Abstract
The glucagon-like peptide 1 (GLP1) receptor is a member of Family B G protein-coupled receptors and represents an important drug target for type 2 diabetes. Despite recent solution of the structure of the amino-terminal domain of this receptor and that of several close family members, understanding of the molecular basis of natural ligand GLP1 binding to its intact receptor remains limited. The goal of this study was to explore spatial approximations between specific receptor residues within the carboxyl terminus of GLP1 and its receptor as normally docked. Therefore, we developed and characterized two high affinity, full-agonist photolabile GLP1 probes having sites for covalent attachment in positions 24 and 35. Both probes labeled the receptor specifically and saturably. Subsequent peptide mapping using chemical and proteinase cleavages of purified wild-type and mutant GLP1 receptor identified that the Arg131–Lys136 segment at the juxtamembrane region of the receptor amino terminus contained the site of labeling for the position 24 probe, and the specific receptor residue labeled by this probe was identified as Glu133 by radiochemical sequencing. Similarly, nearby residue Glu125 within the same region of the receptor amino-terminal domain was identified as the site of labeling by the position 35 probe. These data represent the first direct demonstration of spatial approximation between GLP1 and its intact receptor as docked, providing two important constraints for the modeling of this interaction. This should expand our understanding of the molecular basis of natural agonist ligand binding to the GLP1 receptor and may be relevant to other family members.
Introduction
G protein-coupled receptors (GPCRs)2 are the largest group of membrane receptors with seven transmembrane domains and represent targets for over 30% of approved drugs. Understanding of the molecular basis of ligand binding and activation of these receptors will facilitate the development and refinement of drugs acting at these targets. Currently, the molecular basis of ligand binding and activation of Family B GPCRs is less well understood than that of the larger Family A, where high resolution crystal structures have recently been described for several members (1–4).
A characteristic structural feature of this family is a long and structurally complex extracellular amino-terminal domain containing six conserved cysteine residues that form disulfide bonds important for stabilizing the folded structure. This domain has been suggested to interact with the carboxyl-terminal region of the natural peptide ligands, based on structure-activity relationship, site-directed mutagenesis, and chimeric receptor studies (5–10), and this is a consistent theme throughout their family.
Insights into the structure of the predominant ligand binding domain, the amino terminus of the Family B GPCRs, have substantially advanced with the solution of NMR and crystal structures of the isolated ligand-bound amino terminus of the receptors for corticotrophin-releasing factor (11–13), pituitary adenylate cyclase-activating peptide (14), gastric inhibitory polypeptide (GIP) (15), glucagon-like peptide 1 (GLP1) (16), and parathyroid hormone (PTH) (17). Most of these ligands represented amino-terminally truncated hormones or analogues of these hormones. The GIP receptor structure included its natural agonist peptide (15). In all these structures, the carboxyl-terminal regions of the ligands were present within a binding cleft of the amino terminus of the receptor. However, inconsistencies are present in these structures, both in the absolute site of ligand binding and in the positioning of the ligand in place in the binding cleft. This suggests that the mechanisms proposed for natural peptide ligand binding to these receptors may be distinct, with no consistent experimental basis for the proposed intact receptor structures.
Photoaffinity labeling is a technique for directly establishing residue-residue spatial approximation between a docked ligand and its receptor. It is particularly powerful for study of the sparse and hydrophobic G protein-coupled receptors, which continue to be extremely challenging to study by more direct, high-resolution physicochemical methods. The GLP1 receptor is a Family B GPCR that has a natural agonist ligand that is a gluco-incretin hormone. GLP1 plays an important physiological role in maintaining blood glucose homeostasis, and it holds promise as a very effective therapeutic drug for the treatment of type 2 diabetes. Therefore, we performed photoaffinity labeling on the intact GLP1 receptor with probes incorporating photolabile residues in the carboxyl-terminal region of the GLP1 peptide to test, confirm, and refine the molecular basis of agonist ligand binding to their receptor.
In this work, we used two GLP1 probes incorporating a photolabile p-benzoyl-l-phenylalanine (Bpa) in the carboxyl-terminal region of the peptide, in positions 24 and 35. Both probes were full agonists and bound to and labeled the receptor specifically and saturably. The labeled receptor was purified and submitted to chemical and proteinase cleavages and radiochemical Edman degradation sequencing. This identified residues Glu133 and Glu125 within the amino-terminal domain of the receptor as the sites of labeling for the position 24 and 35 probes, respectively. These represent the first demonstration of direct residue-residue approximations between GLP1 and its receptor and provide two important constraints for future molecular modeling. This should contribute substantially to our understanding of the molecular basis of ligand binding and activation of the clinically important GLP1 receptor.
EXPERIMENTAL PROCEDURES
Materials
The solid-phase oxidant, N-chlorobenzenesulfonamide (IODO-BEAD), cyanogen bromide (CNBr), and m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester (Sulfo-MBS) were purchased from Pierce Chemical Co. 3-Isobutyl-1-methylxanthine, and N-(2-aminoethyl-1)-3-aminopropyl glass beads were from Sigma. Endoproteinase Lys-C (Lys-C) was from Roche Applied Science (Indianapolis, IN) and endoproteinase Asp-N (Asp-N) was from New England Biolabs Inc. (Ipswich, MA). Fetal Clone II and tissue culture medium were from Invitrogen (Grand Island, NY). Human GLP1-(7–36)-amide was purchased from Bachem (Torrance, CA). All other reagents were of analytical grade.
Peptide Synthesis and Radioiodination
The photolabile GLP1 probes, [Arg26,34,Bpa24]GLP1-(7–36) (Bpa24 probe) and [Arg26,34,Bpa35]GLP1-(7–36) (Bpa35 probe), were designed to incorporate a photolabile Bpa to replace Ala24 and Gly35 in the carboxyl-terminal region of the ligand, respectively. Both probes contain a naturally occurring Tyr residue in position 19 as a site for radioiodination. Lysine residues in positions 26 and 34 were replaced by arginines to prevent Lys-C cleavage, and this has previously been shown to be well tolerated (18). Peptides were synthesized by manual solid-phase techniques and were purified by reverse-phase HPLC as we described previously (19). They were radioiodinated oxidatively using 1 mCi Na125I and exposure to the solid-phase oxidant, iodobead for 15 s, and were purified using reversed-phase HPLC to yield specific radioactivities of 2,000 Ci/mmol (20).
Site-directed Mutagenesis
Development of new GLP1 receptor mutants that incorporated additional sites for Lys-C and CNBr cleavage was necessary for the current work. Three GLP1 receptor mutants (S136K, R121K, and S124M) were generated using the QuikChange Site-directed Mutagenesis kit from Stratagene (La Jolla, CA). Additionally, two new GLP1 receptor mutants representing alanine replacement of the sites of labeling by the Bpa24 and Bpa35 probes (E133A and E125A, respectively) were prepared in analogous manner and characterized. Verification of the desired mutations was performed by direct DNA sequencing.
