Abstract
Regulated proteolysis by ATP-dependent proteases is universal in all living cells. In Bacillus subtilis, the degradation of the competence transcription factor ComK is mediated by a ternary complex involving the adaptor protein MecA and the ATP-dependent protease ClpCP. Here we demonstrate that a C-terminal, 98-amino acid domain of MecA (residues 121–218) serves as a non-recycling, degradation tag and targets a variety of fusion proteins to the ClpCP protease for degradation. MecA-(121–218) facilitates productive oligomerization of ClpC, stimulates the ATPase activity of ClpC, and allows the activated ClpC complex to stably associate with ClpP. Importantly, the ClpCP protease undergoes dynamic cycles of assembly and disassembly, which are triggered by association with MecA and the degradation of MecA, respectively.
Introduction
Regulated proteolysis by ATP-dependent proteases is universal in all living cells and is responsible for the timely turnover of many intracellular proteins (1–4). These proteases ensure the welfare of a cell by removing toxic proteins, which may be misfolded or mistranslated, and regulatory proteins, whose degradation may be pivotal to cell fate under specific cues. Representative examples of the proteases include the Clp family of bacterial proteases, such as ClpAP, ClpCP, and ClpXP (1, 5), and the eukaryotic and prokaryotic proteasomes (6–10). A shared feature of these proteases is that the proteolytic active sites are sequestered within a closed chamber, which is formed by oligomeric complexes of the protease protomer. Such a topological arrangement necessitates the unfolding and translocation of substrate proteins, both of which are mediated by the hexameric, AAA+ ATPase complexes.
ClpA, ClpC, and ClpX belong to the conserved Hsp100/Clp family of AAA+ ATPases, each of which forms a hexameric assembly in its active form (11). The ClpA or ClpX hexamer unfolds and translocates substrate protein to the lumen of two heptameric rings of ClpP for degradation. To perform this function, ClpA or ClpX directly associates with the ClpP peptidase. One way of ensuring the degradation specificity is provided by an 11-residue peptide sequence known as the SsrA tag, which is added to the C terminus of a nascent polypeptide chain when translation is stalled (12). The SsrA tag, initially identified in Escherichia coli, is recognized not only by ClpA or ClpX from E. coli (13) but also by the PAN complex from the thermophilic archaebacterium Methanocaldococcus jannaschii) (14). Recent investigations revealed significant insights into the molecular mechanisms by which the ClpXP protease and the PAN proteasome degrade the SsrA-tagged substrate protein (15–21).
When compared with ClpXP and other ATP-dependent proteases, the ClpCP protease from Bacillus subtilis is unique. Two genes, mecA and mecB, were initially identified as the negative regulators of the competence transcription factor ComK in B. subtilis (22–24). ComK activates the transcription of a set of late competence genes, which encode proteins that bind and transport exogenous DNA (25). MecB was quickly recognized to be the heat shock protein ClpC in B. subtilis (26). ComK was found to form a ternary complex with MecA and ClpC (27), which targets ComK for ClpP-mediated degradation (28). In contrast to ClpA and ClpX, activation of ClpC and its association with ClpP are both mediated by MecA (29). Thus, MecA appears to be an obligatory activator of the ClpCP protease as well as an essential determinant of substrate specificity. Consistent with this conclusion, an N-terminal fragment of MecA was found to recognize ComK, whereas a C-terminal domain of MecA directly interacted with ClpC (30).
Paradoxically, although MecA is required for activation of the ClpCP protease, it is degraded along with the substrate protein ComK (28). Notably, even in the absence of ComK, the full-length MecA protein or a C-terminal fragment of MecA was also degraded efficiently (30). These intriguing observations point to a number of important, unanswered questions. First, is the C-terminal fragment of MecA required for assembly of the oligomeric ClpCP protease? Although the available data seem to suggest so, experimental evidence is lacking. Second and more importantly, if the C-terminal fragment of MecA is required for assembly of the ClpCP protease, what happens to the assembled, oligomeric ClpCP protease once MecA is degraded? There are two apparent scenarios here. One is that MecA merely serves as a primer to help overcome the energy barrier for assembly of the ClpCP protease; in this case, the degradation of MecA should have no impact on the integrity of the ClpCP protease. The other scenario is that MecA degradation results in the disassembly of the ClpCP protease. Third, what are the minimal sequences in MecA that are sufficient for assembly of the ClpCP protease? What are the minimal sequences in MecA that are required for subsequent degradation of MecA by the ClpCP protease? Are these two sets of minimal sequences the same? Fourth, can MecA serve as a degradation tag for other proteins in a way that is reminiscent of SsrA? Last but not least, how does the structure of MecA support its function?
In this study, we provide definitive answers to the first four questions. We also report the high resolution structure of the C-terminal domain of MecA (PDB ID:3JTP) in the accompanying article (39), which sheds light on the fifth question. Biochemical characterization reported in this study gives rise to a working model of the MecA-ClpCP system, with significant insights into how the system may work in cells. In particular, we report that a 98-amino acid, C-terminal domain (residues 121–218) of MecA serves as a non-recycling, degradation tag, which represents a minimal domain for assembly of the ClpCP protease. An additional flexible sequence of at least 9 amino acids is required for this degradation tag to be degraded by the assembled ClpCP protease. Heterologous fusion of this degradation tag with other folded proteins leads to their rapid degradation by the ClpCP protease. The function of MecA-(121–218) to serve as a degradation tag resides in its ability to oligomerize and activate the ATPase activity of ClpC. Our biochemical analysis suggests a highly dynamic model for the ClpCP protease in cells, where its assembly and disassembly are constantly ongoing and synchronized with MecA association and MecA degradation, respectively.
