Abstract
Irregular facets (If) is a dominant mutation of Drosophila that results in small eyes with fused ommatidia. Previous results showed that the gene Krüppel (Kr), which is best known for its early segmentation function, is expressed ectopically in If mutant eye discs. However, it was not known whether ectopic Kr activity is either the cause or the result of the If mutation. Here, we show that If is a gain-of-function allele of Kr. We then used the If mutation in a genetic screen to identify dominant enhancers and suppressors of Kr activity on the third chromosome. Of 30 identified Kr-interacting loci, two were cloned, and we examined whether they also represent components of a natural Kr-dependent developmental pathway of the embryo. We show that the two genes, eyelid (eld) and extramacrochaetae (emc), which encode a Bright family-type DNA binding protein and a helix-loop-helix factor, respectively, are necessary to achieve the singling-out of a unique Kr-expressing cell during the development of the Malpighian tubules, the excretory organs of the fly. The results indicate that the Kr gain-of-function mutation If provides a tool to identify genes that are active during eye development and that a number of them function also in the control of Kr-dependent developmental processes.
The gene Krüppel (Kr) encodes a zinc finger-type transcription factor (1, 2) expressed in spatially and temporally restricted patterns throughout Drosophila embryogenesis (3). It functions as a gap gene required for the proper segmentation of the central region of the embryo during early blastoderm stage (reviewed in refs. 4–6). Within the segmentation gene cascade, Kr protein functions mainly as a repressor of other gap genes and pair-rule genes (6–8), restricting their localized activities along the anterior–posterior axis of the embryo (9). After blastoderm stage, Kr is expressed functionally during the following development of the larval visual system (10), formation of the central and stomatogastric nervous system (11, 12), muscle differentiation (13, 14), and generation of the kidney-like Malpighian tubules (15–17).
In addition to the multiple activity patterns of Kr during embryogenesis, Kr mis-expression had been observed in the eye imaginal discs of the dominant Irregular factes (If) mutation (18), which results in reduced adult eyes lacking the regular array of ommatidia (19). However, the question of whether the notable ectopic Kr expression is the cause of the If mutant eye phenotype or just a peripheral consequence of the mutation has not been addressed. Here, we show that If is a dominant gain-of-function allele of Kr that causes mis-expression of the gene in the developing eye imaginal disc. We used the dosage-sensitive If mutation in a genetic screen to isolate dominant enhancers and suppressors of Kr activity that are located on the third chromosome. We identified 30 loci that modulate the eye phenotype generated by the ectopic Kr activity. A more detailed analysis of two genes demonstrates that the modifier screen involving the If mutation provides a tool to isolate factors that act, in addition to being expressed and possibly required during eye development, in a Kr-dependent developmental pathway during embryogenesis. The two genes described here, eyelid (eld) and extramacrochaetae (emc), code for a member of the Bright family of DNA binding proteins and for a helix-loop-helix protein, respectively (20–22). They control specific aspects of Kr expression required for the proper allocation of a unique Kr-dependent cell fate within the Malpighian tubule primordium.
MATERIALS AND METHODS
Drosophila Strains, Mutagenesis, and Mutant Embryos.
Drosophila strains were kept under standard conditions. The ethylmethane sulfonate (EMS) screen on If mutants was done as described (1). The F2 progeny was inspected for embryonic segmentation defects. Twelve Kr alleles were obtained, all of which suppressed the If phenotype to some degree. In the modifier screen, If homozygous females were crossed to males bearing a mutant third chromosome in trans over balancer chromosomes TM3 or TM6B. F1 progeny carrying a mutant third chromosome in the If mutant background were scored for an altered If/+ eye phenotype. The screen involved 335 lethal P-element insertion lines of the Berkeley Genome Project (23) and deficiency chromosomes (Umea and Bloomington stock collection) that uncover the regions of the third chromosome as shown in Fig. 2 (black bars; details in the legend).
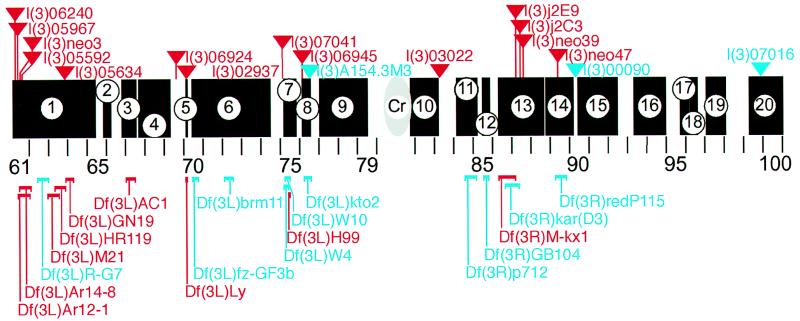
Figure 2.
