Abstract
Exosomes secreted by normal and cancer cells carry and deliver a variety of molecules. To date, mechanisms referring to tumor exosome trafficking, including release and cell-cell transmission, have not been described. To gain insight into this, exosomes purified from metastatic melanoma cell medium were labeled with a lipid fluorescent probe, R18, and analyzed by spectrofluorometry and confocal microscopy. A low pH condition is a hallmark of tumor malignancy, potentially influencing exosome release and uptake by cancer cells. Using different pH conditions as a modifier of exosome traffic, we showed (i) an increased exosome release and uptake at low pH when compared with a buffered condition and (ii) exosome uptake by melanoma cells occurred by fusion. Membrane biophysical analysis, such as fluidity and lipid composition, indicated a high rigidity and sphingomyelin/ganglioside GM3 (N-acetylneuraminylgalactosylglucosylceramide) content in exosomes released at low pH. This was likely responsible for the increased fusion efficiency. Consistent with these results, pretreatment with proton pump inhibitors led to an inhibition of exosome uptake by melanoma cells. Fusion efficiency of tumor exosomes resulted in being higher in cells of metastatic origin than in those derived from primary tumors or normal cells. Furthermore, we found that caveolin-1, a protein involved in melanoma progression, is highly delivered through exosomes released in an acidic condition. The results of our study provide the evidence that exosomes may be used as a delivery system for paracrine diffusion of tumor malignancy, in turn supporting the importance of both exosomes and tumor pH as key targets for future anti-cancer strategies.
Introduction
Cells of different histotypes release intact organelles known as microvesicles or exosomes (1–3). Exosomes have been studied for their molecular composition and biological functions and indicate a specific protein profile (1, 2, 4–6). Much less is known on the regulatory role of these microvesicles. Several hypotheses suggest that exosomes may represent a vehicle for intercellular communication. In fact, exosomes may deliver proteins, soluble factors, and most importantly, RNA and microRNAs which modulate the protein expression of recipient cells (7). This appears of great importance for both the normal homeostasis of the body and the pathogenesis of various diseases, including tumors. As an example, human cancer cells produce large amounts of microvesicles bearing proapoptotic molecules (1–3, 8). These in turn are able to induce apoptosis of activated tumor-specific T cells, thus impairing the ability of effector lymphocytes to exert their cytolytic activity against tumor targets. As a consequence, exosomes may create an immuno-privileged environment within the tumors, similar to some physiologic conditions needing immunotolerance (9). Moreover, exosomes released by normal and tumor cells seem to differ in both functional and structural properties (10). It is also conceivable that exosomes are actively released within tumor tissues or directly spilled into the blood stream but apparently without any specific commitment to tissue targeting (2, 3, 8).
However, there are some unsolved mysteries regarding the mechanism(s) regulating exosome traffic between the outside and the inside of cells, including internalization of exosomes in target cells. It has been proposed that the interaction of exosomes and recipient cells may occur through receptor-ligand binding (11). Alternatively, exosomes enter into normal dendritic cells by an endocytic pathway or they may fuse with platelets (12, 13). Studies looking at the nature of the interaction of tumor exosomes with cells are inexistent. To address the hypothesis that exosome internalization takes place in tumor cells through membrane fusion mechanism, we set up a fusion assay based on R18 lipid probe dequenching, previously used in viral fusion tests (14).
Some features of tumor microenvironment may represent key factors in the regulation of exosome traffic within the tumor mass. In particular, the tumor microenvironment is acidic, and we have demonstrated that acidity is involved in the regulation of some vesicle-mediated malignant tumor cell functions, such as cannibalism and drug resistance (15–17). Malignant melanoma cells survive in an acidic microenvironment, a condition that does not allow survival of normal cells (17). This survival option of malignant tumor cells is conceivably due to hyperfunctional proton pumps that do not allow acidification of cytosol. In fact, a specific inhibition of H+ release (through proton pump inhibitors (PPI)2) can induce both acidification of the tumor cell cytosol (15) and acidic vesicle retention within tumor cells (16). On the basis of these findings we tested whether microenvironmental acidity might be involved in the traffic of tumor exosomes in regulating both their release and uptake by tumor cells. Accordingly, we set up a suitable in vitro model to study the role of low pH in favoring exosome uptake in human metastatic and primary melanoma cell culture. These melanoma cells, which are capable of bearing a low pH condition (17), allowed us to show that melanoma cell acidity was involved in both the exosome release and the uptake by fusion with cell membranes. This was consistent with a different lipid composition of exosomes released at different pH values and with a clear inhibition of exosome uptake after pretreatment with PPI (16). Finally, to investigate the possible physiologic role of acidity in exosome uptake, we used a caveolin-1 (cav-1)-bearing exosome model, previously reported to play a role in melanoma progression (18). By using this model, we were able to demonstrate that in an acidic condition cav-1 associated to melanoma exosome is delivered more efficiently to less aggressive tumor cells. In conclusion, in our study we provide evidence that low pH, which is a hallmark of malignant tumors, plays a key role in aberrantly regulating exosome traffic within the tumor mass.
EXPERIMENTAL PROCEDURES
Melanoma and Peripheral Blood Mononuclear Cells (PBMCs)
Melanoma cell lines were supplied by Istituto Nazionale dei Tumori, Milan, Italy, and were obtained from metastatic lesions (Mel1-Mel3, Me665/1) and from primary lesions (MelP1-MelP3, WM983A). PBMC were purified from buffy coat by Ficoll-Hypaque following manufacturer's instructions (Amersham Biosciences).
Cell Culture and Exosome Isolation
Melanoma cells were cultured in RPMI 1640 medium with 10% fetal bovine serum previously deprived of bovine microvesicles by ultracentrifugation (60 min at 100,000 × g) and antibiotics in a humidified 5% CO2 incubator. Experiments were performed with cells in exponential growth in acidic (pH 6.0) (Mel1ac) or buffered (pH 7.4) (Mel1) media. Microvesicles were isolated from supernatant of Mel1 (exoMel1) or Mel1ac (exoMel1ac) cells by sequential centrifugations as previously described (1, 11) and then purified on a sucrose gradient (11). The density of each fraction was determined by refractometry. Gradient fractions were collected and analyzed by Western blotting with antibodies directed to: Lamp-2 (monoclonal, BD Pharmingen), Rab 5B (polyclonal, Santa Cruz, Santa Cruz, CA), CD81 (monoclonal, Ancell, Bayport, MN). Fractions positive for Lamp-2, CD81, and Rab 5B were pooled, pelleted, and used in all experiments.
Western Blotting
For biotin-NHS labeling, exoMel1 was first incubated with water-soluble Sulfo-Biotin-NHS (Calbiochem) (0.5 mg/ml; 30 min at 4 °C), then washed at 100,000 × g for 1 h at 4 °C to remove unbound biotin and purified on sucrose gradient. Equal volumes of each fraction were resolved by 10% SDS-PAGE under reducing and denaturing conditions. The blots were analyzed by blotting with horseradish peroxidase-conjugated streptavidin (Pierce) according to the manufacturer's instructions. For the experiments with PPI, the cells were incubated overnight with 10 μg/ml PPI (AstraZeneca R&D, Mölndal, Sweden), and then biotinylated exosomes (15 μg) were incubated with unlabeled Mel1 cells (1 × 106) for 1 h. Cells were washed and lysed with buffer (150 mm NaCl, 20 mm Tris (pH 7.4), 1% Nonidet P-40, 10% glycerol) supplemented with protease inhibitors mixture (Roche Applied Science). Cell lysates were resolved by 10% SDS-PAGE and analyzed by blotting with horseradish peroxidase-streptavidin. In other experiments the blots were blocked using 5% nonfat dry milk in TBST buffer (10 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1% Tween 20) for 1 h at room temperature followed by incubation with primary antibodies anti-nucleolin (Clone C23, Santa Cruz) and anti-cav-1 polyclonal antibodies (Santa Cruz). After washing, the filters were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Bio-Rad) for 1 h at room temperature, and reactivity was detected by the enhanced chemiluminescence kit (Pierce).
