Abstract
Thyroid hormone (T3) is essential for normal development and organ function throughout vertebrates. Its effects are mainly mediated through transcriptional regulation by T3 receptor (TR). The identification and characterization of the immediate early, direct target genes are thus of critical importance in understanding the molecular pathways induced by T3. Unfortunately, this has been hampered by the difficulty to study gene regulation by T3 in uterus-enclosed mammalian embryos. Here we used Xenopus metamorphosis as a model for vertebrate postembryonic development to identify direct T3 response genes in vivo. We took advantage of the ability to easily induce metamorphosis with physiological levels of T3 and to carry out microarray analysis in Xenopus laevis and genome-wide sequence analysis in Xenopus tropicalis. This allowed us to identify 188 up-regulated and 249 down-regulated genes by T3 in the absence of new protein synthesis in whole animals. We further provide evidence to show that these genes contain functional TREs that are bound by TR in tadpoles and that their promoters are regulated by TR in vivo. More importantly, gene ontology analysis showed that the direct up-regulated genes are enriched in categories important for transcriptional regulation and protein degradation-dependent signaling processes but not DNA replication. Our findings thus revealed the existence of interesting pathways induced by T3 at the earliest step of metamorphosis.
Introduction
Thyroid hormone (T3)2 is critical for adult organ homeostasis and function and also essential for vertebrate development (1–9). T3 deficiency during development leads to severe developmental defects in mammals, including cretinism in human, which is characterized by severe short stature and mental retardation (5). During early mammalian development, there is little T3 in the fetus, although some maternal T3 reaches the embryo. High levels of T3 are present only during the so-called postembryonic period, which spans from several months prior to birth to several months after birth in human (3, 10). This is a critical period of organ growth and maturation, and thus, not surprisingly, T3 deficiency during this period causes severe developmental defects (5, 11, 12). Interestingly, appropriate levels of maternal T3 are also important to ensure proper mammalian development (13–15). These observations suggest that proper levels of T3 in both the mother and fetus are critical for mammalian development, which makes it difficult to separate the direct effects of T3 on the development of the fetus versus indirect effects through maternal influence. Furthermore, there are only a limited number of known direct T3 response genes in different model systems, and no systematic analysis has been carried out to isolate such genes in development. All these have hampered our understanding of how T3 regulates development in vivo.
Amphibian metamorphosis is a postembryonic process that is also dependent on T3 (4, 16). Studies in anurans such as Xenopus laevis in the past century have shown that T3 controls every aspect of metamorphosis and is both necessary and sufficient (9, 17–19). During metamorphosis, different tissues have different fates (4, 17). For example, the tail and gills undergo resorption, the brain, skin, intestine, and other visceral organs undergo remodeling while the limbs are generated de novo. All these changes are controlled by T3 in mostly organ-autonomous manner.
T3 action is primarily mediated through thyroid receptors (TRs), which are transcription factors and are members of nuclear receptor superfamily (1, 2, 9, 19–21). TRs bind to chromosomal sites in the promoter regions (known as thyroid response elements (TRE)) of direct response genes of T3. For T3-inducible genes, these binding sites are often composed of two direct repeats of the consensus sequence AGGTCA with a 4-bp spacer sequence. TR functions mainly as a heterodimer with 9-cis-retinoic acid receptor (RXR) at these TREs and represses and activates these genes by recruiting corepressor and coactivator complexes in the absence or presence of T3, respectively.
We have previously proposed a dual function model for TR function during anuran metamorphosis (22, 23). According to this model, in the absence of T3, TR/RXR heterodimers repress direct target genes to ensure proper growth of the premetamorphic tadpoles and prevent premature metamorphic organ transformations. In the presence of T3, TR/RXR heterodimers activate these target genes to initiate metamorphosis. Over the years, studies by us and others have shown that TRs are both necessary and sufficient to mediate the metamorphic effects of T3 in X. laevis (9, 19). Thus all the remarkable morphological and developmental changes in the different tissues are effected through gene expression cascades initiated by TRs. Furthermore, TRs indeed have dual functions during metamorphosis. They recruit corepressor complexes in premetamorphic tadpoles to control metamorphic timing and recruit coactivator complexes to initiate metamorphosis (24–28). In addition, the levels of coactivator complexes also regulate the rate of metamorphic progression (29).
Using subtractive hybridization screening and microarrays, we and others have previously documented the differentially expressed genes in different organs during both T3-induced and natural metamorphosis (4, 17, 30–33). These studies have provided us with an understanding of the global gene expression changes and signaling pathways involved in metamorphosis. How these genes are regulated remains largely unknown, although a few have been shown to be direct target genes of TRs (34–38). Here we have made use of the recent advances in microarray analysis of gene expression in X. laevis and genomic sequencing in the highly related species Xenopus tropicalis to systematically identify direct target genes of T3, which are likely key players in propagating the effects of T3 in regulating metamorphosis. This is made possible in part by the fact that X. laevis and X. tropicalis undergo essentially identical T3-dependent metamorphosis. In addition, TR and RXR genes are regulated and function similarly in both species during development (39). As the regulation of immediate early, direct TR target genes by T3 should be independent of new protein synthesis, to identify these direct targets, we treated premetamorphic X. laevis tadpoles with T3 to induce metamorphosis and included protein synthesis inhibitors to block the synthesis of new proteins. We identified the resulting T3 response genes by using an X. laevis microarray. Gene ontology (GO) analysis revealed that these genes are enriched in categories important for the earliest steps of cellular transformations during metamorphosis. To determine whether these genes are regulated by TR directly at the transcriptional level, we then carried out bioinformatics analysis of the homologous genes in X. tropicalis followed by in vitro DNA binding studies. These results showed that essentially all of the genes had one or more functional TREs in and around their promoters. More importantly, we demonstrated that TR was bound to these TREs in tadpoles and regulated their promoters in vivo. Thus, our studies not only identified many direct TR target genes in vivo but also revealed a number of signaling transduction pathways that are regulated by T3 as the first step toward inducing metamorphosis.
MATERIALS AND METHODS
RNA Sample Collection, Probe Preparation, and Microarray Hybridization
X. laevis tadpoles (stage 45/46, 10 days old, Nasco Sci., Fort Atkinson, WI) were treated in 0.1× Marc's modified Ringer solution alone, with 100 nm 3,5,3′-triiodo-l-thyronine (T3), with protein synthesis inhibitors (referred to as cycloheximide (CHX, 20 μg/ml) and anisomycin (25 μg/ml), which is known to inhibit over 95% of new protein synthesis in vivo (40)) or with T3 and CHX. Each treatment included 12 tadpoles and was replicated 3 times. For tadpoles treated with both T3 and CHX, the tadpoles were treated with CHX for 1 h before T3 addition. The tadpoles were treated in T3 for 14 h and/or CHX for 15 h. They were sacrificed, and total RNA was isolated using TRIzol reagent (Invitrogen). The RNA samples were further purified using LiCl precipitation according manufacturer's recommendations (Ambion, Austin, TX). The RNA samples were analyzed using Bioanalyzer (Agilent Technologies, Santa Clara, CA) to check RNA quality. 2 μg RNA from each sample was used to prepare cDNA, and then Cy3-labeled cRNA using a Low RNA Input Linear Amplification Kit from Agilent Technologies was used. The Cy3-labeled cRNA and Cy5-labeled X. laevis Universal Reference cRNA (made from mRNA isolated from tadpoles at multiple stages of development) (31, 32) were hybridized together onto the X. laevis 60-mer oligonucleotide microarray from Agilent Technologies (AMADID# 013665) using the Microarray Hybridization kit (Agilent Technologies) with a two-color reference design system as described (31, 32). After 17-h hybridization at 65 °C, microarray slides were washed according to manufacturer's instructions and scanned with an Agilent Technologies Microarray scanner. Intensity value of each feature was obtained by the Feature Extraction software from Agilent, and statistically significant differentially expressed genes were obtained using Genespring GX software by performing one-way analysis of variance without assuming that all variances were equal and by setting the Benjamini and Hochberg false discovery rate to 5% for multivariate correction (41, 42).
Real-time qRT-PCR Assays Using SYBR Green Dye and TaqMan Probes
cDNA was prepared from 2 μg of total RNA using the Applied Biosystems' High Capacity cDNA Archive kit according to the manufacturer's instructions in a total reaction volume of 50 μl. For quantitative reverse transcription (qRT)-PCR using TaqMan probes, 4 μl of cDNA was used in each reaction using Applied Biosystems 2× PCR Master mix and 20× or 60× TaqMan probe mix. Ribosomal protein L8 was analyzed as a normalization control (43). For qRT-PCR based on SYBR Green detection, 2 μl of cDNA was used for each reaction, and EF1α was used as the control. The primer sequences are listed in Table 1 or as published previously (31, 43).