Cell Culture and Membrane Preparation
A receptor-bearing CHO cell line stably expressing the human GLP1 receptor (CHO-GLP1R) was established as described previously (21). The GLP1 receptor mutants descried above were expressed transiently in COS-1 cells (American Type Culture Collection, Manassas, VA) after transfection using a modification of the diethylaminoethyl-dextran method. Cells were cultured in Ham's F-12 medium (for CHO-GLP1R) or DMEM medium (for COS-1) supplemented with 5% Fetal Clone II, 100 units of penicillin, and 100 μg of streptomycin in a humidified atmosphere containing 5% CO2 at 37 °C. Cells were passaged approximately twice a week.
After grown to confluency, cells were harvested mechanically and plasma membranes were prepared from these cells using discontinuous sucrose gradient centrifugation (22). Membranes were then suspended in Krebs-Ringers/HEPES (KRH) buffer (25 mm HEPES, pH 7.4, 104 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm KH2PO4, 1.2 mm MgSO4) containing 0.01% STI and 1 mm PMSF and stored at −80 °C until use.
Receptor Binding Assay
This assay was used for characterization of the newly synthesized photolabile probes and receptor mutants that were used in the current study. Briefly, ∼200,000 CHO-GLP1R cells were incubated at room temperature for 1 h with a constant amount of radioligand, 125I-GLP1 (5–10 pm), in the absence and presence of varied concentrations of nonradiolabeled ligand (0–1 μm GLP1 or Bpa24 or Bpa35 probe) in KRH buffer containing 0.01% STI and 0.2% bovine serum albumin. Bound and free radioligands were separated by washing the cells twice with ice-cold KRH buffer with STI and bovine serum albumin. Cells were lysed with 0.5 m NaOH, and bound radioactivity was quantified using a γ-counter. Nonspecific binding was determined in the presence of 1 μm unlabeled GLP1 and represented less than 15% of total radioligand binding. The same assay was also utilized to characterize the binding activity of COS-1 cells transiently expressing the GLP1 receptor mutant constructs. Binding curves were analyzed and plotted using the non-linear regression analysis program in the Prism software suite v3.02 (GraphPad Software, San Diego, CA). Binding kinetics was determined by analysis with the LIGAND program of Munson and Rodbard (23). Data are reported as the means ± S.E. of duplicate determinations from a minimum of three independent experiments.
Biological Activity Assay
The biological activity of the Bpa24 and Bpa35 probes to stimulate receptor-bearing cell line CHO-GLP1R to signal was assessed by measuring cAMP responses in these cells. Approximately 8,000 cells were placed in each well of 96-well plates. Two days after plating, cells were washed with phosphate-buffered saline and stimulated for 30 min at 37 °C with increasing concentrations of individual peptides (ranging from 0 to 1 μm) in KRH buffer containing 0.01% STI, 0.2% bovine serum albumin, 0.1% bacitracin, and 1 mm 3-isobutyl-1-methylxanthine. The peptide solution was then removed and 6% ice-cold perchloric acid was added to lyse the cells. After adjusting the pH to 6 with 30% KHCO3, the lysates were introduced directly into an assay of cAMP levels using a LANCE™ cAMP-384 kit from PerkinElmer (Boston, MA). The assay was performed as per manufacturer's instructions and repeated in at least three independent experiments. Analogous procedures were used to characterize the GLP1 mutant receptors expressed in COS-1 cells.
Receptor Photoaffinity Labeling
Photoaffinity labeling experiments were conducted as we described previously (24). Briefly, enriched receptor-bearing plasma membranes (50–100 μg) were incubated in dark for 1 h at room temperature with 0.1 nm 125I-labeled Bpa24 or Bpa35 probe in KRH buffer containing 0.01% STI and 1 mm PMSF in the presence of increasing amounts of competing GLP1 (reaction volume, 500 μl). The reactions were then exposed to photolysis for 30 min at 4 °C using a Rayonet photochemical reactor (Southern New England Ultraviolet Co., Bradford, CT) equipped with 3500-Å lamps. The efficiency of incorporation of each of the photolabile probes into receptor-bearing membranes after photolysis was ∼10%. After being washed with ice-cold KRH buffer, membranes were solubilized in SDS sample buffer and separated on 10% SDS-polyacrylamide gels. The bands of interest were visualized by exposure to Kodak x-ray film at −80 °C, and band densitometric analysis was performed by the NIH ImageJ software. The apparent molecular weights of the radioactive bands were determined by interpolation on a plot of the mobility of the appropriate ProSieve protein Markers (Cambrex, Rockland, ME) versus the log values of their apparent masses.
Peptide Mapping
Radiolabeled receptor bands were cut out from the gel, eluted in water, and lyophilized under vacuum. For selected experiments, receptor samples were deglycosylated. This was achieved by treatment with N-glycosidase F (PNGase F) following the protocol described in the manual (Proenzyme, San Leandro, CA). Cleavage of labeled GLP1 receptor or mutants with CNBr, Lys-C, or Asp-N was performed as described (24, 25). Products of cleavage were analyzed on 10% Bis-Tris NuPAGE gels (Invitrogen, Carlsbad, CA) using MES running buffer system under reducing conditions, and the radiolabeled bands were detected by autoradiography. The apparent molecular weights of the radioactive bands were determined by interpolation on a plot of the mobility of the appropriate Multimark multicolored or Seeblue plus2 prestained standards (Invitrogen) versus the log values of their apparent masses.
Radiochemical Sequencing
Identification of specific receptor residues labeled with the Bpa24 and Bpa35 probes was performed by radiochemical Edman degradation sequencing (26). Briefly, the labeled receptor fragments (Arg131–Lys197 for the Bpa24 probe and Glu125–Lys130 for the Bpa35 probe) were purified and covalently coupled through the cysteine residue (Cys174 for both Bpa24 and Bpa35 probes) to m-maleimidobenzoyl-N-succinimide-activated N-(2-aminoethyl-1)-3-aminopropyl glass beads. Immobilized fragments were then subjected to manual Edman degradation sequencing as described previously (26). Radioactivity eluted in each cycle was quantified using a γ-spectrometer.
RESULTS
Characterization of Photolabile Probes
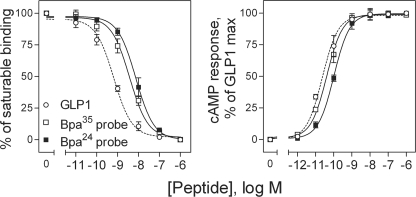
The Bpa24 and Bpa35 GLP1 probes were synthesized by manual solid phase techniques and purified by reversed-phase HPLC, and their identities were verified by mass spectrometry. They were characterized functionally, exploring their ability to bind to the GLP1 receptor and to stimulate biological responses in receptor-bearing CHO-GLP1R cells. As shown in Fig. 1, both probes bound the GLP1 receptor specifically and saturably, with slightly lower affinity than natural GLP1 (GLP1, Ki = 0.7 ± 0.1 nm; Bpa24 probe, Ki = 6.4 ± 2.5 nm; Bpa35 probe, Ki = 4.6 ± 0.5 nm). They were full agonists (GLP1, EC50 = 19 ± 1 pm; Bpa24 probe, EC50 = 86 ± 10 pm; Bpa35 probe, EC50 = 40 ± 7 pm), stimulating cAMP accumulation in CHO-GLP1R cells that were not different from that elicited by natural GLP1.
