Abstract
LapB is a non-heme Fe(II)-dependent 2,3-dioxygenase that catalyzes the second step of a long-chain alkylphenol (lap) degradation pathway in Pseudomonas sp. KL28 and belongs to the superfamily of type I extradiol dioxygenases. In this study, the crystal structures of substrate-free LapB and its complexes with a substrate or product were determined, along with a functional analysis of the active site residues. Structural features of the homotetramer are similar to those of other type I extradiol dioxygenases. In particular, the active site is located in the C-domain of each monomer, with a 2-His-1-carboxylate motif as the first coordination shell to iron ion. A comparison of three different structures in the catalytic cycle indicated catalysis-related local conformational changes in the active site. Specifically, the active site loop containing His-248 exhibits positional changes upon binding of the substrate and establishes a hydrogen-bonding network with Tyr-257, which is near the hydroxyl group of the substrate. Kinetic analysis of the mutant enzymes H248A, H248N, and Y257F showed that these three mutant enzymes are inactive, suggesting that this hydrogen-bonding network plays a crucial role in catalysis by deprotonating the incoming substrate and leaving it in a monoanionic state. Additional functional analysis of His-201, by using H201A and H201N mutants, near the dioxygen-binding site also supports its role as base and acid catalyst in the late stage of catalysis. We also noticed a disordered-to-ordered structural transition in the C-terminal region, resulting in the opening or closing of the active site. These results provide detailed insights into the structural and functional features of an extradiol dioxygenase that can accommodate a wide range of alkylcatechols.
Introduction
Non-heme iron-dependent dioxygenases constitute one group of enzymes involved in bacterial biodegradation pathways of aromatic compounds (1). In general, chemically stable aromatic compounds are initially subject to hydroxylation by enzymes in the pathway, producing aromatic compounds with vicinal diols. Subsequently, under aerobic conditions, dioxygenase catalyzes a ring cleavage of the catecholic compound by activating and incorporating molecular dioxygen into the substrate. The ring-opened product is further degraded in the pathway, and the resulting final compound is used as the nutrition source for microorganisms via the citric acid cycle. Enzymes are classified according to their mode of ring cleavage as intradiol and extradiol dioxygenases, which are distinct both in their sequences and structures as well as in their reaction mechanisms (1, 2).
Although some enzymes utilize Mn(II), extradiol dioxygenases primarily utilize Fe(II) as a cofactor without changes in its redox state during catalysis (3) and cleave the meta-position to the hydroxyl groups to produce muconic semialdehyde adducts (Scheme 1) (4, 5). Structural classification indicates that extradiol dioxygenases comprise three superfamilies (2). Among these, type I enzymes contain a common structural domain that is composed of two repeated βαβββ motifs; structural analysis revealed that these enzymes exhibit various oligomeric states (1). In addition, type I enzymes share similar structural features in the active site, with two histidines and one glutamate conserved in the metal-binding site. Various spectroscopic and computational analyses have suggested the identity of the putative intermediates in the catalytic cycle (6, 7). In particular, an alkylperoxo intermediate was proposed to yield a lactone intermediate via Criegee rearrangement, but previous crystallographic studies of extradiol dioxygenases revealed only the binding mode of a substrate and/or O2 molecule in the active site. Only recently, the first structural evidence for the long searched for alkylperoxo intermediate (8), the O–O bond cleavage (9), and the binding mode of the product (8) has been visualized in structural analyses of homoprotocatechuate 2,3-dioxygenase (HPCD)2 from Brevibacterium fuscum. In this homotetrameric dioxygenase, four independent subunits form one asymmetric unit of crystal and thus have different crystal packings in each subunit. Due to these unique crystalline environments, each subunit contains different intermediates in the reaction mechanism when a slow substrate and a limiting amount of O2 are introduced into a crystalline enzyme.
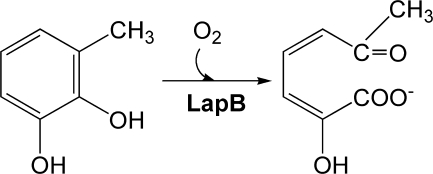
SCHEME 1.
Schematic representation of a ring cleavage of 3-methylcatechol by LapB dioxygenase.
LapB is a catechol 2,3-dioxygenase that catalyzes the second step of a long-chain alkylphenol (lap) degradation pathway in Pseudomonas sp. KL28, which can utilize 4-n-alkylphenols (C1–C5) as growth substrates (10). Phenols with bulky alkyl group(s) are commonly found in aquatic environments and have been known to cause adverse effects such as developmental and reproductive toxicity in animals (11, 12). The product analysis of LapB and its sequence alignment with other dioxygenases (see Fig. 1A) suggest that LapB belongs to the type I superfamily of extradiol dioxygenases, with a substrate preference for 4- or 3-methylcatechol (4MC and 3MC, respectively) (2, 10). To better understand the structural and functional features of LapB, we determined the crystal structure of LapB in a substrate-free form, a substrate-bound form, and a product-bound form, and we conducted a functional analysis of the active site residues. These results provide insight into the ligand-induced conformational changes in the active site and the functional role of the hydrogen-bonding networks involving Tyr-257 and His-248, which are conserved among type I enzymes, in the active site.
FIGURE 1.