TABLE 1.
Primer pairs used for SYBR Green qRT-PCR
| Gene | GenBankTM accession | Strand | Sequence (5′–3′) |
|---|---|---|---|
| Cyclin J | BC076737 | Forward | AGATACAAAGAACTGAAACTTCC |
| Reverse | TAAAGAGATCCAGCAGGTAAAC | ||
| Cyclin F | BC056134 | Forward | ATGAAGGGAGGAGCCTTGC |
| Reverse | CAAATGTGGATGAACCTCTCG | ||
| REV1 | BC070743 | Forward | ATCCGTTGGCATAGGTTC |
| Reverse | ACTCTATAGTCCTGCCAACTCC | ||
| RCOR2 | BC070565 | Forward | CACTGAAACGCCAGGTTC |
| Reverse | CAGGAGTTGTTCGTCTGTAGTC | ||
| EF1α | M25504 | Forward | CTATCCCCGCCAAACATCT |
| Reverse | CCATCTCAGCAGCTTCCTTC |
Bioinformatic Search for TREs in X. tropicalis Genes
Expressed sequence tag or cDNA sequences of CHX-resistant T3-regulated genes were obtained from NCBI Entrez. These sequences were searched against the protein data base for X. tropicalis from Ensembl release 48 (October, 2007, available on-line) using the blastx algorithm. The results were cross-checked for consistency against the Joint Genome Institute website to confirm that the correct gene model was obtained. 4000-bp sequence (2000 bp upstream and 2000 bp downstream) around the putative transcriptional start site (either the 5′-end of the Joint Genome Institute annotated cDNA or the homologous position of the 5′-end of the X. laevis cDNA sequence) was extracted. To search these promoter regions for TRE, the fuzznuc program in the EMBOSS suite of programs was used (44). The pattern used was (A/G)(A/G)GT(C/T)ANNNN(A/G)(A/G)GT(C/T)A with two mismatches allowed.
Gel Mobility Shift Assay
TRα and RXRα proteins were obtained by coupled transcription/translation from cDNA cloned in pSP64 (poly(A)) vector (45) using the TnT® SP6 Quick Coupled Transcription/Translation System from Promega (Madison, WI). For each in vitro translation reaction, 1.5 μg of plasmid DNA was used in a 50-μl reaction. TRα and RXRα reactions were mixed after the in vitro translation to yield 100 μl of TRα/RXRα solution. IR700-labeled TREs from the promoter region of Xenopus TRβA gene (34, 46) were custom made by Integrated DNA Technologies (Coralville, IA). Both sense and antisense oligonucleotides were 5′ IR700-labeled. They were annealed together, and 50 fmol was used in each reaction. The oligonucleotides (sense and antisense) for different TREs or the mutant TRE from TRβA gene (34) were annealed and diluted to 200 fmol/μl, 1 pmol/μl, and 5 pmol/μl to obtain 4×, 20×, and 100× unlabeled oligonucleotides for competition. The total volume of each gel mobility shift reaction was 15 μl. All gel mobility shift assays were done by incubating the mixture of 1 μl of TRα/RXRα protein mix with 1 μl of 50 fmol/μl, 3 μl of 5× gel shift binding buffer (Promega), and 1 μl of appropriate dilution of unlabeled competitor oligonucleotides at room temperature for 20 min, followed by the addition of 1.5 μl of Orange loading dye (LI-COR Corp.) and analysis on 6% DNA retardation gels (Invitrogen). Gels were run at 100 V for 1 h and then scanned using a LI-COR Odyssey Infrared scanner.
ChIP Assay
Stage 54 X. tropicalis tadpoles (Nasco Sci.) were treated with 10 nm T3 for 48 h in 0.1× Marc's modified Ringer solution (or in 0.1× Marc's modified Ringer solution only for control tadpoles), and chromatin immunoprecipitation (ChIP) was done with antibodies for TR and ID14, an extracellular protein as a negative control (47), using the protocol described before (46). All the treatment and control groups had three replicates, and each replicate consisted of three tadpoles. For TR immunoprecipitation, the new anti-TR antibody (PB) was used (39). Primers designed around the putative TREs or a downstream exon region as a negative control (listed in supplemental Table 10A) were used in real-time quantitative PCR using SYBR Green for detection. All the enrichment data are represented as the means ± S.E. of the percentage of input chromatin, and a p value ≤ 0.05 was considered to be statistically significant using Student's t test.
Cloning of Promoter Regions
Promoter regions of cyclin J, collagenase-3, and deiodinase-3 genes were PCR-amplified from X. tropicalis genomic DNA with appropriate primers (listed in supplemental Table 10B). These regions were chosen to include the putative TRE regions and the promoters. The PCR-amplified products from cyclin J and collagenase-3 promoter regions were cloned into the KpnI and XhoI sites of pGL4.10 vector (Promega). Deiodinase-3 promoter region was cloned into the XhoI and HindIII sites of pGL4.10. The pGL-TRE luciferase reporter vector (TRE-Luc) containing the T3-dependent promoter of the X. laevis TRβA gene driving the expression of the firefly luciferase has been described before (48).
Transcription Assay in the X. laevis Oocyte System
The pSP64 plasmids containing TR and RXR, which encode X. laevis TRβA and RXRα, were linearized and transcribed in vitro using a SP6 kit (Ambion) as previously described (45). The cytoplasm of stage VI oocytes from X. laevis was injected with 5.75 ng/oocyte of the TR and RXR mRNAs. Four hours later, the luciferase reporter TRE-Luc plasmid DNA (0.33 ng/oocyte), serving as a positive control, and the control vector, phRG-TK (0.03 ng/oocyte), which contained the herpes simplex virus thymidine kinase promoter driving the expression of the Renilla luciferase, were co-injected into the nucleus. In parallel, the firefly luciferase reporter under the control of the cyclin J, collagenase-3, or deiodinase-3 promoter (0.33 ng/oocyte) and the control vector phRG-TK (0.03 ng/oocyte) were co-injected similarly into the nucleus. After incubation at 18 °C overnight in the presence or absence of 100 nm T3 (45), the injected oocytes were prepared for luciferase assay using the Dual-Luciferase-Reporter Assay system according to the manufacturer's protocol (Promega). Five oocytes per sample were lysed in 75 μl of 1× lysis buffer (Promega), and 10 μl of the lysate was used for luciferase assay. Five independent samples were done for each injection at the same time, and the experiments were repeated three times. The relative expression of firefly luciferase from the reporter plasmid to Renilla luciferase from the control plasmid was determined and is reported here. Each data point represents the average from each group. Data are represented as the means ± S.E.; a p value ≤ 0.05 was considered to be statistically significant using Student's t test, and T3 regulation for all promoters were found to be significant.
RESULTS
Identification of the Direct Response Genes of Thyroid Hormone in X. laevis
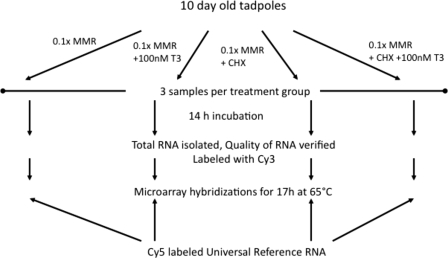
To identify immediate early, direct T3 response genes, we treated 10-day-old X. laevis tadpoles, which were approximately at stage 45/46 and competent to respond to exogenous T3 due to the expression of high levels of TRα (4, 49, 50), with four different solutions: control (0.1× Marc's modified Ringer solution, group 1), 100 nm T3 (group 2), protein synthesis inhibitors (CHX group, group 3), and CHX plus 100 nm T3 (group 4) (Fig. 1). Each treatment was done in three-independent repeats simultaneously. Total RNA was isolated from whole tadpoles and subjected to microarray analysis with a 60-mer oligonucleotide microarray. For microarray analysis, we used a two-color labeling system, with Cy3-labeled experimental sample and Cy5-labeled universal control made of RNA isolated from whole animals of different metamorphic stages as the internal reference (Fig. 1). Quality control of the data were performed as previously described (31) and demonstrated that the three replicas of each treatment group had highly reproducible gene expression profiles (data not shown).
FIGURE 1.
Schematic representation of the experimental design for the X. laevis microarray analysis.