Schematic representation of the third chromosome of Drosophila (section 61–100) showing the regions that were screened with deficiencies (black bars; nos. 1–20). Deficiencies that enhance (blue) or suppress (red) are indicated below the sections; P-element-tagged loci that enhance (blue) or suppress (red) the If phenotype are indicated by triangles above the sections. The following deficiencies were tested: (1) region 61A to 65C: Df(3L)emc-E12, Df(3L)Ar12–1, Df(3L)Ar14–8, Df(3L)R-G5, Df(3L)R-G7, Df(3L)Aprt-1, Df(3L)GN19, Df(3L)GN34, Df(3L)GN24, Df(3L)M21, Df(3L)HR370, Df(3L)HR232, Df(3L)HR119, Df(3L)GN50, Df(3L)ZM47; (2) region 65F3 to 66B10: Df(3L)pbl-X1; (3) 66F5–67D7–13: Df(3L)29A6, Df(3L)AC1; (4) region 67F2–3 to 69B4–5: Df(3L)BK9, Df(3L)lxd6, Df(3L)vin2, Df(3L)vin6, Df(3L)vin7; (5) region 70A2–3 to A5–6: Df(3L)Ly; (6) region 70C1 to 74C: Df(3L)fzGF3b, Df(3L)fzD21, Df(3L)st-f13, Df(3L)st7; (7) region 75B3–6 to 75F1: Df(3L)W10, Df(3L)Cat, Df(3L)W4, Df(3L)H99; (8) region 76A3-B2 to76D5: Df(3L)VW3, Df(3L)kto2; (9) region 77A1 to 79C9: Df(3L)rdgC, Df(3L)ri79c, Df(3L)Pc-Mk; (10) region 81F to 83A: Df(3R)ME15, Df(3R)4–75, Df(3R)P-93, Df(3R)2–2; (11) region 84A1–2 to 85B6: Df(3R)Scr, Df(3R)Antp17, Df(3R)p712, Df(3R)pXT103, Df(3R)p819; (12) region 85D8 to 85F6: Df(3R)by10, Df(3R)GB104, Df(3R)by62; (13) 86C1;88E5–6: Df(3R)M-Kx1, Df(3R)karD3, Df(3R)ry506–85c, Df(3R)red1; (14) 88F;90A: Df(3R)Po4, Df(3R)redP115, Df(3R)C4; (15) region 90C2-D1 to 92D3–6: Df(3R)P14, Df(3R)ChaM7, Df(3R)D1-BX12; (16) region 93B3–5 to 94: Df(3R)e-R1, Df(3R)e-N19; (17) region 95E8-F1 to 96A17–18: Df(3R)crbS87–4, Df(3R)crbS87–5, Df(3R)XS; (18) region 96B-D: Df(3R)XTAI; (19) region 97A to 98A1–2: Df(3R)TI-P; and (20) region 99B to 100F: Df(3R)L127, Df(3R)B81, Df(3R)awd-KRB. Cr: position of centromer. Details in Materials and Methods.
The following fly stocks were used in this work: Oregon R, l(3)00090, l(3)04539, l(3)05592 (23), emcE12 (24), E(Spl)X1, E(Spl)8D06, da1, Kr9, Kr25 (19), brm2 (25), and b pr cn wx If (obtained from the Tübingen stock collection). The two eld alleles (elddust55 and elddust31) were recovered in a standard EMS mutagenesis screen (26). eld alleles were used in trans over the l(3)00090 P-element insertion, shown to cause an eld mutation (20). Complementation analysis showed that l(3)00090 and l(3)04539 are allelic. The Malpighian tubules were examined in trans-heterozygotes of the respective EMS alleles and the l(3)00090 P-insertion. Ectopic expression of Kr in the eye imaginal disc was induced by using the Gal4/UAS system (27) comprised of the sevenless heatshock promoter (gift of K. Basler) or the eyeless-Gal4 (U.W., unpublished work) and UAS-Kr (M.H. and H.J., unpublished work) transgenes.
SEM, Immunocytochemistry, and Molecular Procedures.
Flies were prepared for SEM as described (20), and micrographs were obtained by using a Philipps XL 20 electron microscope. Immunological stainings of whole-mount embryos were carried out as described (28) by using the Vectastain ABC Elite horseradish peroxidase kit (Vectastain, Vector). After staining, embryos were examined as described (29). Antibody dilutions were: rabbit anti-Krüppel 1:20 (3); Mab22C10 1:20 (Hybridoma Bank); mouse anti-β-galactosidase 1:1000 (Cappel). Whole-mount in situ hybridizations were performed with digoxigenin-labeled DNA probes (30) or digoxigenin-labeled RNA probes (8). As probes, we used lacZ, emc (obtained from J. Modolell), and eld cDNA fragments. Stained embryos were examined and photographed by using a Zeiss Axiophot microscope.