EXPERIMENTAL PROCEDURES
Protein Preparation
All clones were generated using a standard PCR-based cloning strategy, and the identities of individual clones were verified through double-strand plasmid sequencing. ClpC and MecA variants were overexpressed in E. coli strain BL21(DE3) at 22 °C using pGEX-6P-1 vectors (GE Healthcare) with an N-terminal glutathione S-transferase (GST)3 tag or PKTG vectors with a C-terminal GST tag. The soluble fraction of the E. coli lysate was purified over a glutathione-Sepharose column and cleaved by PreScissionTM Protease (GE Healthcare) if necessary. ClpP was overexpressed in E. coli strain BL21 (DE3) as a C-terminal His6 tag protein using a pET-21b vector (Novagen). The soluble fraction of ClpP-His6 in the E. coli lysate was purified over a nickel-nitrilotriacetic acid column (Qiagen). ComK was overexpressed in E. coli strain BL21 (DE3) using a PLM303 vector (a derivative of pET-27a). The soluble fraction of maltose-binding protein fusion ComK in the E. coli extracts was purified over amylose resin (New England Biolabs) and then cleaved by PreScission protease (GE Healthcare). After affinity purification, all proteins were further purified by anion-exchange chromatography (Source-15Q, GE Healthcare) and gel filtration chromatography (Superdex 200, GE Healthcare). The concentrations of proteins were determined by spectroscopic measurement at 280 nm. All recombinant proteins were characterized by gel filtration.
ClpCP Degradation Assay
In vitro degradation assays were carried out as described previously (27, 28). ClpC and ClpP were added at 4 μm final concentration. All MecA variants and ComK were added at 6 μm final concentration. All protein concentrations reported in this study refer to those of the monomers, regardless of the oligomerization states of the proteins.
ATPase Assay
Enzyme activity of ClpC was determined with MecA or MecA121–218 as an activator using a coupled spectrophotometric assay (31). All reactions were carried out at 25 °C in 1.8 ml of reaction mixture containing 100 mm Hepes buffer, pH 7.5, 100 mm KCl, 10 mm MgCl2, 200 μm NADH, 1 mm phosphoenolpyruvate, 10 units/ml lactate dehydrogenase, 7.5 units/ml pyruvate kinase. In these reactions, ClpC was fixed at 0.3 μm, and MecA was added from 0 to 1.2 μm. The progress of the reaction was monitored continuously by following the formation of NAD+ at 340 nm on a PerkinElmer Life Sciences Lambda 45 spectrophotometer equipped with a magnetic stirrer in the cuvette holder. The concentrations of ADP formed in the ClpC-catalyzed reaction were determined using an extinction coefficient for NADH of 6220 cm−1 m−1 at 340 nm. The concentrations of ClpC, MecA, and MecA121–218 were determined spectrophotometrically based on their extinction coefficients calculated from their primary amino acid sequences and verified by the method of Bradford with bovine serum albumin as a standard.
Unfolding Assay
The unfolding assay was modified from a published protocol (32). Green fluorescent protein (GFP) fluorescence (excitation wavelength 400 nm and emission wavelength 510 nm) measurements were made on a Hitachi fluorescence spectrometer F4600. Fluorescence measurement reactions were carried out at 37 °C in 100 mm Hepes buffer, pH 7.5, 100 mm KCl, 20 mm MgCl2, 2 mm phosphoenolpyruvate, 50 units/ml pyruvate kinase, with or without 20 mm ATP. In these reactions, ClpC and MecA96–218-GFP were added at 2 and 3 μm, respectively, in the presence or absence of 15 μm GroEL trap (D87K). ClpP was used at 2 μm.
Interaction Assay by Gel Filtration
Size exclusion chromatography, using a Superdex 200 column (10/30; GE Healthcare), was employed to examine protein interaction. In all cases, proteins are incubated at 4 °C for at least 45 min to allow equilibrium to be reached. The flow rate was 0.5 ml/min, and the buffer contained 10 mm Tris, pH 8.0, 150 mm NaCl, and 2 mm dithiothreitol. All fractions were collected at 0.5 ml each. Aliquots of relevant fractions were mixed with SDS sample buffer and subjected to SDS-polyacrylamide gel electrophoresis. The proteins were visualized by Coomassie Blue staining. The column was calibrated with molecular mass standards.
RESULTS
Sequence Determinants of MecA for ClpCP Degradation
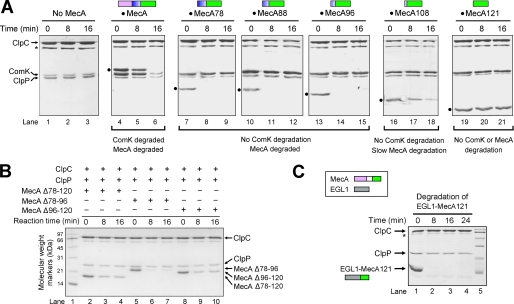
The full-length MecA protein is required for ComK degradation in cells. To determine the sequence requirement of MecA for ComK degradation, we reconstituted an in vitro ComK degradation assay on the basis of a published protocol (30). This assay relies on highly purified, recombinant proteins ClpC, ClpP, MecA, and ComK. Sufficient quantities of pyruvate kinase and phosphoenolpyruvate were included in the assay to ensure timely regeneration of ATP. In the absence of MecA, incubation of ClpC and ClpP with the substrate protein ComK did not lead to its degradation (Fig. 1A, lanes 1–3). Inclusion of the full-length MecA protein in this reaction resulted in the gradual degradation of ComK (lanes 4–6). Consistent with previous studies (30), the full-length MecA protein was also degraded (lanes 4–6).
FIGURE 1.