To identify significantly regulated genes, we performed analysis of variance across all treatment groups with statistical significance of 5% false discovery rate with the -fold change cut-off value set at ≥1.3 for the regulated genes. Because of the reproducibility of the microarray and the use of three biological replicates/treatment, it was possible to obtain statistically significant changes at this fold change cut-off. (It is worth pointing out that due to the very short treatment time, the induction observed under the current conditions was much lower than that after longer treatment for well characterized genes such as TRβ and stromelysin-3, which is expected. Thus, the T3 induction in gene expression will likely be more dramatic in development in vivo. Thus, we set the cut-off at the relatively low level of 1.3). This analysis led to the identification of 188 genes up-regulated and 249 genes down-regulated by T3 in the presence of CHX (group 4 compared with group 3) (Table 2, supplemental Tables 1 and 3, and Fig. 2, A and B). In addition, 311 genes were up-regulated and 162 genes were down-regulated by T3 alone (in the absence of CHX, i.e. group 2 compared with group 1) (supplemental Tables 2 and 4). The genes regulated by T3 alone likely included both immediate early, direct response and late response genes, with the regulation of the latter sensitive to CHX. Thus, one might expect that all genes regulated by T3 plus CHX were among the genes regulated by T3 alone. Surprisingly, only 71 genes were commonly up-regulated and 29 genes commonly down-regulated by T3 or T3 plus CHX (Fig. 2). Although most CHX-resistant T3 response genes were not found as T3 response genes in the absence of CHX during the 15-h treatment, they were likely true T3 response genes, but their regulation by T3 alone was not detected during the relatively short treatment period under our experimental conditions (see “Discussion”).
TABLE 2.
List of CHX-resistant genes up-regulated by T3 (>1.5-fold)
| GenBankTM accession number | Gene symbol | Gene name | -fold change |
|
|---|---|---|---|---|
| CHX+T3 versus CHX | T3 versus control | |||
| CB943171 | EST, transcribed locus | 8.75 | 4.34 | |
| BC079830 | BXDC1 | brix domain containing 1 | 8.18 | 4.44 |
| AF170337 | EST, transcribed locus (IM28) | 5.20 | 1.46 | |
| U37375 | NFIL3 | TH/bZIP (E4BP4) | 3.62 | 2.52 |
| M35362 | THRB | Thyroid hormone receptor β | 3.23 | 2.75 |
| BC073008 | DNASE1L3 | Deoxyribonuclease I-like 3 | 3.07 | 1.96 |
| U35408 | KLF9 | Kruppel-like factor 9 | 2.98 | 2.41 |
| BX853054 | EST, transcribed locus | 2.80 | 1.73 | |
| BC088671 | PAAF1 | Proteasomal ATPase-associated factor 1 | 2.70 | 2.29 |
| BC081152 | NFIX | NFI-X2 transcription factor | 2.60 | 2.82 |
| BC076737 | CCNJ | Cyclin J | 2.49 | 2.33 |
| BC070743 | REV1 | REV1 homolog | 2.40 | 1.98 |
| BC060423 | TIMP3 | Tissue inhibitor of metalloproteinases-3 (TIMP3) | 2.36 | 2.28 |
| BQ399739 | EST, transcribed locus | 2.33 | 1.80 | |
| CD255175 | HIGD1B | HIG1 domain family, member 1B | 2.31 | 0.65 |
| Z27093 | MMP11 | Stromelysin-3 | 2.30 | 3.75 |
| U41855 | Gene 12b-1, no open reading frame | 2.30 | 3.46 | |
| BC091709 | MGAT4B | Mannosyl (α-1,3-)-glycoprotein β-1,4-N-acetylglucosaminyltransferase, isozyme B | 2.16 | 1.46 |
| BC056134 | CCNF | Cyclin F | 2.15 | 1.44 |
| BQ733202 | EST, transcribed locus | 2.12 | 1.44 | |
| U41824 | MMP13 | Matrix metallopeptidase 13 (collagenase 3) | 2.12 | 1.44 |
| BX844667 | EST, transcribed locus | 2.11 | 1.43 | |
| CO389665 | ANKRD42 | Ankyrin repeat domain 42 | 2.10 | 1.00 |
| BX844877 | EPPK1 | Epiplakin 1 | 2.07 | 1.00 |
| BQ728188 | EST, transcribed locus | 2.02 | 1.00 | |
| L28111 | DIO3 | Deiodinase, iodothyronine, type III | 1.97 | 1.95 |
| L29495 | PIM1 | X. laevis Pim-1 oncogene | 1.96 | 2.18 |
| BG264771 | EST, transcribed locus | 1.94 | 1.49 | |
| BC084073 | FAM139A | Family with sequence similarity 139, member A | 1.91 | 1.37 |
| BJ032419 | EST, transcribed locus | 1.89 | 1.00 | |
| BC060754 | CORO1C | Coronin, actin binding protein, 1C | 1.84 | 1.51 |
| CD254756 | EST, transcribed locus | 1.77 | 1.00 | |
| BG346626 | FHOD1 | Formin homology 2 domain containing 1 | 1.76 | 1.00 |
| BX855084 | EST, transcribed locus | 1.74 | 1.47 | |
| CB559498 | EST, transcribed locus | 1.74 | 1.37 | |
| BJ628593 | NAALAD2 | N-Acetylated α-linked acidic dipeptidase 2 | 1.72 | 1.32 |
| BC081091 | GBP3 | Guanylate-binding protein 3 | 1.72 | 1.00 |
| AF351126 | EZH2 | Enhancer of zeste homolog 2 | 1.70 | 1.61 |
| BC071004 | SULT2B1 | Sulfotransferase family, cytosolic, 2B, member 1 | 1.70 | 1.62 |
| BJ039688 | CASR | Calcium-sensing receptor | 1.70 | 1.37 |
| BC068899 | XPNPEP2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound | 1.70 | 1.33 |
| BJ035117 | EST, transcribed locus | 1.68 | 1.50 | |
| BG017896 | SEH1L | SEH1-like | 1.68 | 1.00 |
| BG022981 | EST, transcribed locus | 1.67 | 1.00 | |
| BC056049 | MAPKSP1 | Mitogen-activated protein kinase scaffold protein 1-B | 1.67 | 1.50 |
| BE507589 | HAL | Histidine ammonia-lyase | 1.65 | 2.36 |
| BJ065503 | GJB1 | Gap junction protein, β1, 32 kDa | 1.63 | 1.30 |
| BC072927 | FKBP2 | FK506-binding protein 2, 13 kDa | 1.62 | 1.42 |
| AY685227 | GDF6 | Growth differentiation factor 6 | 1.61 | 1.70 |
| AJ009285 | HES5 | Hairy and enhancer of split 5 (Drosophila) | 1.61 | 1.00 |
| CD300904 | EST, transcribed locus | 1.60 | 1.79 | |
| BC084912 | BCL6 | B-cell CLL/lymphoma 6 | 1.60 | 1.70 |
| BQ731946 | APOH | Apolipoprotein H (β2-glycoprotein I) | 1.58 | 1.00 |
| BC071014 | CCNB1 | Cyclin B1 | 1.58 | 1.00 |
| BC093541 | AMDHD1 | Amidohydrolase domain containing 1 | 1.56 | 1.32 |
| BJ630946 | TMCO4 | Transmembrane and coiled-coil domains 4 | 1.56 | 1.92 |
| U76636 | CALB1 | Calbindin 1, 28 kDa | 1.55 | 1.00 |
| BC070565 | RCOR2 | REST corepressor 2 | 1.55 | 1.77 |
| BC081124 | IPMK | Inositol polyphosphate multikinase | 1.55 | 1.00 |
| CD256754 | MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 1.54 | 1.36 |
| BC077863 | EIF2C2 | Eukaryotic translation initiation factor 2C, 2 | 1.53 | 1.00 |
| BC081082 | NR3C2 | X. laevis mineralocorticoid receptor mRNA | 1.52 | 1.48 |
| BQ731635 | EST, transcribed locus | 1.52 | 1.00 | |
| CA973474 | SPINT1 | Serine peptidase inhibitor, Kunitz type 1 | 1.51 | 1.00 |
| BC044272 | KDELR2 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 | 1.51 | 1.00 |
| BE509179 | EST, transcribed locus | 1.50 | 1.00 | |
| CA971171 | SIGLEC12 | Sialic acid binding Ig-like lectin 12 | 1.50 | 0.73 |
| BI441787 | EST, transcribed locus | 1.50 | 1.00 | |
FIGURE 2.
Venn diagrams showing that only a small fraction of genes are commonly regulated by T3 or T3 in the presence of CHX. A, 311 genes were up-regulated by T3 alone (T3 group compared with control group), whereas 188 were up-regulated by T3 in the presence of CHX (T3 plus CHX group compared with CHX group). Among them, 71 genes were commonly up-regulated by T3 or T3 plus CHX. B, 162 genes were down-regulated by T3 alone (T3 group compared with control group), whereas 249 were down-regulated by T3 in the presence of CHX (T3 plus CHX group compared with CHX group). Among them, 29 genes were commonly down-regulated by T3 or T3 plus CHX.