Membrane Fusion Assay
Exosome fusion activity was followed by the increase in fluorescence resulting from dilution of the self-quenched probe R18 (19). Exosomes (20 μg of protein/ml) and large unilamellar vesicles (10 μg protein/ml) were labeled, respectively, with 1 and 0.5 μl of an ethanolic solution of the fluorescent lipophilic probe octadecyl rhodamine B chloride (R18) (Molecular Probes, Eugene, OR) (1 mm) for 30 min at room temperature in 1 ml of MES buffer (10 mm MES (pH 6.0 or pH 7.4), 145 mm NaCl, 5 mm KCl). The unincorporated R18 was removed by using a Sephadex G-75 column (20 × 1 cm) equilibrated with saline buffer. Ten μg of labeled exosomes were added to MES fusion buffer in a thermostatted spectrofluorometer FluoroMax-2 (Spex), and fluorescence was measured continuously at 560-nm excitation and 590-nm emission wavelengths (slits 1.5 nm). After an equilibration time of 10 min, unlabeled cells (1 × 106) were added to the exosomes, and fluorescence was monitored for a further 30 min. The fusion reaction was stopped by the addition of Triton X-100 plus octylglucoside at a final concentration of 0.3% and 60 mm respectively, which resulted in maximal probe dilution. The fluorescence increase was measured as the difference with respect to initial fluorescence of labeled exosomes and expressed as % of maximal fluorescence dequenching (FD) according to the equation, % FD = ((F − Fi)/(Fmax − Fi)) × 100, where F is the fluorescence intensity after 30 min of incubation exosomes/cells, Fi is the initial fluorescence value of labeled exosomes, and Fmax is the fluorescence intensity after detergent membrane disruption. R18-labeled exosomes were fixed with 0.5% paraformaldehyde (PAF) for 20 min at room temperature, then centrifuged at 100,000 × g for 1 h and used for fusion experiment. In other experiments exosomes were solubilized with 60 mm octylglucoside (Invitrogen) then dialyzed against MES buffer (pH 7.4) overnight at 4 °C. After dialysis, exosomes were recovered and labeled with R18 probe. Fusion activity was tested for 30 min at 37 °C on 1 × 106 Mel1 cells. For cell treatment, 1 × 106 Mel1 cells were incubated with 10 μg/ml filipin (Sigma) for 20 min at 37 °C, then washed and subjected to fusion.
Large Unilamellar Vesicle Preparation
Large unilamellar vesicles were prepared by filter exclusion using an extrusion apparatus. Briefly, appropriate amounts of lipids dissolved in chloroform were mixed, and the solvent was evaporated under nitrogen. The resulting lipid films were kept overnight under vacuum. The dry lipids were dispersed by vortex-mixing in 2 mm l-histidine, 2 mm TES, 150 mm NaCl, 1 mm EDTA (pH 7.4). The suspensions were submitted to 10 cycles of freezing and thawing, then passed 21 times through two stacked 0.1-μm-diameter pore polycarbonate membranes in a Liposofast-Miniextruder (Avestin, Ottawa, Canada). All vesicles were supplemented with trace amounts of radiolabeled lipids, and their concentration was determined by radioactivity measurements.
Confocal Microscopy
To analyze the intracellular fate of exosomes after their entry into tumor cells, the cells were plated on coverslips in WillCo dishes (WillCo Wells, BV, Amsterdam, NH) and fixed with methanol for 10 min at −20 °C. After 2 washes in PBS, cells were probed for 30 min with the following antibodies: Rab 5B (Santa Cruz) for early endosomes, Lamp-1 (clone H4A3, BD Pharmingen) for lysosomes, Golgina (clone CDF4, Molecular Probes) for Golgi apparatus. Then cells were incubated with appropriate Alexa Fluor 488-conjugated secondary antibodies (Molecular Probes). Coverslips were mounted in Vectashield (Vector Laboratories, Burlingame, CA). Exosomes were labeled with exosome proteins/NHS-rhodamine (10 μg/μl NHS-rhodamine, Pierce) 1:3 (w/w) for 30 min at 4 °C in PBS followed by quenching with serum-free RPMI and washing by centrifugation at 100,000 × g for 1 h. In lipid colocalization experiments of exoMel1 with parental cells or PBMC, exoMel1 were stained with R18 as previously described, and the cells were labeled with PKH67 dye (green fluorescent cell linker kit, Sigma). Briefly, the cells were incubated in diluent C containing PKH67 for 5 min followed by quenching with fetal bovine serum and then washed with PBS. PKH67-labeled PBMC were plated on poly-l-lysine-coated glass coverslips. Samples were observed in vivo without fixation with a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems, Mannheim, Germany). Observations were performed with an objective HCX PL APO CS 63 × 1.40 oil in sequential double fluorescence mode (between lines). The excitation wavelengths used were 488 and 543 nm for fluorescein (PKH67) and rhodamine (R18), respectively. Fluorescence emissions were collected after passage through excitation beam splitter FW DD 488/543 filter in a spectral range of 510–555 or 650–700 nm for fluorescein and rhodamine, respectively. The images represent optical sections of double-stained samples.
Electron Microscopy
Melanoma cells, cultured in buffered and acidic conditions, were fixed with 2.5% glutaraldehyde, post-fixed in 1% OsO4, and then dehydrated in alcohol and embedded in epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate and finally examined with a Philips 208 transmission electron microscope (FEI Co.).
Membrane Fluidity Assay
To analyze membrane fluidity, the exosomes released from cells cultured at acidic or buffered pH values and parental cells were loaded with the fluorescent probe Laurdan (20) as previously described (21). Briefly, Laurdan from a DMSO stock solution (Laurdan and DMSO final concentrations of 0.11 μm and 0.06%, respectively) was added to Mel1 cells (1 × 106) or exoMel1 (50 μg) resuspended in 2.5 ml of MES buffer, and the probe loading was allowed to proceed in the dark at 37 °C for 45 min. At the end of the loading procedure, a thermal step gradient (40 to 5.5 °C, 0.5 °C min−1) was applied, and fluorescence excitation spectra (320 to 420 nm at both 435- and 490-nm emission wavelengths) were obtained at the selected temperatures. Fluorescence measurements were carried out with a computer-driven L-type spectrofluorometer (FluoroMax-2) equipped with a stirring accessory and thermostatted (±0.1 °C) by a circulating water bath. The monochromator band pass was set at 5 nm for both the excitation and emission path. Spectroscopic data were elaborated in terms of generalized polarization (GP) according to the equation GP = (I435 − I490)/(I435 + I490), where I435 and I490 are the intensity at each excitation wavelength using emission wavelengths of 435 and 490 nm, respectively. GP values were calculated after subtraction of cell or exosome background signal. The signal of unpartitioned probe was always <4% and did not need to be subtracted. The GP value is an index of the rate and extent of membrane water dipolar relaxation processes, which have been found to reliably mirror the lipid packing order irrespective of phospholipid polar head composition and charge in a wide pH range (22). The slope of the GP excitation spectrum can be exploited to ascertain the coexistence of lipid phases. A negative or positive slope indicates a single liquid-crystalline phase or coexisting gel and liquid-crystalline phase domains, respectively (20).
Cholesterol Measurement and Lipid Analysis
To determine free cholesterol amounts, exosomes and cells were lysed with PBS containing 5 mm EDTA, 1.5% CHAPS (w/w), and a proteases inhibitor mixture (Roche Applied Science) for 30 min at 4 °C. Cholesterol was determined using the cholesterol oxidase-based Amplex Red Cholesterol assay kit (Molecular Probes) according to the manufacturer's protocol. The Amplex Red Fluorescence was excited at 530 nm and detected at 590-nm wavelengths using a Wallac Victor2 plate reader. The amount of free cholesterol was normalized to the total protein content for each sample. Protein amount was detected by protein assay (BCA, Pierce).