Sequence determinants of MecA for ClpCP-mediated degradation of ComK and MecA. A, characterization of deletion variants of MecA. The full-length MecA and the substrate protein ComK were degraded by the ClpCP protease (lanes 4–6). Removal of the N-terminal 77 amino acids in MecA led to complete abrogation of ComK degradation but had no impact on MecA degradation. Further removal of the linker sequence 78–120 resulted in abolishment of MecA degradation. All SDS-PAGE gels in this study were visualized by Coomassie Brilliant Blue staining. The asterisks indicate the position of pyruvate kinase. The MecA variants are indicated by black dots. MecA78 denotes MecA-(78–218), MecA121 denotes MecA-(121–218), and so on. B, the linker sequence 78–120 is not required for MecA degradation. Removal of amino acids 78–96, 96–120, or 78–120 from the full-length MecA protein still allowed the degradation of the cognate MecA variants. C, replacement of the linker sequence 78–120 by a flexible sequence from EGL-1 allowed efficient degradation of the fusion protein between EGL-1 and MecA-(121–218).
Next, we generated a number of MecA deletion variants and evaluated their abilities to mediate ComK degradation as well as degradation of themselves. Removal of the N-terminal 77 or more residues in MecA led to complete abrogation of ComK degradation (Fig. 1A, lanes 7–21), demonstrating that the N-terminal domain is required for substrate degradation. Consistent with this observation, the N-terminal residues 1–92 of MecA were shown to interact with ComK (30), but none of the deletion variants retained the interaction with ComK (data not shown).
Intriguingly, deletion of the N-terminal 77, 87, or 95 amino acids in MecA allowed rapid degradation of these MecA variants (lanes 7–15). When compared with these variants, MecA-(108–218) exhibited a reduced ability to be degraded (lanes 16–18). By contrast, MecA-(121–218) completely failed to be degraded by the ClpCP protease (lanes 19–21). One potential explanation for these observations is that the linker sequence 78–120 of MecA may play an indispensable role in the degradation of MecA. Contrary to this notion, deletion of residues 78–96, 96–120, or 78–120 had relatively minor impact on the degradation of the resulting MecA variants (Fig. 1B). To further examine this scenario, we generated a chimeric protein in which the N terminus of MecA-(121–218) was fused with a peptide fragment (residues 45–87) derived from a reference protein EGL-1 (33). Despite the lack of sequence similarity between MecA-(78–120) and EGL-1-(45–87), the EGL-1-MecA fusion protein was rapidly degraded by the ClpCP protease (Fig. 1C).
MecA-(121–218) as a Degradation Tag
These experimental observations demonstrate that neither the linker sequence 78–120 nor the N-terminal domain of MecA is required for the degradation of MecA-(121–218). Importantly, these results engendered two testable hypotheses. First, the C-terminal 98-amino acid fragment of MecA (residues 121–218) may serve as a signal sequence for the degradation of itself and other proteins attached. Second, MecA-(121–218) may have a requirement for the length, but not the identity, of the peptide sequence at its N terminus to function as a degradation tag.
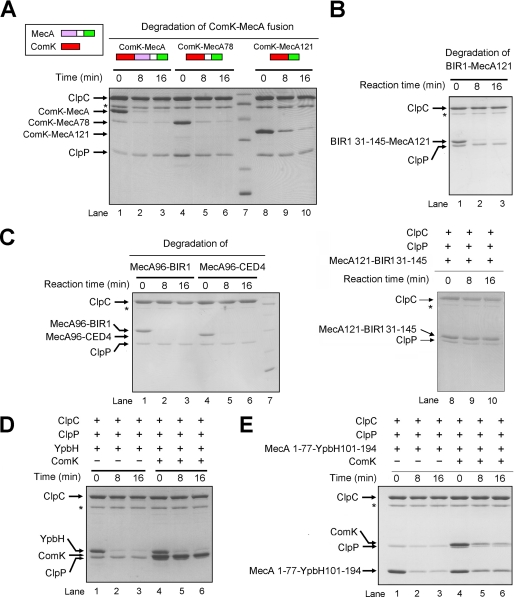
First, we examined the hypothesis that MecA-(121–218) may serve as a degradation tag. If this were true, MecA-(121–218) should be able to target other unrelated proteins for ClpCP-mediated degradation when fused together. In agreement with this prediction, fusion of the full-length ComK protein with MecA, MecA-(78–218), or MecA-(121–218), led to efficient degradation of the chimeric protein (Fig. 2A). Similarly, the BIR1 domain (residues 31–145) of DIAP1 (34) was rapidly degraded when it was fused to MecA-(121–218) (Fig. 2B). In these chimeric proteins, MecA-(121–218) was fused at the C terminus of the target substrate protein. Next, we examined whether the putative degradation tag MecA-(121–218) could be placed at the N terminus of a substrate protein. Both the BIR1 domain (residues 31–145) of DIAP1 and the CARD domain (residues 1–99) of CED-4 were readily degraded when their N termini were fused to the C terminus of MecA-(96–218) (Fig. 2C, lanes 1–6) but remained intact when fused to MecA-(121–218) (Fig. 2C, lanes 8–10). These observations strongly support the notion that MecA-(121–218) serves as a degradation tag but may require an N-terminal flexible sequence to function properly.
FIGURE 2.
MecA-(121–218) functions as a degradation tag. A, MecA-(121–218) targets ComK for degradation by the ClpCP protease. The full-length ComK protein was efficiently degraded when it was covalently fused to the N terminus of the full-length MecA, MecA-(78–218), or MecA-(121–218). B, MecA-(121–218) targets the BIR1 domain (residues 131–145) of DIAP1 for degradation by the ClpCP protease. The BIR1 domain was fused to the N terminus of MecA-(121–218). C, MecA-(96–218) targets the BIR1 domain of DIAP1 and the CARD domain of CED4 for degradation by the ClpCP protease. The BIR1 domain and the CARD domain were fused to the C terminus of MecA-(96–218). The flexible linker sequence 96–120 preceding MecA-(121–218) is required for degradation. D, the MecA paralog YpbH was degraded by ClpCP but failed to target ComK for degradation. E, the degradation tag YpbH-(101–194) targeted both MecA-(1–77) and ComK for ClpCP-mediated degradation. MecA-(1–77) was fused to the N terminus of YpbH-(101–194), which corresponds to MecA-(121–218).