Plasmid rescue for obtaining genomic DNA from the lines l(3)00090, l(3)04539, and l(3)05592 as well as for the other P-element modifier lines identified was performed as described (32). The screening of genomic and cDNA libraries, the handling of DNA, and the preparation of probes were as described in ref. 33. The sequences of the genomic DNA of If, the If revertant alleles, and the cDNAs as well as the DNA surrounding the P-element insertion sites were determined by using the dideoxynucleotide method (34). Sequence comparison and database searches [National Center for Biotechnology Information database (25)] revealed that the P-elements were inserted in the first intron of the previously identified eld gene and in the 5′-untranscribed region of the emc gene, respectively. A 3.5-kb HindIII plasmid rescued of line l(3)00090 (see Fig. 2) was used to screen for eld cDNAs.
RESULTS AND DISCUSSION
Irregular Facets Is a Dominant Krüppel Gain-of-Function Mutation.
If was identified as a spontaneous dominant mutation that causes a severe adult eye phenotype (19). Heterozygous adults bearing the dominant If mutation develop small and narrow eyes that are pointed ventrally; the facets are irregularly arranged, sometimes fused or absent in ventral portions (Fig. 1 A and B). In homozygous If mutants, eyes are further reduced in size and form narrow slits with a glossy surface lacking most of the ommatidia (Fig. 1C). The eye defects can be traced back to an irregular arrangement of differentiating photoreceptor cells during eye imaginal disc development (35) and correlate with ectopic expression of Kr in the If mutant discs, as has been described (18).
Figure 1.
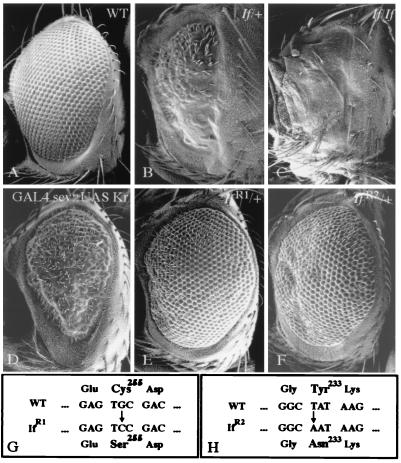
SEM of wild-type and If mutant Drosophila eyes. (A) Wild-type eye. (B) If/+ mutant eye. Note a reduction of the size of eyes; the ommatidia are fused or absent in ventral portions of the eye (Upper) and irregularly arranged dorsally (Lower). (C) Eye remnant of a homozygous If mutant fly. (D) Eye phenotype caused by ectopic Kr expression provided by the sevenless/heatshock-GAL4/UAS-Kr transgene combination (see Materials and Methods). Note the If-like symptoms such as a reduction of the eye and the irregular arrangement of the ommatidia. A similar although less severe eye phenotype was caused by eyeless -GAL4. The difference in the severity is likely to be due to differences between the ectopic promoter in If and the eyeless or sevenless/heat shock promoter used to misexpress Kr (see Materials and Methods). (E) IfR1/+ mutant eye indicating that the If mutant phenotype is almost reversed to wild type. (F) IfR2/+ mutant eye indicating that the If mutant phenotype is weakened significantly. (G) Molecular lesion generated in the IfR1 DNA. The G/C transversion causes a replacement of cysteine255 by serine within the second zinc finger motif of Krüppel (37). This lesion causes a strong Kr segmentation phenotype (38). (H) Molecular lesion of the IfR2 allele; T/A transversion resulting in a replacement of tyrosine233 by asparagine within the first zinc finger motif of Krüppel that causes a weak Kr segmentation phenotype (39). For details see text.
We reasoned that the ectopic Kr expression de-regulates gene activities during eye imaginal disc development. We tested this proposal by misexpression of Kr by the UAS/GAL4 system (27). Misexpression of Kr during eye development resulted in an If mutant-like phenotype (Fig. 1D; details in the legend). Furthermore, we also found the If phenotype with flies that carry the If-bearing chromosome in trans to a Kr deficiency or to a chromosome bearing a Kr lack-of-function allele (data not shown). These results suggest that If is a neomorphic allele of Kr. This proposal is supported by the identical cytological location of both If and Kr at 60F3 and by the lack of recombinants between If and Kr (36).