FIGURE 1.
Binding and biological activity of the Bpa24 and Bpa35 probes. Left, competition-binding curves of increasing concentrations of GLP1 and Bpa24 and Bpa35 probes to displace the binding of radioligand 125I-GLP1 to CHO-GLP1R membranes. Values illustrated represent saturable binding as percentages of maximal binding observed in the absence of the competing peptide. They are expressed as the means ± S.E. of duplicate values from a minimum of three independent experiments. Right, intracellular cAMP responses to increasing concentrations of GLP1 and the Bpa24 and Bpa35 probes in CHO-GLP1R cells. Data points represent the means ± S.E. of three independent experiments performed in duplicate, normalized relative to the maximal response to GLP1.
Photoaffinity Labeling of the GLP1 Receptor
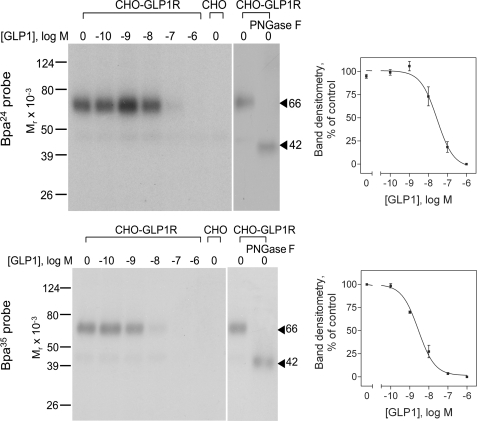
The ability of the Bpa24 and Bpa35 probes to covalently label the GLP1 receptor was explored with CHO-GLP1R cell membranes. As shown in Fig. 2, both probes specifically and saturably labeled the GLP1 receptor, with the labeled protein bands migrating at approximate Mr = 66,000. This labeling was competed by GLP1 in a concentration-dependent manner (Bpa24 probe, IC50 = 37 ± 11 nm; Bpa35 probe, IC50 = 2.3 ± 0.1 nm). No radioactive band was observed in the affinity-labeled non-receptor-bearing CHO cell membranes. The labeled receptor bands shifted to Mr = 42,000 after deglycosylation with PNGase F. These data indicated that both probes were efficiently able to label the GLP1 receptor. Thus, we proceeded with identification of their regions of labeling by peptide mapping.
FIGURE 2.
Photoaffinity labeling of the GLP1 receptor. Left, typical autoradiographs of 10% SDS-PAGE gels used to separate the products of affinity labeling of CHO-GLP1R cell membranes by the Bpa24 (top) and Bpa35 (bottom) probes in the absence and presence of increasing concentrations of competing unlabeled GLP1. The labeled receptor with each probe migrated at Mr = 66,000 and shifted to Mr = 42,000 after deglycosylation with PNGase F. No bands were detected in affinity-labeled non-receptor-bearing CHO cell membranes. Right, densitometric analyses of data from three similar experiments, with data points expressed as means ± S.E.
Identification of the Site of Labeling with the Bpa35 Probe
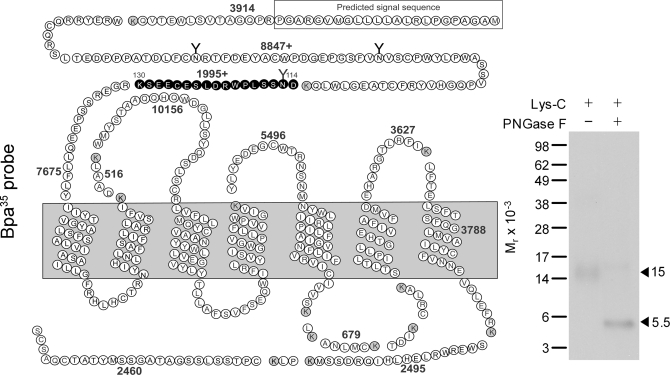
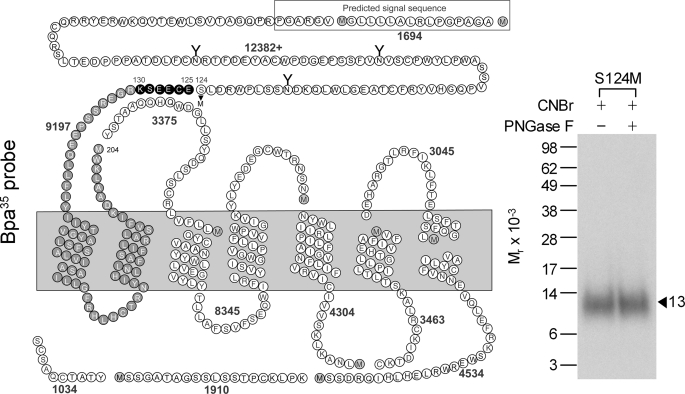
The GLP1 receptor photolabeled with the Bpa35 probe was first cleaved by Lys-C to gain an initial indication of the domain that was labeled. Theoretically, this proteinase cleaves a peptide at the carboxyl terminus of lysine residues, and the cleavage of the GLP1 receptor would result in 16 receptor fragments, with two of these containing sites of glycosylation. As shown in Fig. 3, the cleaved, labeled band migrated at approximate Mr = 15,000 and shifted to approximate Mr = 5,500 after PNGase F treatment. Given the molecular mass of the probe (3,672 Da) and glycosylated nature of the labeled fragment, the receptor fragment labeled could be limited to only one candidate. This was the fragment within the amino-terminal domain that contains the third glycosylation site (Asp114–Lys130).
FIGURE 3.
Endoproteinase Lys-C cleavage of the Bpa35 probe labeled GLP1 receptor. Left, diagram of the predicted sites of Lys-C cleavage of the human GLP1 receptor along with the masses. Right, a representative autoradiograph of 10% NuPAGE gel used to separate the products of Lys-C cleavage of the GLP1 receptor labeled with the Bpa35 probe. Cleavage of the labeled native receptor resulted in a band migrating at approximate Mr = 15,000 that shifted to Mr = 5,500 after deglycosylation. The glycosylated fragment (in bold circles) within the receptor amino terminus was the best candidate to represent the domain of labeling by the Bpa35 probe. Data are representative of three independent experiments.