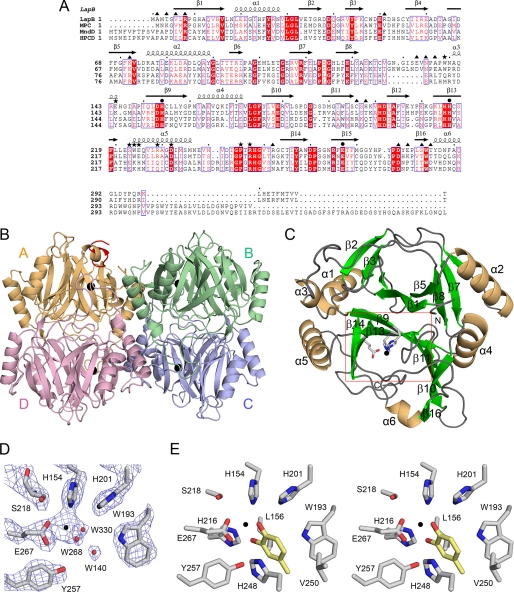
Sequence alignment of LapB and its structure. A, the amino acid sequence of LapB is aligned with other members of type I extradiol dioxygenases. Sequences used in this alignment are LapB from Pseudomonas sp. strain KL28 (GenBankTM accession number AAP92388), MPC for metapyrocatechase dioxygenase from P. putida mt-2 (V01161), MndD for homoprotocatechuate 2,3-dioxygenase from A. globiformis (33356869), and HPCD for homoprotocatechuate 2,3-dioxygenase from B. fuscum (146386887). Note that highly conserved residues are shown in red type and boxed in blue; strictly conserved residues are shown on a red background. Residues that interact with iron ion in the active site are indicated by a black filled circle, whereas the filled triangle and asterisks represent the residues involved in the dimeric and tetrameric interfaces, respectively. The secondary structural elements are shown for the corresponding sequences. This figure was prepared using ESPript (25). B, the quaternary structure of the four subunits, A, B, C, and D, in substrate-free LapB is shown in different colors. In each subunit, the iron ion is represented by a black sphere. The residues Ala-290 to Arg-298 corresponding to the ordered C terminus in subunit A are indicated in red. C, the monomeric structure of LapB is shown in an orientation that places the N-domain in the upper part and the C-domain in the bottom part. The iron-coordinating residues in the C-domain are indicated with a ball-and-stick model and a black sphere for iron ion. D, zoom-in view of the active site in the absence of a substrate is shown, with 2Fo − Fc map at 1 σ. This view is indicated by a rectangular box in red in C. Red and black spheres represent water molecules and bound iron ion, respectively. For clarity, the iron-coordinating residue His-216 is not included in this figure. E, a stereo view of the active site in the LapB-4MC complex is shown for a better view of the active site. This is an orientation identical with that of Fig. 3B. For a more detailed description, see the legend for Fig. 3.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization of LapB
For the expression of the C-terminal His-tagged LapB protein, the gene for LapB (927 bp) from Pseudomonas sp. KL28 (Korean Collection for Type Cultures 22206) was generated by PCR using a pair of primers (forward primer, 5′-GCAGATATACATATGGCGATGACAGGTGTATTGCG-3′; reverse primer, 5′-AATTCGGATCCGGTCACAACGGTCATGAAGG-3′) including an NdeI site in the forward primer and a BamHI site in the reverse primer (underlined). The resulting amplified PCR product was subcloned into the corresponding restriction sites of the pET28 vector (Novagen) containing six histidine residues at the C terminus, and the construct verified by DNA sequencing was transformed into Escherichia coli BL21(DE3) (Stratagene). Transformed cells were grown at 37 °C in Luria-Bertani medium containing 50 μm kanamycin to an A600 of 0.8–1.0 and induced for 7 h at 28 °C with the addition of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside. Cells were harvested by centrifugation at 30,000 × g for 10 min at 4 °C and sonicated in buffer A (50 mm NaH2PO4, 500 mm NaCl, pH 7.5). The resulting His-tagged LapB protein was purified using immobilized metal affinity chromatography with a solution of buffer A plus 500 mm imidazole. The eluted protein was pooled based on SDS-PAGE analysis and dialyzed against buffer B (20 mm HEPES, pH 7.4). Gel filtration chromatography using a Superdex 200 column (GE Healthcare) was subsequently performed with buffer B. In this step, LapB was estimated to form a tetramer in solution and concentrated to ∼10 mg/ml for crystallization screening.
Initial crystallization using the purified C-terminal His-tagged LapB was carried out using a sitting-drop vapor-diffusion method at 22 °C. Typically, each drop was prepared by mixing 2 μl of the protein solution containing an additional 1 mm Fe(II)(NH4)2(SO4)2 with 2 μl of the reservoir solution. Crystals of LapB with a long rod-like appearance were produced in a crystallization buffer of 0.2 m ammonium fluoride and 20% polyethylene glycol 3350. Subsequently, crystallization conditions were optimized to 0.2 m ammonium fluoride and 12–14% polyethylene glycol 3350. The complex of LapB with substrate was achieved by a soaking experiment, in which crystals of LapB were soaked with 5 mm concentrations of the ligands 3MC or 4MC.
Although the presence of Fe(II) is crucial to forming the crystal, we were not able to assess the redox state of the iron ion in the crystal structures. Given that crystallization and soaking experiments were carried out under aerobic conditions, iron ion is likely to be substantially oxidized into the ferric state.
Data Collection and Structure Determination
Single wavelength data for the native crystal of LapB and its complexes with 3MC and 4MC were collected at resolutions of 2.3, 2.0, and 1.94 Å, respectively, at the Pohang Accelerator Laboratory (Pohang, Korea) on beamlines 4A and 6C. Crystals were soaked in a cryoprotectant solution containing a crystallization solution, 20% (v/v) glycerol, and if necessary, additional 3MC or 4MC for formation of the complex. The collected data were indexed and scaled using HKL2000 (13). Detailed data for the native crystal and the complexes are shown in Table 1. The space groups of the native and complex structures were P212121, with four monomers in an asymmetric unit.
TABLE 1.
