknown at this time.
The requirement of GRK2 association with Gβγ subunits, as exemplified by the R587Q mutant, to suppress receptor signaling was also of significant interest. This regulation does not appear to involve targeting of GRK2 to the receptor as the R587Q mutant was similarly associated with the receptor as wild-type GRK2 as revealed with the co-immunoprecipitation experiments. Moreover, the R587Q GRK2 mutant was able to fully support agonist-induced receptor phosphorylation, indicating that Gβγ interactions are not required for receptor phosphorylation. One possibility is that the requirements for GRK2-mediated phosphorylation and receptor uncoupling are different. Alternatively, Gβγ interactions may be required for GRK2-mediated phosphorylation of the yet-to-be-identified phosphoprotein that is involved in the uncoupling response.
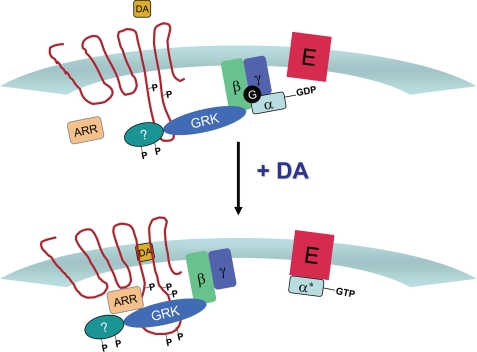
Based on our current and previously published data (15), we propose a model for how DA and GRK2 regulate the expression and activity of the D2 receptor (Fig. 13). Under basal conditions (Fig. 13, top panel), GRK2 is constitutively associated with the D2 receptor, most probably within a multiprotein complex. The association of GRK2 with the D2 DAR complex negatively regulates receptor signaling through two different mechanisms. First, GRK2 promotes a loss of cell surface expression and down-regulation in a phosphorylation-independent fashion. Second, GRK2 attenuates receptor-G protein coupling though a mechanism that also does not involve receptor phosphorylation. Rather, this regulation appears to be dependent on the ability of GRK2 to phosphorylate an additional, unknown protein that regulates receptor-G protein coupling. This latter regulatory activity of GRK2 is dependent on its ability to bind Gβγ, but these interactions are not required to target GRK2 to the receptor. In Fig. 13, bottom panel, DA binding to the receptor promotes a conformationally induced active signaling state, which recruits arrestin to the receptor complex, resulting in desensitization and receptor internalization. Neither of these events involves GRK2-mediated phosphorylation of the receptor. The alteration in receptor conformation, however, does lead to profound GRK2 phosphorylation, which regulates receptor trafficking and recycling to the cell surface subsequent to receptor internalization. Overall, these findings provide a vivid example of the diversity of mechanisms underlying GPCR regulation and clearly show that a unitary or canonical model of agonist-induced regulation does not exist.
FIGURE 13.
Model for GRK2-mediated regulation of the D2 DAR. ARR, arrestin; E, effector.
Supplementary Material
Acknowledgments
We thank Dr. Vanitha Ramakrishnan for the HEK293tsa201 cells and Dr. Lisa Hazelwood for assistance with data analysis. We also thank the NINDS DNA sequencing facility for assistance.
This work was supported, in whole or in part, by the National Institutes of Health through the intramural research program of NINDS (to D. R. S.), Grants MH54137 and DA022413 (to J. A. J.), and a NINDS competitive fellowship (to Y. N.). This work was also supported by the Lieber Center for Schizophrenia Research and Treatment (to J. A. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- DAR
- dopamine receptor
- DA
- dopamine
- GPCR
- G protein-coupled receptor
- DMEM
- Dulbecco's modified Eagle's medium
- EBSS
- Earle's balanced salt solution
- GRK
- G protein-coupled receptor kinase
- siRNA
- short interfering RNA
- PH
- pleckstrin homology
- RH
- regulator of G-protein signaling (RGS) homology
- mGluR1
- metabotropic glutamate receptor-1
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- BRET
- bioluminescence resonance energy transfer
- WT
- wild-type.
