Abstract
The great success of therapeutic monoclonal antibodies has fueled research toward mimicry of their binding sites and the development of new strategies for peptide-based mimetics production. Here, we describe a new combinatorial approach for the production of peptidomimetics using the complementarity-determining regions (CDRs) from gastrin17 (pyroEGPWLEEEEEAYGWMDF-NH2) antibodies as starting material for cyclic peptide synthesis in a microarray format. Gastrin17 is a trophic factor in gastrointestinal tumors, including pancreatic cancer, which makes it an interesting target for development of therapeutic antibodies. Screening of microarrays containing bicyclic peptidomimetics identified a high number of gastrin binders. A strong correlation was observed between gastrin binding and overall charge of the peptidomimetic. Most of the best gastrin binders proceeded from CDRs containing charged residues. In contrast, CDRs from high affinity antibodies containing mostly neutral residues failed to yield good binders. Our experiments revealed essential differences in the mode of antigen binding between CDR-derived peptidomimetics (Kd values in micromolar range) and the parental monoclonal antibodies (Kd values in nanomolar range). However, chemically derived peptidomimetics from gastrin binders were very effective in gastrin neutralization studies using cell-based assays, yielding a neutralizing activity in pancreatic tumoral cell lines comparable with that of gastrin-specific monoclonal antibodies. These data support the use of combinatorial CDR-peptide microarrays as a tool for the development of a new generation of chemically synthesized cyclic peptidomimetics with functional activity.
Introduction
Antibody-based therapeutics have emerged as important components of therapies for an increasing number of debilitating and life-threatening diseases (1–3). The unique properties of antibodies provide a source of inspiration for active research in antibody engineering. Over the years, a wide range of antibody fragments (Fab, scFv)8 and variants (dia-, tria-, tetra-, mini-bodies, single-domain antibodies, intramers, etc.) have been developed (4–8), some of which are used today in clinical therapies (9, 10). One step further in downsizing the antibody molecule is to use peptides derived from one or more of the six hypervariable loops, or “complementarity-determining regions” (CDRs; Fig. 1A) (11). Mutational analysis of antibody-combining sites suggests that only a subset of interface contact residues is essential for binding (12, 13). Several publications have appeared since the first report on CDR-derived peptides (14), with bioactivities even approaching those of the parent antibodies in a few cases. Heap et al. (15) reported a cyclic 17-mer peptide derived from the H3 CDR of an anti-gp120 mAb with only 37-fold lower affinity (KD = 7.5 nm versus 0.2 nm for the mAb) and 32-fold lower HIV-1 neutralizing capacity. Some studies also use a rational design-based approach to make antibody-like binders, with remarkably high in vivo activities (16, 17).
FIGURE 1.
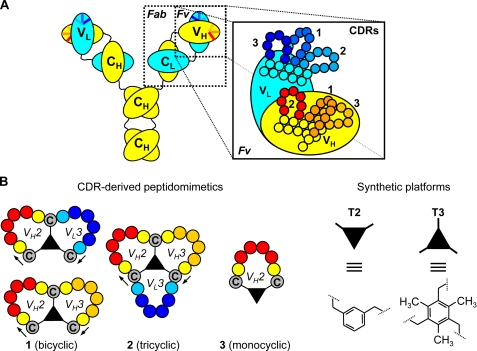
Structure of antibody and CDR-derived peptidomimetics. A, schematic representation of the protein domain structure in antibodies (constant heavy chain (CH), constant light chain (CL), variable heavy chain (VH), and variable light chain (VL)), with amplified structural details for the variable fragment (Fv: CDR1, CDR2, and CDR3 of VH and VL chain). B, monocyclic, bicyclic, and tricyclic peptidomimetics are depicted together with molecular details for the scaffolds (T2 and T3) used in Chemical LInkage of Peptides onto Scaffolds methodology.
In most cases, however, CDR-derived peptides showed a much lower activity than the parental antibodies, even though the 1:1 affinities were reported to be in the same range (18–20). For example, IC50 values for the inhibition of viral reproduction were 350-fold higher (∼7 μm) for a peptide derived from the anti-CD4 mAb ST40 (∼20 nm), whereas the reported affinity of the peptide for CD4 was only <3-fold lower than the antibody (KD = 900 pm versus 370 pm) (18). Similarly, partial inhibition of formation of an idiotypic mAb1·mAb2 complex (KD ∼1 nm) occurred only at 6.6 μm for the best peptide, whereas the reported difference in affinities was only ∼10 (19). Obviously, this raises concerns about potential differences in the antigen-binding mechanism between antibodies and corresponding mimics.
The peptide hormone gastrin is an important growth factor for gastric, pancreatic, and other gastrointestinal malignancies (21–25) through autocrine, paracrine, and endocrine mechanisms (26). Recently, gastrin has been described as an essential cofactor for gastric corpus carcinogenesis (27). Due to this fact, gastrin is considered an important therapeutic target for gastrointestinal cancers (28, 29). In fact, an anti-G17 vaccine, which is producing a significant increase in the survival time of patients, is being used in phase III clinical trials for pancreatic cancer and in phase II for colorectal and gastric cancer patients (30).
Here, we report the use of a synthetic combinatorial strategy for the production of CDR-derived peptidomimetics targeting the tumor antigen G17 (pyroEGPWLEEEEEAYGWMDF-NH2). We describe synthesis and high throughput screening of >10,000 mimetics from five anti-G17 antibodies with KD values ranging from 500 pm to >1 μm. The most active peptidomimetics neutralized G17 in an effective manner (IC50 ∼50 μm) in cell-based proliferation assays using colorectal Colo320 WT and pancreatic BxPc3 tumoral cells (31, 32).
EXPERIMENTAL PROCEDURES
Peptides and CDR Peptidomimetics
G17, G17 variants, and CDR peptidomimetics were provided by Pepscan Therapeutics (Lelystad, The Netherlands). T2 (α,α-dibromoxylene) and T3 (2,4,6-tris(bromomethyl)mesitylene) were purchased from Sigma.