Lipids were extracted from cells and exosomes as described (23) and were analyzed by thin layer chromatography (TLC). Lipid extracts (normalized on 100 μg of protein/sample) were resolved on silica gel plates (Merck), and lipid analysis was performed using a solvent system of chloroform/methanol/water (65:25:4, v/v/v) (24). The areas containing individual lipids were identified by comigration of standards and were visualized by staining with 3% copper acetate solution in 8% phosphoric acid and subsequent heating at 180 °C for 10 min. The relative amount of lipids was obtained through densitometry analysis by using the Quantity One Bio-Rad software program.
Flow Cytometry
Purified exoMel1 and exoMel1ac (10 μg) were stained with R18 as described above and added at different times to melanoma cells (3 × 104 cells/ml) at the corresponding pH values. After PBS washes the cells were sorted on a FACScan machine (BD Biosciences), and at least 1 × 104 events were collected and analyzed using Cell Quest software (BD Biosciences).
Cell Membrane Isolation
To evaluate the uptake of exoMe665/1 and exoMe665/1ac into cell membrane compartments, membranes from WM983A cells (1 × 106) treated or not with exoMe665/1 and exoMe665/1ac (65 μg for 2 h at 37 °C followed by PBS washes) were compared. To purify membrane proteins from WM983A, cells were lysed in 0.1 ml of buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA) supplemented with a protease inhibitor mixture for 30 min at 4 °C and then pelleted down at 1000 × g to remove nuclei. The supernatants were then centrifuged at 30,000 × g for 60 min. The whole amount of membranes recovered from untreated or exosome-treated WM983A cells was analyzed by Western blotting for cav-1 and Lamp-2 expression.
Cell Cycle Analysis
Me665/1 cells were fixed and permeabilized in 70% ethanol in PBS at 4 °C for 30 min then PBS-washed and treated with RNase (40 units/μl, Roche Applied Science) for 1 h at room temperature and stained with propidium iodide (50 μg/ml). Flow cytometry experiments were carried out using a FACS Canto (BD Biosciences). Acquisition and analysis were performed using FACS Diva software (BD Biosciences).
Statistical Analysis
Results are expressed as the means ± S.D. Comparisons between means were performed by two-tail unpaired Student's t test, and p < 0.05 was regarded as significant.
RESULTS
Exosome Release Is Increased by Acidic pH
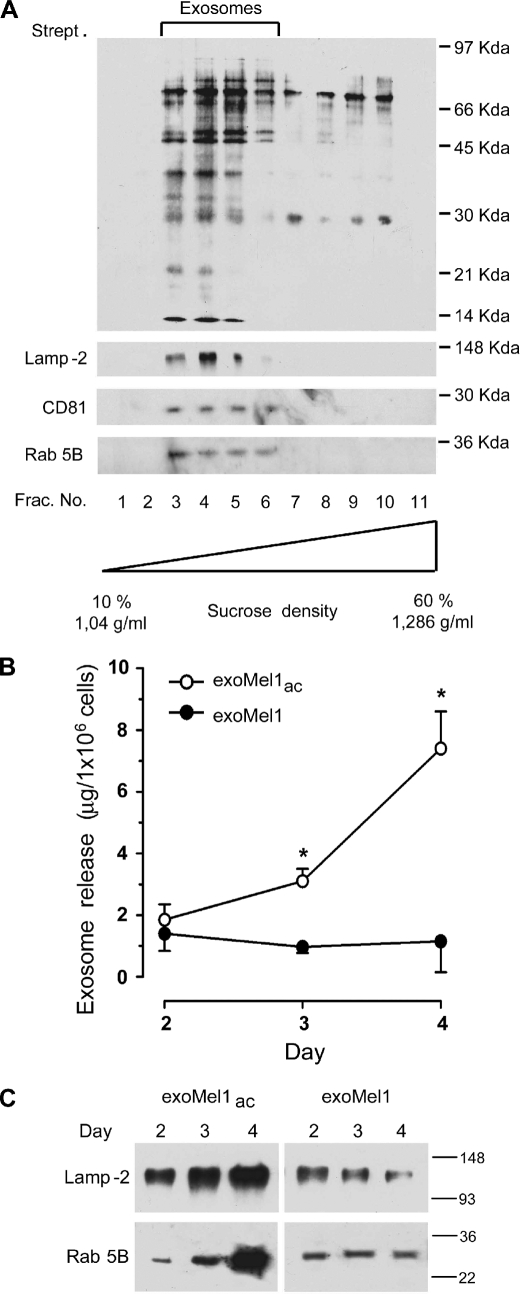
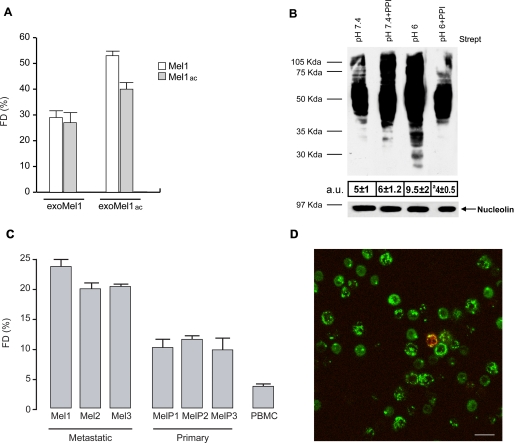
As a model for our studies, we used human metastatic melanoma cells (Mel1), which produce constant exosome levels (1, 25) and are able to sustain low pH conditions without showing levels of cytotoxicity (17). Exosomes were, thus, purified from Mel1 supernatants, labeled with NHS-biotin, and layered on a sucrose gradient to estimate the purity of isolated exosomes (11, 26). A selective and conspicuous protein pattern was detectable by streptavidin blotting in lighter gradient fractions (fractions 3–6) (Fig. 1A), corresponding to exosome density (11). Exosome identity was confirmed by Western blotting for some housekeeping exosome markers, such as Lamp-2, Rab 5B, and CD81 (27, 28) (Fig. 1A). However, the level of exosome release in low pH conditions, which is the physiologic pH condition of tumors (16), was unknown. Our cellular model was fully able to grow in an acidic medium (17). In fact, cell cycle analysis indicated that cell viability was not affected by low pH (supplemental Fig. 1A). These data were further confirmed by electron microscopy analysis showing the absence of ultrastructural damage in cells cultured in acidic conditions (supplemental Fig. 1B). Thus, we cultured Mel1 cells in either an acidic (pH 6.0) or buffered (pH 7.4) medium. After 2, 3, and 4 days, cell culture supernatants were collected, and exosomes exoMel1ac and exoMel1, respectively, were purified as described (1, 25). Results showed that melanoma cells secreted more exosomes in an acidic than in a buffered condition, as analyzed by protein assay (Fig. 1B) and confirmed by Western blot analysis of exosome markers, such as Lamp-2 and Rab 5B (Fig. 1C).
FIGURE 1.
Exosome release in acidic and buffered pH conditions. A, exosomes (100 μg) isolated from Mel1 cells culture medium were NHS-biotin-labeled and loaded on a 10–60% continuous sucrose gradient. Eleven fractions were analyzed by Western blotting with horseradish peroxidase-streptavidin. Results indicate a molecules profile with molecular mass ranging between 97 and 21 kDa enriched in fractions 3–6 corresponding to density 1.11–1.17 g/ml. These fractions were also found positive for exosome markers such as Lamp-2, CD81, and Rab 5B by Western blotting. B, Mel1 cells (3 × 106 cell/ml) were cultured in buffered (pH 7.4) or acidic (pH 6.0) media. At the indicated days exosomes were isolated, quantified by protein assay, and normalized on 1 × 106 viable cells. Points, means (n = 4); bars, S.D. *, p < 0.05. C, Western blotting of Lamp-2 and Rab 5B in exoMel1ac and exoMel1 released from parental cells at 2, 3, and 4 days. Note that Lamp-2 and Rab 5B expressions increased with time in the acidic exosomes. A representative Western blotting of three independent experiments is shown.