REFERENCES
- 1.Missale C., Nash S. R., Robinson S. W., Jaber M., Caron M. G. (1998) Physiol. Rev. 78, 189–225 [DOI] [PubMed] [Google Scholar]
- 2.Sibley D. R., Monsma F. J., Jr. (1992) Trends Pharmacol. Sci. 13, 61–69 [DOI] [PubMed] [Google Scholar]
- 3.Neve K. A., Seamans J. K., Trantham-Davidson H. (2004) J. Recept. Signal Transduct. Res. 24, 165–205 [DOI] [PubMed] [Google Scholar]
- 4.Seeman P. (2007) Synapse 61, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 5.Kapur S., Remington G. (2001) Biol. Psychiatry 50, 873–883 [DOI] [PubMed] [Google Scholar]
- 6.Tandon R., Keshavan M. S., Nasrallah H. A. (2008) Schizophr. Res. 100, 4–19 [DOI] [PubMed] [Google Scholar]
- 7.Ferguson S. S., Barak L. S., Zhang J., Caron M. G. (1996) Can. J. Physiol. Pharmacol. 74, 1095–1110 [DOI] [PubMed] [Google Scholar]
- 8.Gainetdinov R. R., Premont R. T., Bohn L. M., Lefkowitz R. J., Caron M. G. (2004) Annu. Rev. Neurosci. 27, 107–144 [DOI] [PubMed] [Google Scholar]
- 9.Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 10.Marchese A., Paing M. M., Temple B. R., Trejo J. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 601–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore C. A., Milano S. K., Benovic J. L. (2007) Annu. Rev. Physiol. 69, 451–482 [DOI] [PubMed] [Google Scholar]
- 12.Dhami G. K., Ferguson S. S. (2006) Pharmacol. Ther. 111, 260–271 [DOI] [PubMed] [Google Scholar]
- 13.Ferguson S. S. (2007) Trends Pharmacol. Sci. 28, 173–179 [DOI] [PubMed] [Google Scholar]
- 14.Kim O. J., Gardner B. R., Williams D. B., Marinec P. S., Cabrera D. M., Peters J. D., Mak C. C., Kim K. M., Sibley D. R. (2004) J. Biol. Chem. 279, 7999–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Namkung Y., Dipace C., Javitch J. A., Sibley D. R. (2009) J. Biol. Chem. 284, 15038–15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pao C. S., Benovic J. L. (2002) Sci. STKE 2002, PE42. [DOI] [PubMed] [Google Scholar]
- 17.Willets J. M., Challiss R. A., Nahorski S. R. (2003) Trends Pharmacol. Sci. 24, 626–633 [DOI] [PubMed] [Google Scholar]
- 18.Monsma F. J., Jr., McVittie L. D., Gerfen C. R., Mahan L. C., Sibley D. R. (1989) Nature 342, 926–929 [DOI] [PubMed] [Google Scholar]
- 19.Namkung Y., Sibley D. R. (2004) J. Biol. Chem. 279, 49533–49541 [DOI] [PubMed] [Google Scholar]
- 20.Watts V. J., Neve K. A. (1996) Mol. Pharmacol. 50, 966–976 [PubMed] [Google Scholar]
- 21.Gardner B., Hall D. A., Strange P. G. (1996) Br. J. Pharmacol. 118, 1544–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong G., Penn R., Benovic J. L. (1994) J. Biol. Chem. 269, 13084–13087 [PubMed] [Google Scholar]
- 23.Tesmer V. M., Kawano T., Shankaranarayanan A., Kozasa T., Tesmer J. J. (2005) Science 310, 1686–1690 [DOI] [PubMed] [Google Scholar]
- 24.Sterne-Marr R., Tesmer J. J., Day P. W., Stracquatanio R. P., Cilente J. A., O'Connor K. E., Pronin A. N., Benovic J. L., Wedegaertner P. B. (2003) J. Biol. Chem. 278, 6050–6058 [DOI] [PubMed] [Google Scholar]
- 25.Carman C. V., Barak L. S., Chen C., Liu-Chen L. Y., Onorato J. J., Kennedy S. P., Caron M. G., Benovic J. L. (2000) J. Biol. Chem. 275, 10443–10452 [DOI] [PubMed] [Google Scholar]
- 26.Koch W. J., Hawes B. E., Inglese J., Luttrell L. M., Lefkowitz R. J. (1994) J. Biol. Chem. 269, 6193–6197 [PubMed] [Google Scholar]
- 27.Watts V. J., Neve K. A. (2005) Pharmacol. Ther. 106, 405–421 [DOI] [PubMed] [Google Scholar]
- 28.Reiter E., Marion S., Robert F., Troispoux C., Boulay F., Guillou F., Crepieux P. (2001) Biochem. Biophys. Res. Commun. 282, 71–78 [DOI] [PubMed] [Google Scholar]
- 29.Perroy J., Adam L., Qanbar R., Chénier S., Bouvier M. (2003) EMBO J. 22, 3816–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cant S. H., Pitcher J. A. (2005) Mol. Biol. Cell 16, 3088–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman J. L., Gonzalo P., Pitcher J. A., Claing A., Lavergne J. P., Reboud J. P., Lefkowitz R. J. (2002) Biochemistry 41, 12850–12857 [DOI] [PubMed] [Google Scholar]
- 32.Pitcher J. A., Freedman N. J., Lefkowitz R. J. (1998) Annu. Rev. Biochem. 67, 653–692 [DOI] [PubMed] [Google Scholar]
- 33.Stanasila L., Abuin L., Diviani D., Cotecchia S. (2006) J. Biol. Chem. 281, 4354–4363 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.