Synthesis of Bicyclic Peptidomimetic for High Throughput Screening Studies
Synthesis of peptide microarrays on polypropylene support was performed as described previously (33, 34). After side chain deprotection using trifluoroacetic acid and scavengers, the microarrays were washed with excess of milliQ/H2O (five times for 10 min) and treated with a 0.5 mm solution of T3 in a 1:1 mixture of acetonitrile/NH4HCO3 (20 mm, pH 7.8) for 45–60 min to afford the corresponding chemical linkage of peptides onto scaffolds-peptides (format *CT(X)nCT(X)nCT-resin, where n = 4–6 and “CT” represents cysteines that are chemically linked via the T3 scaffold to two other CT values). Finally, the microarrays were washed with excess of acetonitrile/H2O, 1:1 (three times for 10 min), and sonicated in disrupt-buffer (1% SDS, 0.1% β-mercaptoethanol in PBS at 70 °C for 30 min, followed by sonication in milliQ/H2O for another 45 min). Binding studies with G17 were performed as described before (33, 34). Complete methods of bulk synthesis of linear peptides, backbone-cyclized peptides, and CDR-derived peptidomimics are shown as supplemental material.
Liquid Chromatography-Mass Spectrometry Characterization of 1–5 Mimetics
Chromatographic analysis of the peptides was performed on a C18 reversed-phase high performance liquid chromatography column, either using a DeltaPack C18 (3.9 × 150 mm (dxl), 5-μm particle size, 100-Å pore size; Waters) or an Atlantis C18 column (4.6 × 50 mm (dxl), 3-μm particle size; Waters) with a linear gradient of 5–65% solvent B in solvent A over 10 min (6%/min) (solvent A = 0.05% trifluoroacetic acid in H2O; solvent B = 0.05% trifluoroacetic acid in acetonitrile). Alternatively, analysis was performed on an Acquity UPLC (Waters) using an RP-18 preparative “BEH” column (2.1 × 50 inner diameter, 1.7-mm particle size, Waters) with a linear AB gradient (5–55% solvent B, 25% solvent B/min), where solvent A was 0.05% trifluoroacetic acid in water and solvent B was 0.05% trifluoroacetic acid in acetonitrile. The molecular weight of the peptides was determined by electrospray ionization on a Waters ZQ or a Quattro II SG (Micromass, UK) mass spectrometer. In all cases, the experimental and the calculated molecular mass (MM) of the peptides was equivalent (maximum variation <0.9 Da).
Preparation and Screening of Peptide/CDR Mimic Microarrays
Microarrays displaying CDR mimetics were prepared as described previously (33, 34). Screening for mAb binding was performed using known methodologies (34). G17 binding studies with CDR mimic arrays were performed using biotinylated G17 (pyroEGPWLEEEEEAYGWMDFK(biotin); 10 μg/ml) in PBS/Tween 80, followed by conjugation with streptavidin peroxidase (Southern Biotech; 1:1000 dilution in 5% bovine serum albumin/horse serum) following the standard procedure (34).
Surface Plasmon Resonance (SPR) Studies
HBS-EP (10 mm HEPES, pH 7.4, 150 mm NaCl, 3.4 mm NaEDTA, 0.005% surfactant P-20) was used as running and dilution buffer for experiments involving CDR mimics and PBS containing 0.005% surfactant P-20 for experiments with the mAbs/scFvs. CM5 sensor surfaces (BR-1000-14, Biacore AB) were used, and interactions were monitored at 25 °C. Cysteine-extended G17 was immobilized using thiol-coupling chemistry. The immobilization level of G17 and variants was adjusted to avoid mass transfer limitations as follows: less than 10 response units for analyzing interactions with the mAbs and above 1000 response units for the CDR mimics. The reference surface was treated similarly to the ligand surfaces, except that peptide injection was omitted. SPR responses of the peptidomimetics were expressed as percent of the theoretical binding capacity of the surfaces (%Rmax) assuming a 1:1 interaction (Rmax = Rligand·MManalyte/MMligand). This allowed us to normalize responses for differences in ligand immobilization levels (Rligand) and in MM of injected analytes (MManalyte).
Screening of CDR Mimics
Stock solutions of CDR mimics in H2O (1.0 mm) were diluted with HBS-EP to 1–100 μm. In case of solubility problems, stock solutions were prepared in 10–20 mm NaOH. Before injection, solutions were centrifuged and filtered through a 0.22-μm MILLEX GP Millipore filter. CDR mimics were injected at 20 μl/min flow rate. Sensor surfaces were regenerated with 10 μl of 50–100 mm HCl.
Determination of Binding Affinities
Analytes (mAb and CDR mimics) were injected over G17 and corresponding reference surfaces at 5–7 different concentrations with a 30-μl/min flow rate for 60 s (CDR mimics) and 120 s(mAb). The surfaces were regenerated by a 10-μl injection of 100 mm HCl. BIAevaluation 3.2 software was used for data evaluation.
G17-dependent Colo320 WT and BxPc3 Cell Proliferation Assays
Experiments were carried out following established procedures (35, 36). Briefly, Colo320 WT cells were plated onto 96-well tissue culture plates at a density of 104 cells/well in RPMI 1640 medium supplemented with 10% fetal bovine serum, antibiotics, and geneticin in duplicate. Once attached, cells were washed with PBS and cultured with different concentrations of peptidomimetics at five different concentrations (ranging 200 to 12.5 μm) in RPMI 1640 medium containing 5 × 10−10 mol/liter of G17, geneticin, and antibiotics for 72 h at 37 °C in 5% CO2. After removing the medium, cell proliferation was scored by staining cells with 100 μl of the chromogenic dye 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma) at a final concentration of 1 mg/ml in RPMI 1640 medium. The cells were further incubated for 1 h at 37 °C and 5% CO2. Then, 100 μl of DMSO (Sigma) were added to each well. Absorbance was read at 570 nm. Cell viability is represented as the ratio of absorbance between peptidomimetics-, scFv-, or mAb-treated cells and nontreated cells, expressed as a percentage. The same assay was used for BxPc3 pancreatic adenocarcinoma cell line with minor modifications. BxPc3 cells were cultured at a density of 5 × 103 cells per well in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. The next day, the cells were washed with PBS and cultured in Dulbecco's modified Eagle's medium containing antibiotics and the peptidomimetics, scFvs or mAbs, in the same conditions as the Colo320 WT cells. All the experiments were performed in duplicate and repeated three times.