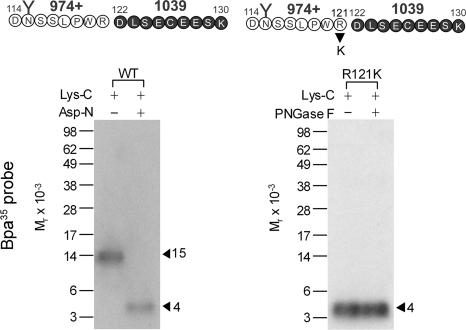
To further define the region of labeling by the Bpa35 probe, the Lys-C cleaved fragment was subsequently submitted to Asp-N cleavage. This proteinase cleaves a peptide at the amino terminus of aspartic acid residues. It should be noted that this enzyme also cleaves the Bpa35 probe, resulting in truncation of eight amino acids at the amino terminus, while keeping the sites of radioiodination and covalent attachment intact. As shown in Fig. 4, Asp-N cleavage of the glycosylated Mr = 15,000 fragment yielded a labeled fragment migrating at approximate Mr = 4,000, suggesting that the site of labeling was within the carboxyl-terminal half (Asp122–Lys130) of the Lys-C-cleaved fragment. This was further confirmed by Lys-C cleavage of the labeled R121K receptor mutant. This mutant bound GLP1 and signaled normally (Ki = 2.5 ± 0.5 nm; EC50 = 64 ± 16 pm), and it was labeled by the Bpa35 probe specifically and saturably (data not shown). Theoretically, Lys-C cleavage of the labeled mutant would result in exactly the same product as did the Asp-N cleavage of the labeled wide-type receptor as described above (highlighted in bold circles in Fig. 4). As expected, a band migrating at approximate Mr = 4,000 that did not further shift after deglycosylation was observed (right panel of Fig. 4). These data further confirmed that the segment between Asp122 and Lys130 contained the site of labeling with the Bpa35 probe.
FIGURE 4.
Further localization of the domain of Bpa35 probe labeling. Left shows that endoproteinase Asp-N cleavage of the Mr = 15,000 fragment resulting from Lys-C cleavage of the wild-type receptor yielded a band migrating at approximate Mr = 4,000. Right shows that Lys-C cleavage of the labeled receptor mutant R121K also resulted in the same size band and did not further shift after deglycosylation. These identified that the segment between Asp122 and Lys130 contained the site of labeling with the Bpa35 probe.
To further narrow down the domain of labeling, we generated another mutant, S124M, introducing an additional methionine residue within the Asp122–Lys130 candidate fragment labeled by the Bpa35 probe. This mutant bound and signaled similarly to that of the wild-type receptor (Ki = 1.4 ± 0.1 nm; EC50 = 40 ± 9 pm). It was labeled with the Bpa35 probe specifically and saturably (data not shown). CNBr cleavage of the labeled intact S124M receptor mutant yielded a non-glycosylated fragment migrating at approximate Mr = 13,000 (Fig. 5), representing the site of labeling by the Bpa35 probe being within the segment between Glu125 and Met204. Taking into account the insight from the Lys-C cleavage described above, the Glu125–Lys130 segment contained the site of labeling for the Bpa35 probe.
FIGURE 5.
CNBr cleavage of the S124M GLP1 receptor mutant labeled with the Bpa35 probe. Left shows the diagram illustrating the theoretical fragments resulting from CNBr cleavage of the S124M GLP1 receptor mutant. Right shows that CNBr cleavage of the labeled receptor mutant yielded a band migrating at approximate Mr = 13,000 that did not further shift after deglycosylation. Taking into account the data in Fig. 4, this identified the fragment between Glu125 and Lys130 containing the site of labeling with the Bpa35 probe.
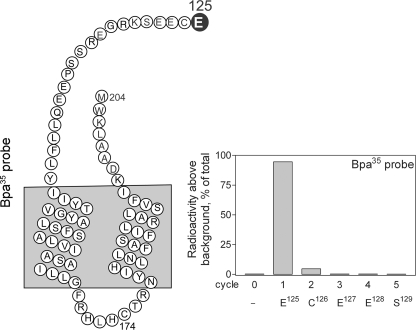
The specific residue labeled with the Bpa35 probe was identified by manual radiochemical Edman degradation sequencing of the CNBr fragment (Glu125–Met204) from the labeled GLP1 receptor mutant S124M. As shown in Fig. 6, a radioactive peak was eluted in the first cycle, corresponding to the covalent labeling of receptor residue Glu125 within the amino-terminal domain of the GLP1 receptor.
FIGURE 6.
Identification of the receptor residue labeled with the Bpa35 probe by radiochemical sequencing. Left, diagram of the CNBr fragment (Glu125–Met204) resulting from cleavage of the S124M receptor mutant labeled with the Bpa35 probe. Right, a representative radioactivity elution profile of Edman degradation sequencing of this fragment. A radioactive peak consistently eluted in cycle 1 represents covalent labeling of the receptor residue Glu125 with the Bpa35 probe.
Identification of the Site of Labeling with the Bpa24 Probe
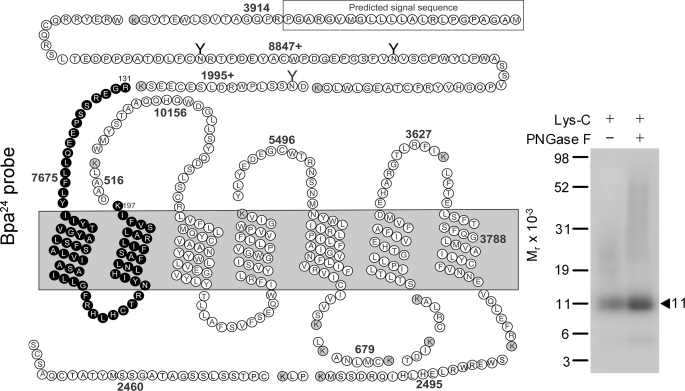
Lys-C was used as the first indication of the domain of labeling with the Bpa24 probe. As shown in Fig. 7, Lys-C cleavage of the labeled GLP1 receptor yielded a band migrating at approximate Mr = 11,000 that did not further shift after PNGase F treatment. Considering the non-glycosylated nature and the mass of the Bpa24 probe (3,658 Da), the receptor fragment labeled could be limited to only one candidate. This was the fragment containing the carboxyl terminus of the amino-terminal domain, the first and second transmembrane domains, and the first intracellular loop (the Arg131–Lys197 fragment highlighted in bold circles).
FIGURE 7.
Endoproteinase Lys-C cleavage of the Bpa24 probe-labeled GLP1 receptor. Left shows the diagram of the predicted sites of Lys-C cleavage of the GLP1 receptor. Right shows that Lys-C cleavage of the Bpa24 probe-labeled GLP1 receptor yielded a band migrating at approximate Mr = 11,000 that did not further shift after PNGase F treatment. Given the molecular mass of the probe (3,658 Da) and non-glycosylated nature of the labeled fragment, the site of labeling with the Bpa24 probe was within the Arg131-Lys197 fragment within the amino terminus of the GLP1 receptor. The molecular weight standards used in this figure were the discontinued Multimark multicolored standards. The Seeblue plus2 prestained standards were used in all other figures.