Another adaptor protein YpbH from B. subtilis shares extensive sequence similarity with MecA, particularly in their respective N- and C-terminal domains (35). Similar to MecA, both the full-length YpbH protein and its C-terminal fragment (residues 92–194) were rapidly degraded by the ClpCP protease (Fig. 2D, lanes 1–3; and data not shown). These results suggest that the C-terminal residues 101–194 of YpbH, which correspond to MecA-(121–218), may also function as a degradation tag. Supporting this analysis, MecA-(1–77) was degraded when it was fused to the N terminus of YpbH-(101–194) (Fig. 2E, lanes 1–3). In addition, although the full-length YpbH failed to target ComK for degradation (Fig. 2D, lanes 4–6), the chimeric protein between MecA-(1–77) and YpbH-(101–194) successfully accomplished this task (Fig. 2E, lanes 4–6).
Requirement of a Flexible Sequence for Degradation
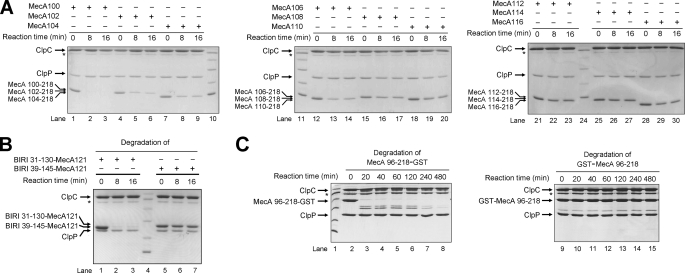
Next, we examined the length requirement of an N-terminal extension for MecA-(121–218) to function as a degradation tag. Because MecA-(108–218), but not MecA-(96–218), exhibited a compromised ability to be degraded by ClpCP (Fig. 1A), we used MecA-(96–218) as the starting point and generated a series of deletion variants, each involving removal of 2 amino acids from the previous variant. Then, we purified these deletion variants to homogeneity and examined their abilities to undergo ClpCP-mediated degradation. Under identical experimental conditions, MecA-(100–218) was completely degraded by ClpCP (Fig. 3A, lanes 1–3), whereas MecA-(116–218) was largely unscathed (lanes 28–30), with the other variants in between showing a decreasing extent of degradation as the deletion proceeded toward the C terminus (lanes 4–9, 12–23, and 25–27). We estimated that MecA-(112–218) had nearly lost the ability to be degraded by ClpCP. This analysis suggests that an N-terminal, flexible sequence of at least 9 amino acids is needed for MecA-(121–218) to function properly as a degradation tag.
FIGURE 3.
MecA-(121–218) requires an N-terminal flexible sequence for ClpCP degradation. A, systematic deletion analysis of MecA. MecA-(121–218) appears to require extra 9 amino acids at its N terminus in order for itself to be degraded by ClpCP. B, the degradation of the chimeric protein between the BIR1 domain and MecA-(121–218) requires an N-terminal flexible sequence. Deletion of 8 amino acids from the N terminus of the BIR1 domain (residues 31–145) led to abrogation of degradation (lanes 5–7). By contrast, removal of internal sequences in BIR1 had no impact on degradation (lanes 1–3). C, the degradation of GST requires an N-terminal flexible sequence. GST was degraded only when it was fused C-terminal to MecA-(96–218) (left panel). GST fused N-terminal to MecA-(96–218) resisted degradation, presumably due to the lack of flexible sequences at its N terminus.
Our experimental evidence is fully consistent with the requirement of an N-terminal flexible sequence for MecA-mediated degradation. This conclusion predicts that a flexible sequence of appropriate length should be present for the degradation of any folded domain that is fused to MecA-(121–218). Supporting this prediction, the BIR1 domain (residues 31–145), which was degraded when fused to the N terminus of MecA-(121–218) (Fig. 2B), contains a flexible sequence comprising residues 31–39 (34). Deletion of 8 amino acids from the N terminus of this BIR1-MecA fusion led to complete abrogation of degradation by the ClpCP protease (Fig. 3B, lanes 5–7). By contrast, deletion of 15 amino acids from the intervening sequences between the two folded domains BIR1 and MecA-(121–218) did not affect the degradation of the chimeric protein (Fig. 3B, lanes 1–3).
In addition to the BIR1 and CARD domains, MecA-(121–218) was also able to target all other tested proteins for ClpCP-mediated degradation. For example, GST fused at the C terminus of MecA-(96–218) was degraded to completion (Fig. 3C, lanes 2–8), albeit at a slower rate when compared with BIR1 or CARD. The presence of the flexible sequences 96–120 was presumably required for the degradation. By contrast, GST fused at the N terminus of MecA-(96–218) resisted degradation by ClpCP (Fig. 3C, lanes 9–15), presumably due to the absence of flexible sequences at the N terminus of GST (36). Intriguingly, the degradation of the fusion protein between MecA-(96–218) and GST exhibited two phases (Fig. 3C, lanes 2–8): a rapid phase of 20 min, during which the fusion protein was reduced to the intermediate degradation product, and a slow phase of up to 480 min, during which the intermediate degradation products were completely degraded. Analysis by N-terminal peptide sequencing and mass spectroscopy revealed that the intermediate degradation products contained the entire GST moiety and residues 184–218 of MecA. These observations demonstrate that the degradation of the fusion protein began from its N terminus.