Confirmation of the Regulated Genes by qRT-PCR Assay in X. laevis
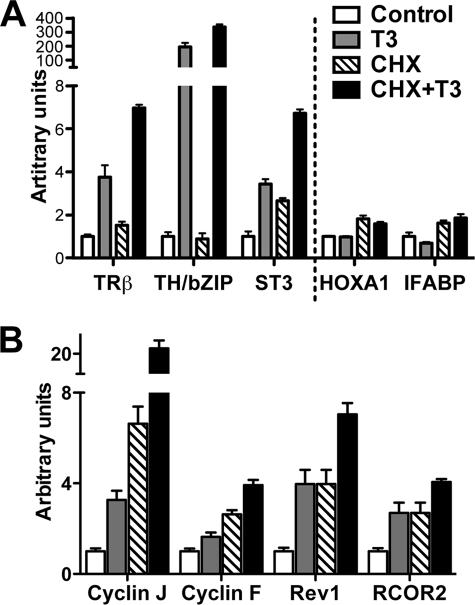
Having identified the regulated genes by microarray analysis, we next validated the regulation of these genes by T3 and or T3 plus CHX. We analyzed the expression of a number of genes regulated by T3 in the array in independently isolated total RNA samples using either TaqMan probe-based or SYBR Green detection-based qRT-PCR. We tested eight genes with TaqMan probes in qRT-PCR assay: three previously known direct up-regulated genes, TRβA (34, 51), TH/bZIP (35), and stromelysin-3 (36), four genes known to be late T3 response genes, BMP-4 (52), gelatinase A (53), IFABP (54), and TIMP2 (55), and one gene independent of T3, HOXA1 (56) (Fig. 3A and data not shown). The results were all consistent with the data obtained from microarray analysis. In addition, we used SYBR Green detection-based qRT-PCR to analyze four newly identified genes induced by T3 plus CHX: cyclin J, cyclin F, REV1, and RCOR2. Again, the qRT-PCR results confirmed the microarray findings (Fig. 3B).
FIGURE 3.
Verification of microarray data by qRT-PCR. Primer sets for indicated genes were used for qRT-PCR analysis with independently isolated total RNA from control, T3-treated, CHX-treated, and T3 plus CHX-treated X. laevis tadpoles (n = 4 in each treatment group). A, three previously known T3 direct-regulated genes, one late T3-down-regulated gene (IFABP), and one non-regulated gene (HOXA1) were analyzed by TaqMan qRT-PCR with rpL8 (ribosomal protein L8) as the control gene (non-regulated). Note that IFABP was down-regulated by T3 alone but not by T3 plus CHX. B, four newly identified T3 direct response genes were analyzed by SYBR Green qRT-PCR with EF1α (elongation factor 1α) as the control gene (non-regulated). For all genes, the signal was normalized with the control gene and the mean of the control group was set to 1. Student's t test was carried out between pairs of control and T3-treated groups and pairs of CHX-treated and CHX plus T3-treated groups, and all pairwise comparisons had p values <0.05.
Functional Classification of the Up-regulated Genes
We next used GO classification to identify significantly enriched functional categories in the genes up-regulated by T3 alone or by T3 in the presence of CHX. The gene symbols were derived from the homologous human genes as described before (31, 32). GoMiner software (57) was used to find these enriched GO categories with p value set at <0.05. Among GO categories with three or more genes up-regulated by either T3 or T3 in the presence of CHX, many categories related to metabolism/catabolism or cell proliferation (cell cycle/DNA replication) were found to be common between the T3 and T3 plus CHX groups, suggesting that the genes in these categories are involved in the early events of metamorphosis (Table 3A and supplemental Tables 5 and 6). Interestingly, careful comparisons of the two lists of enriched GO categories revealed that some of the categories enriched in T3 up-regulated genes were not enriched in CHX-resistant T3 up-regulated genes (Table 3A). For example, DNA replication genes (GO: 0006260) were highly enriched in the T3 up-regulated group (3.0-fold enrichment, p = 0.00) but not in CHX-resistant T3 up-regulated group (1.51-fold enrichment, p = 0.32). Similarly, ribosomal biogenesis and assembly genes (GO: 0042254) were highly enriched in T3-up-regulated genes (3.9-fold enrichment; p = 0.00) but not significantly enriched in the CHX-resistant up-regulated genes (0.0-fold enrichment; p = 1.00). These results suggest that, although cell cycle progression was induced by both T3 and T3 plus CHX, some late response genes, sensitive to CHX, are required for the cells to progress to DNA replication. Such late genes might be among the GO categories for DNA replication and ribosomal biogenesis and assembly. Likewise, apoptosis, another major event occurring during metamorphosis in many tissues (4), seemed to require genes that are sensitive to CHX, because genes in the apoptotic program were enriched in the T3 group but not in the T3 plus CHX group (Table 3A).
TABLE 3.
Selected GO categories significantly enriched in CHX-resistant T3 genes
| GO ID | GO category name | Total genes | Changed genes (p value/enrichment)a |
|
|---|---|---|---|---|
| CHX-resistant regulated genes | T3-regulated genes | |||
| A) Selected GO categories significantly enriched in CHX-resistant up-regulated genes and T3 up-regulated genes | ||||
| 0009310 | Amine catabolic process | 33 | 5 (0.00/8.3) | 5 (0.00/5.9) |
| 0042254 | Ribosome biogenesis and assembly | 58 | 0 (1.00/0.0) | 7 (0.00/3.9) |
| 0007154 | Cell communication | 1237 | 21 (0.03/1.37) | 41 (0.33/1.07) |
| 0007049 | Cell cycle | 385 | 12 (0.04/1.7) | 21 (0.01/1.8) |
| 0006260 | DNA replication | 109 | 3 (0.32/1.5) | 10 (0.00/3.0) |
| 0006261 | DNA-dependent DNA replication | 44 | 0 (1.00/0.0) | 5 (0.01/3.7) |
| 0008632 | Apoptotic program | 48 | 2 (0.22/2.28) | 5 (0.02/3.4) |
| 0042592 | Homeostatic process | 177 | 9 (0.00/2.8) | 6 (0.47/1.09) |
| 0003707 | Steroid hormone receptor activity | 23 | 4 (0.00/9.5) | 2 (0.16/2.8) |
| 0003700 | Transcription factor activity | 373 | 14 (0.00/2.1) | 13 (0.37/1.1) |
| 0008237 | Metallopeptidase activity | 65 | 5 (0.01/4.2) | 7 (0.00/3.5) |
| B) Selected GO categories significantly enriched in CHX-resistant T3 down-regulated genes and T3 down-regulated genes | ||||
| 0030154 | Cell differentiation | 739 | 21 (0.048/1.5) | 8 (0.83/0.76) |
| 0007010 | Cytoskeleton organization and biogenesis | 223 | 10 (0.01/2.3) | 5 (0.2/1.6) |
| 0051301 | Cell division | 138 | 7 (0.02/2.6) | 1 (0.86/0.5) |
| 0005819 | Spindle | 46 | 5 (0.00/5.6) | 2 (0.14/3.06) |
| 0048513 | Organ development | 460 | 12 (0.19/1.4) | 13 (0.01/2.0) |
| 0001501 | Skeletal development | 87 | 2 (0.51/1.2) | 5 (0.01/4.1) |
| 0006811 | Ion transport | 246 | 9 (0.01/2.6) | 9 (0.051/1.9) |
a Numbers of changed genes and enrichment factors are shown in bold in statistically significant GO categories (p < 0.05 as determined by the GoMiner software).
Conversely, GO categories related to transcriptional regulation, such as the transcription factor activity category (GO: 0003700), were significantly enriched in the CHX-resistant T3-up-regulated genes, but not in the T3 up-regulated genes (Table 3A and supplemental Tables 5 and 6). This may not be surprising as it is expected that transcriptional regulation should be an important initial event of many signaling pathways. Taken together, the gene ontology analysis indicates that the T3 direct-response genes (i.e. CHX-resistant T3 up-regulated genes) may not be sufficient for DNA replication or ribosomal biogenesis by themselves and hence will not drive cell proliferation. They, however, are likely critical to initiate cell cycle progression and/or other signaling processes by regulating the transcription of downstream genes.