Exosome Uptake by Fusion with the Cell Membrane
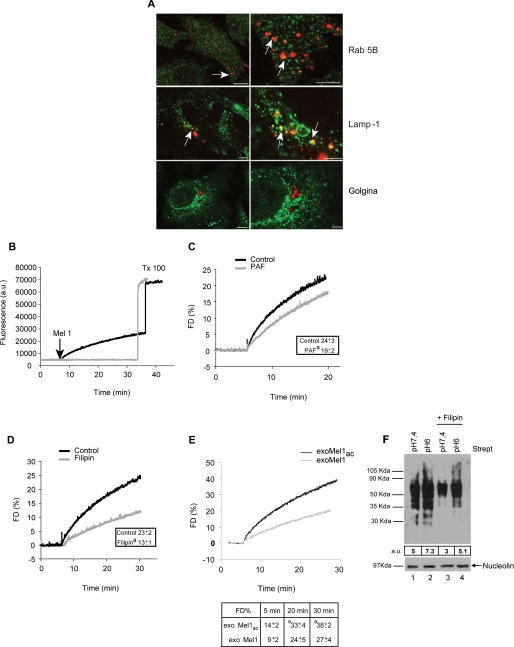
This set of experiments was aimed at investigating the exosome uptake by tumor cells. In fact, the fate of exosomes once released outside the cells is mostly entirely unknown. To demonstrate that purified exosomes added to the cell culture might be internalized by melanoma cells, exosomes were stained with NHS-rhodamine (NHS-ROD-exoMel1) and incubated with tumor cells. Then cells were fixed and analyzed by confocal microscopy to evaluate the interaction of exosomes with intracellular structures such as endosomes, lysosomes, and Golgi apparatus. Results clearly showed areas of exosome/Rab 5B and exosome/Lamp-1 colocalization but not Golgi apparatus (Fig. 2A), suggesting a specific interaction of internalized exosomes with acidic vesicles of melanoma cells.
FIGURE 2.
Exosome fusion with parental cells. A, a panel of confocal laser-scanning microscopy images of a human metastatic melanoma is shown. NHS-rhodamine-labeled exosomes (red) were incubated with cells for 4 h followed by fixation and labeling with green fluorescent antibodies directed to Rab 5B (early endosomes), Lamp-1 (lysosomes), and Golgina (Golgi apparatus). Arrows indicate the events of colocalization (yellow) in all samples, with the exception of Golgi compartment. Images in the right column represent magnification of the images on the left column. Bars: left panels, 10 μm; right panels, 4 μm. B, R18-exoMel1 were left untreated or mixed with 1 × 106 Mel1 cells. Note that a fluorescence dequenching (FD) curve was observed only after the addition of the cells. Tx 100, Triton X-100. C, R18-exoMel1 were left untreated or pretreated with 0.5% PAF before the addition to Mel1 cells, and fusion activity was tested. A representative fluorescence dequenching curve is shown. Inset, statistical analysis obtained by 20 min of kinetic experiments is shown. Values are the means ± S.D. a = p < 0.05 versus control (n = 3). D, Mel1 cells (1 × 106) were left untreated or treated with filipin, then subjected to a fusion test with R18-exoMel1. A representative fluorescence dequenching curve is shown. Inset, statistical analysis obtained by 30 min kinetic experiments is shown. Values are the means ± S.D. a = p < 0.01 versus control (n = 3). E, R18-exoMel1ac and R18-exoMel1 (10 μg) were mixed with parental cells at the corresponding pH, and fusion was monitored. A representative fluorescence dequenching curve is shown. Statistical analysis on 5-, 20-, and 30-min kinetic experiments is represented. Values are the means ± S.D. a, p < 0.05 (n = 3). F, streptavidin blotting is shown. 20 μg of biotinylated exosomes were incubated with parental cells (1 × 106) at the corresponding pH (lane 1, exoMel1 on Mel1; lane 2, exoMel1ac on Mel1ac) or with filipin-treated cells (lane 3, exoMel1 on Mel1; lane 4, exoMel1ac on Mel1ac) for 1 h at 37 °C. Immunoblotting of nucleolin protein expression represents a control for protein equal loading. A representative Western blot of three independent experiments is shown. Numbers expressed in arbitrary units (a.u.) represent streptavidin densitometry analysis.
With these results we had the evidence that exosomes externally administered to melanoma cells are internalized and interact with cytoplasmic vesicles. However, we did not have evidence for the mechanism that exosomes use for cell internalization. Our hypothesis was that exosomes might fuse with the cell membrane through a lipid-lipid interaction, possibly influenced by the pH condition of the culture medium. To verify this hypothesis, we first tested the ability of exosomes to fuse with parental-producing cells through a fusion assay based on lipid mixing (19, 29). Exosomes were labeled with lipophilic fluorescent dye R18 then added to the cell cultures, and the fluorescence emission was monitored up to 30 min after exosome addition. The results showed that lipid mixing continuously occurred (Fig. 2B), reaching about 30% of the maximum fluorescence obtained with Triton X-100 plus octylglucoside at 30 min of incubation. No spontaneous dequenching was observed in R18-labeled exosomes before Triton X-100 addition, thus indicating the specificity of the reported dequenching (Fig. 2B). A fusion efficiency curve of 2.5 up to 70 μg of exosome obtained on the same number of Mel1 cells (1 × 106) is shown in supplemental Fig. 2A. Given that the fluorescence signal rose with the increasing exosome amount, we observed that 10 μg of exosomes represented the best compromise between the signal/noise fluorescence ratio and fusion efficiency. For this reason we used 10 μg of exosomes in all the subsequent experiments. We next tested the influence of stimuli in fusion events, such as temperature and calcium addition, previously described as specific requirements for fusion between biological membranes (29). Our results showed that exosome fusion increased as a function of temperature, reaching the higher values at physiological temperature (supplemental Fig. 2B). However, it was only weakly CaCl2-dependent (supplemental Fig. 2C), in accordance with other reports (29).
To evaluate a specific role of lipids and/or proteins in fusion events, we pretreated R18-labeled exosomes with 0.5% PAF (14). The results showed that fluorescence dequenching was only slightly affected by the addition of PAF-treated exosomes (about 20%) (Fig. 2C), suggesting a minor role of proteins in exosome fusion. Fully comparable results were obtained by pretreating cells with 2% PAF (data not shown), further suggesting a reciprocal lipid role in fusion mechanism. These results were further supported by experiments showing that by pretreating cells with filipin, a molecule known to perturb the composition of cell membranes through its association with cholesterol, we obtained a 50% reduction of fusion activity (Fig. 2D). Altogether these data indicate that the lipid composition of tumor cell membranes is a key factor in determining exosome-tumor cell fusion.
However, a protein role in fusion process cannot be completely ruled out. To test such a hypothesis, we prepared two vesicles populations, (i) lipid vesicles (large unilamellar vesicles) made as a mixture of 10% sphingolipids, 10% acid phospholipids, 60% neutral phospholipid, and 20% cholesterol (a composition resembling cell plasma membrane) and (ii) exosomes solubilized with octylglucoside, then reconstructed by dialysis and subjected to fusion test on Mel1 cells (supplemental Fig. 2D). Interestingly, both microvesicles types did not show fusion activity when compared with untreated exosomes. This result suggests that, although lipids are directly involved in membrane-membrane fusion, proteins may exert a structural role that is key for fusion process.
Exosome Uptake and Fusion under Different pH Conditions
This set of experiments was aimed at analyzing whether the acidic tumor microenvironment might have a role in increasing the exosome uptake by tumor cells. We, thus, used the cellular model and the methods described in the previous paragraphs to assess exosome fusion efficiency in different pH conditions. The fluorescence of R18 probe was unaffected by acidic pH per se (data not shown). Consequently, the fusion ability of R18-labeled exosomes collected from Mel1 and Mel1ac cells was tested on the parental-producing cells under different pH conditions. The results showed that the fusogenic activity of exoMel1ac was higher than exoMel1 (Fig. 2E). This was appreciable within 5 min and became statistically significant at 20–30 min (Fig. 2E). In a parallel set of flow cytometry time-course experiments, we confirmed that R18-labeled exoMel1ac uptake in Mel1ac cells was higher than exoMel1 uptake in Mel1 cells as early as 15 min after the addition of exosomes to the cell cultures (supplemental Fig. 3).