MecA-(121–218) Facilitates Oligomerization of ClpC and Interaction with ClpP
Our observations indicate that MecA-(121–218) serves as a signal for the degradation of a number of chimeric proteins. However, how does MecA-(121–218) carry out this activity? On the basis of available experimental data, we made three inter-related hypotheses. First, MecA-(121–218) may facilitate the activation of ClpC by promoting the formation of a functional, oligomeric complex. Second, MecA-(121–218) is likely to be indispensable for the formation of the oligomeric ClpCP protease. Third, upon complex formation, MecA-(121–218) may markedly stimulate the ATPase activity of ClpC. In this and the next several paragraphs of this study, we provide strong evidence in support of all three hypotheses.
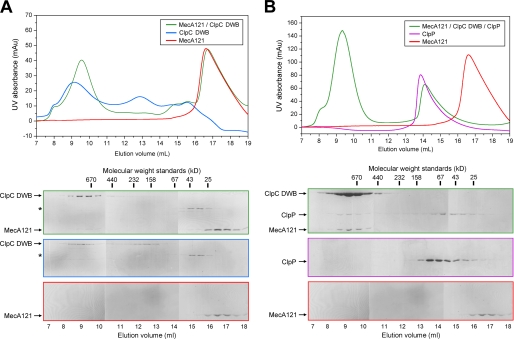
First, we examined the interaction of MecA-(121–218) with ClpC. All protein concentrations reported in this study refer to those of the monomers, regardless of the oligomerization states of the proteins. At 10 μm concentration, the full-length ClpC (with double mutations E280A/E618A in the walker B motifs, referred to as DWB) was eluted from gel filtration in two distinct peaks (Fig. 4A). One peak corresponds to an apparent molecular mass of over 670 kDa, likely representing an inactive oligomer, and the other peak corresponds to ∼180 kDa (Fig. 4A). Incubation of MecA-(121–218) with ClpC DWB, in the absence of any exogenous nucleotide, led to the formation of a stable complex, which was co-eluted from gel filtration with an apparent molecular mass of ∼700 kDa (Fig. 4A), consistent with that of a hexameric MecA-ClpC complex. The elution volume for the MecA-ClpC complex was slightly but reproducibly smaller than that of the inactive ClpC oligomer (Fig. 4A), suggesting that MecA binding may have induced conformation changes in ClpC. Incubation with exogenous ATP or ADP had no apparent impact on the elution volume of the complex between MecA-(121–218) and ClpC DWB. Identical results were obtained for the full-length MecA protein (data not shown). Analysis by liquid chromatography-mass spectroscopy revealed, however, that the recombinant ClpC protein contained a substoichiometric amount of bound ADP.
FIGURE 4.
MecA-(121–218) facilitates oligomerization of ClpC and interaction with ClpP. A, the formation of a stable binary complex between MecA-(121–218) and ClpC in the absence of exogenous ATP or ADP. MecA-(121–218) was co-eluted from gel filtration together with ClpC DWB, indicating the formation of a stable binary complex. The asterisk denotes the position of a contaminating protein. mAU, milliabsorbance units. B, assembly of a stable ternary complex among MecA-(121–218), ClpC, and ClpP. ClpP and ClpC DWB only formed a stable ternary complex in the presence of MecA and ATP.
Then, we examined the role of MecA-(121–218) in the assembly of an oligomeric ClpC-ClpP complex. ClpP alone was eluted from gel filtration over a broad range of molecular mass, from 160 to 25 kDa (Fig. 4B), suggesting the unstable nature of the heptameric ClpP complex from B. subtilis. In the absence of MecA-(121–218), ClpC DWB and ClpP failed to form a stable complex in the presence of ATP, as judged by gel filtration (data not shown). This result is consistent with a previous study (29). In the presence of MecA-(121–218) and ATP, ClpP was co-eluted with ClpC DWB and MecA, indicating the formation of a stable ternary complex with an apparent molecular mass in excess of 670 kDa (Fig. 4B). Thus, we conclude that MecA-(121–218) is required for the formation of the ClpCP protease.
MecA-(121–218) Stimulates the ATPase Activity of ClpC
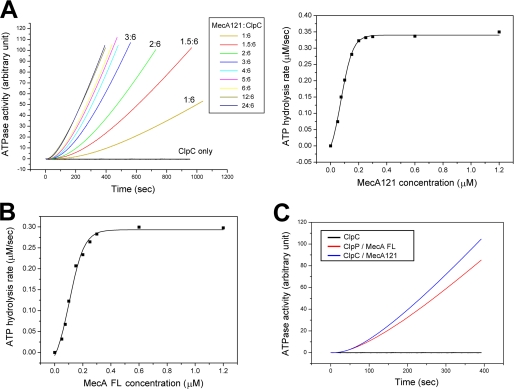
Next, we examined whether MecA-(121–218) is able to stimulate the ATPase activity of ClpC upon complex formation. In the absence of MecA, the wild-type ClpC protein exhibited an undetectable level of ATPase activity (Fig. 5A). Incubation of MecA-(121–218) with ClpC, even at a substoichiometric molar ratio of 1:6, markedly stimulated the ATPase activity. Increasing molar ratios of MecA-(121–218) over ClpC resulted in progressive, but saturable, increase of the ATPase activity, with the maximal activity approaching 0.34 μm/s for 0.3 μm ClpC (Fig. 5A).
FIGURE 5.
MecA-(121–218) stimulates the ATPase activity of ClpC. A, MecA-(121–218) stimulated the ATPase activity of ClpC in a concentration-dependent manner. At 0.3 μm ClpC, the ATPase activity reached a maximum of 0.34 μm/s with a saturating amount of MecA-(121–218). B, the full-length MecA protein stimulated the ATPase activity of ClpC in a concentration-dependent manner. At 0.3 μm ClpC, the maximal ATPase activity was 0.3 μm/s. C, MecA-(121–218) stimulated the ATPase activity of ClpC to a similar extent as the full-length MecA protein.