If If is a neomorphic allele of Kr, point mutations within the coding region of the gain-of-function allele If should cause a reversion of the dominant eye mutant phenotype and generate a Kr segmentation phenotype in homozygousity. To obtain If revertants, we performed an EMS mutagenesis screen and examined whether the sequence of the Kr gene is affected in such If revertants (details in Materials and Methods). One If reversion, termed “IfR1,” was left with subtle eye defects, meaning that the severeness of the If mutation is substantially reduced, almost to wild type (Fig. 1E). We also obtained a series of weaker revertants of the If mutant phenotype, indicating that the reversion was less effective than in the first case. One such weaker revertant, termed “IfR2,” is shown in Fig. 1F. All of the If revertants are recessive embryonic lethals and cause a typical Kr mutant segmentation phenotype (not shown). Moreover, although If complements Kr mutations (ref. 36; unpublished work), the If revertants fail to do so. Thus, the If revertants are alleles of Kr; the dominant If mutation is a Kr gain-of-function allele whereas the reversions cause Kr loss-of-function alleles. To establish this link between If and Kr firmly, we determined the molecular lesions generated in IfR1 and IfR2.
Sequencing and comparison of If and IfR1 DNA revealed a Kr wild-type coding region in If DNA and a single missense mutation causing a replacement of cysteine 255 by a serine residue of the IfR1 protein (Fig. 1G). This replacement causes a disruption of the second zinc finger motif of Krüppel, explaining the strong Kr segmentation phenotype in mutant embryos (37, 38). The weak eye defects that were left with the revertant IfR1 (Fig. 1E) argue for some residual Kr activity that may interfere with eye development. The Kr sequence of the revertant IfR2 shows a different single base pair exchange that results in a replacement of tyrosine 233 within the first Krüppel zinc finger motif by asparagine (Fig. 1H). This finding is in agreement with a weak Kr mutant segmentation defect generated by such a replacement (39), and it argues that Kr function is not as strongly impaired as in the first case. The molecular analyses of two of several EMS-induced Kr revertants establish unequivocally allelism between If and Kr.
The results establish that the phenotype of the spontaneous If mutation is caused by a dominant gain-of-function of Kr activity in eye imaginal discs. One possibility that would explain the mutational event is that If may have acquired additional cis-acting enhancer sequence elements due to a small chromosomal rearrangement or a transposition event that conducts a new Kr expression domain during the eye imaginal disc development. This proposal is consistent with the notion that DNA fragments outside the 18-kb cis-acting Kr control region (40) of If and wild-type flies are different in a sense that If DNA, but not Kr wild-type DNA, contains repetitive DNA (41). This suggests that the spontaneous If mutation could be caused by a transposon insertion resulting in the misexpression of the Kr gene.
Dominant Modifiers of Ectopic Kr Activity in the Eye Imaginal Disc.
Homozygous If eyes are more severely affected than heterozygous If eyes. Furthermore, a small reduction of Kr activity during eye development, as exemplified with the IfR2 mutation, reduces the strength of the If phenotype. These observations suggest that the If mutant phenotype is sensitive to the dose and activity of Kr in the eye disc and may thereby provide a means to identify modifiers of Kr activity. We therefore conducted a genetic screen, searching for mutations on the third chromosome that dominantly increase (enhance) or decrease (suppress) the severity of the If mutant eye phenotype. For this, we examined the eye phenotypes of heterozygous If mutants in combination with chromosomal deficiencies or lethal P-element enhancer trap insertions (Berkeley collection; see Materials and Methods). The chromosome combinations examined cover ≈75% of the third chromosome, and 30 modifier loci (12 enhancers, 18 suppressors) were identified. The results of the screen and the by now few molecularly characterized candidate genes are summarized in Fig. 2. Here, we will focus on two modifiers of ectopic Kr activity, eld and emc (Fig. 3), showing that their activities also are required in a natural context during Kr-dependent embryonic organ development.
Figure 3.