RESULTS
In previous studies, we described the production of a wide collection of scFvs and monoclonal antibodies with gastrin neutralizing activity (35, 36). Here, microarrays of bicyclic peptidomimetics (Fig. 1B and Table 1) were prepared by using the CDR sequences of three anti-G17 murine mAbs (189DB3, 243BA5, and 23CA8), which bind G17 with Kd,1 between 0.54 and 11.3 nm, as measured by SPR (Fig. 2A), as starting material. Chemical synthesis of the peptidomimetics at the microarray surface was performed as follows. First, we synthesized linear peptides composed of two CDR fragments, separated by three cysteines (format, C-CDRA-C-CDRB-C-resin, where C represents cysteines interconnected via either a T3 or T2 scaffold; Fig. 2B). Subsequently, all three cysteines were chemically linked to a synthetic “platform” using enzyme-linked immunosorbent assay technology (Fig. 1B) (37), whereby the CDR fragments become cyclized to structurally mimic the CDRs as closely as possible. The microarrays cover every possible combination of peptide fragments (4–6-mers) derived from the H2, H3, and L3 CDR sequences (Fig. 2B). The total number of peptidomimetics generated in this way was 400–800 for each mAb.
TABLE 1.
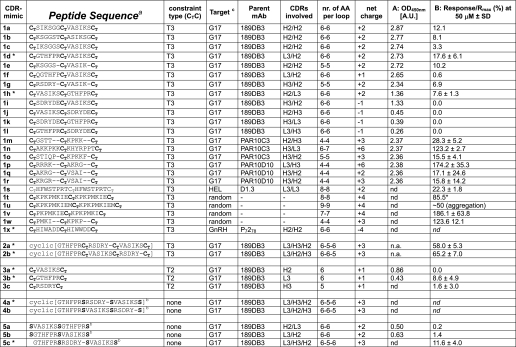
Amino acid sequence, PEPSCAN, and SPR data for CDR peptidomimetics
Sequence information and binding data for G17 CDR 1–5 peptidomimetics are provided. Column A, PEPSCAN, shows peptide screening, and column B shows SPR. nr indicates number.
a CT represents cysteines interconnected via either a T3 or a T2 scaffold; all linear peptides were acetylated at the N terminus and have a C-terminal amide (CONH2); “c[XXXX]” represents backbone-cyclized peptides.
b S represents serines substituting (T3-connected) cysteines in order to study the influence of T3/T2 on G17 binding.
c G17 indicates pyro-EGPWLEEEEEAYGWMDF; GnRH indicates pyro-EHWSYGLRPG (gonadotropin-releasing hormone); HEL indicates hen egg white lysozyme; nd indicates not determined; na indicates not applicable. *, peptidomimetics tested in in vitro neutralization assays.
FIGURE 2.
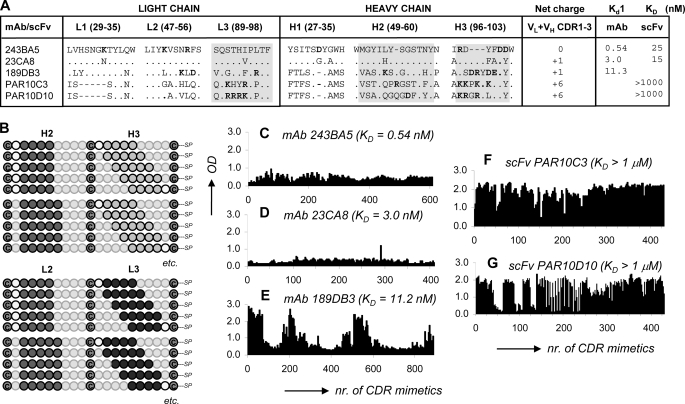
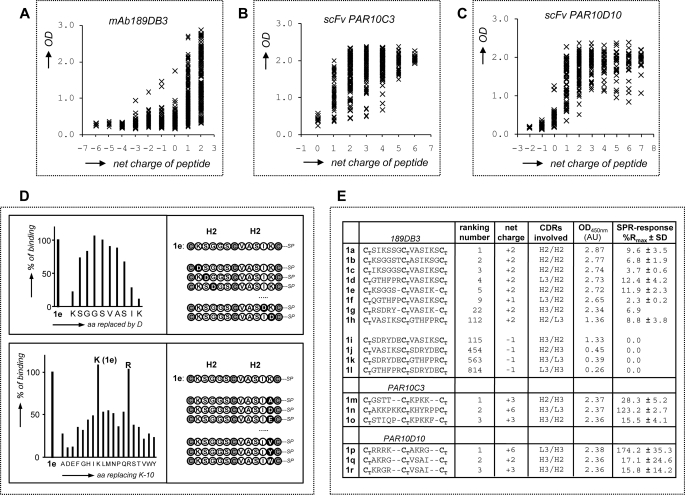
High throughput G17 binding data for microarrays of overlapping CDR-derived bicyclic peptidomimetics. A, amino acid sequences for the CDRs of five anti-G17 mAbs/scFvs + G17 affinity data as determined by SPR or enzyme-linked immunosorbent assay (only PAR10C3); gray areas indicate CDR regions covered in microarray synthesis. B, experimental design of microarrays with overlapping bicyclic peptidomimetics (surface-immobilized) derived from the H2, H3, and L3 CDRs of the five anti-G17 antibodies. C–G, A450 nm values for G17 binding to bicyclic peptidomimetics derived from mAb 243BA5 (C), mAb 23CA8 (D), mAb 189DB3 (E), scFv PAR10C3 (F), and scFv PAR10D10 (G). Binding studies were performed with 10 μg/ml of biotinylated G17 (pyroEGPWLEEEEEAYGWMDFK(biotin), followed by conjugation using peroxidase-labeled streptavidin at 1:1000 dilution. nr, number.