Data collection and refinement for LapB and its complex with a substrate or product
| LapBa | LapB-4-methylcatechola | LapB-3-methylcatechola | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 1.23983 | 1.23985 | 1.00000 |
| Ligand (mm) | 5 | 5 | |
| Cell dimensions | |||
| a, b, c (Å) | 94.9, 97.0, 133.4 | 96.1, 97.6, 133.7 | 95.9, 97.4, 133.5 |
| α, β, γ (deg) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0-2.3 (2.38-2.30)b | 50.0-1.94 (2.01-1.94) | 50.0-2.0 (2.07-2.00) |
| Unique reflections | 55,660 | 93,774 | 85,226 |
| Completeness (%) | 99.9 (99.4) | 99.9 (100.0) | 100.0 (100.0) |
| Redundancy | 13.0 | 11.5 | 7.7 |
| I/σ (I) | 23.2 (3.6) | 35.9 (2.7) | 18.8 (3.2) |
| Rsym (%)c | 11.9 (65.2) | 7.1 (75.4) | 11.1 (57.6) |
| Refinement | |||
| Resolution (Å) | 50.0-2.3 | 50.0-1.94 | 50.0-2.0 |
| No. of reflections | |||
| Working set/test set | 42,020/4,721 | 73,387/8,169 | 64,010/7,191 |
| Rwork (%)d | 20.1 | 24.4 | 22.2 |
| Rfree (%)d | 26.9 | 27.8 | 26.9 |
| r.m.s.d. from ideal geometry | |||
| Bond lengths (Å) | 0.009305 | 0.012134 | 0.006015 |
| Bond angles (degrees) | 1.61074 | 1.88504 | 1.39185 |
| Average B-factor (Å2) | 30.4 | 30.4 | 26.9 |
| No. of molecules | |||
| Protein atoms | 9,277 | 9,328 | 9,399 |
| Water atoms | 332 | 351 | 409 |
| Ligand atoms | 38 | 33 | |
| Ramachandran analysis | |||
| Most favored/allowed (%) | 85.2/14.8 | 87.3/12.7 | 87.8/12.2 |
| Disallowed (%) | 0 | 0 | 0 |
a Residue ranges: substrate-free LapB, subunit A (Ala-2–Arg-298) and subunits B, C and D (Ala-2–Phe-289); LapB-4-methylcatechol, subunits A and B (Ala-2–Phe-289), and subunits C and D (Ala-2–Gln-297); LapB-3-methylcatechol, subunit A (Ala-2–Arg-298), subunits B and D (Ala-2–Phe-289), and subunit C (Ala-2–Gln-297).
b Numbers in parentheses refer to data in the highest resolution shell.
cRsym = Σ|Ih − <Ih>|/ΣIh, where Ih is the observed intensity and <Ih> is the average intensity.
d Rwork = Σ‖Fobs|-k|Fcal‖/Σ|Fobs|; Rfree was calculated using 10% of the data excluded from refinement.
The native structure was determined by the molecular replacement method using CNS (14) with a monomer of metapyrocatechase dioxygenase (Protein Data Bank accession 1MPY (15)) from Pseudomonas putida as a search model. The initial model generated through molecular replacement was further improved by a series of simulated annealing omit maps using CNS, and the resulting map was sufficiently clear to trace most of the residues in four monomers except for those in the disordered C-terminal region (Table 1). Manual model building was accomplished using the model-building program O (16), and refinement was carried out with CNS.
During refinement, we noticed in each subunit a single outstanding density in the Fo − Fc map greater than 5 σ. Based on the distance and geometry of this density relative to the nearby residues, we concluded that this density corresponds to the binding site for the iron ion.
The structures of LapB complexed with a ligand were determined using the final model of native LapB as the starting model and subjecting it to refinement with CNS. For the complex with 4MC, a density for 4MC was observed in three subunits (subunits A, B, and C); a model for 4MC was not included in subunit D due to its highly disordered density. In the structure of the 3MC complex, its product, 2-hydroxy-6-oxohepta-2,4-dienoic acid, was observed in the active site of three subunits (subunits B, C, and D), whereas no density was observed in subunit A. Therefore, LapB complexed with 4MC or 3MC represents a substrate- or product-bound enzyme, respectively. An additional structural feature of the complex is the structural transition of the C-terminal residues from a disordered to an ordered state. In particular, residues 290–298, which were disordered in the substrate-free form, became ordered in subunits C and D of the LapB-4MC complex and in subunit C of the LapB-product complex (Table 1).
The stereochemistry of each final model was checked using the PROCHECK program (17). Neither the native LapB nor its complexes had residues in the disallowed region of the Ramachandran plot. The figures were prepared using PyMOL (27), and the structural analysis was executed using the CCP4 program suite (17).
Site-directed Mutagenesis and Enzyme Assay
To investigate the functional role of the active site residues, three residues, His-201, His-248, and Tyr-257 (see below), were mutated by site-directed mutagenesis. Five mutant enzymes, H201A, H201N, H248A, H248N, and Y257F, were generated using two rounds of PCR. In the first round of PCR, the mutagenic oligonucleotide for the forward primer was 5′- CTCGCACAAGGTTGCTGACATCGCCTTCG-3′ for the mutant H201A, 5′-CTCGCACAAGGTTAATGACATCGCCTTCG-3′ for the mutant H201N, 5′-CGGTCCAACCCGTGCCGGCGTGACTCGCG-3′ for the mutant H248A, 5′- AACGTGGACATCGGTCCAACCCGTAATGGCGTG-3′ for the mutant H248N, and 5′-GGCGTGACTCGCGGTTGCACCATCTTTGCGTGG-3′ for the mutant Y257F. The underlined sequence indicates the mutation site. These forward primers for each mutation were used with the reverse primer (5′-CGCGGATCCGGTCACAACGGTCATGAAGG-3′) for the C terminus, which included a BamHI site (underlined). The resulting PCR product from the first round of PCR was used as the reverse primer in the second round of PCR, in which an oligonucleotide of 5′-GGAATTCCATATGGCGATGACAGGTGTATTG-3′ for the N terminus, which included an NdeI site (underlined), was used as a forward primer. Each mutant LapB was expressed as described above for the wild-type enzyme, and the purified enzyme was concentrated to ∼1.0 mg/ml in 20 mm HEPES (pH 7.5). All mutant enzymes were essentially identical with the wild-type enzyme in their folding and tetramerization (data not shown).