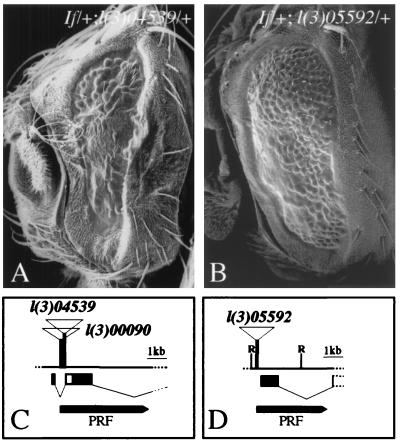
SEM showing the effect of an enhancer (eld) and a suppressor (emc) of the If mutant eye phenotype and the molecular characterization of the genes. (A) If/+; l(3)04539/+ flies develop smaller eyes and fewer ommatidia than If/+ flies, indicating an enhancement of the If phenotype (compare with Fig. 1B). (B) The If phenotype is significantly suppressed in If/+; l(3)05592/+ flies (compare with Fig. 1 A and B). (C) Partial physical map of the eld gene (for details see ref. 20), which was identified by complementation analysis of the P-element insertions l(3)04539 and l(3)00090 previously shown to be an allele of eld. Genomic DNA adjacent to the P-element insertion sites was isolated by plasmid rescue (PRF) and used to screen for cDNAs (see bottom of the physical map). Both P-elements were inserted in the first intron of the previously identified eld gene (20). Exons are indicated by black bars below the physical map. (D) P-element insertion site of l(3)05592 in the 5′ region of emc gene. l(3)05592 fails to complement previously identified emc mutations indicating that it is an allele of emc. Genomic DNA of the emc locus was isolated by plasmid rescue (see Materials and Methods); the diagnostic sequence 3′ adjacent to the P-element insertion is ACTCCGCCTATCGGATTC. Part of the translated region is shown (black bar; for details, see refs. 21 and 22). Diagnostic restriction site: R, EcoRI.
Identification of eld and emc as Kr Interacting Genes.
We molecularly characterized two modifiers of If that were generated by the P-element insertions l(3)00090 and l(3)05592, respectively (23). After plasmid rescue (32), we cloned genomic DNA fragments adjacent to the P-element insertion sites, determined its sequence and identified the transcription units affected by the P-element insertion. The results showed that the P-element insertions were localized in the 5′ regions of the previously characterized genes eld (20) (Fig. 3C) and emc (21, 22) (Fig. 3D), respectively. Transcript mapping combined with cDNA sequencing and complementation analysis with known eld and emc alleles confirmed that the If enhancer is eld (Fig. 3A) whereas the If suppressor is emc (Fig. 3B). Furthermore, previously characterized alleles of the two loci were found to modify the If mutant phenotype, indicating that the effects on ectopic Kr activity in the eye disc were not allele-specific (not shown).
Both modifier genes code for transcription factors. As shown recently, eld encodes a Bright family-type DNA binding protein (20), whereas emc codes for a helix-loop-helix transcription factor (21, 22) required for the proper specification of many cell types in the embryo (24, 42). As expected from their interference with ectopic Kr activity (see above), both genes are expressed in the developing eye imaginal discs (23) (data not shown). In addition, they are expressed at multiple other sites during embryogenesis, including the Malpighian tubules, which develop in a Kr-dependent manner (see below). This allowed us to examine whether the two modifiers of ectopic Kr activity act also in a natural Kr-dependent developmental pathway.
emc-Dependent Singling-Out of the Kr-Expressing Mother Tip Cell.
Kr expression defines the Malpighian tubule anlage at late blastoderm stage and becomes restricted to a ring of cells at the midgut/hindgut boundary from where Kr-expressing Malpighian tubule precursors evert (17). Previous studies have shown that the specification of Malpighian tubule fate and the segregation of the cells depend on Kr expression in the Malpighian tubule anlage (17, 43). In Kr-deficient embryos, the respective cells become part of the hindgut epithelium (44).
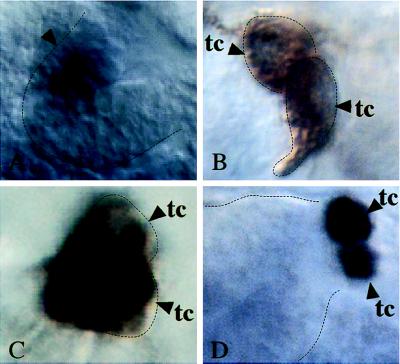
Once the tubules evert, Kr expression becomes restricted to a single cell, termed the “tip mother cell” (Fig. 4A). The singling-out process of this cell from an equivalence group of Malpighian tubule precursors involves the activated Notch pathway (15), which restricts the proneural bHLH proteins encoded by the achaete-scute-complex (ASC) genes (45) to the tip mother cell. In this cell, the ASC proteins act in concert with bHLH protein encoded by daughterless (da) (45–47) to maintain Kr expression (M.H. and H.J., unpublished work). The tip mother cell divides once, and the daughters give rise to the tip cell, which controls proliferation during tubule elongation (44) and differentiates neuronal characteristics (15), and an excretory cell, termed “satellite cell” (Fig. 4B). The satellite cell loses Kr expression in a Notch-dependent manner (15), whereas Kr expression is maintained in the tip cell until the end of embryogenesis (Fig. 4C).
Figure 4.