G17 Binding of Peptidomimetics Derived from Anti-G17 mAbs
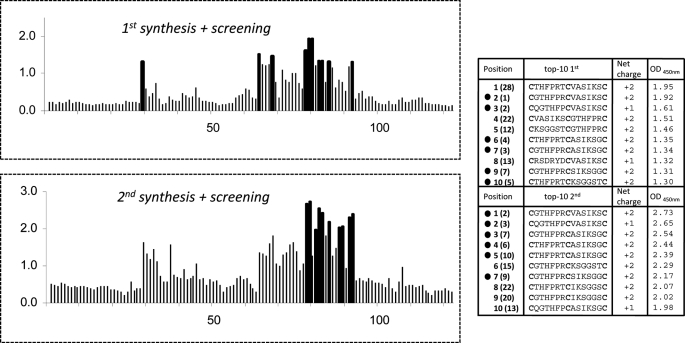
Screening of the microarrays for G17 binding showed that the number of hits differed substantially among the parent antibodies (Fig. 2, C–E). Little to no binding (A450 nm <1.0) was observed for peptidomimetics derived from the CDRs of mAbs 23CA8 and 243BA5 (Fig. 2, C and D), most notably, those with the highest affinities for G17 (Kd1 = 3.0 and 0.54 nm). Instead, 15–20% of the peptidomimetics derived from mAb 189DB3 (Kd1 = 11.3 nm) showed strong binding of G17 (A450 nm >2.0; Fig. 2E). Sequence analysis showed that strong binders were mostly derived from the H2 (VASIKSGGST) and L3 (VQGTHFPRTF) CDRs, whereas H3 CDR (RSDRYDEDY) mainly gave rise to nonbinders. Reproducibility of the chemical synthesis and G17 screening procedure was high. Resynthesis and screening of a subset (∼125) of the 189DB3-library showed excellent correlation with the original data (Fig. 3). Six out of 10 of the best binders of the duplicated dataset were also present in the top 10 of the original set and vice versa. Moreover, 10 of 10 best binders in each set were among the 28 best binders of the other dataset. Control binding studies with linear (no T3 present, cysteines replaced by serines) or single loop mimetics (C-CDR-C-resin; T2 instead of T3) clearly showed that G17 binding was significantly weakened (A450 nm <0.5 and <1.0, respectively).
FIGURE 3.
Sequences, overall charges, and A450 nm values for a subset (125 peptides) of the 189DB3 microarray that was synthesized in duplicate to address the reproducibility of the microarray data. Bars corresponding to the top 10 binders in each set are in black to emphasize their position. Peptides indicated with ● (6 of 10) were also present in the top 10 binders of the duplicate set. Numbers next to ● indicate the top 10 binders according to their A450 nm; parentheses indicate their position in the other assay.
Correlation between Gastrin Binding and Overall Charge of the Peptidomimetics
Further analysis of the 189DB3 dataset revealed a strong correlation between binding and the overall charge of the peptidomimetics (Fig. 4A). The majority of strong binders (99% with A450 nm >1.0) carry an overall positive charge (+1 or +2). The five strongest G17 binders of the 189DB3 dataset all carry a +2 charge (Fig. 4E), the highest possible charge in this library (885 compounds of which 18.1% have +2, 29.6% +1, 18.6% have 0, and 33.8% have −1 to −6 net charge). In sharp contrast to this, the worst five binders in this set were either negatively charged (3 of 5) or neutral (2 of 5). The importance of charge in G17 binding also explains why peptidomimetics derived from the CDRs of either 243BA5 or 23CA8 did not bind G17 (Fig. 2, C and D). In these libraries, not a single mimetic from the >1000 peptidomimetics investigated carries a net positive charge.
FIGURE 4.
Analysis of G17 binding strength of CDR-derived bicyclic peptidomimetics. A–C, correlation of G17 binding levels (A450 nm values) to overall charge of bicyclic peptidomimetics derived from mAb 189DB3 (A), scFv PAR10C3 (B), and scFv PAR10D10 (C). D, complete Asp scan and Lys-10 replacement analysis of bicyclic peptidomimetic 1e (CTKSGGSCTVASIKCT-T3) (derived from mAb 189DB3), showing the crucial role of charge in G17 recognition. E, comparison of G17 binding levels (A450 nm) in PEPSCAN microarrays and SPR (%Rmax, being 100% Rmax when Req = Rmax,calc) in relation to overall charge for some of the best and worst G17 binders derived from mAb 189DB3, scFv PAR10C3, and scFv PAR10D10.
Efforts to correlate G17 binding to the presence of single amino acid types clearly showed that A450 nm values increased with the number of lysines and decreased with the number of aspartic acids, with little influence of noncharged amino acids. However, the number of arginines failed to correlate well with G17 binding (data not shown), which seems due to the fact that two of three of the arginines in the 189DB3 dataset are located within H3 CDR (97RSDRYDEDY105) including four acidic (Asp/Glu) residues. To address this issue in more detail, we synthesized a microarray of ∼1000 peptidomimetics derived from scFvs PAR10C3 and PAR10D10 that have only weak affinity for G17 (KD for G17 ≥1 μm), but the H3 and L3 CDRs of which contain 5–8 Lys/Arg residues and no Asp/Glu residues (Fig. 2A) (38). As expected, >90% of the peptidomimetics showed strong binding to G17 (Fig. 2, F and G) with a clearly positive correlation between A450 nm values, overall charges, and number of Lys/Arg residues present (Fig. 4, B and C).
Replacement studies for peptidomimetics with high affinity for G17 (top 5, see Fig. 4D) also confirm the importance of charge for G17 binding. For example, replacement of each residue in CTKSGGSCTVASIKCT-resin (1e) by Asp strongly impeded binding for Lys-1, Lys-10, and Ile-9 (>90, 80, and 70% binding decrease, respectively), but little for amino acids more remote (i.e. >2 residues) from these (<20% decrease in binding for Gly-4 to Ala-7). Replacement of Lys-10 in 1e by the other 19 natural amino acids showed the strongest effect for replacement by Asp and Glu (∼90% reduction), a lesser effect for other amino acids (50–80%), and none for Arg.