Enzyme assays were performed at 25 °C using a UV-visible spectrophotometer (Jasco V-560). The 2-ml reaction mixture contained 20 mm HEPES (pH 8.0), 80 mm NaCl, 4 μm dithiothreitol, 0.4 μm Fe(II)(NH4)2(SO4)2, 0.114 μm LapB protein, and the substrate 4MC or 3MC (0.5–100 μm). Specifically, enzymes were first incubated for 3 min in the absence of substrate, and the reaction was then initiated by the addition of the substrate. Product formation was monitored as a function of time at its characteristic absorption wavelength of 382 nm with its molar extinction coefficient of 28,100 m−1cm−1 for 4MC as a substrate and of 384 nm with its molar extinction coefficient 13,400 m−1cm−1 for 3MC as a substrate (18, 19). The initial velocity of the reaction was measured during the first 30 s, and kinetic parameters were determined using a Lineweaver-Burk plot constructed using SigmaPlot. The kcat was calculated using a monomer of 35,080 daltons as the molecular mass of the enzyme.
RESULTS
Overall Structure of Substrate-free LapB
The catechol 2,3-dioxygenase LapB is composed of 309 amino acids. The overall structure of LapB in the absence of ligand shows a tetrameric arrangement into an asymmetric unit with non-crystallographic 222 symmetry (Fig. 1B), which is consistent with the chromatographic observation that LapB exists as a tetramer in solution. Therefore, this arrangement of tetramers forming an asymmetric unit is likely to be the functional tetrameric unit in solution. Superposition of all Cα atoms between monomers exhibits root mean square deviation (r.m.s.d.) less than 0.5 Å, suggesting that the four subunits are structurally identical to one another.
Each subunit consists of six α-helices and 16 β-strands that fold into the four repeated βαβββ motifs (Fig. 1, A and C). A short helix α3 and the C-terminal helix α6 are the exception to this motif. From a structural perspective, the monomeric structure can be divided into two domains that are highly homologous in structure: an N-domain (residues 1–149) and a C-domain (residues 150–309). Specifically, there are two repeated βαβββ motifs in each domain: β1-α1-β2-β3-β4 and β5-α2-β6-β7-β8 in the N-domain and β9-α4-β10-β11-β12 and β13-α5-β14-β15-β16 in the C-domain. The two motifs in each domain are arranged in a manner such that the first β-strands in each βαβββ motif (β1 and β5 in the N-domain, β9 and β13 in the C-domain) are antiparallel to each other, resulting in an extended eight-stranded, concave β-sheet. These structural features are observed in both domains, which are related by a r.m.s.d. of 1.61 Å for 116 matched Cα atoms.
The eight β-strands and nearby loops are arranged in such a way that they generate a funnel-like cavity in the concave side of each domain (Fig. 1C). Unlike the N-domain, in which the funnel is sealed off by the side chains, there are two openings in the C-domain. Specifically, one end of the funnel remains open toward the intersubunit interface, but the tetramerization of four subunits essentially blocks this opening. The other end of the funnel connects the active site (see below) to the surface of the molecule.
This putative substrate-binding channel adopts a closed conformation in subunit A and an open conformation in the other three subunits. These different structural features are due to the orientation of nine extra residues, Ala-290 to Arg-298, in the C terminus, which are ordered only in subunit A (Fig. 1B). Crystal contact analysis using the program CONTACT in the CCP4 suite indicated that residues Tyr-295, Pro-296, and Gln-297 in the C-terminal region of subunit A are involved in intermolecular crystal packing with residues Glu-210, Pro-209, and Tyr-208 in a loop between β12 and β13 of the crystallographic symmetry-related subunit C, at a distance of 3.5–5.0 Å, whereas no crystal contacts were observed in the C-terminal residues of the other three subunits. Therefore, the closed conformation of the active site in subunit A in substrate-free LapB is mainly due to crystal packing.
The location of the active site was unambiguously suggested by the presence of a metal-binding site in the C-domain (Fig. 1, C and D). The active site is situated deep in the interior of the cavity of the C-domain. Specifically, the metal-binding site possibly occupied by the iron ion was located at the concave β-sheet and ligated with His-154, His-216 (not shown in this orientation of Fig. 1D), Glu-267, and a water molecule, all within a distance of 2.3 Å.
Dimeric and Tetrameric Interface
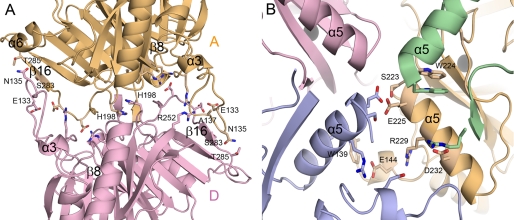
Each subunit contacts the other three subunits via two different interfaces, the dimeric and the tetrameric (Fig. 2). Interface residues are identified as those interacting with each other within 4.0 Å via hydrogen bonds or van der Waals interactions using the program CONTACT, and their surface area was calculated using the AREAIMOL program within the CCP4 suite (17).
FIGURE 2.
Dimeric and tetrameric interfaces. A, the dimeric interface between subunits A and D are shown, with the interface residues defined using the program CONTACT (17). B, the tetrameric interface was formed mainly by residues belonging to α5.
Fig. 2A shows the dimeric interface between subunits A and D, which are related by a non-crystallographic two-fold symmetry. Dimerization of A and D or B and C resulted in buried surface areas of 3,967 and 4,090 Å2, respectively. Most of the residues in the dimeric interface are located in the loop regions (Fig. 1A, filled triangles) and mediate a total of 27 hydrogen bonds and a number of van der Waals interactions at each interface. In particular, the protruding long loop between β8 and α3 in the N-domain has extensive intersubunit interactions with the loop connecting the last β-strand β16 and α6 of the C-domain in the adjacent subunit. Specifically, those contacts are mediated by the main-chain carbonyl oxygen atoms and side chains of Glu-133, Asn-135, and Ala-137 in the protruding loop of one particular subunit and the backbone nitrogen atoms and side chains of Ser-283, Thr-285, and Arg-252 in the interacting subunit. The His-198 residues from both subunits are located at the center of the dimeric interface, with their side chains stacked against each other. The structural environment of the dimeric interface between B and C is essentially identical to those described here for the dimer A and D.