The full-length MecA protein was shown to stimulate the ATPase activity of ClpC (28). To investigate whether MecA-(121–218) retains the full activity of the full-length protein, we measured the ATPase activity of 0.3 μm ClpC in the presence of varying molar ratios of the full-length MecA. As anticipated, the full-length MecA protein stimulated the ATPase activity of ClpC in a concentration-dependent manner, with a maximal activity of ∼0.3 μm/s (Fig. 5B). This analysis demonstrates that within the margin of errors, MecA-(121–218) stimulated the ATPase activity of ClpC to the same extent as the full-length MecA protein (Fig. 5C).
Constant Unfolding and Degradation of MecA by the ClpCP Protease
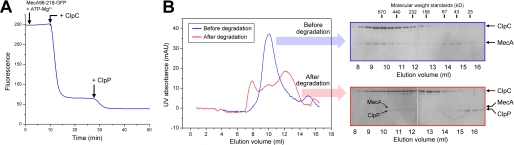
Taken together, our experimental observations suggest a highly dynamic scenario in which MecA undergoes unfolding and degradation by the ClpCP protease as soon as the MecA-ClpCP ternary complex is assembled. To directly examine this scenario, we engineered a chimeric protein between MecA-(96–218) and GFP and purified the chimeric protein to homogeneity. As anticipated, the MecA-GFP chimeric protein formed a stable, oligomeric complex with ClpC in the absence of ATP (data not shown). The intensity of fluorescence remained steady for the MecA-(96–218)-GFP protein, at a concentration of 3 μm, in the presence of 20 mm ATP at 37 °C (Fig. 6A). Then, we monitored the change of fluorescence intensity upon the addition of ClpC and ClpP. The addition of ClpC, at a concentration of 2 μm, led to rapid decrease of fluorescence intensity by ∼74% within 7 min (Fig. 6A). This result demonstrates that the vast majority of MecA-(96–218)-GFP had been unfolded by the activated ClpC complex within 7 min.
FIGURE 6.
Dynamic nature of the MecA-ClpCP system. A, the constant unfolding and degradation of MecA by the ClpCP protease following MecA-mediated assembly of the ClpCP protease. MecA-(96–218) was fused with GFP to create a substrate protein. Fluorescence intensity was monitored as a convenient indicator of unfolding and degradation. B, the degradation of MecA results in the disassembly of the functional ClpCP protease. mAU, milliabsorbance units.
Interestingly, the intensity of fluorescence, which corresponds to the fraction of folded MecA-(96–218)-GFP, remained relatively steady after 7 min (supplemental Fig. 1). One potential explanation for this observation is that the unfolded MecA-(96–218)-GFP could spontaneously refold back to the native conformation and, under the experimental conditions, the refolding rate was similar to the unfolding rate after 7 min. To examine this possibility, we introduced a mutant GroEL protein, which is known to trap the unfolded substrate protein (32, 37), into the reaction and compared the rates of unfolding. The result indicates that, partially consistent with this explanation, the presence of the mutant GroEL, at a concentration of 15 μm, accelerated the unfolding reaction (supplemental Fig. 1). However, the presence of GroEL only reduced the fraction of folded MecA-(96–218)-GFP by ∼2% (from 26 to 24%) after 7 min (supplemental Fig. 1). Moreover, the addition of the ClpP peptidase to the reaction led to further decrease of fluorescence intensity, but the remaining fluorescence still represented 18% of the initial value (Fig. 6A), indicating that there must be an additional explanation to account for the observation. We propose that the remaining fluorescence is a direct consequence of binding equilibrium among ClpP, ClpC, and MecA-(96–218)-GFP. Under the experimental conditions, the leftover, folded MecA-(96–218)-GFP could no longer form a stable complex with the ClpCP protease, hence escaping degradation.
Disassembly of the MecA-ClpCP Complex upon MecA Degradation
Our experimental observations demonstrate that MecA undergoes unfolding and degradation by the ClpCP protease as soon as the MecA-ClpCP ternary complex is assembled in the presence of ATP. What happens to the assembled, oligomeric ClpCP protease once MecA is degraded? One possibility is that MecA is continuously required for maintenance of the oligomeric ClpCP protease; in this case, the degradation of MecA is predicted to trigger the disassembly of the ClpCP protease. Another possibility is that MecA is only required for formation, but not maintenance, of the ClpCP protease; in this case, the degradation of MecA should have no impact on the assembled ClpCP protease. To differentiate between these two possibilities, we incubated the full-length ClpC with an excess of full-length MecA in the absence of ATP. The sample was split into two equal aliquots, one incubated with ClpP in the presence of ATP and the other with buffer as a control. The reaction was allowed to proceed to near completion. Then, both aliquots were individually analyzed by gel filtration for signs of complex formation. For the control aliquot, MecA and ClpC formed a stable binary complex (Fig. 6B). For the aliquot incubated with ClpP and ATP, the vast majority (>80%) of MecA had been degraded by the ClpCP protease, as evidenced by the much decreased intensity of MecA on SDS-PAGE (Fig. 6B). Consequently, the elution volume of ClpP corresponds to a molecular weight that is considerably smaller than that of ClpP from a functional ClpCP complex (Fig. 6B). In addition, the vast majority (>80%) of ClpC was eluted from gel filtration with molecular weights that are considerably smaller than that of ClpC from the MecA-ClpC complex (Fig. 6B). These observations clearly indicate the disassembly of the MecA-ClpCP complex.
DISCUSSION
In this study, we report a number of interconnected findings. First, MecA-(121–218) was identified as a degradation tag for ClpCP-mediated degradation. However, the degradation of a fusion protein between MecA-(121–218) and a folded domain requires the presence of a flexible sequence of at least 9 amino acids at the N terminus. Second, the mechanisms by which MecA-(121–218) functions as a degradation tag were attributed to assembly and activation of the ClpCP protease mediated by MecA-(121–218). MecA-(121–218) stimulated the ATPase activity of ClpC to the same extent as the full-length MecA protein. Then, using an in vitro unfolding and degradation assay, MecA was shown to undergo unfolding and degradation by the ClpCP protease as soon as the ClpCP protease was assembled (Fig. 6). The degradation of MecA, in turn, led to the disassembly of the ClpCP protease (Fig. 6B).