In contrast to the up-regulated categories, there was little overlap among the significantly enriched GO categories between the genes down-regulated by T3 and the genes down-regulated by T3 in the presence of CHX (Fig. 3B and supplemental Tables 7 and 8). This is consistent with the fact that a lot fewer genes were found to be commonly down-regulated by T3 and T3 plus CHX compared with the up-regulated genes (Fig. 2). Interestingly, a number of categories involved in cellular processes, such as cell differentiation and cytoskeletal changes, were significantly enriched in genes down-regulated by T3 in the presence of CHX, whereas a number of categories at the organismal level, such as organ development and multicellular organismal processes, were significantly enriched in genes down-regulated by T3 alone (Table 3B). Such changes suggest that down-regulation of cellular processes occurs prior to the changes at the organ levels, which would be expected, and that the latter requires late/indirect T3 response genes.
Bioinformatic Search for Putative TREs in the X. tropicalis Homologs of the CHX-Resistant T3-up-regulated Genes Found in X. laevis
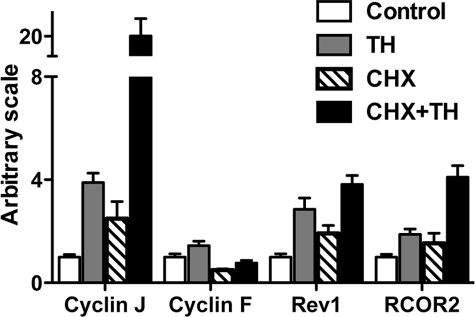
Although immediate early, direct TR target genes are expected to be regulated by TR without a requirement for new protein synthesis, it is possible that some of the genes whose regulation by T3 is resistant to CHX may not be direct target genes of TR, e.g. through non-genomic effects of T3 (58). (Note there should also be direct T3 response genes that are sensitive to CHX. The regulation of such genes by T3 occurs later and requires the synthesis of other proteins even though TR can bind directly to their regulatory regions. Such genes are outside of the scope of this study). Thus, to investigate whether the CHX-resistant T3 response genes were direct TR target genes, we needed to determine whether the genes contained functional TREs. The lack of genomic sequence information in X. laevis made it difficult to carry out such analysis. On the other hand, a highly related species, X. tropicalis, had a nearly completely sequenced genome and an essentially identical developmental profile, although a shorter life cycle. More importantly, its TR and RXR genes are regulated and function similarly during development as in X. laevis and the homologs of several known TR target genes in X. laevis have been shown to be regulated by T3 identically in X. tropicalis (39). To investigate whether the newly identified CHX-resistant T3 response genes are similarly regulated by T3 in X. tropicalis, we carried out qRT-PCR analysis for four homologous genes in X. tropicalis, cyclin J, REV1, cyclin F, and RCOR2, by using total RNA isolated from X. tropicalis tadpoles treated with T3 and/or CHX. All of them were found to be regulated in X. tropicalis just like in X. laevis (Fig. 4, compared with Fig. 3B), suggesting that it is likely that most, if not all, of the newly identified target genes are regulated similarly in X. tropicalis as in X. laevis.
FIGURE 4.
Conservation of the T3-regulation of newly identified genes in X. tropicalis. Selected up-regulated genes were analyzed by SYBR Green qRT-PCR in X. tropicalis tadpoles. Primers sets of four selected T3 direct response genes (same genes as in Fig. 3B) were used to analyze the expression of the genes in total RNA from control, T3-treated, CHX-treated, and T3 plus CHX-treated X. tropicalis tadpole samples (n = 4 in each treatment group). EF1α (elongation factor 1α) was also analyzed as the control gene (non-regulated). For all genes, the signal was normalized with the control gene and the mean of the control group was set to 1. Student's t test was carried out between pairs of control and T3-treated groups, and pairs of CHX-treated and CHX+T3-treated groups and all pairwise comparisons had p values <0.05.
The conservation in their regulation makes it possible to carry out a bioinformatic search for TREs in the target genes. For this purpose, we focused on the CHX-resistant T3 up-regulated genes, because much less is known about how TR down-regulates gene expression in the presence of T3. To do this, we chose the 68 genes whose regulation by T3 in the presence of CHX in X. laevis was 1.5-fold or higher (Table 2) to reduce the likelihood of a gene being a false positive from the microarray analysis. We identified X. tropicalis homologs of these genes by using the blastx algorithm (ftp.ncbi.nlm.nih.gov/blast/) (59). This allowed us to map 46 genes out of the 68 X. laevis CHX-resistant T3 up-regulated genes to the X. tropicalis genome-annotated protein data base (the other 22 genes were not found most likely due to incomplete genome sequence and/or annotation). We then extracted a 2000-bp sequence from either side of the putative transcriptional start site (4000-bp total) and used the fuzznuc program within the EMBOSS suite of programs to search for TRE in these 4000- bp promoter regions of the CHX-resistant genes in the X. tropicalis genome. We searched for the pattern 5′-(A/G)(A/G)GT(C/T)ANNNN(A/G)(A/G)GT(C/T)A-3′. The positive alignments were allowed up to two mismatches. This search yields TRE-like element in all but one of the promoters of CHX-resistant genes identified (supplemental Table 9).
In Vitro Binding of TR/RXR Heterodimers to the Putative TREs
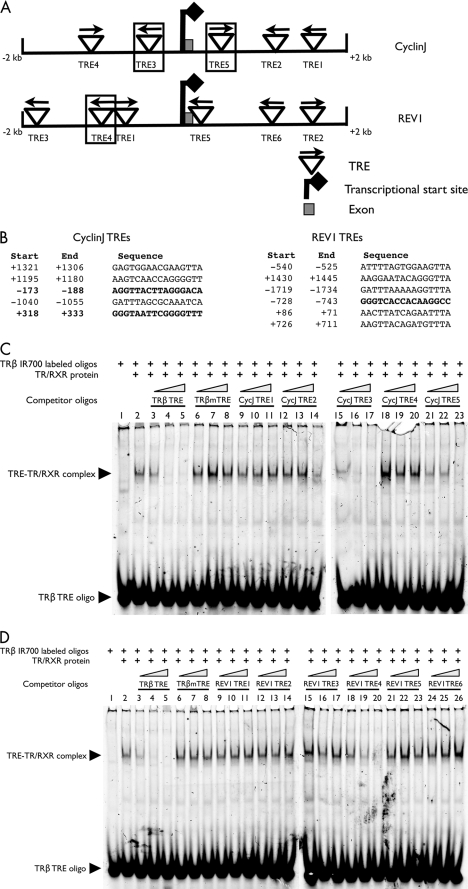
We used a gel-mobility assay to identify the TREs that can bind to TR biochemically. We used the TRE located in the T3-dependent X. laevis TRβA gene promoter as the positive control (34, 51). For T3-induced genes, TR normally functions as a heterodimer with RXR. Thus, to investigate the ability of the putative TREs in the newly identified target genes to bind to TR/RXR heterodimers, we labeled the double-stranded TRβ TRE oligonucleotide and used it for in vitro binding with in vitro translated TR and RXR. We competed against the complex formation between the TRβ TRE and the TR/RXR with the double-stranded oligonucleotides made of the putative TREs in the promoter regions of eight direct target genes (PAAF1, NFIX, collagenase-3, DNaseIL3, deiodinase-3, cyclin J, gene 12b1, and REV1) (supplemental Table 9). The results for two of the genes, cyclin J and REV1, are shown in Fig. 5. As shown in Fig. 5 (C and D), the TRβ TRE formed a complex with TR/RXR (lane 2), and this complex was effectively competed away by the unlabeled TRβ TRE itself (lanes 3–5) but not by a mutated version of the TRβ TRE (TRβ mTRE) (lanes 6–8), demonstrating the specificity of the binding reaction in vitro. Among the unlabeled double-stranded oligonucleotides from cyclin J (Fig. 5C) and REV1 (Fig. 5D) promoter regions, cyclin J TRE3 (Fig. 5C, lanes 15–17) and TRE5 (Fig. 5C, lanes 21–23), and REV1 TRE4 (Fig. 5D, lanes 18–20) competed effectively, whereas the other putative TREs did not, suggesting that these three TREs can bind to TR/RXR at least biochemically in vitro. Using this assay, at least one functional TRE was confirmed for each of the eight genes tested (Fig. 6A).
FIGURE 5.
The binding of TR/RXR heterodimers to the TREs identified in the direct response genes. A, schematic diagram of the putative TREs in the cyclin J and REV1 promoter regions (−2000 to +2000 bp around the transcriptional start sites). The TREs are shown as up-side-down triangles with arrows indicating the orientation of each TRE. Each confirmed TRE is shown with a box drawn around it. B, the sequences of the putative TREs are shown with the positions (relative to the transcriptional start sites). The confirmed TREs are in bold letters. C and D, in vitro recognition of the putative TREs by TR/RXR heterodimers. The binding activity of cyclin J (CycJ) and REV1 TREs to in vitro translated TRα/RXRα heterodimers was analyzed by gel retardation assay to determine their ability to compete away the formation of the complex between the end-labeled X. laevis TRβ promoter TRE and in vitro translated TRα/RXRα. The competition was done with 4-, 20-, or 100-fold of indicated, unlabeled competitor TRE oligonucleotides: the TRβ TRE itself, mutated TRβ TRE (TRβ mTRE), or the putative TREs from the CycJ promoter region (C) or REV1 promoter region (D). The resulting binding mixture was analyzed on native polyacrylamide gels.