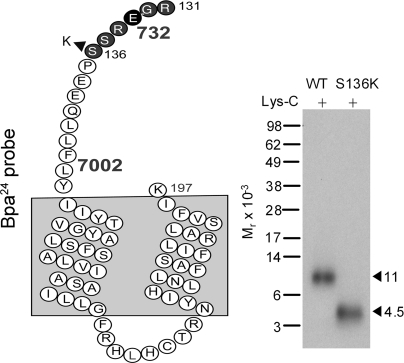
To definitively define the region of labeling by the Bpa24 probe, we generated a GLP1 receptor mutant construct, S136K, to introduce an additional lysine residue within the Arg131–Lys197 candidate fragment. This mutant bound GLP1 and signaled normally (Ki = 1.8 ± 0.2 nm; EC50 = 8 ± 2 pm). It was specifically and saturably labeled with the Bpa24 probe (data not shown). Lys-C cleavage resulted in shifting the Mr = 11,000 band in the wild-type receptor to Mr = 4,500 in the S136K receptor (Fig. 8). This established that the site of labeling for the Bpa24 probe was within the small segment between Arg131 and Lys136 at the amino-terminal domain of the GLP1 receptor adjacent to the first transmembrane domain.
FIGURE 8.
Endoproteinase Lys-C cleavage of the S136K GLP1 receptor mutant labeled with the Bpa24 probe. Shown in the left is a diagram illustrating the theoretical fragment resulting from Lys-C cleavage of the S136K GLP1 receptor mutant. Compared with that of the wild-type receptor, Lys-C cleavage of the labeled S136K receptor mutant yielded a much smaller fragment migrating at approximate Mr = 4,500 (right), representing the fragment Arg131–Lys136 that contained the site of labeling with the Bpa24 probe.
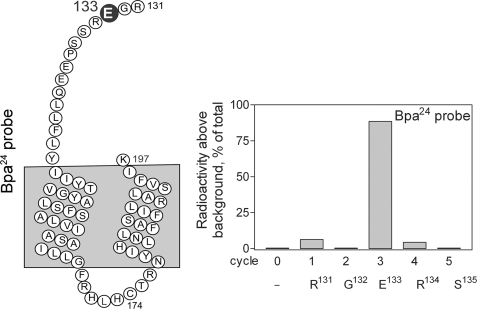
The radiochemically pure Lys-C-cleaved fragment (Arg131–Lys197) from the affinity-labeled wild-type GLP1 receptor was utilized for manual radiochemical Edman degradation sequencing to identify the specific residue labeled with the Bpa24 probe. As shown in Fig. 9, a peak of eluted radioactivity was observed in the third cycle, indicating that the receptor residue Glu133 was the site of covalent attachment of this probe.
FIGURE 9.
Identification of the receptor residue labeled with the Bpa24 probe by radiochemical sequencing. Left, diagram of the Lys-C fragment (Arg131–Lys197) from cleavage of the Bpa24 probe-labeled wild-type GLP1 receptor. Right, a representative profile of eluted radioactivity in which a peak was found in cycle 3, corresponding to the labeling of receptor residue Glu133 within the amino terminus adjacent to the top of the first transmembrane segment.
Characterization of Receptor Photoaffinity Labeling Site Mutants
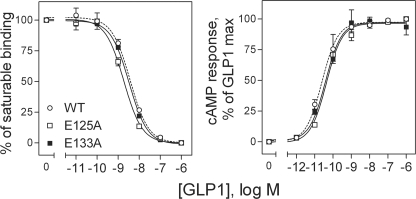
The E125A and E133A GLP1 receptor mutants were expressed transiently in COS-1 cells. These site mutants were studied in this cell system for impact on the binding and biological activity of GLP1 (Fig. 10). Mutation of the residues covalently labeled by the Bpa24 and Bpa35 probes (Glu133 and Glu125, respectively) had no significant effect on the binding and biological effects of GLP1.
FIGURE 10.
Characterization of the E125A and E133A GLP1 receptor mutants. Left, competition-binding curves of increasing concentrations of GLP1 to displace the binding of radioligand 125I-GLP1 to COS-1 cells expressing wild-type (WT), E125A and E133A GLP1 receptors. Values illustrated represent saturable binding as percentages of maximal binding observed in the absence of the competing peptide. They are expressed as the means ± S.E. of duplicate values from a minimum of three independent experiments. Right, intracellular cAMP responses to increasing concentrations of GLP1 in these cells. Data points represent the means ± S.E. of three independent experiments performed in duplicate, normalized relative to the maximal response to GLP1.
DISCUSSION
Detailed molecular insights into the active conformation of a receptor and into the molecular basis of its binding of agonist ligands will facilitate rational design and refinement of receptor-active drugs. The GLP1 receptor, a member of the Family B GPCRs, is a potential target for glucose-dependent therapeutic agents designed to treat hyperglycemia resulting from type 2 diabetes. However, the mechanisms underlying binding and activation of this receptor and other members of this family are poorly understood and the development of small molecule ligands is problematic. This has been approached by previous structure-activity, mutagenesis and chimeric studies (18, 27–35). In this study, we used photoaffinity labeling to demonstrate that the carboxyl terminus of GLP1 is in approximation with the amino terminus of the receptor when docked.
Because of the indirect nature of mutagenesis studies, most of which involve loss of function, and the paucity of examples of successful gain-of-function and complementary mutagenesis experiments, we have used the approach of photoaffinity labeling to identify unambiguous constraints that can be used to dock the natural ligand GLP1 to its receptor. This approach is dependent on the spatial approximation between a photolabile group incorporated into a ligand as it resides in its binding site and a portion of the receptor molecule. Constraints obtained with photoaffinity labeling are particularly powerful, because they are direct and clearly establish spatial approximations between distinct residues within a ligand and a receptor that are extremely useful for molecular modeling.
The current study focused on the carboxyl-terminal region of the GLP1 peptide, using photolabile Bpa24 and Bpa35 probes. It is important to note that both of these GLP1 analogues bound to the GLP1 receptor specifically and with high affinity, retaining the determinants of binding and activation intrinsic to the natural hormone. Additionally, both probes were shown to represent full agonists at the GLP1 receptor. This ensures that the molecular mechanisms of binding to the receptor and inducing conformational changes in that target remain intact. This further supports the physiological significance of the residue-residue approximations that have been experimentally determined in these studies. The current report provided direct spatial approximations between the carboxyl-terminal residues 24 and 35 of GLP1 and receptor residues Glu133 and Glu125 within the amino terminus, respectively. These represent the first demonstration of direct residue-residue approximations between the natural agonist GLP1 and its receptor.