The tetrameric interface, which is defined as the interface formed by the two dimers A-D and B-C, exhibits more extensive interactions. Its buried surface area was calculated to be about 11,786 Å2, providing the structural stability of the tetrameric LapB. Fig. 2B shows the tetrameric interface, along with the interacting residues. For example, the residues participating in tetrameric interactions are those in α5, including residues Ser-223, Trp-224, Glu-225, Arg-229, and Asp-232, and additional residues Trp-139, Glu-144, and Arg-247 (Fig. 1A, asterisks). These residues interact by hydrogen bonds with Asp-232 and Arg-229 in another dimer. None of the dimeric or tetrameric interface residues had dual roles.
Structural Features in the Active Site
As described under “Experimental Procedures,” the substrate 4MC was bound in the active site of the LapB-4MC complex, whereas 2-hydroxy-6-oxohepta-2,4-dienoic acid, a 3MC product generated via a proximal cleavage by LapB, was identified in the active site of the complex with 3MC. Therefore, three different intermediates in the catalytic cycle of extradiol dioxygenation are described in this work, including a substrate-free LapB, the LapB-substrate complex, and the LapB-product complex. Structural superposition using CCP4 Molecular Graphics (17) indicated that the tetrameric structures of the substrate-free form and the complex are essentially identical, with r.m.s.d. values of 0.43 and 0.32 Å for all Cα atoms in the complex with 4MC and product, respectively. Even in the absence of large scale structural changes, some noticeable structural changes in the vicinity of the active site occur with the binding of a substrate or a product.
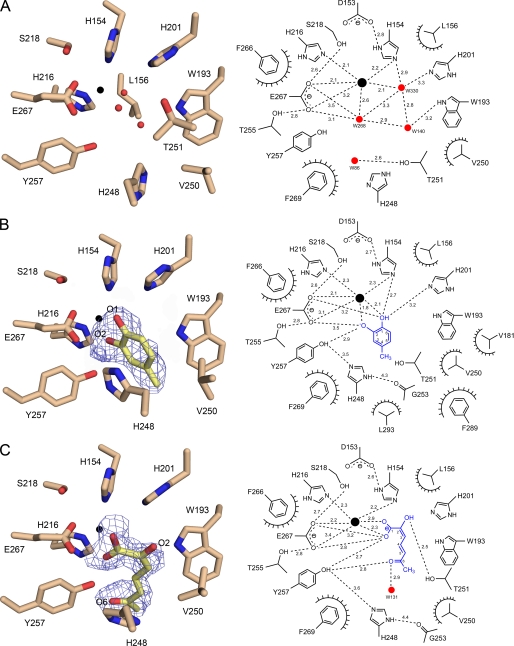
Fig. 3 shows structural features of the active site for the substrate-free LapB and its complexes. In the absence of substrate, cofactor iron ion forms a square pyramidal coordination with two histidine residues (His-154 and His-216), one glutamate (Glu-267), and 2–3 water molecules, depending on subunit (Fig. 3A). In particular, all of the coordinating ligands in the first coordination shell are within 2.2 Å of iron ion, with Glu-267 in the axial position. The possible sixth coordination site at the axial position across from Glu-267 remains vacant or occupied with water molecules in substrate-free LapB and is likely the dioxygen-binding site. These metal-ligating residues (i.e. His-154 and Glu-267) also form hydrogen bonds to adjacent residues, including Asp-153, Ser-218, and Thr-255.
FIGURE 3.
Active site of native LapB and its complexes. The active site for substrate-free LapB and its complex with 4MC or product is shown with a stick model, with a schematic representation of the interactions shown in the right panel. The dashed lines indicate putative hydrogen bonds, which are labeled with the average interatomic distance (in Å), and the decorated arcs represent van der Waals interactions of less than 4.0 Å. A, the active site for substrate-free LapB in subunit B is shown. Three water molecules depicted as red spheres occupy the first coordination shell and interact with iron ion, a black sphere. B, a 2Fo − Fc electron density map contoured at 1 σ is overlaid on the model of 4MC in the LapB-4MC complex (subunit C). For clarity, Leu-156, which is also a part of the active site, is not shown in this figure and also not shown in C but is displayed in Fig. 1E. C, same as B except that 2-hydroxy-6-oxohepta-2,4-dienoic acid, the product of 3MC produced via proximal cleavage by LapB, is shown with a 2Fo − Fc electron density map contoured at 0.8 σ in subunit B.
In the complex with 4MC, we found a density corresponding to 4MC, but none corresponding to dioxygen (Fig. 3B, and see Fig. 1E for a stereo view of the active site). In subunit C, the unambiguous density for the methyl group at C4 allowed the correct localization of the two hydroxyl groups O1 and O2 of the substrate. Overall, 4MC remains in a planar group and is bound in such a manner that the two hydroxyl groups chelate iron ion within 2.1 Å, and the 4-methyl group points toward the opening to the surface of the enzyme in the substrate-binding channel. The two hydroxyl groups displace the two water molecules that previously occupied the first coordination shell in the substrate-free enzyme. The binding of 4MC was further stabilized by various interactions. Among these, the side chain of His-248 was observed to be in a stacking orientation within 3.5 Å of the aromatic ring of 4MC. In addition, the methyl group at C4 is bound in the hydrophobic environment of the active site, which consists of Val-181, Trp-193, Val-250, Phe-289, and Leu-293. In particular, the 4-methyl group is within 3.1–4.0 Å of Trp-193 and Val-250. These hydrophobic residues are a part of a wall of residues in the channel connecting the active site to the surface of the enzyme. This hydrophobic environment may be the key structural contributor to the substrate specificity of LapB for 4-alkylcatechol (10). In addition to these hydrophobic interactions, there is a possible hydrogen-bonding network connecting O2 of 4MC-Oη of Tyr-257-Nϵ2 (His-248) Nδ1-main-chain oxygen of Gly-253, with average distances of 2.9, 3.5, and 4.3 Å, respectively. It is noteworthy that this suggested hydrogen-bonding network was not observed in the substrate-free LapB and that the loop containing His-248 was relatively flexible, as indicated by the high average B-factors of 52.2 Å2 for the main-chain atoms from His-248 to Thr-251 when compared with B-factors of 38.3 Å2 for the corresponding region in the complex. That particular loop became ordered in this complex, together with positional shifts (see below), suggesting that the observed hydrogen-bonding network is established by the binding of a substrate.