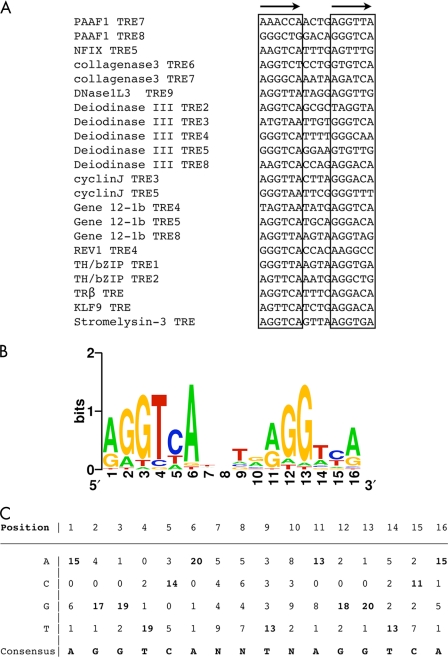
FIGURE 6.
A consensus TRE for Xenopus TR target genes. A, sequences of the 17 new TREs identified in X. tropicalis genes from this study along with the five known TREs for X. laevis genes (the bottom five). B, graphic representation of the consensus TRE generated from the 22 TREs above. The positions of residues in the TRE are shown on the horizontal axis, and the relative frequencies of the four bases at each position are shown in color as bits in the vertical axis. C, the frequencies of the four bases at each position in the 22 Xenopus TREs. The consensus TRE is shown at the bottom.
Consensus TRE Motif for the Xenopus T3-up-regulated Genes
By using a position-specific probability matrix (MEME, available on-line) (60), we generated a consensus TRE for Xenopus TR target genes (Fig. 6, B and C) from the 17 newly identified X. Tropicalis TREs and the 5 previously identified TREs from X. laevis genes TRβA (34), TH/bZIP (two TREs) (35), stromelysin-3 (36), and KLF9 (35) (Fig. 6A). The results from this rather larger set of biochemically characterized naturally occurring TREs indicate that AGGTCAnnTnAGGTCA is the optimal target sequence of endogenous target genes for TR/RXR heterodimers, consistent with previous findings from in vitro and mammalian cell culture studies (61, 62).
In Vivo Regulation of Direct Response Gene Promoters by T3
Having shown that the newly identified direct T3 response genes contain TREs capable of binding to TR/RXR heterodimers in vitro, we next investigated whether the corresponding promoters can be regulated by TR/RXR in vivo in the context of chromatin. For this purpose, we PCR-amplified the promoter regions, including the TREs of the cyclin J, collagenase-3, and deiodinase-3 genes from X. tropicalis. These genes were chosen in part due the relatively small sizes of the regions that contain both the promoter and the TREs. The PCR products were placed in front of the firefly luciferase reporter gene in pGL4.10 vector. To study their regulation by TR, we use the X. laevis oocyte transcription system, where we can study promoter regulation in the context of chromatin (63). We first introduced TRα and RXRα into frog oocyte by microinjecting their mRNAs into the cytoplasm. Four hours later, the firefly luciferase reporter plasmid and an internal control plasmid, which contained the Renilla luciferase under the control of the T3-independent TK promoter, were co-injected into the oocyte nucleus. After overnight incubation in the presence or absence of T3, the oocytes were collected to assay for the activities of the firefly luciferase over those of the Renilla luciferase. The results showed that the expression of TR/RXR in the absence of T3 led to a reduction in the activity of the promoters, and the addition of T3 caused strong activation of these promoters, similar to that observed for the X. laevis TRβA promoter (Fig. 7A), a well characterized direct T3-responsive promoter (34, 51). These results indicate that TR/RXR regulates cyclin J, collagenase-3, and deiodinase-3 promoters in vivo.
FIGURE 7.
TR regulates the T3-direct response genes in vivo. A, TR/RXR heterodimer activates cyclin J, collagenase-3, and deiodinase-3 promoters in the presence of T3. The indicated promoter constructs for firefly luciferase were co-injected with the internal control plasmid phRG-TK driving the expression of Renilla luciferase into the nuclei of the oocytes with or without prior microinjection of mRNAs for TRα and RXRα (TR/RXR) into the cytoplasm. The oocytes were incubated at 18 °C overnight in the presence or absence of 100 nm T3 and then subjected to dual luciferase assays. The normalized firefly luciferase activities of the reporters were plotted. The bars represent the means ± S.E. of one independent experiment performed in quintuple (p < 0.05 was found for all samples); the experiment was performed three times with similar results. B, TR is bound to the TREs of the newly identified target genes in tadpoles. Chromatin was isolated from three whole premetamorphic tadpoles treated in either the presence or absence of 10 nm T3 for 48 h and used in the ChIP assay. Nine TRE regions in four T3-responsive promoters (cyclin J, REV1, collagenase-3, and deiodinase-3) and a control DNA region, REV1 exon 5, were analyzed for TR binding after immunoprecipitation with an anti-TR antibody (new PB-1) by using quantitative PCR with the eluted DNA. The data are represented as percentage of input chromatin. The bars represent mean ± S.E. from two to three replicate experiments performed. Statistically significant (p < 0.05) increases in TR binding in response to T3 at some TREs are marked by an asterisk. Note that all TRE regions have statistically significant enrichment (p value < 0.05) compared with REV1 exon 5, although the ChIP signal varied considerably among different TREs (see text for more details).
In Vivo Binding of TR to the TRE Regions in Direct T3 response Gene Promoters
To determine whether these newly discovered direct T3 response genes are indeed regulated by TR directly in developing tadpoles, we then carried out ChIP assay to analyze the binding of TR to the TRE regions in premetamorphic X. tropicalis tadpoles treated with or without T3 for 2 days. The genomic DNA immunoprecipitated with an anti-TR antibody was analyzed by quantitative PCR for the presence of the TRE regions and of the exon 5 of REV1 gene, which served as a negative control. As shown in Fig. 7B, in the absence of T3 treatment, all the TRE regions were significantly enriched in the anti-TR ChIP DNA compared with the exon 5 region of REV1 gene, suggesting that TR is associated with the TREs in premetamorphic tadpoles. Upon T3 treatment, significant increases in TR binding were observed at the TRE3 of cyclin J TRE3, TRE4 of REV1, and TRE6 of collagenase-3, whereas no significant changes were found at the other TREs, similar the findings at TREs of several known T3 response genes in X. laevis (46, 64), where the T3-induced increase in TR binding has been attributed to the increased TR levels after T3 treatment, thereby leading to more binding of TR to some of the sites not yet full bound by TR. It should be pointed out that, although ChIP signals at different TREs varied considerably, such differences might not indicate the absolute binding of TR to the TREs differed similarly. This is because many factors could influence the ChIP assay outcome, e.g. other proteins at the TRE regions could affect the cross-linking or immunoprecipitation efficiencies during the ChIP assay. Thus, the valid conclusions to be drawn here were whether there was TR binding and if it changed upon T3 treatment. Our results here indicate that TR is bound to the newly identified direct T3 response genes in developing tadpoles.
DISCUSSION
T3 regulates diverse developmental processes and organ function and metabolisms in different vertebrate species. Although non-genomic actions of T3 undoubtedly contribute to some of the biological effects of T3 (58), TR is generally believed to be the main mediator of T3 action through transcriptional regulation. The immediate early, direct target genes of TR likely play critical roles to transduce the biological effects of T3. While a number of microarrays have been carried out to identify T3 response genes in different species, the T3-resposne genes identified from such studies include both immediate early, direct target genes of TR as well as downstream, late T3 response genes. Amphibian metamorphosis is an ideal model system to study T3 action in development because of its total dependence on T3 and the ability to easily manipulate this process without any potential complications of maternal effects as in mammals (3, 4). Furthermore, TR has been shown to be both necessary and sufficient for the metamorphic effects of T3, suggesting an essential role of direct TR target genes in this process (9, 17, 19). Here, we have combined the advantages of two highly related frog species, microarray analysis in X. laevis and genome sequence information in X. tropicalis, to not only identify immediate early, direct target genes of T3 but also provide evidence to support their regulation by TR in developing tadpoles in vivo. Furthermore, our gene ontology analysis has revealed interesting pathways induced by T3 at the earliest stages of the metamorphic process.