Soluble CDR-derived Peptidomimetics Bind Gastrin 17
We also studied binding of a representative set of soluble CDR-derived peptidomimetics to surface-immobilized G17 using SPR (Fig. 4E). A total of 12 soluble peptidomimetics derived from 189DB3 was synthesized (supplemental material), including the top 5 binders in microarrays (1a–e) plus 7 different CDR combinations (1f–1l). Measurable binding at 50 μm was clearly observed for all mimetics with +2 charge (1a–e and 1g–h; %Rmax between 3.3 and 12.4). Mimetic 1f with +1 charge showed only weak binding (%Rmax = 2.3), even though it bound strongly to G17 (A450 nm = 2.65) when immobilized at the microarray surface. The requirement for a positive charge would also explain the absence of binding to G17 in SPR (%Rmax = 0) for mimic 1i, one of the few mimics with charge <0 that showed G17 binding in microarrays. Like 1i, none of the other three mimetics (1j–l) with −1 charge showed any binding to surface-immobilized G17 in SPR. Together, these data confirm G17 binding for 7 of 9 peptidomimetics derived from 189DB3, whereas 2 of 9 binders investigated appeared to be false positives.
Binding to surface-immobilized G17 in SPR was also confirmed for the top 3 binders of the PAR10C3 and PAR10D10 library (1m–1r; Fig. 4E). These data again highlighted the importance of overall charge, but the data also visualized large differences in G17 binding strength among the best G17 binding mimics, most likely as a result of solid-phase effects (multivalency). Moreover, it was found that this binding was decreased by 15–50% upon increasing the NaCl concentration from 150 to 400 mm, which hints at significant electrostatic interactions.
G17 Binding Affinity Evaluation by SPR
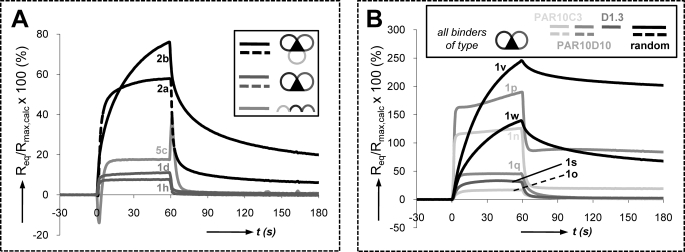
In theory, 1:1 binding affinities (KD values) can be evaluated by SPR only if the injected compounds are homogeneous and monomeric. However, most binding curves typically showed complex kinetics (multiphasic and slow injection/post-injection phases, Req >100% Rmax), indicative of the presence of heterogeneous material and/or multimeric interactions (Fig. 5A and B, and Table 1). Such binding curves were observed for bicyclic mimetic 1 (n/p/v/w; Fig. 5B) and tricyclic mimetics 2a and 2b (Fig. 5A). The latter cover H2, H3, and L3 CDR loop sequences of 189DB3 and have a cyclic backbone rather than a linear backbone (for 1d/1h; Fig. 1B).
FIGURE 5.
SPR binding studies with CDR-derived peptidomimetics. A, SPR curves (%Rmax versus time, being 100% Rmax when Req = Rmax,calc) for G17 binding of 50 μm bicyclic peptidomimetics 1d and 1h, tricyclic mimetics 2a and 2b, and linear control 5c, all derived from mAb 189DB3. B, SPR curves for G17 binding of bicyclic peptidomimetics derived from scFv PAR10C3 (1n/1o), scFv PAR10D10 (1p/1q), anti-egg white lysozyme mAb D1.3 (1s), and random bicyclic mimetics 1v and 1w.
Binding curves for the +2 charged mimetics 1d and 1h (189DB3), 1q (PAR10D10), and 1s (D1.3) were typical of low affinity 1:1 interaction/fast complex formation and dissociation with stable equilibrium responses (Req). Theoretically, KD values can be calculated from the dependence of Req and [bycyclic peptidomimetics], but data fitting approaches like these are tentative when Req is still far below surface saturation (<25% Rmax) at [bycyclic peptidomimetics]max. Therefore, KD values in the 100–500 μm range were estimated for 1d and 1h by comparing experimental and simulated responses (data not shown). SPR evaluation of the linear peptides 5a–c and monocycles 3a–c (linear and single loop controls of 1d/h and 2a/2b) revealed that only 5c (H3/H2/L3 CDRs; +3 charge) and 3c (L3 CDR only; +1 charge) showed measurable responses, with KD values in the 100 μm (5c) to 1 mm (3a) range (Table 1). Interaction of the other controls with G17 was hardly detectable (<1.5% Rmax).
Evaluation of G17 Neutralizing Activity in BxPc3 and Colo320 WT Cell-based Assays
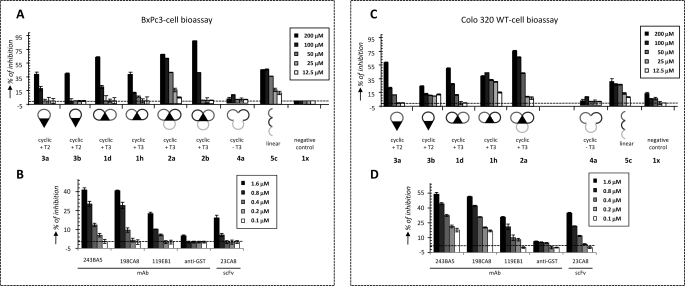
Neutralizing activities of peptidomimetics 1–5 for G17 were evaluated in two different G17-dependent cell proliferation assays (Colo320 WT and BxPc3 cell-based assay; Fig. 6) and compared with previous data obtained with scFv and mAbs (35, 36). Experiments were performed at five different concentrations, ranging from 200 to 12.5 μm. Of all peptidomimetics 1–5, the tricyclic mimetic 2a (+3 charge) showed the highest neutralizing activity in both assays (Fig. 6, A and C). The observed dose response was close to perfect, and the IC50 values of ∼50 μm in both assays were only 30-fold higher than for the high affinity mAbs 243BA5 and 198CA8 (IC50 = 1.6 μm) (Fig. 6, B and D). Most striking was the fact that removal of the T3 central core in 2a, completely abolished the activity (see 4a in Fig. 6). The observed effect was equally strong in both assays. Interestingly, mimic 2b displayed a higher tendency for aggregate formation by SPR than 2a (Fig. 5A). Also, peptide 5c, the linear variant of 2a, showed activity in both assays, despite the absence of T3. However, the dose responses for 5c were poorer than 2a, for example.