Contrary to the complex with 4MC, a product of 3MC, 2-hydroxy-6-oxohepta-2,4-dienoic acid, was observed in the LapB-3MC complex. In subunit B, in which the density of the ring-opened structure was relatively well defined except for the C4 region, the product remains in the active site in such a manner that the two oxygen atoms in the carboxylic group are a part of the first coordination shell, with distances of 2.3 and 2.9 Å from iron, and the rest of the molecule faces toward the opening to the surface of the enzyme, with its extended configuration stacked against His-248 (Fig. 3C). In particular, the hydroxyl group at C2 is stabilized by a hydrogen bond to the hydroxyl group of Thr-251, and the Oη of Tyr-257 remains within hydrogen-bonding distance of the carbonyl oxygen atom at C6 and one of the carboxylic oxygen atoms at C1, with distances of 2.8 and 2.7 Å, respectively. Interestingly, the loop region including His-248 became disordered again in the complex, with a relatively weak density for the side chain and average B-factors of 42.8 Å2 for the main-chain atoms His-248 to Thr-251. Therefore, the validity of the hydrogen bond between Oη of Tyr-257 and Nϵ2 of His-248, separated by a distance of 3.6 Å, is questionable in the LapB-product complex.
Other Structural Differences between Substrate-free LapB and Its Complexes
We carried out a structural comparison with identify catalysis-related conformational changes in the complex. The comparison was performed first between the substrate-free LapB and its complex with substrate 4MC. Although the overall r.m.s.d. for the tetramer is only 0.43 Å for 1,152 equivalent Cα atoms, there are localized ligand-induced structural changes in the vicinity of the active site, including the active site loop between α5 and β14 containing His-248. Positional changes in Cα were as large as 0.9–1.6 Å for the loop residues Arg-247 to Thr-251. In particular, the side chain of His-248 in the loop was in the middle of the substrate-binding pocket in the absence of substrate but shifted its position as much as 2.0 Å for the side chain to accommodate the incoming 4MC in the LapB-4MC complex (Fig. 4A). Because of these movements, the loop was displaced away from the active site in the complex and formed a suggested hydrogen-bonding network connecting the substrate, Tyr-257, His-248 and Gly-253, which are conserved residues in other 2,3-dioxygenases (Fig. 3B) (18). Subsequently, structural comparison between LapB-4MC and the complex with a product did not reveal any noticeable changes. Therefore, conformational features of the enzyme in the presence of product are essentially identical to those of the complex with 4MC.
FIGURE 4.
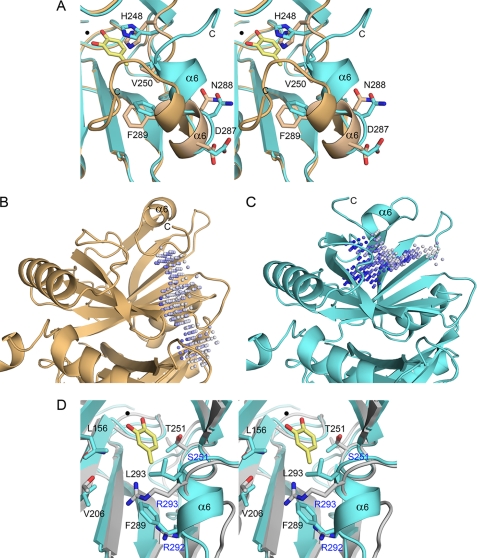
Structural comparison of the active site and a substrate-binding channel. A, a stereo view of the active site between substrate-free LapB (subunit A; brown) and the LapB-4MC complex (subunit C; cyan) is shown. Due to crystal packing, the C-terminal region is ordered in the absence of ligand, but its orientation is different from that in the complex. The bound 4MC and iron ion are indicated with a stick model and a black sphere, respectively. B and C, the shape of the substrate-binding channel is represented for substrate-free LapB (subunit A) and the LapB-4MC complex (subunit C), respectively. The putative channel was predicted using the program PocketPicker (26). The two structures are in identical orientations, and the active site is located at the center of the molecule. Unlike substrate-free LapB, in which the channel is blocked, the LapB-4MC complex contains a channel from the active site to the surface of the molecule. D, a stereo view of the active site between the LapB-4MC complex in cyan and HPCD complexed with 3,4-dihydroxyphenylacetate in gray is shown. The substrate 4MC and iron ion in LapB are indicated with a yellow stick molecule and black circle, respectively. The substrate 3,4-dihydroxyphenylacetate is not shown for clarity. Residues in LapB and HPCD are shown in black and blue, respectively.
Additional structural differences are associated with the C-terminal region, which is located next to the loop containing His-248. This region, including helix α6 and the following C-terminal loop, exhibits a structural transition from a disordered to an ordered conformation depending on ligand binding. In particular, the C-terminal region is disordered in the absence of ligand, except in subunit A in which crystal packing appears to stabilize the region. This region became ordered in the LapB-4MC complex (Fig. 4A). However, this observation was characterized only in subunit C, not in subunits A and B in which 4MC was also bound to the active site. In the complex with product, this C-terminal region remained ordered in subunit C, with an identical conformation to that of the LapB-4MC complex. Although not all subunits containing 4MC or product have an ordered C-terminal region, our observations suggest that the ordering of the C-terminal loop in subunit C is likely related to ligand binding. These conformational changes are associated with the opening or closing of the substrate-binding channel. Specifically, the active site adopts an open conformation with the binding of 4MC (Fig. 4C), possibly for the binding of oxygen, whereas the C-terminal region of subunit A adopts a closed conformation in the substrate-free form (Fig. 4B).