It should be noted that the replacement of each of the residues that were covalently labeled in the current photoaffinity labeling studies (Glu125 and Glu133) with alanine did not interfere with the normal binding and biological activity of natural GLP1. This is consistent with the accommodation of a photolabile residue in these positions within the docked peptide, suggesting that the natural peptide-receptor residues in these positions do not represent critical interactions. This is quite typical of photoaffinity labeling, often providing spatial approximation data rather than identifying critical interactions.
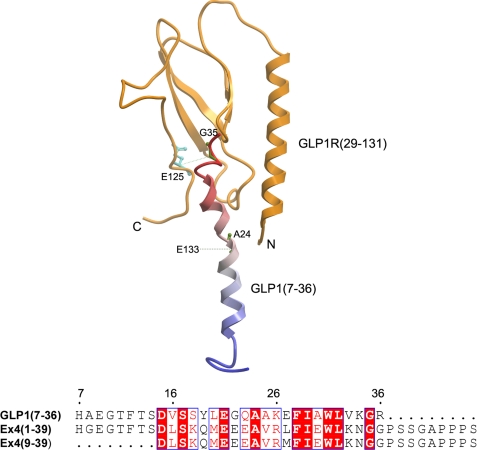
Recently, the crystal structure of the antagonist Extendin-4-(9–39) (Ex4-(9–39))-bound amino-terminal domain of the GLP1 receptor was reported (16). The structure used truncated region between residues 29 and 131 of the amino-terminal domain of the GLP1 receptor, and it did not include both the distal amino- and carboxyl-terminal regions of this domain. To examine whether our data with the natural ligand GLP1 are accommodated in this structure, we replaced the docked antagonist Ex4-(9–39) with the agonist GLP1 (Fig. 11). The receptor residue Glu133 that was labeled by the Bpa24 GLP1 probe is not included in the structure (Fig. 11), whereas the analogous position of Ex4-(9–39) (Ala18) was shown to be in proximity with the receptor residue Glu128 with a Cα-Cα distance of 14.3 Å (16). The receptor residue Glu125 that was labeled by the Bpa35 GLP1 probe was shown to be in proximity with the Gly35 of GLP1 with a Cα-Cα distance of 14.6 Å (Fig. 11), while the analogous position of Ex4-(9–39) (Gly29) is in close proximity with receptor residues Glu68 (Cα-Cα, 5.9 Å) and Tyr69 (Cα-Cα, 7.5 Å) (16). Our data from labeling of the GLP1 receptor through carboxyl-terminal residues of GLP1 (i.e. positions 24 and 35) can be reasonably accommodated in this crystal structure, but they are not consistent with a recent homology model of the GLP1 receptor in which Ala24 and Gly35 of GLP1 were quite distant from their respective labeling sites, Glu133 and Glu125 (36). This may reflect the fact that the model was built to accommodate several ligands based on limited experimental data derived from indirect mutagenesis analysis. In addition, our data suggest differences in the structural details of binding agonist GLP1 from that of binding antagonist Ex4-(9–39). This is not surprising because differences in binding determinants for GLP1 and Ex4 have been previously observed (37).
FIGURE 11.
Homology modeling structure of GLP1-occupied GLP1 receptor amino-terminal domain. Top, schematic representation of the molecular replacement by aligning the agonist Ex4 NMR structure (PDB code: 1JRJ) with the antagonist Ex4-(9–39)-bound GLP1 receptor amino-terminal domain crystal structure (PDB code: 3C59) that followed by replacing with the agonist GLP1 NMR structure (PDB code: 1D0R). The GLP1 receptor amino terminus is colored in orange, and the GLP1 peptide is in blue-red from the amino terminus to the carboxyl terminus. Residue labels are in green on the GLP1 peptide and cyan on the receptor. The side chains of peptide residues Ala24 and Gly35, and receptor residue Glu125 are highlighted by a sticks representation. The distance restraints are represented in green-dotted lines. The site of labeling for the Bpa24 probe, Glu133, is not in the structure. The site of labeling for the Bpa35 probe, Glu125, is in the structure, but its side chain exposes to solvent instead of pointing toward the interaction surface. This figure was made with the Internal Coordinate Mechanics program from Molsoft (55). Bottom, the sequence alignment of agonist GLP1-(7–36), Ex4-(1–39), and antagonist Ex4-(9–39). The residues Ala24 and Gly35 are conserved in GLP1 and Ex4. This alignment was performed with the ClustalW program, and the figure was generated using ESPript.
Ligand binding and activation of the GLP1 receptor have been studied by structure-activity relationship studies. Alanine-scanning of GLP1 has implicated positions 7, 10, 12, 13, 15, 26, 28, and 29 as being critical for receptor binding (18, 30), whereas substitutions at positions 24 and 35 have minimal effects on GLP1 receptor binding and activation (18). This is consistent with our incorporation of a Bpa in these positions. Like natural ligands for other members of Family B GPCRs, GLP1 is a long peptide having diffuse pharmacophoric domains, with its amino-terminal region containing key determinants for receptor selectivity and activation and its carboxyl-terminal region containing determinants for high affinity binding. NMR analysis of the GLP1 peptide reveals that the residues 7–30 adopt a helical structure (38–41). It is assumed that this helical region binds to the amino-terminal domain of the GLP1R while the amino terminus of the peptide interacts with the receptor core (27).
Previous receptor chimeric and mutagenesis studies have suggested the importance of the amino terminus of the GLP1 receptor in ligand binding (27–29, 34), and this domain determines the specificity for the carboxyl terminus of GLP1 (33). This is a theme that is now well established for ligand binding of Family B GPCRs (5–10). However, the precise residues within the amino terminus of the GLP1 receptor that interact with GLP1 have not been mapped. Mutagenesis and chimeric analyses have also demonstrated that the first extracellular loop, and the first and fourth transmembrane domains of the GLP1 receptor are important for ligand binding and receptor activation (31, 32, 35). Based on these studies, a “two-step” or “tethering” mechanism has been proposed, where the carboxyl-terminal region of the peptide ligand initially interacts with the amino-terminal domain of the receptor, and in the second step, the amino-terminal region of the ligand interacts with the core domain of the receptor, which leads to receptor activation (27). This mechanism has also been proposed for other members of Family B GPCRs (42–44). Our current data show that both the position 24 and 35 residues of GLP1 are in proximity with the residues in the amino-terminal domain of its receptor, which is consistent with this mechanism.
The most extensive data for docking a natural peptide ligand in this family to its receptor come from photoaffinity labeling with analogues of secretin where spatial approximations have been established between residues scattered throughout the pharmacophore of secretin in positions 1, 5, 6, 12, 13, 14, 18, 21, 22, 23, and 26 (45, 46). Interestingly, the carboxyl-terminal (positions 18, 21, 22, 23, and 26), mid-region (position 12 and 14), and amino-terminal (position 6) probes have been demonstrated to label the distal amino terminus of the secretin receptor, all within the first 38 residues (45, 46). The only secretin analogue that labels a residue (Val103) within the amino-terminal domain adjacent to the transmembrane domain, in positions analogous to the GLP1 receptor residues labeled by the carboxyl-terminal position 24 and 35 GLP1 probes in this work, is the midregion position 13 probe (25). Another two amino-terminal probes (position 1 and 5) have been shown to label residues within the top of the sixth transmembrane domain and third extracellular loop (43, 46). These data also support the proposed mechanism for activation of Family B GPCRs.