FIGURE 6.
G17 neutralization studies with CDR-derived peptidomimetics in comparison with anti-G17 scFvs and mAbs. A and C, G17-neutralizing activities of 189DB3-derived bicyclic mimetics 1d/1h, tricyclic mimetics 2a/2b, monocyclic mimetics 3a/3b, cyclic T3-less mimetic 4a, and linear mimetic 5 at five different concentrations (200–12.5 μm) using Gonadotropin-releasing hormone-derived bicyclic peptidomimetic 1x (−4 charge) as negative control. B and D, G17 neutralizing activities of anti-G17 scFv and mAbs at five different concentrations (1.6–0.1 μm) using anti-GST mAb as negative control. G17 neutralization studies BxPc3 (A and B) and Colo320 WT cell-based bioassays (C and D). Media and standard deviation of the assays are represented in the figure. B and D, data were adapted from Refs. 35, 36.
Monocyclic peptidomimetics 3a and 3b showed neutralizing activities only emerging at 200 μm. Bicyclic 1h exhibited activity in the Colo320 WT cell assay down to 25 μm, but a typical dose response was not seen. In any case, the difference in activity between mono-cycles and bi-cycles peptidomimetics was marginal. T3-less cyclic peptidomimetics 4a/b were not active either in Colo320 WT or BxPc3 cell assays. Negative control peptide 1x (−4 charge) was totally silent in both assays, confirming the importance of positive charge also for G17 neutralizing activity.
DISCUSSION
Peptidomimetics consisting of one or more CDRs represent the ultimate example of a miniaturized antibody (20, 39–42). Antibodies are depicted as being oversized for the mere purpose of antigen binding, and it has been suggested that the antibody-binding site could be downsized significantly without major loss of binding affinity. This view is supported strongly by virus-neutralizing data and tumor mouse models, which highlighted the potential therapeutic and diagnostic value of this new class of drugs (15, 16, 43, 44). However, little is known about the exact mechanism of production or whether CDR-derived peptidomimetics bind the antigen in a similar or totally different mode as compared with the parent antibody. The high throughput synthetic approach (>10,000 compounds) in combination with the use of a small peptide antigen (G17) and systematic amino acid replacements facilitated the binding analysis to the level of individual amino acids.
Screening of microarrays with bicyclic peptidomimetics showed that binding of G17 occurred exclusively for mimics with multiple cationic residues (Arg/Lys) and a net charge >0. It seemed likely at first sight that these mimics recognize the acidic G17 (−6 charge) mainly via electrostatic attraction. A very similar mode of binding was observed for scFvs with low affinity for G17 (KD >1 μm) (38). The high affinity antibodies seem to bind G17 via a totally different mechanism. Electrostatic interactions do not play a major role here, as judged from the fact that residues most essential to binding (pyroEGPWL) were all neutral (35). Moreover, the antibody CDRs include an almost equal number of basic and acidic residues.
This mode of binding is not easily mimicked with CDR-derived peptidomimetics, as deduced from the fact that none of the peptidomimetics derived from mAbs 23CA8 and 243BA5 showed appreciable affinity for G17 (Fig. 2C). It is expected that shape complementarity and specific H-bonding patterns require a higher level of structural organization, where even small changes will have high impact on binding. In this sense, the grafting of CDR loops onto a synthetic scaffold can be compared with conventional CDR grafting for antibody humanization purposes. The latter often leads to a reduction in affinity (45), which has been associated with subtle conformational effects within framework regions in the periphery of the antibody-combining site (46). Transplantation of CDR fragments onto a synthetic scaffold is clearly more radical than grafting onto a structurally homologous framework. It is therefore obvious that highly specific, short range interactions will be lost and that, at best, only the less specific electrostatic and/or hydrophobic driving forces will be preserved. The latter can nevertheless provide a basic level of affinity that can be enhanced by high throughput screening and selection. The intrinsic conformational flexibility of peptidic loops may even act favorably in this regard.
The CDR-derived peptidomimetics can be envisaged as an exponent of primarily charge-driven binders. They seem to recognize essentially the presence and number of certain amino acid types (charged, aromatic, and hydrophobic) rather than well defined amino acid combinations. The multimeric nature of the peptidomimetic·G17 complexes also suggests a clear difference in binding mechanism. In microarrays, G17 binding is likely the result of multivalent interactions, as judged from slow dissociations (G17 binding was detectable after three 30-s washes). In case of low affinity 1:1 complexes, binding should be lost completely within seconds. Additional evidence comes from SPR studies, where the observed slow dissociation kinetics (kd ≤10−2 s−1) could suggest high affinity 1:1 binding (Fig. 5B). However, when high affinity complexes are formed, surface saturation is expected within seconds upon injection of bicyclic peptidomimetics at 50 μm. The fact that this was not observed further supports the occurrence of multimeric aggregates with poorly defined structures and stoichiometries.
The results of our studies show interesting parallels with previously reported data, suggesting a general mechanism that can be used for other antigens. For example, the HIV-neutralizing “microantibody” (CDLIYYDYEEDYYFDYC; 7 Trp/Tyr/Phe, 6 Asp/Glu, 0 Arg/Lys, and 2 Leu/Ile, 17 residues in total) described by Heap et al. (15) shows remarkably close resemblance to our target antigen G17 (4 Trp/Tyr/Phe, 6 Asp/Glu, 0 Arg/Lys, and 2 Leu/Met, 17 in total). Moreover, the target epitope sequence on gp120 of HIV-1 (KRXXXIGPGR) is highly reminiscent of some of the top 5 G17 binders (Fig. 4E). Similarly, peptides reported by Greene and co-workers (41, 42) to block gp120 binding to CD4 (e.g. FCYICEVEDQCY; (3 Tyr/Phe, 3 Asp/Glu, 0 Arg/Lys, and 1 Ile) or inhibit proliferation of overexpressing tumor cells (e.g. “aromatically modified” FCDGFYACYMDV; (4 Tyr/Phe, 2 Asp, 0 Arg/Lys, and 2 Val/Met) also have similar constitutions. Finally, a series of potent synthetic receptors for platelet-derived growth factor and vascular endothelial growth factor displaying very strong activity in tumor suppression were described (16, 17, 43). The composition of the peptide loops (GDGY in GFB-111; GKGK in GFB-116) in combination with the calix[4]arene scaffold used here (four “Tyr-like” aromatic rings) highlights the crucial presence of aromatic and acidic or basic residues (16, 17). Moreover, many of these systems may also involve structurally ill defined complexes that are multivalent in nature. So far, the molecular structure of complexes resolved by x-ray or NMR has not been reported, which supports this view. Regarding the close analogy between these systems and the one described here, it is likely that similar binding mechanisms apply. The assumption that stable complexes reflect high affinity 1:1 interactions might lead to the incorrect conclusion that such mimics are indeed very good representatives of the parent antibodies (18).