Our microarray analysis in X. laevis identified 188 genes up-regulated and 249 genes down-regulated by T3 in the absence of new protein synthesis. Interestingly, most of these genes were not found as T3 response genes in the absence of CHX during the 15-h treatment. Although the exact reasons remain to be determined, some possibilities include the following. First, CHX can stabilize some mRNAs (Fig. 3). This might have altered the magnitude of the changes for some genes. Some genes with the extent of T3 regulation near the 1.3-fold cut-off might change from being a regulated gene to being a non-regulated gene and vice versa in the presence of CHX. In addition, some of the T3-regulated genes might be expressed at very low levels and thus not detectable or with more variable signals on the microarray in the absence of CHX by the microarray. Upon stabilization by CHX, they were now detectable with more consistent signals as T3-regulated genes. Second, some of the genes regulated by T3 treatment alone were late response genes and were thus not regulated by T3 in the presence of CHX. Finally, undoubtedly some genes with a fold of regulation by T3 around the 1.3-fold cut-off might be missed in the T3 alone group or T3 plus CHX group due to some expected experimental variations. Thus, it is quite likely that most of the genes regulated by T3 in the presence of CHX are true T3 response genes during metamorphosis.
The above conclusion is also supported by our studies in X. tropicalis, a closely related diploid species of X. laevis within the Xenopodinae subfamily of anurans. We have shown previously that TR and RXR genes are regulated and function similarly in X. tropicalis and X. laevis (39). In addition, of the a few direct TR target genes in X. laevis that have been analyzed in X. tropicalis, all are regulated by T3 similarly in X. tropicalis (39).3 Our studies here also indicate that the corresponding genes of the newly identified CHX-resistant T3 response genes in X. laevis are similarly regulated in X. tropicalis, supporting the idea that the T3-dependent gene regulation program is conserved between X. tropicalis and X. laevis.
Furthermore, our bioinformatic search showed that of the 46 CHX-resistant T3-induced genes with clear homologs between X. laevis and X. tropicalis, 45 have candidate TREs within 2000 bp of their putative transcription start site. In addition, biochemical studies showed that, of the genes analyzed, each has at least one TRE capable of binding to the TR/RXR heterodimer strongly in vitro. More importantly, our in vivo ChIP assay has shown that TR is bound to the TREs of newly identified target genes in premetamorphic X. tropicalis tadpoles and that the binding to some of the TREs increases upon T3 treatment, just like that in X. laevis. By using the oocyte transcription system, we have further demonstrated that TR regulates the promoters of the newly identified genes in vivo in the context of chromatin. Thus, most, if not all, of the genes discovered here are likely directly regulated by TR during Xenopus development. These findings allowed us for the first time to generate a Xenopus consensus TRE based on a large number of biochemically characterized, natural occurring TREs. The consensus TRE and the degeneracy identified here should help to refine future searches for TREs in other TR target genes by using position-specific probability matrix (MEME) (60). Of particular interest in this regard is the identification of direct target genes of T3 in different metamorphosing tissues, which are likely important to dictate tissue-specific transformations, an area that we are currently pursuing.
Our microarray analysis showed that only a small fraction of genes was commonly regulated by T3 in the absence or presence of CHX. One reason is that the genes regulated by T3 alone likely include both immediate early, direct T3 response genes, which are CHX-resistant T3 response genes, and late, downstream T3 response genes. Thus, one would expect that the biological functions of the genes regulated by T3 in the absence of CHX and those in the presence of CHX would also only partially overlap. Indeed, our GO categories analysis revealed that there are a number of GO categories overlapping between the genes induced by T3 alone and those induced by T3 in the presence of CHX. On the other hand, there are few common GO categories between the down-regulated genes by T3 alone and those by T3 in the presence of CHX, consistent with the much fewer common down-regulated genes under the two conditions (Fig. 2).
The direct T3 response genes (regulated by T3 in the presence of CHX) are enriched in GO categories related to transcriptional regulation, such as transcription factors (Table 3 and supplemental Table 5). Some of these transcription factors have been reported previously as direct targets of T3, such as TRβ and TH/bZIP (34, 35, 65). Interestingly, GO categories related to transcriptional regulation are not significantly enriched in the genes regulated by T3 alone, likely because of the presence of many late, downstream response genes in this case. These results support the idea that a transcription factor-mediated gene regulation cascade plays an important role in metamorphosis and suggest that the earliest members of the gene expression pathways leading to subsequent morphological changes involve a disproportionately larger fraction of transcription factors.
Another GO category specifically enriched in the direct T3 target genes is metallopeptidases (Table 3 and supplemental Table 5). This suggests that the remodeling of the extracellular matrix and other protein degradation events are important early signaling events induced by T3 during metamorphosis. In fact, one of the first T3 direct response genes found during metamorphosis is stromelysin-3 (MMP-11) (36, 66–68), which has been shown to be essential for T3-induced cell death during metamorphosis (43, 69–71). Thus, signaling processes triggered through the action of metallopeptidases are another important early event toward regulating cell fate and behavior during metamorphosis.
In addition, our GO analysis revealed the enrichment of categories related to cell cycle and DNA replication in the genes induced by T3 alone and/or by T3 in the presence of CHX. Interestingly, whereas cell cycle categories were enriched in the direct target genes, DNA replication categories were enriched in the genes induced by T3 alone. These suggest that, in the absence of protein synthesis, T3 may induce cell cycle changes. The direct target genes by themselves, however, are not sufficient for DNA replication but rather prepare the cells to enter the S-phase. This is strongly supported by the enrichment of G1/S phase transition genes but the absence of DNA replication genes, e.g. the MCM genes involved in replication fork (72), in the direct T3 target genes (Table 3 and supplemental Table 5). Likewise, the absence of the GO categories, related to ribosomal biogenesis and assembly in the CHX-resistant T3 response genes, and their presence in the genes induced by T3 alone, suggest the possibility that downstream genes are required to sustain cell proliferation. Undoubtedly, it would be of importance in the future to investigate the function of the direct target genes to test these predictions.
Supplementary Material
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of NICHD.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–10.