Other Family B GPCRs that have been studied by photoaffinity labeling are PTH, calcitonin and VPAC1 receptors. Like the secretin receptor, while the midregion and carboxyl-terminal PTH or PTH-related peptide probes have been shown to label residues within the amino terminus of the PTH receptor, the amino-terminal probe has been shown to label residues within the sixth transmembrane domain (42, 47, 48). The carboxyl-terminal position 33 PTH probe has been demonstrated to label the receptor amino terminus adjacent to the first transmembrane domain (49), consistent with labeling the GLP1 receptor with the carboxyl-terminal probes in this study. However, another two carboxyl-terminal position 23 and 28 probes were shown to label the distal amino terminus (49, 50). The labeling of the calcitonin receptor is quite consistent with the data from the secretin and PTH receptors (44, 51), but the labeling of the VPAC1 receptor suggests a different mechanism. All the vasoactive intestinal polypeptide (VIP) probes tested so far label the amino-terminal domain adjacent to the transmembrane domain. These include the amino-terminal position 0 and 6, and carboxyl-terminal position 22, 24, and 28 VIP probes that all label residues between residues 107 and 137 (52–54), a region that the current carboxyl-terminal GLP1 probes label. However, whether ligand binding to the GLP1 receptor follows the themes observed with the secretin/PTH receptors or the VPAC1 receptor is not clear, and this can be tested with probes incorporating photolabile residues in other positions, in particular the mid- and amino-terminal regions of the peptide ligand.
In summary, we have identified two residue-residue approximations between the carboxyl-terminal residues of the natural agonist ligand GLP1 and its receptor that are potentially useful for the molecular modeling of the agonist-occupied GLP1 receptor. Although high-resolution crystal or NMR structures of the amino terminus of several Family B GPCRs have recently become available, there are differences in docking the ligands to their receptors. Furthermore, these structures did not include the entire receptor amino-terminal domain, and, in all but one of these structures, an antagonist was used as the docked ligand. In addition, the relationship between the amino-terminal domain and the core domains of the receptor is not clear. Therefore, insights into the residue-residue approximations between the natural ligand GLP1 and its docked intact receptor become very important for constructing a meaningful and testable molecular model for agonist ligand binding of the GLP1 receptor. This should become possible as we expand the number of such experimental constraints, and our insights into the molecular basis of ligand binding of this important receptor will become clearer.
This work was supported, in whole or in part, by National Institutes of Health Grant DK46577. This work was also supported by American Diabetes Association Grant 1-08-RA-42.
- GPCR
- G protein-coupled receptor
- Asp-N
- endoproteinase Asp-N
- Bpa
- p-benzoyl-l-phenylalanine
- CHO
- Chinese hamster ovary
- CHO-GLP1R
- human GLP1 receptor-bearing CHO cells
- CNBr
- cyanogen bromide
- Ex4
- extendin-4
- GIP
- gastric inhibitory polypeptide
- GLP1
- glucagon-like peptide-1
- KRH
- Krebs-Ringers-HEPES
- Lys-C
- endoproteinase Lys-C
- PNGase F
- N-glycosidase F
- PMSF
- phenylmethylsulfonyl fluoride
- PTH
- parathyroid hormone
- STI
- soybean trypsin inhibitor
- VIP
- vasoactive intestinal polypeptide
- MES
- 4-morpholineethanesulfonic acid
- PDB
- Protein Data Bank.
REFERENCES
- 1.Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 4.Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y. J., Gimpl G., Fahrenholz F. (1995) Biochem. Biophys. Res. Commun. 212, 673–680 [DOI] [PubMed] [Google Scholar]
- 6.Gourlet P., Vilardaga J. P., De Neef P., Waelbroeck M., Vandermeers A., Robberecht P. (1996) Peptides 17, 825–829 [DOI] [PubMed] [Google Scholar]
- 7.Graziano M. P., Hey P. J., Strader C. D. (1996) Receptors Channels 4, 9–17 [PubMed] [Google Scholar]
- 8.Holtmann M. H., Hadac E. M., Miller L. J. (1995) J. Biol. Chem. 270, 14394–14398 [DOI] [PubMed] [Google Scholar]
- 9.Jüppner H., Schipani E., Bringhurst F. R., McClure I., Keutmann H. T., Potts J. T., Jr., Kronenberg H. M., Abou-Samra A. B., Segre G. V., Gardella T. J. (1994) Endocrinology 134, 879–884 [DOI] [PubMed] [Google Scholar]
- 10.Stroop S. D., Nakamuta H., Kuestner R. E., Moore E. E., Epand R. M. (1996) Endocrinology 137, 4752–4756 [DOI] [PubMed] [Google Scholar]
- 11.Grace C. R., Perrin M. H., DiGruccio M. R., Miller C. L., Rivier J. E., Vale W. W., Riek R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12836–12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grace C. R., Perrin M. H., Gulyas J., Digruccio M. R., Cantle J. P., Rivier J. E., Vale W. W., Riek R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pioszak A. A., Parker N. R., Suino-Powell K., Xu H. E. (2008) J. Biol. Chem. 283, 32900–32912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C., Song D., Davis-Taber R. A., Barrett L. W., Scott V. E., Richardson P. L., Pereda-Lopez A., Uchic M. E., Solomon L. R., Lake M. R., Walter K. A., Hajduk P. J., Olejniczak E. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7875–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parthier C., Kleinschmidt M., Neumann P., Rudolph R., Manhart S., Schlenzig D., Fanghänel J., Rahfeld J. U., Demuth H. U., Stubbs M. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13942–13947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runge S., Thøgersen H., Madsen K., Lau J., Rudolph R. (2008) J. Biol. Chem. 283, 11340–11347 [DOI] [PubMed] [Google Scholar]