We speculated that the higher fusion efficiency of acidic exosome could be related to a peculiar intrinsic molecular composition. On the basis of the data showing the filipin effect on fusion activity (Fig. 2D), we tested the effects of filipin treatment on exosome uptake under different pH conditions. To this purpose we analyzed the amount of NHS-biotinylated proteins of exoMel1ac or exoMel1 recovered into untreated- or filipin-treated Mel1 cells at the corresponding pH by using Western blotting. The results clearly showed that exosome uptake increased in cells cultured in an acidic condition and that filipin treatment markedly reduced exosome uptake in both acidic and buffered conditions (Fig. 2F).
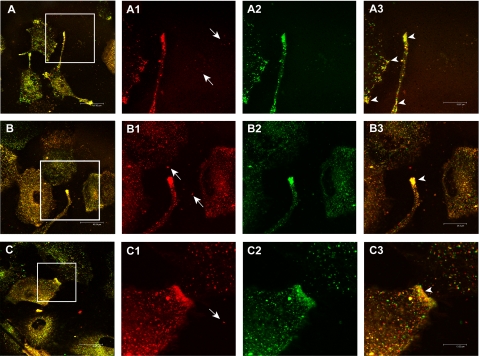
To visualize the intracellular distribution of exosomes, we performed confocal laser-scanning microscopy on cells pre-labeled with PKH67 lipid dye and co-cultured with R18-labeled exosomes. These experiments were performed without fixing the cells inasmuch as both PKH67 and R18 lipid dyes are unstable in fixed cells. Moreover, only the continuous in vivo observation of the unfixed cells permitted the capture of very rapid lipid mixing. By using this approach we showed that there were multiple sites of exosome/cell lipids merging (yellow staining) both at the plasma membrane and intracellular vesicle levels, likely indicating exosome/cell lipid fusion areas (Fig. 3, A–C).
FIGURE 3.
In vivo exosome/melanoma lipids colocalization analysis through confocal laser-scanning microscopy. The panel shows three different fields (A, B, and C) of unfixed cell cultures in which co-cultivation of R18-labeled exosomes (red, magnifications in A1, B1, C1) and PKH67-labeled cells (green, magnifications in A2, B2, C2) is analyzed. In particular, arrows in A1, B1, and C1 images indicate free exosomes in cell culture medium not yet interacting with the cells. The yellow points (A, B, and C and arrows in magnification A3, B3, and C3) correspond to cell/exosome lipid mixing events, both at plasma membrane and intracellular levels. Bars: A, 48 μm; B, 46 μm; C, 42 μm; A3, 20 μm; B3, 24 μm; C3, 13 μm.
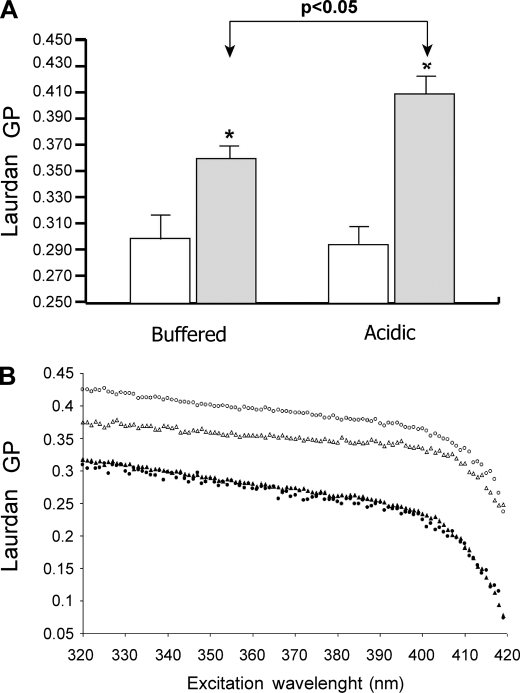
Acidic Exosomes Display High Membrane Rigidity
Once a key role of lipids in the exosome/melanoma cell fusion process was verified, we evaluated if buffered and acidic exosomes displayed different biophysical and biochemical membrane properties, such as membrane fluidity, phase state, and lipid composition. Our experimental approach was also based on the notion that lipid lateral phase separation, i.e. the coexistence of liquid crystalline- and gel-phase state lipid domains, may induce bilayer defects promoting membrane fusion (30). Laurdan fluorescence spectroscopy was applied to assess both membrane fluidity and phase state by evaluating the GP value and its behavior over the entire excitation Laurdan spectrum (20). We found that at physiological temperature, the membranes of exoMel1ac were significantly more rigid than those of exoMel1 and melanoma cells. As shown in Fig. 4A, the ranking order of GP values was exoMel1ac > exoMel1 > Mel1 cells = Mel1ac cells. In all samples, whatever the starting GP value, the GP decreased with increasing excitation wavelength (Fig. 4B). This indicated that the membranes of both cells and exosomes in either buffered and acidic conditions were in the liquid-crystalline phase state and, as a result, ruled out the occurrence of lipid lateral phase separation in our system. Low temperatures decreased the membrane fluidity but did not induce lipid phase separation in any sample (data not shown). Altogether, these data provide evidence of marked differences in membrane fluidity between cells and exosomes and, more importantly, between exosomes released under different pH culture conditions. Moreover, they allow us to exclude a direct role of lipid lateral phase separation in the exosome/cell fusion process.
FIGURE 4.
Exosomes and parental cells membrane fluidity and lipid phase state at different pH conditions. A, shown are Laurdan GP values (340-nm excitation wavelength; 37 °C) of buffered and acidic Mel1 cells (open bars) and exoMel1 and exoMel1ac (filled bars). The higher GP values of exosomes indicate a higher rigidity of such membranes with respect to bulk of the cellular membranes. ExoMel1ac are significantly more rigid than exoMel1. Data are the means ± S.E. of at least three independent experiments. *, p < 0.05 versus the corresponding bulk cellular membranes. B, Laurdan excitation GP spectra of Mel1 (a, filled triangles), Mel1ac cells (b, filled circles), exoMel1 (c, open triangles), and exoMel1ac (d, open circles) at 37 °C. Laurdan GP values decrease by increasing excitation wavelength, indicating that both cell and exosome membranes are in a liquid-crystalline lipid phase in the absence of coexisting gel-phase lipid domains. The curves are representatives of three consistent experiments.
Changes in membrane fluidity might conceivably be related to lipid composition. Therefore, we analyzed the lipid content in both exosome and cell membranes in buffered and acidic pH conditions. A lipid composition analysis was performed through TLC separation, staining, and densitometry quantification. The results showed that the most representative classes of lipids were SM, GM3, phosphatidylserine, phosphatidylinositol, phosphatidylcholine, and phosphatidylethanolamine (Table 1). Comparison between exosomes and cells in both pH conditions showed that exosomes were more enriched in SM+GM3, whereas parental cells contained higher phosphatidylethanolamine and phosphatidylserine plus phosphatidylinositol plus phosphatidylcholine lipid amounts. Interestingly, acidic exosomes were more enriched in SM+GM3 than buffered exosomes (Table 1). The free cholesterol content in both exosomes and melanoma cells was determined with a different technique. The results showed that exosomes contain a higher amount of free cholesterol compared with their parental cells (exoMel1ac 0.25 μg/μg of protein, Mel1ac cells 0.03 μg/μg of protein, exoMel1 0.67 μg/μg of protein, and Mel1 cells 0.06 μg/μg of protein).
TABLE 1.