In summary, we report the first example of CDR-derived peptidomimetics with neutralizing and in vitro anti-tumoral activity. The synthetically derived peptidomimetics were extremely effective in their gastrin neutralizing activity, despite their relative low affinity. IC50 values were only 30-fold higher (50 versus 1.6 μm) than for the most active high affinity mAbs (243BA5 and 198CA8) (35). However, our studies revealed essential differences in their binding. Given that the high throughput screening procedure will likely select affinity contributions of low specificity (hydrophobic and electrostatic interactions), it may be desirable to focus in consecutive rounds on specificity-enhancing modifications. CDR sequences might be regarded as a good starting point but by no means should be considered an end point. The results described in this study provide a useful new tool for future design of antibody mimetics and pave the way to their clinical application in the treatment of pancreatic and other gastric tumors.
Supplementary Material
Acknowledgment
We thank R. Boshuizen for help with analyzing the G17 screening results.
This work was supported in part by European Commission FP6 Cooperative Research Project COOP-CT-2004-512691.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures.”
- scFv
- single-chain variable fragment
- CDR
- complementarity-determining region
- mAb
- monoclonal antibody
- MM
- molecular mass
- G17
- gastrin17
- SPR
- surface plasmon resonance
- PBS
- phosphate-buffered saline
- WT
- wild type
- HIV-1
- human immunodeficiency virus, type 1
- T2
- α,α-dibromoxylene
- T3
- 2,4,6-tris(bromomethyl)mesitylene.
REFERENCES
- 1.Adams G. P., Weiner L. M. (2005) Nat. Biotechnol. 23, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 2.Carter P. J. (2006) Nat. Rev. Immunol. 6, 343–357 [DOI] [PubMed] [Google Scholar]
- 3.Reichert J. M., Rosensweig C. J., Faden L. B., Dewitz M. C. (2005) Nat. Biotechnol. 23, 1073–1078 [DOI] [PubMed] [Google Scholar]
- 4.Dumoulin M., Last A. M., Desmyter A., Decanniere K., Canet D., Larsson G., Spencer A., Archer D. B., Sasse J., Muyldermans S., Wyns L., Redfield C., Matagne A., Robinson C. V., Dobson C. M. (2003) Nature 424, 783–788 [DOI] [PubMed] [Google Scholar]
- 5.Famulok M., Blind M., Mayer G. (2001) Chem. Biol. 8, 931–939 [DOI] [PubMed] [Google Scholar]
- 6.Haupt K., Mosbach K. (1998) Trends Biotechnol. 16, 468–475 [DOI] [PubMed] [Google Scholar]
- 7.Holliger P., Hudson P. J. (2005) Nat. Biotechnol. 23, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 8.Revets H., De Baetselier P., Muyldermans S. (2005) Expert Opin. Biol. Ther. 5, 111–124 [DOI] [PubMed] [Google Scholar]
- 9.Hayden E. C. (2008) Nat. Med. 14, 108–108 [DOI] [PubMed] [Google Scholar]
- 10.Kourlas H., Abrams P. (2007) Clin. Ther. 29, 1850–1861 [DOI] [PubMed] [Google Scholar]
- 11.Kieber-Emmons T., Murali R., Greene M. I. (1997) Curr. Opin. Biotechnol. 8, 435–441 [DOI] [PubMed] [Google Scholar]
- 12.Muller Y. A., Chen Y., Christinger H. W., Li B., Cunningham B. C., Lowman H. B., de Vos A. M. (1998) Structure 6, 1153–1167 [DOI] [PubMed] [Google Scholar]
- 13.Braden B. C., Goldman E. R., Mariuzza R. A., Poljak R. J. (1998) Immunol. Rev. 163, 45–57 [DOI] [PubMed] [Google Scholar]
- 14.Kang C. Y., Brunck T. K., Kieber-Emmons T., Blalock J. E., Kohler H. (1988) Science 240, 1034–1036 [DOI] [PubMed] [Google Scholar]
- 15.Heap C. J., Wang Y., Pinheiro T. J., Reading S. A., Jennings K. R., Dimmock N. J. (2005) J. Gen. Virol. 86, 1791–1800 [DOI] [PubMed] [Google Scholar]
- 16.Blaskovich M. A., Lin Q., Delarue F. L., Sun J., Park H. S., Coppola D., Hamilton A. D., Sebti S. M. (2000) Nat. Biotechnol. 18, 1065–1070 [DOI] [PubMed] [Google Scholar]
- 17.Sun J., Blaskovich M. A., Jain R. K., Delarue F., Paris D., Brem S., Wotoczek-Obadia M., Lin Q., Coppola D., Choi K., Mullan M., Hamilton A. D., Sebti S. M. (2004) Cancer Res. 64, 3586–3592 [DOI] [PubMed] [Google Scholar]
- 18.Casset F., Roux F., Mouchet P., Bes C., Chardes T., Granier C., Mani J. C., Pugnière M., Laune D., Pau B., Kaczorek M., Lahana R., Rees A. (2003) Biochem. Biophys. Res. Commun. 307, 198–205 [DOI] [PubMed] [Google Scholar]
- 19.Laune D., Molina F., Mani J., Del Rio M., Bouanani M., Pau B., Granier C. (2000) J. Immunol. Methods 239, 63–73 [DOI] [PubMed] [Google Scholar]
- 20.Takasaki W., Kajino Y., Kajino K., Murali R., Greene M. I. (1997) Nat. Biotechnol. 15, 1266–1270 [DOI] [PubMed] [Google Scholar]