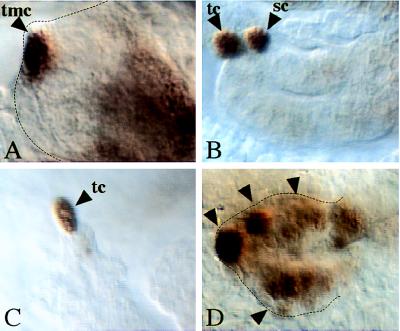
emc represses Kr expression in the Malpighian tubules. (A–C) Wild-type embryos. (D) Homozygous emc mutant embryo (emcE12) stained with anti-Krüppel antibodies. (A) Restricted Kr expression in the tip mother cell (tmc). (B) The tip mother cell divides and gives rise to the tip cell (tc) and the satellite cell (sc) (15). (C) Kr expression is maintained only in the tip cell (tc). (D) In emc mutants, Kr is expressed in many cells along the tubule (arrowheads).
emc expression accompanies Malpighian tubule development in a manner similar to Kr expression. However, once the tip cell is formed, the patterns of expression become complementary, meaning that emc expression continues in all cells of the elongating Malpighian tubules except in the tip cell (ref. 24; unpublished work). To test whether the complementary patterns of Kr and emc expression reflect a regulatory effect of emc on Kr, as indicated during eye development in the If mutant, we examined Kr expression in the Malpighian tubules of emc mutant embryos. Fig. 4D shows multiple Kr-expressing cells in emc mutant Malpighian tubules. This finding is consistent with the previous finding that emc mutant embryos develop multiple tip cells and that each of them continues to express achaete (24). Virtually the same observations have been made previously with Notch mutants, and it was shown that Notch acts toward restricting the activity of the proneural bHLH proteins, which are required to maintain Kr expression first in the tip mother cell and subsequently in the tip cell (15). However, although the activated Notch pathway acts through transcriptional repression of the ASC genes, emc protein antagonizes proneural bHLH activities by sequestering the proteins as heterodimers that are incapable of binding to DNA (48, 49). Our results are therefore consistent with the proposal that emc functions in the control of Kr expression by antagonizing proneural bHLH activities that are required to maintain Kr expression in the tip mother cell.
eld Antagonizes Kr-Dependent Tip Cell Differentiation.
The eld protein shows a nuclear localization, consistent with its suspected function as a transcription factor (20). It appears to act in multiple signaling pathways because it antagonizes wingless activity, suppresses Ras1 activity in the eye (50), and blocks Notch-dependent neuronal differentiation (20). During Malpighian tubule development, eld is expressed in a restriced area of the everting precursors that corresponds to the equivalence group of cells expressing the proneural genes (Fig. 5A).
Figure 5.
eld represses Kr expression in the sibling cell. (A) RNA in situ hybridization of wild-type embryos with an eld probe (see Materials and Methods) showing expression in a group of cells (arrowhead) corresponding to the cluster of cells in the outgrowing tubules from which the tip mother cell is selected. (B and D) Embryos transheterozygous for the eld P-insertion l(3)00090 and elddust55 and (C) for l(3)00090 and elddust31. (B and C) Mab22C10 antibody staining, characteristic for tip cells (15), shows that two instead of a single tip cell (tc) are formed. (D) Formation of two tip cells corresponds to maintained Kr expression in the satellite cell, as revealed by anti-Krüppel antibody stainings (arrowheads). Details in the text.
eld mutant embryos exert a distinct phenotype during Malpighian tubule development that is linked to Kr activity. Whereas the anlage and the four tubules evert normally (data not shown), each tubule develops two instead of the normal one tip cell (Fig. 5 B and C). Tip cell development is under the control of Kr activity (M.H. and H.J., unpublished work), so we next asked whether and when Kr expression is altered in eld mutant embryos. In correspondence with the mutant phenotype, the initial expression of Kr, including its restriction to the tip mother cell, appears to be normal (not shown). However, once the tip mother cell has undergone division, two instead of only one of the daughter cells maintain Kr expression (Fig. 5D). This indicates that eld activity is necessary to prevent Kr expression in the sibling of the tip cell and allows for its differentiation into a satellite cell. Thus, although emc is necessary for the restriction of Kr to the tip mother cell, eld functions specifically at the subsequent step during Malpighian tubule development where an alternative and Kr-dependent cell fate decision is taken between the daughters of the tip mother cell.