Relative lipid composition of exoMel1ac, exoMel1, and parental cells
Lipids extracted as described under “Experimental Procedures” were stained and quantified by densitometry analysis. The numbers represent the relative amount (%) of each class of lipid with respect to the total identified lipids in the same preparation. Results are the means ± S.D. (n = 3). PS, phosphatidylserine; PI, phosphatidylinositol; PC, phosphatidylcholine; PE, phosphatidylethanolamine.
| SM+GM3 | PE | PS+PI+PC | Total | |
|---|---|---|---|---|
| % | % | % | % | |
| exoMel1ac | 29 ± 1a | 28 ± 1a | 43 ± 1 | 100 |
| exoMel1 | 22 ± 1 | 35.5 ± 2 | 42.5 ± 1 | 100 |
| Mel1ac | 15 ± 3 | 38.5 ± 3 | 46.5 ± 1 | 100 |
| Mel1 | 15.5 ± 3 | 37 ± 4 | 47.5 ± 1 | 100 |
ap < 0.05 vs exoMel1.
These data provided a possible explanation for the increased membrane rigidity of acidic exosomes. In fact, sphingolipids hold long, largely saturated acyl chains that make them pack more tightly than glycerophospholipids (31). Moreover, cholesterol and sphingolipids may contribute in stabilizing the formation of highly ordered microdomains, characterized by high values of Laurdan GP (32). Thus, the high rigidity of exosomes is likely due to high cholesterol, GM3, and SM content. More specifically, the higher membrane rigidity of acidic exosomes might correlate with a higher association of SM with cholesterol.
Role of Acidity in Triggering Exosome-to-cell Fusion
A hallmark of malignant tumors is extracellular low pH, which is maintained by highly active proton pumps, therefore justifying the use of both anti-neoplastic drugs (15) and chemosensitizers (16). Consequently, our melanoma cells, capable of growing under low pH culture conditions, were suitable to investigate a possible role of low pH in altering exosome-to-cell fusion efficiency. Accordingly, we tested the fusion activity of exoMel1 and exoMel1ac on both Mel1 and Mel1ac cell populations (Fig. 5A). We found that exoMel1ac fused much better on tumor cells cultured in both acidic and buffered conditions, whereas exoMel1 showed a lower fusion capability independently of culture medium pH (Fig. 5A).
FIGURE 5.
Role of microenvironmental pH in exosome fusion. A, R18-exoMel1 and R18-exoMel1ac were mixed with both acidic and buffered Mel1 cells (1 × 106), and fusion efficiency was tested for 30 min. Points, means ± S.D. FD, fluorescence dequenching. B, NHS-biotin-buffered (pH 7.4) or acidic (pH 6.0) exosomes (25 μg) were incubated for 1 h with untreated or PPI-treated parental cells, then cells were analyzed by streptavidin blotting. Western blotting of nucleolin represents a control for cell protein equal loading. Numbers represent the whole lane densitometry analysis expressed in arbitrary units (a.u.). Points, means ± S.D. a p < 0.01 versus control (pH 6.0) (n = 3). C, R18-exoMel1 were added to metastatic melanoma (Mel1–3), primary melanoma (MelP1–3), and normal donor-derived PBMC, and fusion efficiency was tested for 30 min. Points, means ± S.D., (n = 3). D, R18-exoMel1 (red) were incubated for 3 h with PKH67-PBMC (green) and analyzed by confocal laser-scanning microscopy. The image clearly shows the absence of lipid colocalization areas. The rare event of colocalization observed in PBMC cells is likely due to the presence of monocytes, as recognizable from nuclear morphology. Bar, 20 μm.
Thus, we used the model of acidic exosomes and cells versus buffered exosomes and cells to test whether we could revert exosome fusion efficiency by inhibiting culture acidification through PPI. Moreover, PPI are pro-drugs needing low pH to be transformed in the active molecule sulfenamide (15, 16, 33). In a new set of experiments, we pretreated melanoma cells with PPI in different pH conditions and then co-cultured for 1 h with biotinylated acidic or buffered exosomes, respectively. We then analyzed them by streptavidin incubation. Results clearly indicated that PPI pretreatment of Mel1ac cells significantly decreased exoMel1ac protein uptake, as indicated also by densitometry analysis, but did not affect exoMel1 entry into Mel1 cells (Fig. 5B). This supports our hypothesis that the environmental acidic pH is a key requirement for exosome fusion.
This set of data was supplemented with a series of experiments aimed at evaluating the ability of exosomes to target cells of different origins. We compared the fusion of exoMel1 to either metastatic melanomas (Mel1–3), primary melanomas (MelP1–3), or normal PBMC. The results clearly showed that exosomes fused better with metastatic tumor cells (range 19–23%) compared with primary melanomas (range 9–12%), whereas the exosome fusion activity was only barely detectable in primary PBMC derived from healthy donors (Fig. 5C). The low level of exosome fusion in PBMC was also confirmed by confocal microscopy analysis, showing the absence of lipid-dependent exosome entry in PBMC but some scattered monocytes (Fig. 5D).
Exosome-associated Caveolin-1 Delivery Is Enhanced by Acidic Conditions
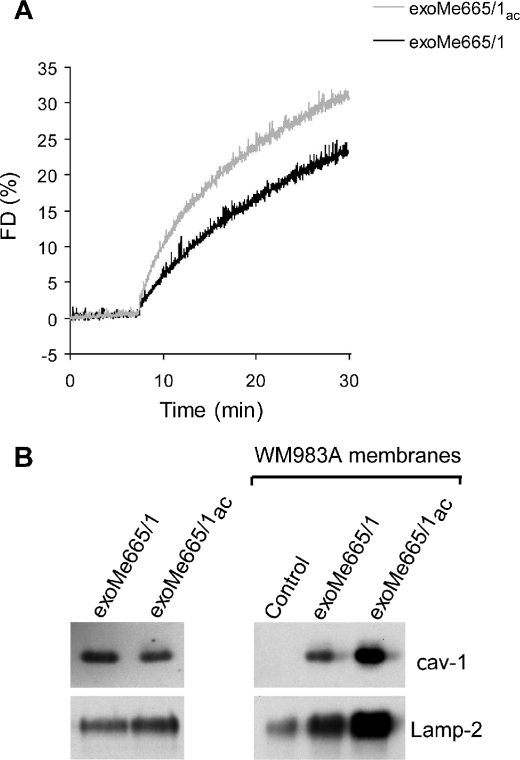
Metastatic melanoma (Me665/1) expresses high levels of endogenous and exosome-associated cav-1 (18), and high levels of plasmatic cav-1-bearing exosomes have been detected in melanoma patients (25). Moreover, it has been shown that cav-1-bearing exosomes may promote tumor progression (18). Therefore, we used the model of cav-1-bearing exosomes and melanoma cells not expressing cav-1 (WM983A) to test whether low pH conditions might increase the intercellular delivery of tumor-associated molecules through exosomes. We first evaluated the fusion efficiency of acidic and buffered exoMe665/1 on the autologous cells. As expected, we found that acidic pH induced a higher fusion activity after 30 min when compared with buffered pH (Fig. 6A). Then we studied whether acidity could influence exoMe665/1 cav-1 delivery to the less aggressive WM983A melanoma lacking cav-1. ExoMe665/1 and exoMe665/1ac were incubated with WM983A cells, and cell membranes were analyzed for the levels of cav-1 and Lamp-2 (Fig. 6B). Results showed a clear increase of cav-1 and Lamp-2 in association with exoMe665/1ac, suggesting effective exoMe665/1 protein incorporation into the WM983A recipient cell membranes. On one hand, these results confirmed that exosomes may serve as a natural delivery system between cells (18). On the other hand, these data suggest that in acidic conditions the delivery activity of exosomes increases with possible dangerous effects, such as the transfer of a more aggressive phenotype to less aggressive sibling cells (18).
FIGURE 6.
Exosomes delivery of cav-1. A, R18-exoMe665/1ac and R18-exoMe665/1 (10 μg) were mixed with parental cells at the corresponding pH and fusion monitored. A representative fluorescence dequenching (FD) curve is shown. B, cav-1 and Lamp-2 immunoblotting on WM983A cell membranes after incubation with exoMe665/1 and exoMe665/1ac. Control represents WM983A membranes in the absence of exoMe665/1 incubation. A representative Western blot of two independent experiments is shown.