Enzyme Assay of LapB Mutants
Our structural analysis identified three residues likely involved in catalysis: His-201, situated near the dioxygen-binding site; Tyr-257, which was near the active site and within hydrogen-bonding distance to bound 4MC or product; and His-248, which maintains a hydrogen-bonding network with Tyr-257 (Figs. 1E and 3). To investigate the functional roles of these residues, we generated five mutants, H201A, H201N, H248A, H248N, and Y257F, and measured their enzyme activities using 4MC or 3MC as a substrate. For the wild-type LapB, the kinetic parameters for 4MC were measured to be 1.5 μm for Km and 45 min−1 for kcat, resulting in a kcat/Km of 29.3 μm−1min−1, whereas Km and kcat for 3MC were 3.8 μm and 85 min−1, respectively, with a kcat/Km of 22.2 μm−1min−1. All five mutants were essentially inactive, even when using a 10-fold higher concentration of mutant enzyme, suggesting that these three residues play a crucial role in the catalysis of extradiol ring cleavage.
DISCUSSION
In this study, we report on the crystal structure of non-heme Fe(II)-dependent catechol 2,3-dioxygenase LapB from Pseudomonas sp. KL28, which belongs to the type I class of extradiol dioxygenases. The crystalline enzyme containing four subunits in an asymmetric unit was catalytically active, and structures of the enzyme in complex with a substrate, 4MC, or product were determined by soaking the crystalline enzyme in an authentic substrate, 4MC or 3MC, to provide structural and functional insights into the catalytic cycle.
From a structural perspective, LapB exhibits high similarity to other extradiol dioxygenases. A structural homology search using the program DALI (20) identified 13 structures with Z-scores greater than 10 and r.m.s.d. values ranging from 1.3 to 3.4 Å in comparison with the LapB monomer. Among these, metapyrocatechase dioxygenase from P. putida (15), HPCD from Arthrobacter globiformis (21), and B. fuscum (8) showed the highest Z-scores of 43.6, 34.1, and 33.2, respectively. Metapyrocatechase dioxygenase, used as a search model for molecular replacement in this study, bears a sequence identity of 49%. Despite a relatively low sequence identity of 24–26%, HPCD also displays high structural similarity to LapB. These four enzymes, which are all in homotetrameric conformation, contain conserved metal-ligating residues, including two histidine residues and one glutamate residue in the active site, but are characterized by differences in the length and sequence of the C-terminal region (Fig. 1A). This C-terminal region was found to form an independent domain in the structure of HPCD and was suggested to be a lid domain on the substrate-binding channel (8, 21). Unlike HPCD, the corresponding region in LapB, which is shorter in length when compared with that of HPCD, displays a structural transition from a disordered to an ordered conformation with the binding of a substrate or a product. These dynamic features of the loop are further reinforced by the observation that the C-terminal region in subunit C is free of crystalline packing. Due to its intrinsic flexibility, the C-terminal loop is likely involved in catalysis by opening or closing the channel according to the ligation state of the active site (Fig. 4).
From a functional perspective, our structure also provides the molecular basis for the substrate specificity of LapB for 3- or 4-alkylcatechol (10). A structural comparison between LapB-4MC and HPCD-3,4-dihydroxyphenylacetate (Protein Data Bank accession 1F1V (21)) from A. globiformis indicates that in LapB, the cavity near C3 or C4 of the substrate is composed of hydrophobic residues, including Leu-156, Val-181, Trp-193, Val-206, Val-250, Phe-289, and Leu-293 (Figs. 3 and 4D). Therefore, the hydrophobic cavity in LapB could accommodate a 3- or 4-alkylcatechol. However, those hydrophobic residues are not present in the HPCD complex. Instead, the cavity near the substrate consists of hydrophilic residues such as Asn-157, Thr-205, Arg-292, and Arg-293 (MndD in Fig. 1A), which are structurally equivalent to Leu-156, Val-206, Phe-289, and Leu-293, respectively, in LapB (Fig. 4D). In particular, the side chain of Arg-293 in the HPCD complex was suggested to mediate a crucial hydrogen bond to the substrate (21). Substrate specificity appears to be determined by these structural differences, which could not be inferred from a simple sequence comparison.
Recent structural studies of various intermediates that form during the catalytic cycle of HPCD-mediated extradiol ring cleavage provided firm evidence for the reaction mechanism (4, 8, 9). In this study, we gained newly defined mechanistic insights, based on a functional study of mutant enzymes and structural determination of substrate-free LapB and its complex with a substrate (4MC) or a product.