B. Das, R. A. Heimeier, D. R. Buchholz, and Y.-B. Shi, unpublished observation.
- T3
- thyroid hormone
- TR
- thyroid receptor
- TRE
- thyroid response element
- RXR
- 9-cis-retinoic acid receptor
- GO
- Gene ontology
- CHX
- cycloheximide
- qRT
- quantitative reverse transcription
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Lazar M. A. (1993) Endocr. Rev. 14, 184–193 [DOI] [PubMed] [Google Scholar]
- 2.Yen P. M. (2001) Physiol. Rev. 81, 1097–1142 [DOI] [PubMed] [Google Scholar]
- 3.Tata J. R. (1993) BioEssays 15, 239–248 [DOI] [PubMed] [Google Scholar]
- 4.Shi Y. B. (1999) Amphibian Metamorphosis: From Morphology to Molecular Biology, John Wiley & Sons, Inc., New York [Google Scholar]
- 5.Hetzel B. S. (1989) The Story of Iodine Deficiency: An International Challenge in Nutrition., Oxford University Press, Oxford [Google Scholar]
- 6.Freake H. C., Oppenheimer J. H. (1995) Annu Rev. Nutr. 15, 263–291 [DOI] [PubMed] [Google Scholar]
- 7.Franklyn J. A., Gammage M. D. (1996) Trends Endocrinol. Metab. 7, 50–54 [DOI] [PubMed] [Google Scholar]
- 8.Silva J. E. (1995) Thyroid 5, 481–492 [DOI] [PubMed] [Google Scholar]
- 9.Shi Y. B. (2009) Thyroid 19, 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howdeshell K. L. (2002) Environ. Health Perspect. 110, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porterfield S. P., Hendrich C. E. (1993) Endocr. Rev. 14, 94–106 [DOI] [PubMed] [Google Scholar]
- 12.Hsu J. H., Brent G. A. (1998) TEM 9, 103–112 [DOI] [PubMed] [Google Scholar]
- 13.de Escobar G. M., Obregón M. J., del Rey F. E. (2004) Best Pract. Res. Clin. Endocrinol. Metab. 18, 225–248 [DOI] [PubMed] [Google Scholar]
- 14.de Escobar G. M., Obregón M. J., del Rey F. E. (2007) Public Health Nutrition 10, 1554–1570 [DOI] [PubMed] [Google Scholar]
- 15.Anselmo J., Cao D., Karrison T., Weiss R. E., Refetoff S. (2004) JAMA 292, 691–695 [DOI] [PubMed] [Google Scholar]
- 16.Gilbert L. I., Tata J. R., Atkinson B. G. (1996) Metamorphosis: Post-embryonic Reprogramming of Gene Expression in Amphibian and Insect Cells, Academic Press, New York [Google Scholar]
- 17.Brown D. D., Cai L. (2007) Dev. Biol. 306, 20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchholz D. R., Tomita A., Fu L., Paul B. D., Shi Y. B. (2004) Mol. Cell. Biol. 24, 9026–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchholz D. R., Paul B. D., Fu L., Shi Y. B. (2006) Gen. Comp. Endocrinol. 145, 1–19 [DOI] [PubMed] [Google Scholar]
- 20.Evans R. M. (1988) Science 240, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai M. J., O'Malley B. W. (1994) Annu. Rev. Biochem. 63, 451–486 [DOI] [PubMed] [Google Scholar]
- 22.Shi Y. B., Wong J., Puzianowska-Kuznicka M., Stolow M. A. (1996) BioEssays 18, 391–399 [DOI] [PubMed] [Google Scholar]
- 23.Sachs L. M., Damjanovski S., Jones P. L., Li Q., Amano T., Ueda S., Shi Y. B., Ishizuya-Oka A. (2000) Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 126, 199–211 [DOI] [PubMed] [Google Scholar]
- 24.Paul B. D., Fu L., Buchholz D. R., Shi Y. B. (2005) Mol. Cell. Biol. 25, 5712–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havis E., Sachs L. M., Demeneix B. A. (2003) EMBO Rep. 4, 883–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato Y., Buchholz D. R., Paul B. D., Shi Y. B. (2007) Mech. Dev. 124, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul B. D., Buchholz D. R., Fu L., Shi Y. B. (2005) J. Biol. Chem. 280, 27165–27172 [DOI] [PubMed] [Google Scholar]
- 28.Paul B. D., Buchholz D. R., Fu L., Shi Y. B. (2007) J. Biol. Chem. 282, 7472–7481 [DOI] [PubMed] [Google Scholar]
- 29.Matsuda H., Paul B. D., Choi C. Y., Hasebe T., Shi Y. B. (2009) Mol. Cell. Biol. 29, 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denver R. J., Pavgi S., Shi Y. B. (1997) J. Biol. Chem. 272, 8179–8188 [DOI] [PubMed] [Google Scholar]
- 31.Buchholz D. R., Heimeier R. A., Das B., Washington T., Shi Y. B. (2007) Dev. Biol. 303, 576–590 [DOI] [PubMed] [Google Scholar]
- 32.Das B., Cai L., Carter M. G., Piao Y. L., Sharov A. A., Ko M. S., Brown D. D. (2006) Dev. Biol. 291, 342–355 [DOI] [PubMed] [Google Scholar]
- 33.Cai L., Das B., Brown D. D. (2007) Dev. Biol. 304, 260–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranjan M., Wong J., Shi Y. B. (1994) J. Biol. Chem. 269, 24699–24705 [PubMed] [Google Scholar]
- 35.Furlow J. D., Kanamori A. (2002) Endocrinology 143, 3295–3305 [DOI] [PubMed] [Google Scholar]
- 36.Fu L., Tomita A., Wang H., Buchholz D. R., Shi Y. B. (2006) J. Biol. Chem. 281, 16870–16878 [DOI] [PubMed] [Google Scholar]
- 37.Puzianowska-Kuznicka M., Shi Y. B. (1996) J. Biol. Chem. 271, 6273–6282 [DOI] [PubMed] [Google Scholar]
- 38.Shi Y. B. (1996) in Metamorphosis: Post-embryonic Reprogramming of Gene Expression in Amphibian and Insect Cells (Gilbert L. I., Tata J. R., Atkinson B. G. eds) Academic Press, New York [Google Scholar]
- 39.Wang X., Matsuda H., Shi Y. B. (2008) Endocrinology 149, 5610–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanamori A., Brown D. D. (1992) J. Biol. Chem. 267, 739–745 [PubMed] [Google Scholar]
- 41.Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. 57, 289–300 [Google Scholar]
- 42.Hamatani T., Carter M. G., Sharov A. A., Ko M. S. (2004) Dev. Cell 6, 117–131 [DOI] [PubMed] [Google Scholar]
- 43.Fu L., Ishizuya-Oka A., Buchholz D. R., Amano T., Matsuda H., Shi Y. B. (2005) J. Biol. Chem. 280, 27856–27865 [DOI] [PubMed] [Google Scholar]
- 44.Rice P., Longden I., Bleasby A. (2000) Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 45.Wong J., Shi Y. B. (1995) J. Biol. Chem. 270, 18479–18483 [DOI] [PubMed] [Google Scholar]
- 46.Buchholz D. R., Paul B. D., Shi Y. B. (2005) J. Biol. Chem. 280, 41222–41228 [DOI] [PubMed] [Google Scholar]
- 47.Buchholz D. R., Ishizuya-Oka A., Shi Y. B. (2004) Gene Expr. Patterns 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 48.Amano T., Leu K., Yoshizato K., Shi Y. B. (2002) Dev. Dyn. 223, 526–535 [DOI] [PubMed] [Google Scholar]
- 49.Tata J. R. (1968) Dev. Biol. 18, 415–440 [DOI] [PubMed] [Google Scholar]
- 50.Yaoita Y., Brown D. D. (1990) Genes Dev. 4, 1917–1924 [DOI] [PubMed] [Google Scholar]
- 51.Wong J., Liang V. C., Sachs L. M., Shi Y. B. (1998) J. Biol. Chem. 273, 14186–14193 [DOI] [PubMed] [Google Scholar]
- 52.Ishizuya-Oka A., Ueda S., Amano T., Shimizu K., Suzuki K., Ueno N., Yoshizato K. (2001) Cell Tissue Res. 303, 187–195 [DOI] [PubMed] [Google Scholar]
- 53.Hasebe T., Hartman R., Matsuda H., Shi Y. B. (2006) Cell Tissue Res. 324, 105–116 [DOI] [PubMed] [Google Scholar]
- 54.Shi Y. B., Hayes W. P. (1994) Dev. Biol. 161, 48–58 [DOI] [PubMed] [Google Scholar]
- 55.Walsh L. A., Carere D. A., Cooper C. A., Damjanovski S. (2007) PLoS ONE 3, e1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puzianowska-Kuznicka M., Damjanovski S., Shi Y. B. (1997) Mol. Cell Biol. 17, 4738–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeeberg B. R., Feng W., Wang G., Wang M. D., Fojo A. T., Sunshine M., Narasimhan S., Kane D. W., Reinhold W. C., Lababidi S., Bussey K. J., Riss J., Barrett J. C., Weinstein J. N. (2003) Genome Biology 4, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis P. J., Davis F. B. (1996) Thyroid 6, 497–504 [DOI] [PubMed] [Google Scholar]
- 59.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 60.Bailey T. L., Elkan C. (1994) in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36, AAAI Press, Menlo Park, CA: [PubMed] [Google Scholar]
- 61.Kurokawa R., Yu V. C., Näär A., Kyakumoto S., Han Z., Silverman S., Rosenfeld M. G., Glass C. K. (1993) Genes Dev. 7, 1423–1435 [DOI] [PubMed] [Google Scholar]
- 62.Umesono K., Murakami K. K., Thompson C. C., Evans R. M. (1991) Cell 65, 1255–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong J., Shi Y. B., Wolffe A. P. (1995) Genes Dev. 9, 2696–2711 [DOI] [PubMed] [Google Scholar]
- 64.Sachs L. M., Shi Y. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13138–13143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishizuya-Oka A., Ueda S., Shi Y. B. (1997) Dev. Genet 20, 329–337 [DOI] [PubMed] [Google Scholar]
- 66.Shi Y. B., Brown D. D. (1993) J. Biol. Chem. 268, 20312–20317 [PubMed] [Google Scholar]
- 67.Wang Z., Brown D. D. (1993) J. Biol. Chem. 268, 16270–16278 [PubMed] [Google Scholar]
- 68.Patterton D., Hayes W. P., Shi Y. B. (1995) Dev. Biol. 167, 252–262 [DOI] [PubMed] [Google Scholar]
- 69.Mathew S., Fu L., Fiorentino M., Matsuda H., Das B., Shi Y. B. (2009) J. Biol. Chem. 284, 18545–18556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ishizuya-Oka A., Li Q., Amano T., Damjanovski S., Ueda S., Shi Y. B. (2000) J. Cell Biol. 150, 1177–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu L., Buchholz D., Shi Y. B. (2002) Mol. Reprod Dev. 62, 470–476 [DOI] [PubMed] [Google Scholar]
- 72.Ryu S., Driever W. (2006) Cell Cycle 5, 1140–1142 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.