DISCUSSION
This study imparts a series of novelties on the mechanism(s) driving exosome release and recovery by human tumor cells. Our study shows that an acidic microenvironment is a key factor in increasing both exosome release and entry into melanoma cells, suggesting that acidity may drive a sort of paracrine traffic of microvesicles within tissues and tumor mass. Moreover, for the first time this study highlights that exosomes are not just internalized by tumor cells through a putative endocytic mechanism. Rather, they go across a lipid-dependent fusion process that is resistant to paraformaldehyde fixation, suggesting that proteins are only marginally involved in the exosome-to-cell membrane fusion. This was also extended to exosomes released from melanoma cells in a low pH condition. Accordingly, the increased fusion capacity of exosomes released in an acidic condition was associated to a change in membrane rigidity that was higher than in a buffered condition, in turn related to an increased amount of SM+GM3 lipids. GM3 is a recognized marker of highly ordered sphingolipid/cholesterol plasma membrane microdomains, known as “lipid rafts” (34, 35). Lipid rafts and, in particular, sphingolipids play a key role in viral fusion with cell membranes by affecting protein binding and by modulating membrane physicochemical and mechanical properties (36). Moreover, sphingomyelin-enriched microdomains have also been found to modulate the efficiency of membrane fusion (37). Hence, it seems conceivable that the high SM+GM3 content of the acidic exosomes positively affect their fusion ability.
Consistent with this finding, we also show that acidic exosomes fuse better with melanoma cells cultured in a buffered condition. A possible explanation is that acidic exosomes, which are intrinsically endowed with negative charge due to high GM3 content, once released in a highly rich H+ medium, are positively charged and fuse better with cells cultured in a buffered condition. This could be due to the fact that there are more negative charges available than in an acidic environment. As a consequence, it might be speculated that exosome-to-melanoma cell fusion may strictly depend on the electrostatic charges present in the close proximity of the cell-to-exosome contact sites. However, low microenvironmental pH is the in vivo steady-state condition of malignant tumor cells. We, therefore, considered co-cultivation of acidic exosomes and acidic cells the most physiologic condition because it realistically mimics exosome-to-cell contact within the tumor mass. Accordingly, we showed that fusion occurs better between acidic exosomes and acidic cells than between buffered exosomes and buffered cells. This suggests that exosome-to-cell fusion is a more frequent event within the tumor mass than in the surrounding normal tissues.
Consistent with this hypothesis, our data show that tumor exosomes can be preferentially delivered to metastatic tumor cells rather than primary (i.e. more differentiated) tumor cells. Because a peculiarity of malignant tumor cells is their ability to grow in an acidic condition (17), depending on the activity of proton pumps, we used a proton pump inhibitor (38, 39) to analyze the role of low pH in favoring exosome traffic in malignant tumor cells. We found that exosome entry into target cells was significantly reduced by preincubation with proton pump inhibitors, suggesting that inhibition of extracellular acidification may interfere with exosome traffic in malignant tumor cells.
This result is also consistent with a clear anti-neoplastic effect of PPI that correlates with the level of acidity of malignant tumors (15, 16). A point to note is that PPI are pro-drugs needing acidity to form the active drug (tetracyclic sulfenamide). Thus, tumor acidity represents a sort of specific target for PPIs, which have been proposed as new anti-cancer compounds (40).
We showed through confocal microscopy analysis that exosome proteins delivered in recipient cells are preferentially targeted to cytoplasmic vesicles expressing either endosome or lysosome markers. Thus, it seems conceivable from our data that exosomes may drive information into the internal structure of the cells but also that exogenous proteins and lipids may be stored for some time into intracellular vesicles. This suggests that exosomes may transfer proteins from one tumor cell to another tumor cell, in turn contributing to a paracrine diffusion of a malignant phenotype within the tumor mass. Thanks to their ability to circulate and transport a broad protein spectrum deriving from different compartments of the cell, exosomes may act as important mediators of intercellular cross-talk and molecule delivery. This hypothesis has recently been supported by data showing that cav-1-bearing microvesicles may transfer cav-1 to less aggressive cells and promote their invasive activity (18). Moreover, we have shown that cav-1 is highly detectable in exosomes purified from the plasma of melanoma patients with poor prognosis (25). In our study we have shown that cav-1-expressing exosomes released by metastatic melanoma cells deliver cav-1 to primary melanoma more efficiently in acidic than in buffered conditions. This result supports the hypothesis that tumor exosomes may represent a natural nano-delivery system for tumor-associated proteins, such as cav-1. Moreover, these data provide the evidence that features of tumor microenvironment, such as acidity, can contribute to increasing malignancy by enhancing exosome-mediated cav-1 intercellular delivery.
Notably, the acidity-mediated entry of exosomes into tumor cells through fusion may be comparable with the entry of retroviruses within cells (41). However, it is also conceivable that both tumors and viruses may hijack the cellular microvesicle traffic to increase their dissemination in the human body but also to interfere with normal communication between cells for their own advantage.
In summary, our study adds much to the knowledge of the biological relevance of exosomes as a nano-device for cell-to-cell communication. However, it is conceivable that in a tumor microenvironment exosomes may participate in increasing tumor malignancy using different mechanisms. In fact, it is known that exosomes deliver functional pro-apoptotic (1, 2) and differentiating molecules (42), requiring protein-protein interaction to trigger stimulation of different signaling pathways. However, it is interesting to note that the same signals that exosomes trigger in human normal cells are not triggered when exosomes interact with tumor cells. One example is the case of Fas-ligand-bearing exosomes that trigger Fas-mediated apoptosis in T cells but do not kill the melanoma cells that normally release them (1). One hypothesis is that the mechanism of interaction between exosomes and malignant tumor cells differs from the interaction between exosomes and normal cells. Here we provide evidence that exosomes preferentially fuse with the membrane of tumor cells and that this interaction is under the control of microenvironmental pH, which in turn modulates the lipid composition of cell and exosome membranes. We also provide evidence that, through fusion, exosomes may transfer tumor-associated proteins (e.g. cav-1) to cells that do not express these proteins in a pH-dependent way. This in turn suggests that acidic exosomes may really represent a delivery system for malignant diffusion within the tumor mass. Nevertheless, these microvesicles may have a key role in diffusing proteins that may be potentially transferred to many types of cells in several body compartments. This may be operative in tumor patients, but exosomes may have a role in cell-to-cell, organ-to-organ communication in both normal and other pathological conditions as well. In fact, some human body compartments, such as bone marrow, gut mucosa, and skin, are normally characterized by proliferation or rapid cell turnover that continuously change the pH of these tissues. However, in inflammatory, autoimmune, and infectious disease states the pH of various organs and compartments may vary due to a great deal of factors. Therefore, it appears conceivable that an acidic microenvironment may favor exosome-to-cell fusion in both normal and pathologic conditions.
Supplementary Material
Acknowledgments
We thank Mauro Biffoni and Mathias Viard for helpful experimental suggestions, Barbara Rossi for technical assistance in Laurdan experiments, and Alfonso Zito and Giuseppe Loreto for graphics.