In the absence of substrate, the active site iron ion has a five-coordination with a 2-His-1-carboxylate motif, including His-154, His-216, Glu-267, and two water molecules that occupy only one face of the active site (Fig. 3A). The asymmetric binding of 4MC displaces iron ion-ligating water molecules with the two hydroxyl groups O1 and O2, leaving the sixth coordination site vacant for dioxygen and causing various effects on the conformation of the active site region. Specifically, substrate binding displaces the side chain of His-248 that occupied a position near the active site in the substrate-free form, resulting in the stabilization of the loop containing His-248 by a stacking interaction (Fig. 4A). As a result of this series of events, a hydrogen-bonding network was established connecting O2 of 4MC-Oη of Tyr-257-Nϵ2 (His-248) Nδ1-main-chain oxygen of Gly-253 (Figs. 1E and 3B). Previous spectroscopic and computational analyses indicated that only one of the two hydroxyl groups in a catecholic compound should be deprotonated upon binding to the active site (6, 22). Both our structural data for the proposed hydrogen-bonding network and our kinetic analysis of the mutant enzymes Y257F, H248A, and H248N strongly support the hypothesis that the hydroxyl group of Tyr-257 has an essential role in catalysis and that His-248 appears to enhance the basicity of Tyr-257 via a hydrogen-bonding network and stabilizes the orientation of Tyr-257 toward O2 of the substrate. These functional assignments are consistent with a previous study in which the highly conserved residues within catechol 2,3-dioxygenase were subjected to mutational analysis, and it was concluded that the histidine equivalent to His-248 in LapB is involved in catechol deprotonation (18). Therefore, Tyr-257 serves as a base catalyst by deprotonating the O2 hydroxyl group of 4MC, leaving the incoming substrate in a monoanionic form. Analogous to Tyr-257 and His-248 in LapB, which are conserved among type I extradiol dioxygenases, the residue pair His-115 and Asp-114 has also been suggested to play a crucial role as an acid-base catalyst in 2,3-dihydroxyphenylpropionate 1,2-dioxygenase, which is classified as a type II dioxygenase (23). His-201 in LapB, which is located near the putative dioxygen-binding site (Figs. 1E and 3), is the invariant residue in extradiol dioxygenase (Fig. 1A). Various mutational studies have indicated that this histidine residue is crucial in forming an alkylperoxo intermediate and a lactone intermediate, possibly by serving as base and acid catalyst, respectively (24). Consistent with these observations, the mutant LapB enzymes H201A and H201N were catalytically inactive.
We report structural and functional evidence that LapB is a type I extradiol dioxygenase. Structural analysis of three different intermediates revealed ligand-induced conformational changes in the active site loop containing His-248 and in the C-terminal region, which have not been described in other extradiol dioxygenases. The residues His-201, His-248, and Tyr-257, which are conserved in type I enzymes, were identified as crucial active site residues. In particular, a hydrogen-bonding network including Tyr-257 and His-248 is likely responsible for deprotonating the incoming substrate. We also identified a hydrophobic environment near the C3 and C4 positions of the substrate, providing the structural determinants for 3- and 4-alkylcatechols as substrates. These studies provide detailed structural and functional insight into an extradiol dioxygenase involved in the degradation of environmentally hazardous alkylphenols by Pseudomonas.
This work was supported by Korea Science and Engineering Foundation (KOSEF) Grants R01-2007-000-21067-0 and R11-2008-062-01002-0 and by a grant from Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology, Republic of Korea.
The atomic coordinates and structure factors (codes 3HPV, 3HPY, and 3HQ0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HPCD
- homoprotocatechuate 2,3-dioxygenase
- lap
- long-chain alkylphenol
- 4MC
- 4-methylcatechol
- 3MC
- 3-methylcatechol
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1.Vaillancourt F. H., Bolin J. T., Eltis L. D. (2006) Crit. Rev. Biochem. Mol. Biol. 41, 241–267 [DOI] [PubMed] [Google Scholar]
- 2.Bugg T. D., Ramaswamy S. (2008) Curr. Opin. Chem. Biol. 12, 134–140 [DOI] [PubMed] [Google Scholar]
- 3.Emerson J. P., Kovaleva E. G., Farquhar E. R., Lipscomb J. D., Que L., Jr. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7347–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipscomb J. D. (2008) Curr. Opin. Struct. Biol. 18, 644–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovaleva E. G., Lipscomb J. D. (2008) Nat. Chem. Biol. 4, 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegbahn P. E., Haeffner F. (2004) J. Am. Chem. Soc. 126, 8919–8932 [DOI] [PubMed] [Google Scholar]
- 7.Kovaleva E. G., Neibergall M. B., Chakrabarty S., Lipscomb J. D. (2007) Acc. Chem. Res. 40, 475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovaleva E. G., Lipscomb J. D. (2007) Science 316, 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovaleva E. G., Lipscomb J. D. (2008) Biochemistry 47, 11168–11170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong J. J., Kim J. H., Kim C. K., Hwang I., Lee K. (2003) Microbiology 149, 3265–3277 [DOI] [PubMed] [Google Scholar]
- 11.Giger W., Brunner P. H., Schaffner C. (1984) Science 225, 623–625 [DOI] [PubMed] [Google Scholar]
- 12.Sharpe R. M., Fisher J. S., Millar M. M., Jobling S., Sumpter J. P. (1995) Environ. Health Perspect. 103, 1136–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 14.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 15.Kita A., Kita S., Fujisawa I., Inaka K., Ishida T., Horiike K., Nozaki M., Miki K. (1999) Structure 7, 25–34 [DOI] [PubMed] [Google Scholar]
- 16.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project, Number 4 ( 1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760– 76315299374 [Google Scholar]
- 18.Viggiani A., Siani L., Notomista E., Birolo L., Pucci P., Di Donato A. (2004) J. Biol. Chem. 279, 48630–48639 [DOI] [PubMed] [Google Scholar]
- 19.Takeo M., Nishimura M., Takahashi H., Kitamura C., Kato D., Negoro S. (2007) J. Biosci. Bioeng. 104, 309–314 [DOI] [PubMed] [Google Scholar]
- 20.Holm L., Sander C. (1998) Nucleic Acids Res. 26, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vetting M. W., Wackett L. P., Que L., Jr., Lipscomb J. D., Ohlendorf D. H. (2004) J. Bacteriol. 186, 1945–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaillancourt F. H., Barbosa C. J., Spiro T. G., Bolin J. T., Blades M. W., Turner R. F., Eltis L. D. (2002) J. Am. Chem. Soc. 124, 2485–2496 [DOI] [PubMed] [Google Scholar]
- 23.Mendel S., Arndt A., Bugg T. D. H. (2004) Biochemistry 43, 13390–13396 [DOI] [PubMed] [Google Scholar]
- 24.Groce S. L., Lipscomb J. D. (2005) Biochemistry 44, 7175–7188 [DOI] [PubMed] [Google Scholar]
- 25.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 26.Weisel M., Proschak E., Schneider G. (2007) Chem. Cent. J. 1, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]