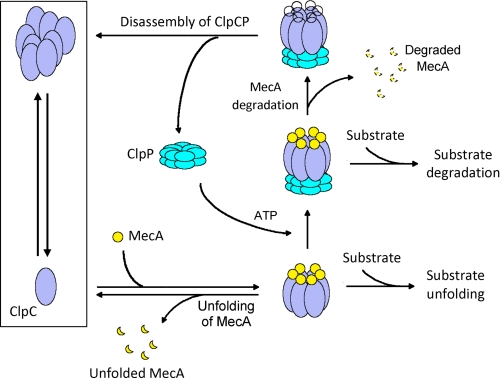
Together, our experimental observations give rise to a working model for the ClpCP protease in cells (Fig. 7). The major elements of this model, such as dynamic cycles of ClpCP assembly, have not been proposed previously and are borne out of direct experimental evidence presented in this study. In this model, assembly of the ClpCP protease strictly depends on the presence of MecA, and the assembled MecA-ClpCP protease is highly dynamic, where the degradation of ComK is accompanied by consumption of MecA and subsequent disassembly of the ClpCP protease. ATP plays a key role in these processes as it is required for assembly of the MecA-ClpCP protease and for ATP hydrolysis-dependent unfolding and translocation of substrate proteins. Given the availability of ATP in bacteria, the dynamic cycles of assembly and disassembly of the ClpCP protease, triggered by the appearance and disappearance of MecA, are likely at work at all times (Fig. 7). This dynamic process likely ensures a high level of sensitivity toward changes of regulator concentrations in the cytoplasm. For example, elevated concentrations of the anti-adaptor protein ComS, which binds competitively to MecA, may immediately liberate ComK from MecA-targeted degradation (27, 38).
FIGURE 7.
A working model of the MecA-ClpCP system. In this highly dynamic system, the assembly and disassembly of the ClpCP protease are triggered by appearance and disappearance of MecA. A key to this system is that MecA itself is rapidly degraded by the ClpCP protease as soon as MecA accomplishes the task of ClpCP assembly. This dynamic system may ensure rapid response to changes in bacteria. The exact molar ratio between MecA and ClpC, although shown here as 1:1, remains to be determined.
Our results demonstrate that the C-terminal domain of MecA, comprising residues 121–218, is a degradation tag for the ClpCP protease. Fusion of folded proteins with MecA-(121–218) results in rapid degradation of the chimeric proteins. Similar to other representative degradation tags such as SsrA in bacteria and polyubiquitin in eukaryotes, MecA-(121–218) needs to be covalently attached to the substrate protein to function as a degradation tag. In analogy to SsrA but different from polyubiquitin, the MecA degradation tag is degraded along with the substrate protein and hence is not recycled. The SsrA tag is placed at the C terminus of the substrate protein, and consequently, the degradation of SsrA-tagged protein proceeds in the C-to-N direction. Evidence presented in this study (Fig. 3C) indicates that the degradation of MecA or the fusion protein occurs in the N-to-C direction.
The observation that MecA is not recycled (MecA was degraded along with the substrate protein ComK) does not necessarily indicate that MecA is non-reusable within the assembled ClpCP protease. For example, it is possible that MecA can target multiple molecules of ComK for degradation and that the degradation of MecA only occurs when there is no alternative substrate remaining. To address this issue, we incubated ComK of increasing concentrations with the same amount of the MecA-ClpCP protease and examined the extent of ComK degradation. Preliminary results show that higher concentrations of ComK failed to increase the extent of ComK degradation (supplemental Fig. 2), suggesting that MecA may not be reusable within the assembled ClpCP protease.
Examination of the N-terminal, flexible sequences in the various MecA fusion proteins revealed no obvious sequence motif for ClpC recognition. However, the length of the peptide sequences was apparently important as shortening of the peptide length led to gradual decrease of MecA degradation (Fig. 3A). These experimental observations suggest that the length, but not the identity, of the N-terminal peptide sequence is needed for MecA-mediated degradation. Because such a recognition sequence could be extremely degenerate and simple, we cannot completely rule out the possibility that it does exist and is hidden at the N termini of the fusion proteins examined in this study.
Given that MecA-(121–218) is sufficient for assembly of the ClpCP protease, why does MecA-(121–218) still need an N-terminal, flexible sequence for degradation? The degradation of MecA-(121–218) requires its unfolding and translocation by ClpC, propelled by cycles of ATP hydrolysis. The initiation of unfolding and translocation likely involves the interaction between the N-terminal residue(s) of MecA and the A-Φ loops of the ATPase domains of ClpC, as suggested from studies of the ClpXP protease (15). The length requirement of the peptide sequence preceding MecA-(121–218) may reflect the need for its N terminus to reach the A-Φ loops of ClpC.
MecA targets substrate protein, such as ComK and ComS, to the ClpCP protease for degradation. This function is reminiscent of ubiquitin in the eukaryotic proteasome, where only polyubiquitinated substrate protein is recognized, unfolded, and translocated into the 20 S core particle for degradation, all by the preassembled 19 S regulatory particle. The polyubiquitin chain is released from the unfolded substrate protein and recycled by the deubiquitinating enzymes within the regulatory particle. By contrast, assembly and activation of the ClpC complex strictly depend on MecA. The MecA degradation tag is degraded by the ClpCP protease, together with the non-covalently attached substrate protein.
Many important questions remain. How does MecA recognize the substrate protein? How does MecA facilitate the oligomerization and activation of ClpC? How is MecA itself degraded by the ClpCP protease? Deciphering these mechanisms requires elucidation of the MecA-ClpC structure and associated biochemical analysis.