This work was supported in part by a grant from Fondo per gli Investimenti della Ricerca di Base (FIRB) year 2003 (RBNE03FMCJ-002) of the Italian Ministry for University and Research (to M. S. and I. P.), by an Italy-to-China partnership project, by the Chemores FP6 European Project, and by a Italy-NIH partnership project supported by the Italian Ministry of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- PPI
- proton pump inhibitor
- cav-1
- caveolin-1
- PBMC
- peripheral blood mononuclear cell(s)
- PAF
- paraformaldehyde
- TES
- N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid
- GP
- generalized polarization
- SM
- sphingomyelin
- GM3
- N-acetylneuraminylgalactosylglucosylceramide
- MES
- 4-morpholineethanesulfonic acid
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Andreola G., Rivoltini L., Castelli C., Huber V., Perego P., Deho P., Squarcina P., Accornero P., Lozupone F., Lugini L., Stringaro A., Molinari A., Arancia G., Gentile M., Parmiani G., Fais S. (2002) J. Exp. Med. 195, 1303–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huber V., Fais S., Iero M., Lugini L., Canese P., Squarcina P., Zaccheddu A., Colone M., Arancia G., Gentile M., Seregni E., Valenti R., Ballabio G., Belli F., Leo E., Parmiani G., Rivoltini L. (2005) Gastroenterology 128, 1796–1804 [DOI] [PubMed] [Google Scholar]
- 3.Valenti R., Huber V., Filipazzi P., Pilla L., Sovena G., Villa A., Corbelli A., Fais S., Parmiani G., Rivoltini L. (2006) Cancer Res. 66, 9290–9298 [DOI] [PubMed] [Google Scholar]
- 4.Bard M. P., Hegmans J. P., Hemmes A., Luider T. M., Willemsen R., Severijnen L. A., van Meerbeeck J. P., Burgers S. A., Hoogsteden H. C., Lambrecht B. N. (2004) Am. J. Respir. Cell Mol. Biol. 31, 114–121 [DOI] [PubMed] [Google Scholar]
- 5.Calzolari A., Raggi C., Deaglio S., Sposi N. M., Stafsnes M., Fecchi K., Parolini I., Malavasi F., Peschle C., Sargiacomo M., Testa U. (2006) J. Cell Sci. 119, 4486–4498 [DOI] [PubMed] [Google Scholar]
- 6.Mears R., Craven R. A., Hanrahan S., Totty N., Upton C., Young S. L., Patel P., Selby P. J., Banks R. E. (2004) Proteomics 4, 4019–4031 [DOI] [PubMed] [Google Scholar]
- 7.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 8.Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., Tursz T., Amigorena S., Raposo G., Angevin E., Zitvogel L. (2002) Lancet 360, 295–305 [DOI] [PubMed] [Google Scholar]
- 9.Kim J. W., Wieckowski E., Taylor D. D., Reichert T. E., Watkins S., Whiteside T. L. (2005) Clin. Cancer Res. 11, 1010–1020 [PubMed] [Google Scholar]
- 10.Taylor D. D., Lyons K. S., Gerçel-Taylor C. (2002) Gynecol. Oncol. 84, 443–448 [DOI] [PubMed] [Google Scholar]
- 11.Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Conde I., Shrimpton C. N., Thiagarajan P., López J. A. (2005) Blood 106, 1604–1611 [DOI] [PubMed] [Google Scholar]
- 13.Morelli A. E., Larregina A. T., Shufesky W. J., Sullivan M. L., Stolz D. B., Papworth G. D., Zahorchak A. F., Logar A. J., Wang Z., Watkins S. C., Falo L. D., Jr., Thomson A. W. (2004) Blood 104, 3257–3266 [DOI] [PubMed] [Google Scholar]
- 14.Chu V. C., McElroy L. J., Chu V., Bauman B. E., Whittaker G. R. (2006) J. Virol. 80, 3180–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Milito A., Iessi E., Logozzi M., Lozupone F., Spada M., Marino M. L., Federici C., Perdicchio M., Matarrese P., Lugini L., Nilsson A., Fais S. (2007) Cancer Res. 67, 5408–5417 [DOI] [PubMed] [Google Scholar]
- 16.Luciani F., Spada M., De Milito A., Molinari A., Rivoltini L., Montinaro A., Marra M., Lugini L., Logozzi M., Lozupone F., Federici C., Iessi E., Parmiani G., Arancia G., Belardelli F., Fais S. (2004) J. Natl. Cancer Inst. 96, 1702–1713 [DOI] [PubMed] [Google Scholar]
- 17.Lugini L., Matarrese P., Tinari A., Lozupone F., Federici C., Iessi E., Gentile M., Luciani F., Parmiani G., Rivoltini L., Malorni W., Fais S. (2006) Cancer Res. 66, 3629–3638 [DOI] [PubMed] [Google Scholar]
- 18.Felicetti F., Parolini I., Bottero L., Fecchi K., Errico M. C., Raggi C., Biffoni M., Spadaro F., Lisanti M. P., Sargiacomo M., Carè A. (2009) Int. J. Cancer 125, 1514–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoekstra D., de Boer T., Klappe K., Wilschut J. (1984) Biochemistry 23, 5675–5681 [DOI] [PubMed] [Google Scholar]
- 20.Parasassi T., Krasnowska E. K., Bagatolli L., Gratton E. (1998) J. Fluoresc. 8, 365–373 [Google Scholar]
- 21.Palleschi S., Silvestroni L. (1996) Biochim. Biophys. Acta 1279, 197–202 [DOI] [PubMed] [Google Scholar]
- 22.Harris F. M., Best K. B., Bell J. D. (2002) Biochim. Biophys. Acta 1565, 123–128 [DOI] [PubMed] [Google Scholar]
- 23.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 24.Wubbolts R., Leckie R. S., Veenhuizen P. T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J. W., Geuze H. J., Stoorvogel W. (2003) J. Biol. Chem. 278, 10963–10972 [DOI] [PubMed] [Google Scholar]
- 25.Logozzi M., De Milito A., Lugini L., Borghi M., Calabrò L., Spada M., Perdicchio M., Marino M. L., Federici C., Iessi E., Brambilla D., Venturi G., Lozupone F., Santinami M., Huber V., Maio M., Rivoltini L., Fais S. (2009) PLoS One 4, e5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caby M. P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. (2005) Int. Immunol. 17, 879–887 [DOI] [PubMed] [Google Scholar]
- 27.Riteau B., Faure F., Menier C., Viel S., Carosella E. D., Amigorena S., Rouas-Freiss N. (2003) Hum. Immunol. 64, 1064–1072 [DOI] [PubMed] [Google Scholar]
- 28.Théry C., Zitvogel L., Amigorena S. (2002) Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 29.Vidal M., Hoekstra D. (1995) J. Biol. Chem. 270, 17823–17829 [DOI] [PubMed] [Google Scholar]
- 30.Cevc G., Richardsen H. (1999) Adv. Drug Deliv. Rev. 38, 207–232 [DOI] [PubMed] [Google Scholar]
- 31.Holthuis J. C., Pomorski T., Raggers R. J., Sprong H., Van Meer G. (2001) Physiol. Rev. 81, 1689–1723 [DOI] [PubMed] [Google Scholar]
- 32.Bagatolli L. A., Gratton E., Fidelio G. D. (1998) Biophys. J. 75, 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsson H., Mattson H., Sundell G., Carlsson E. (1985) Scand. J. Gastroenterol. Suppl.108, 23–35 [DOI] [PubMed] [Google Scholar]
- 34.Hakomori S. (2003) Curr. Opin. Hematol. 10, 16–24 [DOI] [PubMed] [Google Scholar]
- 35.Sorice M., Parolini I., Sansolini T., Garofalo T., Dolo V., Sargiacomo M., Tai T., Peschle C., Torrisi M. R., Pavan A. (1997) J. Lipid Res. 38, 969–980 [PubMed] [Google Scholar]
- 36.Teissier E., Pécheur E. I. (2007) Eur. Biophys. J. 36, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogasevskaia T., Coorssen J. R. (2006) J. Cell Sci. 119, 2688–2694 [DOI] [PubMed] [Google Scholar]
- 38.Lu X., Qin W., Li J., Tan N., Pan D., Zhang H., Xie L., Yao G., Shu H., Yao M., Wan D., Gu J., Yang S. (2005) Cancer Res. 65, 6843–6849 [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Zaguilan R., Lynch R. M., Martinez G. M., Gillies R. J. (1993) Am. J. Physiol. 265, C1015–C1029 [DOI] [PubMed] [Google Scholar]
- 40.Fais S., De Milito A., You H., Qin W. (2007) Cancer Res. 67, 10627–10630 [DOI] [PubMed] [Google Scholar]
- 41.Gould S. J., Booth A. M., Hildreth J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10592–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lakkaraju A., Rodriguez-Boulan E. (2008) Trends Cell Biol. 18, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.