Notch signaling recently was shown to be required first for the selection of the tip mother cell and subsequently for the distinction between its daughters to either develop a tip cell or a satellite cell (15). Consistently, in Notch mutant embryos, all cells of the proneural equivalence group develop first into tip mother cells; these cells divide and subsequently develop into the multiple tip cells that continue Kr expression (15). In contrast, only two tip cells were found in eld mutants. This finding implies that, if eld acts in a Notch-dependent manner and/or mediates Notch signaling (20), its activity is required only for the second of the two Notch-dependent differentiation steps during Malpighian tubule development. Thus, eld participates as an optional component in the Notch-signaling pathway and is needed to prevent, directly or indirectly, the maintenance of Kr expression in the satellite cell that would otherwise develop into a second tip cell.
CONCLUSIONS
The results presented here demonstrate that gene activities that were identified via an artificial experimental situation, namely the ectopic expression of Kr in the developing eye disc, can lead to the identification of integral components of a Kr-dependent developmental pathway during embryogenesis. In the eye imaginal disc, emc suppresses Kr activity whereas eld has an opposite effect, but both act during embryonic Malpighian tubule development as negative regulators of Kr. We have no explanation for this phenomenon. It could mean, in negative terms, that the Kr misexpression screen turned up dosage-sensitive genes affecting cell fate that were several steps downstream from Kr activity and thus have no direct interaction with Kr. Thus, each gene identified in the modifyer screen represents a candidate gene that needs to be evaluated critically through additional criteria as outlined here for eld and emc. The additional screening is essential to distinguish between direct Kr interactors and genes that mediate different read-outs of the Kr pathway in cells that have a different organ or tissue competence. However, in view of the fragmentary information concerning the spatial and temporal control of postblastodermal Kr expression (17, 40) and in view of the fact that the few Kr target genes of Kr were identified by molecular approaches (13, 51), our experimental strategy to assess components of a Kr-dependent regulatory circuitry seems a valid one.
Acknowledgments
We thank Gordon Dowe and Sonja Fellert for technical support and Michael Stauber for various and important contributions. We thank in particular Dieter Kötting and Kurt Müller for help in preparing the SEM data. We thank especially Marcos González-Gaitán for helpful discussions. B.K. was supported by a fellowship of the Boehringer Ingelheim Fonds, and P.C. was supported by a CSIC postdoctoral fellowship. This work was supported by the SFB 271 of the Deutsche Forschungsgemeinschaft (H.J. and M.H.).
ABBREVIATION
- EMS
ethylmethane sulfonate
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Preiss A, Rosenberg U B, Kienlin A, Seifert E, Jäckle H. Nature (London) 1985;313:27–32. doi: 10.1038/313027a0. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg U B, Preiss A, Seifert E, Jäckle H, Knipple D C. Nature (London) 1985;313:703–706. doi: 10.1038/313703a0. [DOI] [PubMed] [Google Scholar]
- 3.Gaul U, Seifert E, Schuh R, Jäckle H. Cell. 1987;50:639–647. doi: 10.1016/0092-8674(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 4.Hoch M, Jäckle H. Curr Opin Genet Dev. 1993;3:566–573. doi: 10.1016/0959-437x(93)90092-4. [DOI] [PubMed] [Google Scholar]
- 5.Niessing D, Rivera-Pomar R, La Rosée A, Häder T, Schöck F, Purnell B A, Jäckle H. J Cell Phys. 1997;173:162–167. doi: 10.1002/(SICI)1097-4652(199711)173:2<162::AID-JCP15>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 6.Pankratz M J, Jäckle H. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1993. pp. 467–516. [Google Scholar]
- 7.Stanojevic D, Small S, Levine M. Science. 1991;254:1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 8.Klingler M, Gergen J P. Mech Dev. 1993;43:3–19. doi: 10.1016/0925-4773(93)90019-t. [DOI] [PubMed] [Google Scholar]
- 9.Rivera-Pomar R, Jäckle H. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- 10.Schmucker D, Taubert H, Jäckle H. Neuron. 1992;9:1025–1039. doi: 10.1016/0896-6273(92)90063-j. [DOI] [PubMed] [Google Scholar]
- 11.Romani S, Jiménez F, Hoch M, Patel N H, Taubert H, Jäckle H. Mech Dev. 1996;60:95–107. doi: 10.1016/s0925-4773(96)00603-x. [DOI] [PubMed] [Google Scholar]