Supplementary Material
This work was supported by grants from the Ministry of Science and Technology (Grant 2009CB918801), Tsinghua University 985 Phase II funds, Project 30888001 supported by the National Natural Science Foundation of China, and Beijing Municipal Commissions of Education and Science and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GST
- glutathione S-transferase
- GFP
- green fluorescent protein
- CARD
- caspase recruitment domain.
REFERENCES
- 1.Baker T. A., Sauer R. T. (2006) Trends Biochem. Sci. 31, 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inobe T., Matouschek A. (2008) Curr. Opin. Struct. Biol. 18, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg A. L. (1992) Eur. J. Biochem. 203, 9–23 [DOI] [PubMed] [Google Scholar]
- 4.Baumeister W., Walz J., Zühl F., Seemüller E. (1998) Cell 92, 367–380 [DOI] [PubMed] [Google Scholar]
- 5.Groll M., Bochtler M., Brandstetter H., Clausen T., Huber R. (2005) Chembiochem. 6, 222–256 [DOI] [PubMed] [Google Scholar]
- 6.Baumeister W., Lupas A. (1997) Curr. Opin. Struct. Biol. 7, 273–278 [DOI] [PubMed] [Google Scholar]
- 7.Goldberg A. L. (2007) Biochem. Soc. Trans. 35, 12–17 [DOI] [PubMed] [Google Scholar]
- 8.Hanna J., Finley D. (2007) FEBS Lett. 581, 2854–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 10.Bochtler M., Ditzel L., Groll M., Hartmann C., Huber R. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 295–317 [DOI] [PubMed] [Google Scholar]
- 11.Sauer R. T., Bolon D. N., Burton B. M., Burton R. E., Flynn J. M., Grant R. A., Hersch G. L., Joshi S. A., Kenniston J. A., Levchenko I., Neher S. B., Oakes E. S., Siddiqui S. M., Wah D. A., Baker T. A. (2004) Cell 119, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiler K. C., Waller P. R., Sauer R. T. (1996) Science 271, 990–993 [DOI] [PubMed] [Google Scholar]
- 13.Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998) Genes Dev. 12, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benaroudj N., Goldberg A. L. (2000) Nat. Cell Biol. 2, 833–839 [DOI] [PubMed] [Google Scholar]
- 15.Martin A., Baker T. A., Sauer R. T. ( 2008) Nat. Struct. Mol. Biol. 15, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin A., Baker T. A., Sauer R. T. (2008) Nat. Struct. Mol. Biol. 15, 139–145 [DOI] [PubMed] [Google Scholar]
- 17.Martin A., Baker T. A., Sauer R. T. (2008) Mol. Cell 29, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin A., Baker T. A., Sauer R. T. (2007) Mol. Cell 27, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin A., Baker T. A., Sauer R. T. (2005) Nature 437, 1115–1120 [DOI] [PubMed] [Google Scholar]
- 20.Zhang F., Wu Z., Zhang P., Tian G., Finley D., Shi Y. (2009) Mol. Cell 34, 485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Mol. Cell 34, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubnau D., Roggiani M. (1990) J. Bacteriol. 172, 4048–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn J., Kong L., Dubnau D. (1994) J. Bacteriol. 176, 5753–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Sinderen D., Venema G. (1994) J. Bacteriol. 176, 5762–5770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen I., Christie P. J., Dubnau D. (2005) Science 310, 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Msadek T., Kunst F., Rapoport G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5788–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turgay K., Hamoen L. W., Venema G., Dubnau D. (1997) Genes Dev. 11, 119–128 [DOI] [PubMed] [Google Scholar]
- 28.Turgay K., Hahn J., Burghoorn J., Dubnau D. (1998) EMBO J. 17, 6730–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirstein J., Schlothauer T., Dougan D. A., Lilie H., Tischendorf G., Mogk A., Bukau B., Turgay K. (2006) EMBO J. 25, 1481–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persuh M., Turgay K., Mandic-Mulec I., Dubnau D. (1999) Mol. Microbiol. 33, 886–894 [DOI] [PubMed] [Google Scholar]
- 31.Cook P. F., Neville M. E., Jr., Vrana K. E., Hartl F. T., Roskoski R., Jr. (1982) Biochemistry 21, 5794–5799 [DOI] [PubMed] [Google Scholar]
- 32.Weber-Ban E. U., Reid B. G., Miranker A. D., Horwich A. L. (1999) Nature 401, 90–93 [DOI] [PubMed] [Google Scholar]
- 33.Yan N., Gu L., Kokel D., Chai J., Li W., Han A., Chen L., Xue D., Shi Y. (2004) Mol. Cell 15, 999–1006 [DOI] [PubMed] [Google Scholar]
- 34.Yan N., Wu J. W., Chai J., Li W., Shi Y. (2004) Nat. Struct. Mol. Biol. 11, 420–428 [DOI] [PubMed] [Google Scholar]
- 35.Persuh M., Mandic-Mulec I., Dubnau D. (2002) J. Bacteriol. 184, 2310–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanaoka Y., Ago H., Inagaki E., Nanayama T., Miyano M., Kikuno R., Fujii Y., Eguchi N., Toh H., Urade Y., Hayaishi O. (1997) Cell 90, 1085–1095 [DOI] [PubMed] [Google Scholar]
- 37.Fenton W. A., Kashi Y., Furtak K., Horwich A. L. (1994) Nature 371, 614–619 [DOI] [PubMed] [Google Scholar]
- 38.Prepiak P., Dubnau D. (2007) Mol. Cell 26, 639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., Mei M., Qi Y., Yan C., Xiang S., Zhou Z., Hu Q., Wang J., Shi Y. (2009) J. Biol. Chem. 284, 34376–34381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.