- 21.Singh P., Owlia A., Varro A., Dai B., Rajaraman S., Wood T. (1996) Cancer Res. 56, 4111–4115 [PubMed] [Google Scholar]
- 22.Singh P., Walker J. P., Townsend C. M., Jr., Thompson J. C. (1986) Cancer Res. 46, 1612–1616 [PubMed] [Google Scholar]
- 23.Hur K., Kwak M. K., Lee H. J., Park D. J., Lee H. K., Lee H. S., Kim W. H., Michaeli D., Yang H. K. (2006) J. Cancer Res. Clin. Oncol. 132, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith J. P., Fantaskey A. P., Liu G., Zagon I. S. (1995) Am. J. Physiol. 268, R135–141 [DOI] [PubMed] [Google Scholar]
- 25.Smith J. P., Shih A., Wu Y., McLaughlin P. J., Zagon I. S. (1996) Am. J. Physiol. 270, R1078–R1084 [DOI] [PubMed] [Google Scholar]
- 26.Watson S. A., Durrant L. G., Morris D. L. (1988) Br. J. Surg. 75, 342–345 [DOI] [PubMed] [Google Scholar]
- 27.Takaishi S., Tu S., Dubeykovskaya Z. A., Whary M. T., Muthupalani S., Rickman B. H., Rogers A. B., Lertkowit N., Varro A., Fox J. G., Wang T. C. (2009) Am. J. Pathol. 175, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilliam A. D., Watson S. A., Henwood M., McKenzie A. J., Humphreys J. E., Elder J., Iftikhar S. Y., Welch N., Fielding J., Broome P., Michaeli D. (2004) Eur. J. Surg. Oncol. 30, 536–543 [DOI] [PubMed] [Google Scholar]
- 29.Brett B. T., Smith S. C., Bouvier C. V., Michaeli D., Hochhauser D., Davidson B. R., Kurzawinski T. R., Watkinson A. F., Van Someren N., Pounder R. E., Caplin M. E. (2002) J. Clin. Oncol. 20, 4225–4231 [DOI] [PubMed] [Google Scholar]
- 30.Gilliam A. D., Watson S. A. (2007) Expert Opin. Biol. Ther. 7, 397–404 [DOI] [PubMed] [Google Scholar]
- 31.Yu H. G., Schrader H., Otte J. M., Schmidt W. E., Schmitz F. (2004) Biochem. Pharmacol. 67, 135–146 [DOI] [PubMed] [Google Scholar]
- 32.Tan M. H., Nowak N. J., Loor R., Ochi H., Sandberg A. A., Lopez C., Pickren J. W., Berjian R., Douglass H. O., Jr., Chu T. M. (1986) Cancer Invest. 4, 15–23 [DOI] [PubMed] [Google Scholar]
- 33.Timmerman P., Puijk W. C., Meloen R. H. (2007) J. Mol Recognit. 20, 283–299 [DOI] [PubMed] [Google Scholar]
- 34.Timmerman P., Van Dijk E., Puijk W., Schaaper W., Slootstra J., Carlisle S. J., Coley J., Eida S., Gani M., Hunt T., Perry P., Piron G., Meloen R. H. (2004) Mol. Divers. 8, 61–77 [DOI] [PubMed] [Google Scholar]
- 35.Barderas R., Shochat S., Timmerman P., Hollestelle M. J., Martínez-Torrecuadrada J. L., Höppener J. W., Altschuh D., Meloen R., Casal J. I. (2008) Int. J. Cancer 122, 2351–2359 [DOI] [PubMed] [Google Scholar]
- 36.Barderas R., Desmet J., Timmerman P., Meloen R., Casal J. I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9029–9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmerman P., Beld J., Puijk W. C., Meloen R. H. (2005) Chembiochem 6, 821–824 [DOI] [PubMed] [Google Scholar]
- 38.Barderas R., Shochat S., Martínez-Torrecuadrada J., Altschuh D., Meloen R., Ignacio Casal J. (2006) J. Immunol. Methods 312, 182–189 [DOI] [PubMed] [Google Scholar]
- 39.Laune D., Molina F., Ferrieres G., Mani J. C., Cohen P., Simon D., Bernardi T., Piechaczyk M., Pau B., Granier C. (1997) J. Biol. Chem. 272, 30937–30944 [DOI] [PubMed] [Google Scholar]
- 40.Monnet C., Laune D., Laroche-Traineau J., Biard-Piechaczyk M., Briant L., Bès C., Pugnière M., Mani J. C., Pau B., Cerutti M., Devauchelle G., Devaux C., Granier C., Chardès T. (1999) J. Biol. Chem. 274, 3789–3796 [DOI] [PubMed] [Google Scholar]
- 41.Park B. W., Zhang H. T., Wu C., Berezov A., Zhang X., Dua R., Wang Q., Kao G., O'Rourke D. M., Greene M. I., Murali R. (2000) Nat. Biotechnol. 18, 194–198 [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Piatier-Tonneau D., Auffray C., Murali R., Mahapatra A., Zhang F., Maier C. C., Saragovi H., Greene M. I. (1996) Nat. Biotechnol. 14, 472–475 [DOI] [PubMed] [Google Scholar]
- 43.Sun J., Wang D. A., Jain R. K., Carie A., Paquette S., Ennis E., Blaskovich M. A., Baldini L., Coppola D., Hamilton A. D., Sebti S. M. (2005) Oncogene 24, 4701–4709 [DOI] [PubMed] [Google Scholar]
- 44.Qiu X. Q., Wang H., Cai B., Wang L. L., Yue S. T. (2007) Nat. Biotechnol. 25, 921–929 [DOI] [PubMed] [Google Scholar]
- 45.Roguska M. A., Pedersen J. T., Henry A. H., Searle S. M., Roja C. M., Avery B., Hoffee M., Cook S., Lambert J. M., Blättler W. A., Rees A. R., Guild B. C. (1996) Protein Eng. 9, 895–904 [DOI] [PubMed] [Google Scholar]
- 46.Foote J., Winter G. (1992) J. Mol. Biol. 224, 487–499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.