- 12.González-Gaitán M, Jäckle H. Development. 1995;121:2313–2325. doi: 10.1242/dev.121.8.2313. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann C, Landgraf M, Bate M, Jäckle H. EMBO J. 1997;16:5299–5309. doi: 10.1093/emboj/16.17.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Gomez M, Romani S, Hartmann C, Jäckle H, Bate M. Development. 1997;124:3407–3414. doi: 10.1242/dev.124.17.3407. [DOI] [PubMed] [Google Scholar]
- 15.Hoch M, Broadie K, Jäckle H, Skaer H. Development. 1994;120:3439–3450. doi: 10.1242/dev.120.12.3439. [DOI] [PubMed] [Google Scholar]
- 16.Gloor H. Arch Jul Klaus Stiftung. 1950;29:277–287. [Google Scholar]
- 17.Gaul U, Weigel D. Mech Dev. 1991;33:57–68. doi: 10.1016/0925-4773(90)90135-9. [DOI] [PubMed] [Google Scholar]
- 18.Baker N E, Moses K, Nakahara D, Ellis M C, Carthew R W, Rubin G M. J Neurogenetics. 1992;8:85–100. doi: 10.3109/01677069209084154. [DOI] [PubMed] [Google Scholar]
- 19.Lindsley D L, Zimm G G. The Genome of Drosophila melanogaster. San Diego, CA: Academic; 1992. [Google Scholar]
- 20.Treisman J E, Luk A, Rubin G M, Heberlein U. Genes Dev. 1997;11:1949–1962. doi: 10.1101/gad.11.15.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis H M, Spann D R, Posakony J W. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- 22.Garrell J, Modolell J. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- 23.Karpen G H, Spradling A C. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cubas P, Modolell J, Ruiz-Gomez M. Development. 1994;120:2555–2566. doi: 10.1242/dev.120.9.2555. [DOI] [PubMed] [Google Scholar]
- 25.Kennison J A, Tamkun J W. Proc Natl Acad Sci USA. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigliatti T. In: Mutagenesis. Roberts D B, editor. Oxford: IRL Press; 1986. pp. 39–48. [Google Scholar]
- 27.Brand A H, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 28.Macdonald P, Struhl G. Nature (London) 1986;324:537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt-Ott U, González-Gaitán M, Jäckle H, Technau G M. Proc Natl Acad Sci USA. 1994;91:2664–2668. doi: 10.1073/pnas.91.18.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 31.González-Gaitán M, Jäckle H. In: In Situ Localization of Proteins in Whole Mount Tissue. Crampton J M, Beard C B, Louis C, editors. New York: Chapman & Hall; 1997. pp. 283–294. [Google Scholar]
- 32.Wilson C, Pearson R K, Bellen H J, O′Kane C J, Grossniklaus U, Gehring W J. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renfranz P J, Benzer S. Dev Biol. 1989;136:411–429. doi: 10.1016/0012-1606(89)90267-4. [DOI] [PubMed] [Google Scholar]
- 36.Wieschaus E, Nüsslein-Volhard C, Kluding H. Dev Biol. 1984;104:172–186. doi: 10.1016/0012-1606(84)90046-0. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg U B, Schröder C, Preiss A, Kienlin A, Côté S, Riede I, Jäckle H. Nature (London) 1986;319:336–339. [Google Scholar]
- 38.Redemann N, Gaul U, Jäckle H. Nature (London) 1988;332:90–92. doi: 10.1038/332090a0. [DOI] [PubMed] [Google Scholar]
- 39.Gaul U, Redemann N, Jäckle H. Proc Natl Acad Sci USA. 1989;86:4599–4603. doi: 10.1073/pnas.86.12.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoch M, Schroeder C, Seifert E, Jäckle H. EMBO J. 1990;9:2587–2596. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preiss A. Ph.D. thesis. Tübingen, Germany: University of Tübingen; 1985. [Google Scholar]
- 42.Ellis H M. Mech Dev. 1994;47:65–72. doi: 10.1016/0925-4773(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 43.Harbecke R, Janning W. Genes Dev. 1989;3:114–122. doi: 10.1101/gad.3.1.114. [DOI] [PubMed] [Google Scholar]
- 44.Skaer H. In: The Development of Drosophila melanogaster. Martinez Arias A, Bate M, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1993. pp. 941–1013. [Google Scholar]
- 45.Campos-Ortega J A. In: The Development of Drosophila melanogaster. Martinez Arias A, Bate M, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1091–1130. [Google Scholar]
- 46.Cronmiller C, Schedl P, Cline T W. Genes Dev. 1988;1988:1666–1676. doi: 10.1101/gad.2.12a.1666. [DOI] [PubMed] [Google Scholar]
- 47.Cabrera C V, Alonso M C. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Doren M, Ellis H M, Posakony J W. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- 49.Cabrera, C. V., Alonso, M. C. & Huikeshoven, H. (1994) Development 120. [DOI] [PubMed]
- 50.Karim F D, Chang H C, Therrien M, Wassarman D A, Laverty T, Rubin G M. Genetics. 1996;143:315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartmann C, Jäckle H. Dev Genes Evol. 1997;207:186–193. doi: 10.1007/s004270050106. [DOI] [PubMed] [Google Scholar]