- 17.Pioszak A. A., Xu H. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adelhorst K., Hedegaard B. B., Knudsen L. B., Kirk O. (1994) J. Biol. Chem. 269, 6275–6278 [PubMed] [Google Scholar]
- 19.Powers S. P., Pinon D. I., Miller L. J. (1988) Int. J. Pept. Protein. Res. 31, 429–434 [DOI] [PubMed] [Google Scholar]
- 20.Ulrich C. D., 2nd, Pinon D. I., Hadac E. M., Holicky E. L., Chang-Miller A., Gates L. K., Miller L. J. (1993) Gastroenterology 105, 1534–1543 [DOI] [PubMed] [Google Scholar]
- 21.Dong M., Gao F., Pinon D. I., Miller L. J. (2008) Mol. Endocrinol 22, 1489–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadac E. M., Ghanekar D. V., Holicky E. L., Pinon D. I., Dougherty R. W., Miller L. J. (1996) Pancreas 13, 130–139 [DOI] [PubMed] [Google Scholar]
- 23.Munson P. J., Rodbard D. (1980) Anal. Biochem. 107, 220–239 [DOI] [PubMed] [Google Scholar]
- 24.Dong M., Wang Y., Pinon D. I., Hadac E. M., Miller L. J. (1999) J. Biol. Chem. 274, 903–909 [DOI] [PubMed] [Google Scholar]
- 25.Zang M., Dong M., Pinon D. I., Ding X. Q., Hadac E. M., Li Z., Lybrand T. P., Miller L. J. (2003) Mol. Pharmacol. 63, 993–1001 [DOI] [PubMed] [Google Scholar]
- 26.Ji Z., Hadac E. M., Henne R. M., Patel S. A., Lybrand T. P., Miller L. J. (1997) J. Biol. Chem. 272, 24393–24401 [DOI] [PubMed] [Google Scholar]
- 27.Al-Sabah S., Donnelly D. (2003) Br. J. Pharmacol. 140, 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buggy J. J., Livingston J. N., Rabin D. U., Yoo-Warren H. (1995) J. Biol. Chem. 270, 7474–7478 [DOI] [PubMed] [Google Scholar]
- 29.Gallwitz B., Witt M., Morys-Wortmann C., Fölsch U. R., Schmidt W. E. (1996) Regul. Pept. 63, 17–22 [DOI] [PubMed] [Google Scholar]
- 30.Gallwitz B., Witt M., Paetzold G., Morys-Wortmann C., Zimmermann B., Eckart K., Fölsch U. R., Schmidt W. E. (1994) Eur. J. Biochem. 225, 1151–1156 [DOI] [PubMed] [Google Scholar]
- 31.Gelling R. W., Wheeler M. B., Xue J., Gyomorey S., Nian C., Pederson R. A., McIntosh C. H. (1997) Endocrinology 138, 2640–2643 [DOI] [PubMed] [Google Scholar]
- 32.López de Maturana R., Treece-Birch J., Abidi F., Findlay J. B., Donnelly D. (2004) Protein Pept. Lett. 11, 15–22 [DOI] [PubMed] [Google Scholar]
- 33.Runge S., Wulff B. S., Madsen K., Bräuner-Osborne H., Knudsen L. B. (2003) Br. J. Pharmacol 138, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilmen A., Van Eyll B., Göke B., Göke R. (1997) Peptides 18, 301–305 [DOI] [PubMed] [Google Scholar]
- 35.Xiao Q., Jeng W., Wheeler M. B. (2000) J. Mol. Endocrinol 25, 321–335 [DOI] [PubMed] [Google Scholar]
- 36.Lin F., Wang R. (2009) J. Mol. Model 15, 53–65 [DOI] [PubMed] [Google Scholar]
- 37.Mann R., Nasr N., Hadden D., Sinfield J., Abidi F., Al-Sabah S., de Maturana R. L., Treece-Birch J., Willshaw A., Donnelly D. (2007) Biochem. Soc. Trans 35, 713–716 [DOI] [PubMed] [Google Scholar]
- 38.Andersen N. H., Brodsky Y., Neidigh J. W., Prickett K. S. (2002) Bioorg Med. Chem. 10, 79–85 [DOI] [PubMed] [Google Scholar]
- 39.Neidigh J. W., Fesinmeyer R. M., Prickett K. S., Andersen N. H. (2001) Biochemistry 40, 13188–13200 [DOI] [PubMed] [Google Scholar]
- 40.Parker J. C., Andrews K. M., Rescek D. M., Massefski W., Jr., Andrews G. C., Contillo L. G., Stevenson R. W., Singleton D. H., Suleske R. T. (1998) J. Pept. Res. 52, 398–409 [DOI] [PubMed] [Google Scholar]
- 41.Thornton K., Gorenstein D. G. (1994) Biochemistry 33, 3532–3539 [DOI] [PubMed] [Google Scholar]
- 42.Bisello A., Adams A. E., Mierke D. F., Pellegrini M., Rosenblatt M., Suva L. J., Chorev M. (1998) J. Biol. Chem. 273, 22498–22505 [DOI] [PubMed] [Google Scholar]
- 43.Dong M., Li Z., Pinon D. I., Lybrand T. P., Miller L. J. (2004) J. Biol. Chem. 279, 2894–2903 [DOI] [PubMed] [Google Scholar]
- 44.Dong M., Pinon D. I., Cox R. F., Miller L. J. (2004) J. Biol. Chem. 279, 31177–31182 [DOI] [PubMed] [Google Scholar]
- 45.Dong M., Lam P. C., Gao F., Hosohata K., Pinon D. I., Sexton P. M., Abagyan R., Miller L. J. (2007) Mol. Pharmacol. 72, 280–290 [DOI] [PubMed] [Google Scholar]
- 46.Dong M., Lam P. C., Pinon D. I., Sexton P. M., Abagyan R., Miller L. J. (2008) Mol. Pharmacol. 74, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behar V., Bisello A., Bitan G., Rosenblatt M., Chorev M. (2000) J. Biol. Chem. 275, 9–17 [DOI] [PubMed] [Google Scholar]
- 48.Gensure R. C., Carter P. H., Petroni B. D., Jüppner H., Gardella T. J. (2001) J. Biol. Chem. 276, 42692–42699 [DOI] [PubMed] [Google Scholar]
- 49.Gensure R. C., Gardella T. J., Jüppner H. (2001) J. Biol. Chem. 276, 28650–28658 [DOI] [PubMed] [Google Scholar]
- 50.Mannstadt M., Luck M. D., Gardella T. J., Jüppner H. (1998) J. Biol. Chem. 273, 16890–16896 [DOI] [PubMed] [Google Scholar]
- 51.Dong M., Pinon D. I., Cox R. F., Miller L. J. (2004) J. Biol. Chem. 279, 1167–1175 [DOI] [PubMed] [Google Scholar]
- 52.Ceraudo E., Murail S., Tan Y. V., Lacapère J. J., Neumann J. M., Couvineau A., Laburthe M. (2008) Mol. Endocrinol. 22, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ceraudo E., Tan Y. V., Nicole P., Couvineau A., Laburthe M. (2008) J. Mol. Neurosci. 36, 245–248 [DOI] [PubMed] [Google Scholar]
- 54.Tan Y. V., Couvineau A., Murail S., Ceraudo E., Neumann J. M., Lacapère J. J., Laburthe M. (2006) J. Biol. Chem. 281, 12792–12798 [DOI] [PubMed] [Google Scholar]
- 55.Abagyan R. A., Totrov M. M., Kuznetsov D. A. (1994) J. Comp. Chem. 15, 488–506 [Google Scholar]